Abstract
Objectives:
Silver sulfide (Ag2S) quantum dots (QDs) are highly promising nanomaterials in bioimaging systems due to their high activities for both imaging and drug/gene delivery. There is insufficient research on the toxicity of Ag2S QDs coated with meso-2,3-dimercaptosuccinic acid (DMSA). In this study, we aimed to determine the cytotoxicity of Ag2S QDs coated with DMSA in Chinese hamster lung fibroblast (V79) cells over a wide range of concentrations (5-2000 μg/mL).
Materials and Methods:
Cell viability was determined by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) and neutral red uptake (NRU) assays. The genotoxic and apoptotic effects of DMSA/Ag2S QDs were also assessed by comet assay and real-time polymerase chain reaction technique, respectively.
Results:
Cell viability was 54.0±4.8% and 65.7±4.1% at the highest dose (2000 μg/mL) of Ag2S QDs using the MTT and NRU assays, respectively. Although cell viability decreased above 400 μg/mL (MTT assay) and 800 μg/mL (NRU assay), DNA damage was not induced by DMSA/Ag2S QDs at the studied concentrations. The mRNA expression levels of p53, caspase-3, caspase-9, Bax, Bcl-2, and survivin genes were altered in the cells exposed to 500 and 1000 μg/mL DMSA/Ag2S QDs.
Conclusion:
The cytotoxic effects of DMSA/Ag2S QDs may occur at high doses through the apoptotic pathways. However, DMSA/Ag2S QDs appear to be biocompatible at low doses, making them well suited for cell labeling applications.
Keywords: Meso-2, 3-dimercaptosuccinic acid coated silver sulfide quantum dots, genotoxicity, apoptosis
INTRODUCTION
The number of commercial products containing nanoparticles (NPs) is rapidly increasing and NPs are already widely distributed in air, cosmetics, medicines, and even food. As one of the leading nanomaterials, engineered NPs are currently the focus of considerable research attention due to their various applications such as drug and gene delivery, biosensors, and diagnostic tools. The use of functional nanomaterials in biology and biomedicine has been extensively explored, and it has become one of the fastest moving and most exciting research directions.1,2
A key issue in evaluating the utility of these materials is assessing their potential toxicity, which may result from either their inherent chemical composition (e.g., heavy metals) or their nanoscale properties (e.g., inhalation of particulate carbon nanotubes).1,2 To date, a variety of nanomaterials, such as carbon nanotubes, silicon nanowires, gold/silver NPs, and quantum dots (QDs), have been studied and used in a wide range of biological applications.3,4,5,6 NPs have unique features such as high surface-to-volume ratios, surface curvatures, and surface reactivities. They can also be produced with different sizes, chemical compositions, shapes, and surface charges, which affect their passage across the cell membranes, biodistribution, and toxicity.7,8,9 Recently, the use of nanomaterials has also attracted considerable interest in biomedical fields.10
QDs are nanometer-scale semiconductor crystals and are defined as particles with physical dimensions smaller than the exciton Bohr radius. QDs, which are composed of group II to VI or III to V elements, are often described as “artificial atoms”. They exhibit discrete energy levels, and their band gaps can be precisely modulated by varying their size.11,12 In 2002, Applied Spectroscopy published its first review on QDs, “Quantum Dots: A Primer,” by Murphy and Coffer.13 The applications of luminescent nanocrystals have evolved tremendously over the last decade, particularly in bioimaging and bioanalysis. Since the first demonstration of QDs for biological imaging in 1998,14,15 thousands of research articles on QDs have been published. Researchers have exploited the brightness, photostability, size-dependent optoelectronic properties, and superior multiplexing capabilities of QDs for a myriad of applications.16,17,18,19,20,21 Some of the prominent applications include in vitro diagnostics, energy transfer-based sensing, cellular and in vivo imaging, and drug delivery and theranostics.18,22,23 In parallel with these advances in bioimaging and bioanalysis, QDs have also evolved to provide greater flexibility and capability.24
QDs are usually synthesized using group II–VI materials, for example, cadmium telluride (CdTe) or cadmium selenide (CdSe).25,26 Structurally, QDs consist of a metalloid crystalline core and a “cap” or “shell” that shields the core and renders the QD bioavailable. QD cores can be fabricated using different materials with different band gaps for luminescence in the visible or near-infrared region (NIR). Cd or Zn chalcogenides such as CdS, CdSe, CdTe, and ZnS are examples of group II–VI series of QDs27,28 with luminescence in the visible range; indium phosphate and indium arsenate are examples of group III–V series QDs with emission in the red to NIR.29,30
A major limitation with respect to the clinical use of QDs is their potential toxicity due to their chemical composition and nanoscale features.29 The most popular QDs for biological applications are still based on CdSe core materials, which offer high quality and control over the spectroscopic properties of the nanocrystal. Despite several demonstrations of relatively nontoxic compositions being delivered to cells, concerns remain regarding the cytotoxicity of released cadmium ions and the associated oxidative stress.31,32,33,34,35,36
Within the last decade, tremendous efforts have been devoted to developing Cd-free QDs. Silver sulfide (Ag2S) QDs emerged recently as new generation QDs satisfying both of these criteria.37,38 Hocaoglu et al.38 reported meso-2,3-dimercaptosuccinic acid (DMSA)-coated Ag2S QDs as one of the most strongly luminescent, anionic, NI-emitting QDs. These particles were significantly internalized by HeLa cells and provided strong intracellular optical signals, suppressing autofluorescence. No reduction in the viability of HeLa cells and only 20% reduction in NIH/3T3 cells at concentrations up to 840 µg/mL were reported, which is quite unusual for a nonpegylated QD. QDs were found quite hemocompatible as well. This composition is of special interest with respect to numerous applications since surface carboxylic acids can be conjugated with target ligands or drugs, producing theranostic NPs.
In the present study, we performed a detailed toxicity analysis to investigate the potential cytotoxicity, genotoxicity, and apoptosis induced by DMSA/Ag2S QDs in Chinese hamster lung fibroblast (V79) cells. To have a relatively thorough toxicity analysis of DMSA/Ag2S NIR QDs, the MTT and neutral red uptake (NRU) assays were performed to evaluate the potential cytotoxicity; the comet assay was performed to assess the potential genotoxicity; the real-time polymerase chain reaction (RT-PCR) technique was used to evaluate the regulation of mRNA expression of tumor suppressor gene (p53), apoptotic genes (caspase-3, caspase-9, and Bax) and anti-apoptotic genes (Bcl-2 and survivin). The data presented here are the first that give the cytotoxic, genotoxic, and apoptotic effects of DMSA/Ag2S QDs in vitro. Since there is insufficient research on their toxicity, this study provides remarkable information for human health.
MATERIALS AND METHODS
Chemicals
The chemicals were purchased from the following suppliers: hydrogen peroxide (35%) (H2O2) from Merck Chemicals (Darmstadt, Germany); 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), acetic acid, dimethyl sulfoxide (DMSO), DMSA, Dulbecco’s modified eagle’s medium (DMEM), ethanol, ethidium bromide (EtBr), fetal bovine serum (FBS), low melting point agarose, L-glutamin, NR, sodium chloride (NaCl), sodium hydroxide (NaOH), N-lauroyl sarcosinate, normal melting point agarose, silver nitrate (AgNO3), trypsin-EDTA, triton X-100, penicillin/streptomycin, and phosphate buffered saline (PBS) from Sigma-Aldrich Chemicals (St. Louis, MO, USA); and sodium sulfide (Na2S) from Alfa-Aesar (Thermo Fisher Scientific, Karlsruhe, Germany). Milli-Q water (18.2 MOhm) was used as the reaction medium.
