Abstract
Objectives:
Enterococcus faecalis is the major reason for biofilm-related infections and it also interacts with Staphylococcus aureus in biofilms. Gelatinase (gelE) enzyme is an important virulence factor of E. faecalis for biofilm formation. This study aimed to compare the biofilm producing E. faecalis isolates from urine and urinary catheters. The influence of S. aureus on the growth of E. faecalis biofilm cells was also investigated in a dual biofilm model in vitro. Another aim was to evaluate E. faecalis gelE gene expression during biofilm formation.
Materials and Methods:
Firstly, crystal violet staining was used to measure the total biofilm biomass of the isolates. Secondly, plate counting was performed to determine the biofilm formation ability of E. faecalis isolates and the effect of S. aureus on E. faecalis biofilm formation. Finally, the gelE expression profile of the isolates was assessed by quantitative real time-polymerase chain reaction.
Results:
According to crystal violet staining and plate counting, all E. faecalis isolates were biofilm producers and the number of E. faecalis sessile cells increased in the presence of S. aureus. Among the 21 E. faecalis isolates, ten expressed high levels of the gelE gene, while eight of them had low expression profiles (p<0.05).
Conclusion:
When they grow together, S. aureus may give some advantages to E. faecalis such as increasing sessile cell growth. The expression of the gelE gene was not affected by E. faecalis biofilm formation of the isolates collected from the patients with urinary tract infections.
Keywords: Dual biofilm, E. faecalis, S. aureus, gelatinase, quantitative reverse transcription polymerase chain reaction
INTRODUCTION
Biofilms are defined as biotic or abiotic surface-attached microbial consortia and have multiple stages such as initial reversible attachment; production of an extracellular polymeric matrix (EPM) including proteins, polysaccharides, and nucleic acids; irreversible attachment; etc.1,2 Biofilm formation is an important problem causing failure in antimicrobial treatment because sessile cells in the biofilm are highly resistant to antimicrobial agents. It has been highlighted that 65%-80% of all infections are biofilm-related. Biofilm cells are phenotypically, physiologically, and genotypically different from nonattached (planktonic) cells. Moreover, high concentrations of antimicrobial agents are necessary to kill sessile cells in a mature biofilm vs planktonic cells.3
It has been recently shown that most diseases are caused by polymicrobial communities.4,5,6,7,8 Although some infections are considered predominantly monomicrobial, they may be influenced by other microorganismal associations during active infection.4 The physiology of microbial cells in the biofilm has been frequently changed by these interactions and leads to various advantages being obtained, such as resistance to antimicrobials or the human immune system, metabolic cooperation, quorum sensing systems, and more productive gene sharing.9,10,11,12
Enterococcus species have been recognized as opportunistic pathogens for many nosocomial infections and are natural inhabitants of the human intestinal and oral flora. Enterococcus faecalis is the most common species leading to many infections among the other enterococcus species.13,14 They can readily form biofilms and keep growing on various medical devices’ surfaces such as urinary catheters despite a serious inflammatory response.15 Staphylococcus aureus has become an important cause of hospital-acquired infection associated with indwelling medical devices and surgical wounds. It may cause chronic infections that cannot be treated with antibiotics because of the ineffective host immune response. Moreover, staphylococci have nonspecific resistance mechanisms such as biofilm formation.16,17,18
Changing expression levels of virulence factors of E. faecalis have been shown whether they formed a biofilm or not. Among the virulence factors, the gelatinase (gelE) enzyme is an important factor that hydrolyzes gelatin, casein, and collagen.19 Although there have been many studies on biofilm formation and gelE expression by E. faecalis, it is still not clear how gelE expression levels change in mono- or polymicrobial biofilms.20,21,22
In the present study, we evaluated the biofilm ability of E. faecalis isolates by quantification assays and then we set up an in vitro dual biofilm model in a repeatable style and determined the influence of the presence of S. aureus on the growth of E. faecalis by plating assay. Finally, the gelE gene expression levels of E. faecalis were measured by quantitative real time-polymerase chain reactions (qRT-PCRs).
MATERIALS AND METHODS
Strains used in the study
A total of 20 E. faecalis clinical isolates and a strain as a positive control (E. faecalis ATCC 29212) were used in this study. These isolates were taken from urinary catheter (n=10) and urine samples (n=10) from hospitalized intensive care unit patients admitted to a University Hospital from 2000 to 2011.
For dual biofilm formation, all the E. faecalis isolates and E. faecalis ATCC 29212 were cultured with S. aureus ATCC 29213.
Mono and dual biofilm formation in microtiter plates
Final inoculum suspensions of all clinical E. faecalis strains were adjusted to approximately 106 colony-forming units (CFU) mL-1. Each experiment included the biofilm-forming E. faecalis ATCC 29212 strain as a positive control. For dual species biofilms, E. faecalis isolates were co-cultured with a laboratory strain of S. aureus (106 CFU/mL) and incubated at 37°C without shaking. Sterile tryptic soy broth (TSB) (Becton Dickinson GmbH, Heidelberg, Germany) with 0.25% glucose was used as a blank. For each test condition, 12 wells of a flat-bottomed polystyrene 96-well microtiter plate were inoculated with 100 µL of the final inoculum suspension. After 4 h of incubation at 37°C without shaking, nonadhered cells were removed and rinsed with 100 µL of 0.9% physiological saline (PS), then 100 µL of fresh TSB with 0.25% glucose was added, and the plates were incubated for an additional 20 h for biofilm maturation. After 24 h, the supernatants were removed and each well was rinsed with PS before the sessile cells were quantified.
Quantification of the biofilms
Crystal violet staining
The biomass quantification of E. faecalis biofilms was performed according to an optimized assay.19 After washing with sterile PBS, the wells were stained with 100 µL of a solution of 0.2% crystal violet for 15 min. The stained biofilms were rinsed again three times with PBS to remove excess dye and dried for 15 min at room temperature. The bound dye was solubilized in 150 µL of acetone/ethanol solution. The optical densities (ODs) of the stained adherent cells were read at 570 nm using a micro-ELISA plate reader. We defined the cut-off OD (0.282) as three standard deviations above mean OD of the negative control. Each isolate was tested in 12 wells in each assay and each assay was carried out in duplicate (n=24).
Plate counting
Quantification of the number of cells in mature biofilms was done via plate counting using tryptic soy agar (TSA) medium. Biofilms were detached by vortexing (5 min) followed by sonication (5 min). The sonicated fluids were serially diluted and plated on TSA to determine the number of CFU per mL of the isolates. Bile esculin azide agar was used for plating of E. faecalis isolates in mature dual biofilms that were formed by E. faecalis and S. aureus.
Expression of the gelE gene in planktonic and biofilm cells of E. faecalis
Total RNA was extracted from the mono- and dual-species biofilm cells with the RNeasy® Mini Kit according to the manufacturer’s recommendations (Qiagen GmbH, Germany). All RNA extracts were prepared as 100 