Preparation and characterization of DMSA/Ag2S NIR QDs
DMSA/Ag2S NIR QDs were prepared in a one-step reaction. A detailed description and characterization were reported previously by Hocaoglu et al.38 Briefly, 42.5 mg of AgNO3 (0.25 mmol) was dissolved in 75 mL of deoxygenated deionized water. Then 113.89 mg of DMSA (0.625 mmol) was dissolved and deoxygenated in 25 mL of deionized water at pH 7.5 and added to the reaction mixture. The pH was adjusted to 7.5 using NaOH and CH3COOH solutions (2 M). The reaction mixture was stirred at 70°C for 4 h. The prepared colloidal DMSA/Ag2S QDs were washed with deionized water using Amicon-Ultra centrifugal filters (3000 Da cut-off) and stored in the dark at 4°C. In order to calculate the concentration of QDs, a few milliliters of the colloidal solution was dried in a freeze-drier. The concentration of the QD solution was determined as 4.6 mg/mL. The absorbance spectrum of QDs was recorded in a Shimadzu 3101 PC UV-vis-NIR spectrometer in the 300-1000 nm range (Figure 1a). The photoluminescence spectrum was obtained as described in detail previously by Hocaoglu et al.38 Samples were excited with a DPSS laser operating at 532 nm and emission was recorded by an amplified silicon detector with femtowatt sensitivity in the range of 400-1100 nm with a lock-in amplifier. The QDs have an emission maximum at 790 nm with about 129 nm full-width at half maximum (Figure 1Sb). A Malvern zetasizer nano ZS was used for the measurement of the hydrodynamic size (2.9 nm) of aqueous QDs and the zeta potential of aqueous QDs (-30 mV). Hydrodynamic size was measured by dynamic light scattering. No agglomeration in the cell culture medium was observed.
Figure 1.
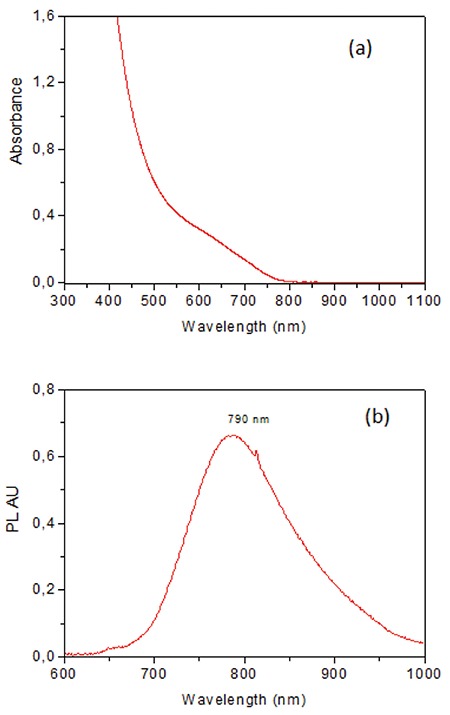
(a) Absorbance spectra, (b) emission spectra of colloidal DMSA/Ag2S QDs
Ag2S: Silver sulfide, QDs: quantum dots
Cell culture
V79 cells were obtained from the American Type Culture Collection (ATCC; Rockville, MD, USA). The cells were grown in DMEM supplemented with 10% heat-inactivated FBS, 1% penicillin/streptomycin solution (10.000 units of penicillin and 10 mg of streptomycin in 0.9% NaCl), and 2 mM L-glutamin at 37°C in a humidified atmosphere of 5% CO2.39 The culture medium was changed every 3 to 4 days. The passage numbers used in our study were between 6 and 10.
Determination of cytotoxicity by MTT assay
The MTT assay by the method described by Mosmann40 with the modifications by Hansen et al.41 and Kuźma et al.42 was carried out. The cells were disaggregated with trypsin/EDTA and then resuspended in the medium. The suspended cells (a total of 105 cells/well) were plated in 96-well tissue-culture plates. The experiment was performed for 12 h, 24 h, and 48 h before and there were no time differences (data not shown). To get a dose range for the further experiments, 24 h incubation was selected. After the incubation for 24 h, the cells were exposed to different concentrations of DMSA/Ag2S QDs (5, 10, 25, 50, 100, 200, 400, 200, 800, 1000, 2000 µg/mL) in the medium for 24 h. Then the medium was removed and MTT solution (5 mg/mL of stock in PBS) was added (10 µL/well in 100 µL of cell suspension). After the incubation of the cells for an additional 4 h with MTT dye, the dye was carefully taken out and 100 µL of DMSO was added to each well. The absorbance of the plate was measured in a microplate reader at 570 nm. The experiment was repeated three times. The results were expressed as the mean percentage of cell growth. IC50 values represent the concentrations that reduced the mean absorbance of 50% of those in the untreated cells.
Determination of cytotoxicity by NRU assay
Determination of the cytotoxicity of DMSA/Ag2S QDs using NRU assay was performed according to the protocols described by Di Virgilio et al.43 and Saquib et al.44 V79 cells were treated with DMSA/Ag2S QDs as described in the MTT assay. After incubation for 24 h, the medium was aspirated. The cells were washed twice with PBS and incubated for an additional 3 h in the medium supplemented with NR (50 µg/mL). The absorbance of the solution in each well was measured in a microplate reader at 540 nm and compared with the wells containing untreated cells. The experiment was repeated three times. The results were expressed as the mean percentage of cell growth inhibition. IC50 values represent the concentrations that reduced the mean absorbance of 50% of those in the untreated cells.
Determination of genotoxicity by comet assay
V79 cells were treated with DMSA/Ag2S QDs as described in the MTT assay. Following the disaggregation of the cells with trypsin/EDTA and the resuspension of the cells in the medium, a total of 2×105 cells/well were plated in 6-well tissue-culture plates. After 24 h of incubation, the cells were incubated with different concentrations of DMSA/Ag2S QDs (5-2000 µg/mL) for an additional 24 h at 37°C. A positive control (50 µM H2O2) was also included in the experiments. The cells were embedded in agarose gel and lysed. Fragmented DNA strands were then drawn out by electrophoresis to form a comet. After electrophoresis, the slides were neutralized and then incubated in 50%, 75%, and 98% alcohol for 5 min. The dried microscopic slides were stained with EtBr (20 µg/mL in distilled water, 60 µL/slide) and were examined with a Leica® fluorescence microscope under green light.
The microscope was connected to a charge-coupled device camera and a personal computer-based analysis system (Comet Analysis Software, version 3.0, Kinetic Imaging Ltd, Liverpool, UK) to determine the extent of DNA damage after electrophoretic migration of the DNA fragments in the agarose gel. In order to visualize DNA damage, 100 nuclei per slide were examined at 400× magnification. The results were expressed as the percent of DNA in the tail, “tail intensity”. The experiment was performed in duplicate and repeated three times.
Determination of apoptotic genes by RT-PCR
V79 cells were treated with DMSA/Ag2S QDs at concentrations of 125, 250, 500, and 1000 µg/mL in 6-well plates for 24 h. After the completion of the exposure time, total RNA was extracted with a Qiagen RNeasy Plus Mini Kit (Valencia, CA, USA) according to the manufacturer’s protocol. The RNA content was estimated using a Nanodrop 8000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA), and the integrity of RNA was visualized on 1% agarose gel using a gel documentation system (Thermo Fisher Scientific, Wilmington, DE, USA). First-strand cDNA was synthesized using an RT2 First Strand Kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions. Quantitative RT-PCR was performed by QuantiTect SYBR Green PCR kit (Qiagen) using a Corbett RotorGene Sequence Detection System (Thermo Fisher Scientific, Wilmington, DE, USA). Two microliters of template cDNA was added to the final volume of 20 µL of reaction mixture. The RT-PCR cycle parameters included 10 min at 95°C followed by 40 cycles involving denaturation at 95°C for 15 s, annealing at 60°C for 20 s, and elongation at 72°C for 20 s. The sequences of the specific sets of primer for p53, caspase-3, caspase-9, Bax, Bcl2, and survivin utilized in the present investigation are given in our previous study.45 Expressions of selected genes were normalized to the gapdh gene and then used as controls. The experiment was performed in duplicate and repeated three times.
Statistical analysis
Statistical analysis was performed with SPSS for Windows 20.0 for the alkaline comet assay. Differences between the means of data were compared by one-way variance analysis and post hoc analysis of group differences by the least significant difference test. The RT-PCR array was analyzed by t-test. Significance in the RT-PCR array was determined based on the fold change from the control ΔΔCt value. The results were expressed as the mean ± standard deviation. A p value of less than 0.05 was considered statistically significant.
RESULTS
Cytotoxicity of DMSA/Ag2S QDs by MTT assay
The V79 cells were treated with DMSA/Ag2S QDs and free DMSA to determine the cytotoxicity of the QDs itself and the coating material over a wide range of concentrations between 0 and 2000 µg/mL for 24 h. The cytotoxicity was then evaluated by MTT assay. The data provided in Figure 2a exhibited no significant cytotoxicity between 5 and 200 µg/mL and a concentration-dependent decline in the survival of cells exposed to DMSA/Ag2S QDs at higher concentrations (400-2000 µg/mL) when compared to the untreated control. IC50 of DMSA/Ag2S QDs was not determined at these concentrations. Cell viability was 54.0±4.8% at the highest doses (2000 µg/mL). As shown in Figure 2b, free DMSA did not cause any significant cytotoxicity in V79 cells within the same concentration range.
Figure 2.

Influence of DMSA/Ag2S QDs (a) and free DMSA solutions (b) on viability of V79 cells using the MTT assay. Cell viability was plotted as percent of negative control (assuming data obtained from untreated cells as 100%). Results were given as the mean ± standard deviation. Differences between the means of data were compared by one-way analysis of variance and post hoc analysis of group differences by least significant difference test. *Significant difference as compared to the negative control (p<0.05). Negative control (1% PBS), positive control (50 μM H2O2). The cell viability of the positive control was 48.5%
Ag2S: Silver sulfide, QDs: Quantum dots, DMSA: Meso-2,3-dimercaptosuccinic acid, MTT: 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide, PBS: Phosphate buffered saline
Cytotoxicity of DMSA/Ag2S QDs by NRU assay
The results for cytotoxicity as evaluated by NRU cell viability indicated no significant cytotoxicity at concentrations between 5 and 400 µg/mL when compared to the untreated control, but a clear dose-dependent toxicity at higher concentrations (800-2000 µg/mL) was observed (Figure 3a). IC50 of DMSA/Ag2S was not determined. Cell viability was 65.7±4.1% at the highest dose (2000 µg/mL) of Ag2S QDs. Similar to the results obtained from the MTT assay, DMSA alone did not show cytotoxicity in V79 cells with the same studied doses (Figure 3b).
Figure 3.
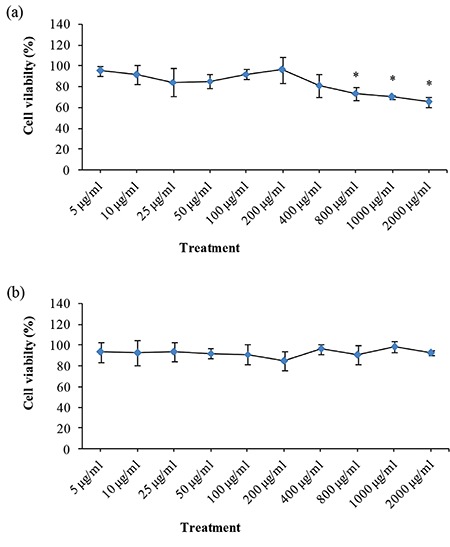
Effects of DMSA/Ag2S QDs (a) and DMSA solutions (b) on viability of V79 cells using the NRU assay. Cell viability was plotted as percent of negative control (assuming data obtained from untreated cells as 100%). Results were given as the mean ± standard deviation. Differences between the means of data were compared by one-way analysis of variance and post hoc analysis of group differences by least significant difference test *Significant difference as compared to the negative control (p<0.05). Negative control (1% PBS), positive control (50 μM H2O2). The cell viability of the positive control was 53.6%
Ag2S: Silver sulfide, QDs: Quantum dots, DMSA: Meso-2,3-dimercaptosuccinic acid, NRU: Neutral red uptake, PBS: Phosphate buffered saline
Genotoxicity of DMSA/Ag2S QDs
Genotoxicity of these QDs was evaluated by comet assay (Figures 4 and 5). DNA damage, expressed as “DNA tail intensity” in V79 cells, is presented in Figure 4. No significant DNA damage was observed, since DMSA/Ag2S QDs treatments (5-2000 µg/mL) for 24 h did not change DNA tail intensity in V79 cells (Figure 5).
Figure 4.
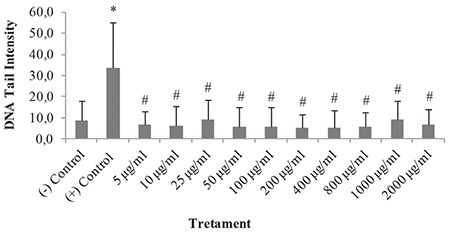
DNA damage expressed as tail intensity in the V79 cells treated with DMSA/Ag2S QDs. Results were given as the mean ± standard deviation. Differences between the means of data were compared by one-way analysis of variance and post hoc analysis of group differences by least significant difference test. *p<0.05, significantly different from the negative control. #p<0.05, significantly different from the positive control. Negative control (1% PBS), positive control (50 μM H2O2)
Ag2S: Silver sulfide, QDs: Quantum dots, DMSA: Meso-2,3-dimercaptosuccinic acid, PBS: Phosphate buffered saline
Figure 5.
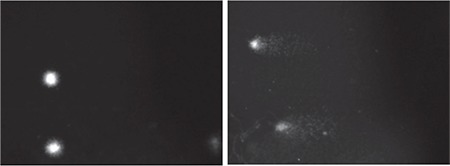
The comet microscopic images of V79 cells. (a) Undamaged cells treated with DMSA/Ag2S QDs and (b) damaged cells treated with 50 μM H2O2 were examined at 400× magnification
Ag2S: Silver sulfide, QDs: Quantum dots, DMSA: Meso-2,3-dimercaptosuccinic acid
Effects of DMSA/Ag2S QDs on the expressions of apoptotic genes
The mRNA expression levels of p53, caspase-3, caspase-9, Bax, Bcl-2, and survivin genes (apoptotic markers) in V79 cells treated with DMSA/Ag2S QDs at concentrations of 125, 250, 500, and 1000 µg/mL for 24 h was analyzed by RT-PCR assay.
The results demonstrated that the mRNA expression levels of apoptotic genes p53, caspase-3, caspase-9, and Bax were up-regulated, while the expressions of anti-apoptotic genes Bcl-2 and survivin were down-regulated in V79 cells treated with the highest concentration of DMSA/Ag2S QDs (1000 µg/mL) (p<0.05) (Figure 6). No significant changes were observed in lower concentrations. The ratio of Bax/Bcl-2 gene expression levels in the cells treated with DMSA/Ag2S QDs (Figure 7) suggests that these two genes may play a significant role in the pathway of DMSA/Ag2S QDs via apoptosis.
Figure 6.
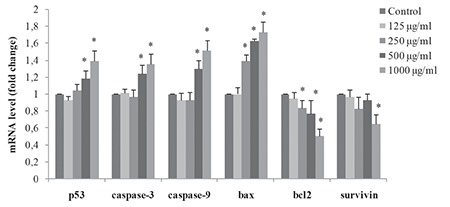
DMSA/Ag2S QDs-induced apoptosis in V79 cells. Cells were exposed to DMSA/Ag2S QDs at the dosages of 0, 125, 250, 500, and 1000 μg/mL for 24 h. At the end of exposure, mRNA levels of the p53, caspase-3, caspase-9, Bax, Bcl2, and survivin genes were measured as described in the Materials and Methods. Results were given as the mean ± standard deviation. The real-time polymerase chain reaction (RT PCR) arrays were analyzed by t-test. Significance in the PCR array was determined based on fold change from the control ΔΔCt value. *Significant difference as compared to the negative control (p<0.05). Negative control (1% PBS)
Ag2S: Silver sulfide, QDs: Quantum dots, DMSA: Meso-2,3-dimercaptosuccinic acid, PCR: Polymerase chain reaction, PBS: Phosphate buffered saline
Figure 7.
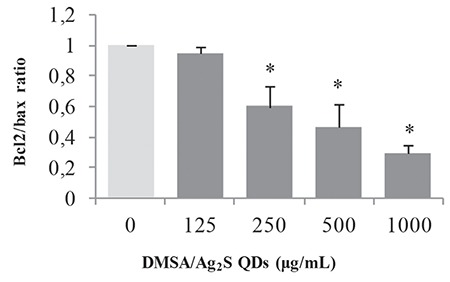
The ratio of Bcl2/Bax mRNA in V79 cells. Cells were exposed to DMSA/Ag2S QDs at the dosages of 0, 125, 250, 500, and 1000 μg/mL for 24 h. *Significant difference as compared to the negative control (p<0.05). Negative control (1% PBS)
Ag2S: Silver sulfide, QDs: Quantum dots, DMSA: Meso-2,3-dimercaptosuccinic acid, PBS: Phosphate buffered saline
DISCUSSION
There has been increasing concern regarding the toxicity of QDs, but further effort is needed to make them safe for biomedical application.46 The toxic effects of different QDs have already been investigated in vitro34,47,48,49,50,51 as well as in vivo.50,52 QDs are suggested to be cytotoxic and/or to change gene expression53 and the cores and coatings of QDs may be responsible for their toxicity.54 Ag2S QDs were considered to be much less toxic than QDs such as PbSe, PbS, and CdHgTe QDs, because of the lack of toxic metals, such as Pb, Hg, and Cd. Ag2S QDs are promising fluorescent probes with both bright photoluminescence in the NIR and high biocompatibility, making them highly selective in in vitro targeting and imaging of different cell lines.55 Ag2S QDs are reported to have no significant effects in altering cell viability, triggering apoptosis or necrosis, forming reactive oxygen species (ROS), or causing DNA damage in in vitro toxicity studies.38,55
In recent years NP applications towards cell apoptosis have been an increasing focus. Unfortunately, such wide use may pose an unwanted threat to human health and so there is a need for a precise analysis of NP cytotoxicity in living cells. An understanding of the exact role their properties (size, shapes, surface charges, dispersion/agglomeration status) play in the decision about NP safety and suitability is necessary. In addition, some aspects of surface modification may be able to reduce the bioreactivity of NPs, thus alleviating their toxicities in certain circumstances. This may provide a way to design even more effective particles of minimum undesired toxicity.
In the present study, it was aimed to evaluate the cytotoxic, genotoxic, and apoptotic potentials of DMSA/Ag2S QDs in the V79 cell line. We performed MTT and NRU cytotoxicity assays, since they are generally used tests to determine the cytotoxicity of NPs in different cell lines.56,57,58,59 These assays differ depending on the different mechanisms leading to cell death. Therefore, it is important to check nanotoxicity with different protocols. The NRU assay is a colorimetric assay measuring the uptake of dye by viable cells and its accumulation in functional lysosomes, while the MTT assay is based on the enzymatic conversion of MTT in the mitochondria.60 The lung fibroblast V79 cell line was used in our experiment. The rationale for choosing this cell line is that it has been widely studied in many nanocytotoxicity and nanogenotoxicity assays, because of its excellent properties in colony formation and also its high sensitivity to many chemicals.61,62,63,64,65 The question of dose becomes important when comparing studies and when developing predictive models of nanoparticle toxicity. This is very important when comparing in vitro and in vivo studies, where physicochemical parameters make simple comparisons difficult. Consistent with the previous studies,61,62,63,64,65 24 h of exposure was selected to be the optimal time for measurements of the effects of NPs on cell viability. It has been reported that rather high concentrations of NM solutions are used in in vitro studies (30 to 400 µg/mL) in the literature.66 There are no cytotoxicity studies for the doses of DMSA/Ag2S in V79 cells, and therefore we used wide concentration ranges of DMSA/Ag2S QDs (0-2000 µg/mL).
In our study, DMSA/Ag2S QDs reduced cell viability above 400 µg/mL using the MTT assay and above 800 µg/mL using the NRU assay, indicating dose-dependent toxicity in both assays. MTT seems to be more sensitive in detecting changes in viability at low concentrations.67 In both the MTT and NRU assays, DMSA alone did not significantly induce cell death in the same concentration range between 5 and 2000 µg/mL. It seems that the coating material may prevent cytotoxicity. The biocompatibility of DMSA coupled with the extremely low solubility of Ag2S core preventing release of high concentration of Ag+ from the core accounts for the biocompatibility of DMSA/Ag2S at least in short-term exposure. Munari et al.54 reported that methyl polyethylene glycol-coated Ag2S (0.01-50 µg/mL) showed neither genotoxic nor cytotoxic effects.
It is important to use the appropriate method to measure the cytotoxicity of interest without false-negative or -positive misconstruction of the result. The MTT and NRU assays may sometimes suffer from severe interferences caused by interaction of metallic NPs with assay reagents. Serious consideration is critical to obtain reliable and realistic data.68 Interference with analytical techniques should be considered in terms of NP intrinsic fluorescence/absorbance and interactions between NPs and assay components. Due to the unique physicochemical properties and increased reactivity of NPs, there is a high potential for these materials to interfere with spectrophotometric and spectrofluorimetric assays. NPs can bind to proteins and dyes and alter their structure and/or function, and it is probable that this process occurs in common toxicity assays. Aluminum NPs showed a strong interaction with the MTT dye, causing significant misreading of the cell viability data.69,70 Some NPs (iron/graphite magnetic particles, super-paramagnetic magnetite/silica NPs, bare and PEGylated silica NPs, and magnetic composites magnetite/FAU zeolite) in culture medium in the absence of cells have the same wavelength used in MTT assays at 525 nm. This absorbance increases with the NP concentration and can greatly interfere with MTT assay results.71 However, in our study DMSA/Ag2S QDs had the emission maximum at 870 nm with broad absorption up to 800 nm. In the MTT and NRU assays the absorbance was 570 nm and 540 nm, respectively. DMSA/Ag2S QDs appear not to interact with MTT reagent, and therefore there is no absorbance interference.
The comet assay is a sensitive method to detect DNA strand breaks as well as oxidatively damaged DNA at single cell level. The effect of NPs to cause DNA damage is an important issue in mutations and carcinogenesis. Oxidative stress but also other mechanisms may also be involved in the genotoxicity of NPs, including direct NP–DNA interactions and disturbance of the mitotic spindle and its components.72,73 In our study, DMSA/Ag2S QD treatments (5-2000 µg/mL) for 24 h did not increase DNA tail intensity in V79 cells, which may indicate no genotoxic effects. The biocompatibility of Ag2S QDs in the mouse fibroblast L929 cell line, including cell proliferation, cell apoptosis/necrosis, production of ROS, and DNA damage using the comet assay, was investigated by Zhang et al.55 in a study comparable with ours. They used different Ag2S QDs with different targeting ligands including dihydrolipoic acid and poly(ethylene glycol) (PEG). The proliferation, ROS production, and DNA damage of L929 cells treated with 6.25, 12.5, 25, 50, and 100 µg/mL Ag2S QDs for 72 h were not significantly different from those of the negative control. The results presenting negligible toxicity of Ag2S QDs at concentrations up to 100 µg/mL show that Ag2S QDs are highly biocompatible in their study. Ag2S QDs did not interfere with cell proliferation, which makes them suitable for use in the labeling of in vitro systems. These observations illustrated the biocompatible nature of Ag2S without side effects on cell proliferation. Previous studies confirmed that some QDs have high biocompatibilities and low toxicities.74,75,76 The coating material may be suggested to reduce cytotoxicity. Consistent with our study, Jebali et al.77 (2014) reported that free fatty acids-coated Ag NPs had less toxicity, higher uptake, and less ROS generation than unbound Ag NPs. Hocaoglu et al.78 showed the biocompatibility of 2-mercaptopropionic acid/Ag2S QDs even at the highest concentration of 600 µg/mL in NIH/3T3 cells after 24 h incubation using the XTT assay. Hocaoglu et al.38 also showed that DMSA/Ag2S QDs did not reduce cell viability up to 200 µg/mL in HeLa cells and showed only 20% reduction in cell viability of 3T3 NIH cells over 24 h.
Apoptosis, via extracellular or intracellular signals, triggers the onset of a signaling cascade with characteristic biochemical and cytological signatures with nuclear condensation and DNA fragmentation.79 Several genes are known to sense DNA damage and apoptosis. In the presence of DNA damage or cellular stress, the p53 protein triggers cell-cycle arrest to provide time for the damage to be repaired or for self-mediated apoptosis.16 The p53 gene maintains genomic stability via activating cell cycle checkpoints, DNA repair, and apoptosis.80 Survivin, described as an inhibitor of caspase-9 and a member of the family of inhibitors of apoptotic proteins, functions as a key regulator of mitosis and programmed cell death. Survivin has been reported to play an important role in both cell proliferation and apoptosis.17 Initially, survivin gene expression is transcriptionally repressed by wild-type p53 and can be deregulated in cancer by several mechanisms, including gene amplification, hypomethylation, increased promoter activity, and loss of p53 function.81 Downregulation of survivin may cause a cell-cycle defect that leads to apoptosis. The Bax and Bcl-2 proteins regulate apoptotic pathways. The Bcl-2 protein has an antiapoptotic activity, while Bax has a pro-apoptotic effect.18 The ratio of Bax/Bcl-2 proteins represents a cell death switch, which determines the life or death of cells in response to an apoptotic stimulus; an increased Bax/Bcl-2 ratio decreases the cellular resistance to apoptotic stimuli, leading to apoptosis. It is crucial in mitochondrial outer-membrane permeabilization and the release of cytochrome C in the cytosol.19,82,83 Moreover, destabilization of mitochondrial integrity by apoptotic stimuli precedes activation of caspases, leading to apoptosis.84,85 Caspases, essential in cellular DNA damage and apoptosis, are known to play a vital role in both the initiation and execution of apoptosis in many cells.86
The transcriptional data on modulation of p53 and Bax/Bcl-2 ratio and release of caspases have strengthened the role of DMSA/Ag2S QDs in inducing mitochondrial dependent apoptotic pathways. The main intrinsic pathway is characterized by mitochondrial dysfunction, with the release of cytochrome c activation of caspase-9, and subsequently of caspase-3 enzyme.87,88 Typically, p53 is activated when DNA damage occurs or cells are stressed; p53 is then translocated to the nucleus, where it can induce pro-apoptotic gene expression on the mitochondrial membrane, activate the effector caspases, and accelerate cell death.88,89 Survivin inhibition induces the activation of caspase-3 and caspase-9 enzymes.89,90,91 Taken together, up-regulation of p53 and down-regulation of survivin lead to activation of pro-apoptotic members of the Bcl-2 family. This includes Bax, inducing permeabilization of the outer mitochondrial membrane, which releases soluble proteins from the intermembrane space into the cytosol, where they promote caspase activation.85,92 The expression of antiapototic protein Bcl-2 was significantly lower, and the expression of pro-apoptotic protein Bax was significantly higher in cells exposed DMSA/Ag2S QDs, suggesting that these genes could be excellent molecular biomarkers to assess the apoptotic response of NPs. In our study, no significant changes in mRNA expression levels were observed between 125 and 500 µg/mL, but a clear effect on apoptotic/antiapoptotic gene expression levels was detected at the dose of 1000 µg/mL. The mRNA expression levels of apoptotic genes p53, caspase-3, caspase-9, and Bax were up-regulated, while the expressions of anti-apoptotic genes Bcl-2 and survivin were down-regulated in V79 cells treated with the highest concentration of 1000 µg/mL of DMSA/Ag2S QDs. The results show that the related gene expression levels may change only at a very high cytotoxic dose, indicating that DMSA/Ag2S QDs may lead to cell death via apoptotic pathways at very high doses.
CONCLUSIONS
In our study, the potential cytotoxic, genotoxic, and apoptotic effects of DMSA/Ag2S QDs in vitro were evaluated. Ag2S QDs coated with DMSA had high biocompatibility and low toxicity, since heavy metal-related cytotoxicity was eliminated by using quite a biocompatible and insoluble Ag2S semiconductor core.
Our data show that DMSA/Ag2S QDs have neither cytotoxic nor genotoxic effects in V79 cells in medically relevant doses. They may induce apoptosis via p53, survivin, Bax/Bcl-2, and caspase pathways at high dose. The underlying mechanisms of DMSA/Ag2S QDs should be confirmed by additional experiments in order to prove our results. Further investigation is needed to determine whether in vivo exposure consequences may exist for DMSA/Ag2S QDs application and also to make QDs safe for widespread use.
Acknowledgments
This work was supported by TÜBİTAK (Project Number: 114S861).
Footnotes
Conflict of Interest: No conflict of interest was declared by the authors.
References
- 1.Rothenfluh DA, Bermudez H, O’Neil CP, Hubbell JA. Biofunctional polymer nanoparticles for intra-articular targeting and retention in cartilage. Nat Mater. 2008;7:248–254. doi: 10.1038/nmat2116. [DOI] [PubMed] [Google Scholar]
- 2.Kostarelos K, Bianco A, Prato M. Promises, facts and challenges for carbon nanotubes in imaging and therapeutics. Nat Nanotechnol. 2009;4:627–633. doi: 10.1038/nnano.2009.241. [DOI] [PubMed] [Google Scholar]
- 3.Michalet X, Pinaud FF, Bentolila LA, Tsay JM, Doose S, Li JJ, Sundaresan G, Wu AM, Gambhir SS, Weiss S. Quantum dots for live cells, in vivo imaging, and diagnostics. Science. 2005;307:538–544. doi: 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Z, Tabakman SM, Goodwin AP, Kattah MG, Daranciang D, Wang X, Zhang G, Li X, Liu Z, Utz PJ, Jiang K, Fan S, Dai H. Protein microarrays with carbon nanotubes as multicolor Raman labels. Nat Biotechnol. 2008;26:1285–1292. doi: 10.1038/nbt.1501. [DOI] [PubMed] [Google Scholar]
- 5.Qian X, Peng XH, Ansari DO, Yin-Goen Q, Chen GZ, Shin DM, Yang L, Young AN, Wang MD, Nie S. In vivo tumor targeting and spectroscopic detection with surface-enhanced Raman nanoparticle tags. Nat Biotechnol. 2008;26:83–90. doi: 10.1038/nbt1377. [DOI] [PubMed] [Google Scholar]
- 6.He Y, Fan C, Lee ST. Silicon nanostructures for bioapplications. Nano Today. 2010;5:282–295. [Google Scholar]
- 7.McAuliffe ME, Perry MJ. Are nanoparticles potential male reproductive toxicants? A literature review. Nanotoxicology. 2007;1:204–210. [Google Scholar]
- 8.Nel A, Xia T, Mädler L, Li N. Toxic potential of materials at the nanolevel. Science. 2006;311:622–627. doi: 10.1126/science.1114397. [DOI] [PubMed] [Google Scholar]
- 9.Oberdörster G, Oberdörster E, Oberdörster J. Nanotoxicology: An emerging discipline evolving from studies of ultrafine particles. Environ Health Perspect. 2005;113:823–839. doi: 10.1289/ehp.7339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singh SK, Kulkarni PP, Dash D. Biomedical Applications of Nanomaterials: An Overview. Bio Nanotechnology: A Revolution in Food. Biomedical and Health Sciences. 2013:1–32. [Google Scholar]
- 11.Chan WC, Maxwell DJ, Gao X, Bailey RE, Han M, Nie S. Luminescent quantum dots for multiplexed biological detection and imaging. Curr Opin Biotechnol. 2002;13:40–46. doi: 10.1016/s0958-1669(02)00282-3. [DOI] [PubMed] [Google Scholar]
- 12.Klimov VI. Spectral and dynamical properties of multiexcitons in semiconductor nanocrystals. Annu Rev Phys Chem. 2007;58:635–673. doi: 10.1146/annurev.physchem.58.032806.104537. [DOI] [PubMed] [Google Scholar]
- 13.Murphy CJ, Coffer JL. Quantum dots: A primer. Appl Spectrosc. 2002;56:16–27. [Google Scholar]
- 14.Bruchez Jr M, Moronne M, Gin P, Weiss S, Alivisatos AP. Semiconductor nanocrystals as fluorescent biological labels. Science. 1998;281:2013–2016. doi: 10.1126/science.281.5385.2013. [DOI] [PubMed] [Google Scholar]
- 15.Chan WCW, Nie S. Quantum dot bioconjugates for ultrasensitive nonisotopic detection. Science. 1998;281:2016–2018. doi: 10.1126/science.281.5385.2016. [DOI] [PubMed] [Google Scholar]
- 16.Medintz I. Universal tools for biomolecular attachment to surfaces. Nat Mater. 2006;5:842. doi: 10.1038/nmat1776. [DOI] [PubMed] [Google Scholar]
- 17.Wagner MK, Li F, Li J, Li XF, Le XC. Use of quantum dots in the development of assays for cancer biomarkers. Anal Bioanal Chem. 2010;397:3213–3224. doi: 10.1007/s00216-010-3847-9. [DOI] [PubMed] [Google Scholar]
- 18.Mattoussi H, Palui G, Na HB. Luminescent quantum dots as platforms for probing in vitro and in vivo biological processes. Adv Drug Deliv. Rev.2012;64:138–166. doi: 10.1016/j.addr.2011.09.011. [DOI] [PubMed] [Google Scholar]
- 19.Frigerio C, Ribeiro DS, Rodrigues SS, Abreu VL, Barbosa JA, Prior JA, Marques KL, Santos JL. Application of quantum dots as analytical tools in automated chemical analysis: A review. Anal Chim Acta. 2012;735:9–22. doi: 10.1016/j.aca.2012.04.042. [DOI] [PubMed] [Google Scholar]
- 20.Frasco MF, Chaniotakis N. Bioconjugated quantum dots as fluorescent probes for bioanalytical applications. Anal Bioanal Chem. 2010;396:229–240. doi: 10.1007/s00216-009-3033-0. [DOI] [PubMed] [Google Scholar]
- 21.Wu X, Liu H, Liu J, Haley KN, Treadway JA, Larson JP, Ge N, Peale F, Bruchez MP. Immunofluorescent labeling of cancer marker Her2 and other cellular targets with semiconductor quantum dots. Nat Biotechnol. 2003;21:41–46. doi: 10.1038/nbt764. [DOI] [PubMed] [Google Scholar]
- 22.Algar WR, Prasuhn DE, Stewart MH, Jennings TL, Blanco-Canosa JB, Dawson PE, Medintz IL. The controlled display of biomolecules on nanoparticles: a challenge suited to bioorthogonal chemistry. Bioconjug Chem. 2011;22:825–858. doi: 10.1021/bc200065z. [DOI] [PubMed] [Google Scholar]
- 23.Rosenthal SJ, Chang JC, Kovtun O, McBride JR, Tomlinson ID. Biocompatible quantum dots for biological applications. Chem Biol. 2011;18:10–24. doi: 10.1016/j.chembiol.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petryayeva E, Algar WR, Medintz IL. Quantum dots in bioanalysis: A review of applications across various platforms for fluorescence spectroscopy and imaging. Appl Spectrosc. 2013;67:215–252. doi: 10.1366/12-06948. [DOI] [PubMed] [Google Scholar]
- 25.Alivisatos AP. Semiconductor clusters, nanocrystals, and quantum dots. Science. 1996;271:933–937. [Google Scholar]
- 26.Weller H. Quantum size colloids: From size-dependent properties of discrete particles to self-organized superstructures. Curr Opin Colloid Interface Sci. 1998;3:194–199. [Google Scholar]
- 27.Dabbousi BO, Rodriguez-Viejo J, Mikulec FV, Heine JR, Mattoussi H, Ober R, Jensen KF, Bawendi MG. (CdSe)ZnS core-shell quantum dots: Synthesis and characterization of a size series of highly luminescent nanocrystallites. J Phys Chem B. 1997;101:9463–9475. [Google Scholar]
- 28.Hines MA, Guyot-Sionnest P. Synthesis and characterization of strongly luminescing ZnS-capped CdSe nanocrystals. J Phy Chem. 1996;100:468–471. [Google Scholar]
- 29.Hardman R. A toxicologic review of quantum dots: Toxicity depends on physicochemical and environmental factors. Environ Health Perspect. 2006;114:165–172. doi: 10.1289/ehp.8284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim S, Lim YT, Soltesz EG, De Grand AM, Lee J, Nakayama A, Parker JA, Mihaljevic T, Laurence RG, Dor DM, Cohn LH, Bawendi MG, Frangioni JV. Near-infrared fluorescent type II quantum dots for sentinel lymph node mapping. Nat Biotechnol. 2004;22:93–97. doi: 10.1038/nbt920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chan WH, Shiao NH, Lu PZ. CdSe quantum dots induce apoptosis in human neuroblastoma cells via mitochondrial-dependent pathways and inhibition of survival signals. Toxicol Let. 2006;167:191–200. doi: 10.1016/j.toxlet.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 32.Chen N, He Y, Su Y, Li X, Huang Q, Wang H, Zhang X, Tai R, Fan C. The cytotoxicity of cadmium-based quantum dots. Biomaterials. 2012;33:1238–1244. doi: 10.1016/j.biomaterials.2011.10.070. [DOI] [PubMed] [Google Scholar]
- 33.Cho SJ, Maysinger D, Jain M, Röder B, Hackbarth S, Winnik FM. Long-term exposure to CdTe quantum dots causes functional impairments in live cells. Langmuir. 2007;23:1974–1980. doi: 10.1021/la060093j. [DOI] [PubMed] [Google Scholar]
- 34.Kirchner C, Liedl T, Kudera S, Pellegrino T, Muñoz Javier A, Gaub HE, Stölzle S, Fertig N, Parak WJ. Cytotoxicity of colloidal CdSe and CdSe/ZnS nanoparticles. Nano Lett. 2005;5:331–338. doi: 10.1021/nl047996m. [DOI] [PubMed] [Google Scholar]
- 35.Li KG, Chen JT, Bai SS, Wen X, Song SY, Yu Q, Li J, Wang YQ. Intracellular oxidative stress and cadmium ions release induce cytotoxicity of unmodified cadmium sulfide quantum dots. Toxicol In Vitro. 2009;23:1007–1013. doi: 10.1016/j.tiv.2009.06.020. [DOI] [PubMed] [Google Scholar]
- 36.Male KB, Lachance B, Hrapovic S, Sunahara G, Luong JHT. Assessment of cytotoxicity of quantum dots and gold nanoparticles using cell-based impedance spectroscopy. Anal Chem. 2008;80:5487–5493. doi: 10.1021/ac8004555. [DOI] [PubMed] [Google Scholar]
- 37.Gao J, Chen X, Cheng Z. Near-infrared quantum dots as optical probes for tumor imaging. Curr Top Med Chem. 2010;10:1147–1157. doi: 10.2174/156802610791384162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hocaoglu I, Demir F, Birer O, Kiraz A, Sevrin C, Grandfils C, Acar HY. Emission tunable, cyto/hemocompatible, near-IR-emitting Ag2S quantum dots by aqueous decomposition of DMSA. Nanoscale. 2014;6:11921–11931. doi: 10.1039/c4nr02935f. [DOI] [PubMed] [Google Scholar]
- 39.Rao BS, Shanbhoge R, Rao BN, Adiga SK, Upadhya D, Aithal BK, Kumar MR. Preventive efficacy of hydroalcoholic extract of Cymbopogon citratus against radiation-induced DNA damage on V79 cells and free radical scavenging ability against radicals generated in vitro. Hum Exp Toxicol. 2009;28:195–202. doi: 10.1177/0960327109104822. [DOI] [PubMed] [Google Scholar]
- 40.Mosmann T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 41.Hansen MB, Nielsen SE, Berg K. Re-examination and further development of a precise and rapid dye method for measuring cell growth/cell kill. J Immunol Methods. 1989;119:203–210. doi: 10.1016/0022-1759(89)90397-9. [DOI] [PubMed] [Google Scholar]
- 42.Kuźma Ł, Wysokińska H, Rózalski M, Krajewska U, Kisiel W. An unusual taxodione derivative from hairy roots of Salvia austriaca. Fitoterapia. 2012;83:770–773. doi: 10.1016/j.fitote.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 43.Di Virgilio AL, Iwami K, Wätjen W, Kahl R, Degen GH. Genotoxicity of the isoflavones genistein, daidzein and equol in V79 cells. Toxicol Lett. 2004;151:1511–1562. doi: 10.1016/j.toxlet.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 44.Saquib Q, Al-Khedhairy AA, Siddiqui MA, Abou-Tarboush FM, Azam A, Musarrat J. Titanium dioxide nanoparticles induced cytotoxicity, oxidative stress and DNA damage in human amnion epithelial (WISH) cells. Toxicol In Vitro. 2012;26:351–561. doi: 10.1016/j.tiv.2011.12.011. [DOI] [PubMed] [Google Scholar]
- 45.Ahamed M, Akhtar MJ, Siddiqui MA, Ahmad J, Musarrat J, Al-Khedhairy AA, AlSalhi MS, Alrokayan SA. Oxidative stress mediated apoptosis induced by nickel ferrite nanoparticles in cultured A549 cells. Toxicology. 2011;283:101–108. doi: 10.1016/j.tox.2011.02.010. [DOI] [PubMed] [Google Scholar]
- 46.Winnik FM, Maysinger D. Quantum dot cytotoxicity and ways to reduce it. Acc Chem Res. 2013;46:672–680. doi: 10.1021/ar3000585. [DOI] [PubMed] [Google Scholar]
- 47.Manshian BB, Soenen SJ, Brown A, Hondow N, Wills J, Jenkins GJ, Doak SH. Genotoxic capacity of Cd/Se semiconductor quantum dots with differing surface chemistries. Mutagenesis. 2016;31:97–106. doi: 10.1093/mutage/gev061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith WE, Brownell J, White CC, Afsharinejad Z, Tsai J, Hu X, Polyak SJ, Gao X, Kavanagh TJ, Eaton DL. In vitro toxicity assessment of amphiphillic polymer-coated CdSe/ZnS quantum dots in two human liver cell models. ACS Nano. 2012;6:9475–9484. doi: 10.1021/nn302288r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smulders S, Luyts K, Brabants G, Golanski L, Martens J, Vanoirbeek J, Hoet PH. Toxicity of nanoparticles embedded in paints compared to pristine nanoparticles, in vitro study. Toxicol Lett. 2015;232:333–339. doi: 10.1016/j.toxlet.2014.11.030. [DOI] [PubMed] [Google Scholar]
- 50.Smulders S, Luyts K, Brabants G, Landuyt KV, Kirschhock C, Smolders E, Golanski L, Vanoirbeek J, Hoet PH. Toxicity of nanoparticles embedded in paints compared with pristine nanoparticles in mice. Toxicol Sci. 2014;141:132–140. doi: 10.1093/toxsci/kfu112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Soenen SJ, Manshian BB, Himmelreich U, Demeester J, Braeckmans K, De Smedt SC. The performance of gradient alloy quantum dots in cell labeling. Biomaterials. 2014;35:7249–7258. doi: 10.1016/j.biomaterials.2014.05.023. [DOI] [PubMed] [Google Scholar]
- 52.Derfus AM, Chan WCW, Bhatia SN. Probing the Cytotoxicity of Semiconductor Quantum Dots. Nano Lett. 2004;4:11–18. doi: 10.1021/nl0347334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Choi O, Clevenger TE, Deng B, Surampalli RY, Ross Jr L, Hu Z. Role of sulfide and ligand strength in controlling nanosilver toxicity. Water Res. 2009;43:1879–1886. doi: 10.1016/j.watres.2009.01.029. [DOI] [PubMed] [Google Scholar]
- 54.Munari M, Sturve J, Frenzilli G, Sanders MB, Brunelli A, Marcomini A, Nigro M, Lyons BP. Genotoxic effects of CdS quantum dots and Ag2S nanoparticles in fish cell lines (RTG-2) Mutat Res Genet Toxicol Environ Mutagen. 2014;776:89–93. doi: 10.1016/j.mrgentox.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 55.Zhang Y, Hong G, Zhang Y, Chen G, Li F, Dai H, Wang Q. Ag2S quantum dot: a bright and biocompatible fluorescent nanoprobe in the second near-infrared window. ACS Nano. 2012;6:3695–3702. doi: 10.1021/nn301218z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Akhtar MJ, Kumar S, Murthy RC, Ashquin M, Khan MI, Patil G, Ahmad I. The primary role of iron-mediated lipid peroxidation in the differential cytotoxicity caused by two varieties of talc nanoparticles on A549 cells and lipid peroxidation inhibitory effect exerted by ascorbic acid. Toxicol In Vitro. 2010;24:1139–1147. doi: 10.1016/j.tiv.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 57.Barillet S, Jugan ML, Laye M, Leconte Y, Herlin-Boime N, Reynaud C, Carrière M. In vitro evaluation of SiC nanoparticles impact on A549 pulmonary cells: cyto-, genotoxicity and oxidative stress. Toxicol Lett. 2010;198:324–330. doi: 10.1016/j.toxlet.2010.07.009. [DOI] [PubMed] [Google Scholar]
- 58.Mahmoudi M, Simchi A, Milani AS, Stroeve P. Cell toxicity of superparamagnetic iron oxide nanoparticles. J Colloid Interface Sci. 2009;336:510–508. doi: 10.1016/j.jcis.2009.04.046. [DOI] [PubMed] [Google Scholar]
- 59.Sharma V, Shukla RK, Saxena N, Parmar D, Das M, Dhawan A. DNA damaging potential of zinc oxide nanoparticles in human epidermal cells. Toxicol Lett. 2009;185:211–218. doi: 10.1016/j.toxlet.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 60.Fotakis G, imbrell JA. In vitro cytotoxicity assays: Comparison of LDH, neutral red, MTT and protein assay in hepatoma cell lines following exposure to cadmium chloride. Toxicol Lett. 2006;160:171–177. doi: 10.1016/j.toxlet.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 61.Chaung W, Mi LJ, Boorstein RJ. The p53 status of Chinese hamster V79 cells frequently used for studies on DNA damage and DNA repair. Nucleic Acids Res. 1997;25:992–994. doi: 10.1093/nar/25.5.992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen Z, Wang Y, Ba T, Li Y, Pu J, Chen T, Song Y, Gu Y, Qian Q, Yang J, Jia G. Genotoxic evaluation of titanium dioxide nanoparticles in vivo and in vitro. Toxicol Lett. 2014;226:314–319. doi: 10.1016/j.toxlet.2014.02.020. [DOI] [PubMed] [Google Scholar]
- 63.Darne C, Terzetti F, Coulais C, Fontana C, Binet S, Gaté L, Guichard Y. Cytotoxicity and genotoxicity of panel of single- and multiwalled carbon nanotubes: in vitro effects on normal Syrian hamster embryo and immortalized v79 hamster lung cells. J Toxicol. 2014;2014:872195. doi: 10.1155/2014/872195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guichard Y, Fontana C, Chavinier E, Terzetti F, Gaté L, Binet S, Darne C. Cytotoxic and genotoxic evaluation of different synthetic amorphous silica nanomaterials in the V79 cell line. Toxicol Ind Health. 2016;32:1639–1650. doi: 10.1177/0748233715572562. [DOI] [PubMed] [Google Scholar]
- 65.Kang SJ, Kim BM, Lee YJ, Chung HW. Titanium dioxide nanoparticles trigger p53-mediated damage response in peripheral blood lymphocytes. Environ Mol Mutagen. 2008;49:399–405. doi: 10.1002/em.20399. [DOI] [PubMed] [Google Scholar]
- 66.Gangwal S, Brown JS, Wang A, Houck KA, Dix DJ, Kavlock RJ, Hubal EA. Informing selection of nanomaterial concentrations for ToxCast in vitro testing based on occupational exposure potential. Environ Health Perspect. 2011;119:1539–1546. doi: 10.1289/ehp.1103750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nogueira DR, Mitjans M, Infante MR, Vinardell MP. Comparative sensitivity of tumor and non-tumor cell lines as a reliable approach for in vitro cytotoxicity screening of lysine-based surfactants with potential pharmaceutical applications. Int J Pharm. 2011;420:51–58. doi: 10.1016/j.ijpharm.2011.08.020. [DOI] [PubMed] [Google Scholar]
- 68.Kong B, Seog JH, Graham LM, Lee SB. Experimental considerations on the cytotoxicity of nanoparticles. Nanomedicine (Lond). 2011;6:929–941. doi: 10.2217/nnm.11.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Maccormack TJ, Clark RJ, Dang MK, Ma G, Kelly JA, Veinot JG, Goss GG. Inhibition of enzyme activity by nanomaterials: Potential mechanisms and implications for nanotoxicity testing. Nanotoxicology. 2012;6:514–525. doi: 10.3109/17435390.2011.587904. [DOI] [PubMed] [Google Scholar]
- 70.Monteiro-Riviere NA, Oldenburg SJ, Inman AO. Interactions of aluminum nanoparticles with human epidermal keratinocytes. J Appl Toxicol. 2010;30:276–285. doi: 10.1002/jat.1494. [DOI] [PubMed] [Google Scholar]
- 71.Díaz B, Sánchez-Espinel C, Arruebo M, Faro J, de Miguel E, Magadán S, Yagüe C, Fernández-Pacheco R, Ibarra MR, Santamaría J, González- Fernández A. Assessing Methods for Blood Cell Cytotoxic Responses to Inorganic Nanoparticles and Nanoparticle Aggregates. Small. 2008;4:2025–2034. doi: 10.1002/smll.200800199. [DOI] [PubMed] [Google Scholar]
- 72.Karlsson HL, Di Bucchianico S, Collins AR, Dusinska M. Can the comet assay be used reliably to detect nanoparticle-induced genotoxicity? Environ Mol Mutagen. 2015;56:82–96. doi: 10.1002/em.21933. [DOI] [PubMed] [Google Scholar]
- 73.Shukla RK, Sharma V, Pandey AK, Singh S, Sultana S, Dhawan A. ROS-mediated genotoxicity induced by titanium dioxide nanoparticles in human epidermal cells. Toxicol In Vitro. 2011;25:231–241. doi: 10.1016/j.tiv.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 74.Dong B, Li C, Chen G, Chen G, Zhang Y, Zhang Y, Deng M, Wang Q. Facile Synthesis of Highly Photoluminescent Ag2Se Quantum Dots as a New Fluorescent Probe in the Second Near-Infrared Window for in vivo Imaging. Chem Mater. 2013;25:2503–2509. [Google Scholar]
- 75.Gu Y-P, Cui R, Zhang Z-L, Xie Z-X, Pang D-W. Ultrasmall Near-Infrared Ag2Se Quantum Dots with Tunable Fluorescence for in vivo Imaging. J Am Chem Soc. 2012;134:79–82. doi: 10.1021/ja2089553. [DOI] [PubMed] [Google Scholar]
- 76.Tang H, Yang ST, Yang YF, Ke DM, Liu JH, Chen X, Wang H, Liu Y. Blood Clearance, Distribution, Transformation, Excretion, and Toxicity of Near-Infrared Quantum Dots Ag2Se in Mice. ACS Appl Mater Interfaces. 2016;8:17859–17869. doi: 10.1021/acsami.6b05057. [DOI] [PubMed] [Google Scholar]
- 77.Jebali A, Hekmatimoghaddam S, Kazemi B. The cytotoxicity of silver nanoparticles coated with different free fatty acids on the Balb/c macrophages: an in vitro study. Drug Chem Toxicol. 2014;37:433–439. doi: 10.3109/01480545.2013.878952. [DOI] [PubMed] [Google Scholar]
- 78.Hocaoglu I, Çizmeciyan MN, Erdem R, Ozen C, Kurt A, Sennaroglu A, Acar HY. Development of highly luminescent and cytocompatible near-IR-emitting aqueous Ag2S quantum dots. J Mater Chem. 2012;22:14674–14681. [Google Scholar]
- 79.Gopinath P, Gogoi SK, Sanpui P, Paul A, Chattopadhyay A, Ghosh SS. Signaling gene cascade in silver nanoparticle induced apoptosis. Colloids Surf B Biointerfaces. 2010;77:240–245. doi: 10.1016/j.colsurfb.2010.01.033. [DOI] [PubMed] [Google Scholar]
- 80.Sherr CJ. Principles of Tumor Suppression. Cell. 2004;116:235–46. doi: 10.1016/s0092-8674(03)01075-4. [DOI] [PubMed] [Google Scholar]
- 81.Ryan BM, O’Donovan N, Duffy MJ. Survivin: A new target for anti-cancer therapy. Cancer Treat Rev. 2009;35:553–562. doi: 10.1016/j.ctrv.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 82.Chougule M, Patel AR, Sachdeva P, Jackson T, Singh M. Anticancer activity of Noscapine, an opioid alkaloid in combination with Cisplatin in human non-small cell lung cancer. Lung Cancer. 2011;71:271–282. doi: 10.1016/j.lungcan.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gao C, Wang AY. Significance of Increased Apoptosis and Bax Expression in Human Small Intestinal Adenocarcinoma. J Histochem Cytochem. 2009;57:1139–1148. doi: 10.1369/jhc.2009.954446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Timmer JC, Salvesen GS. Caspase substrates. Cell Death And Differentiation. Cell Death Differ. 2007;14:66–72. doi: 10.1038/sj.cdd.4402059. [DOI] [PubMed] [Google Scholar]
- 85.Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol. 2008;9:47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
- 86.Jänicke RU, Sprengart ML, Wati MR, Porter AG. Caspase-3 is required for DNA fragmentation and morphological changes associated with apoptosis. J Biol Chem. 1998;273:9357–9360. doi: 10.1074/jbc.273.16.9357. [DOI] [PubMed] [Google Scholar]
- 87.Porter AG, Jänicke RU. Emerging roles of caspase-3 in apoptosis. Cell Death Differ. 1999;6:99–104. doi: 10.1038/sj.cdd.4400476. [DOI] [PubMed] [Google Scholar]
- 88.Saquib Q, Al-Khedhairy AA, Ahmad J, Siddiqui MA, Dwivedi S, Khan ST. Zinc ferrite nanoparticles activate IL-1b, NFKB1, CCL21 and NOS2 signaling to induce mitochondrial dependent intrinsic apoptotic pathway in WISH cells. Toxicol Appl Pharmacol. 2013;273:289–297. doi: 10.1016/j.taap.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 89.Farnebo M, Bykov VJ, Wiman KG. The p53 tumor suppressor: A master regulator of diverse cellular processes and therapeutic target in cancer. Biochem Biophys Res Commun. 2010;396:85–89. doi: 10.1016/j.bbrc.2010.02.152. [DOI] [PubMed] [Google Scholar]
- 90.Blanc-Brude OP, Yu J, Simosa H, Conte MS, Sessa WC, Altieri DC. Inhibitor of apoptosis protein survivin regulates vascular injury. Nat Med. 2002;8:987–994. doi: 10.1038/nm750. [DOI] [PubMed] [Google Scholar]
- 91.Marusawa H, Matsuzawa S, Welsh K, Zou H, Armstrong R, Tamm I, Reed JC. HBXIP functions as a cofactor of survivin in apoptosis suppression. EMBO J. 2003;22:2729–2740. doi: 10.1093/emboj/cdg263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fuentes-Prior P, Salvesen Guy GS. The protein structures that shape caspase activity, specificity, activation and inhibition. Biochem J. 2004;384:201–232. doi: 10.1042/BJ20041142. [DOI] [PMC free article] [PubMed] [Google Scholar]


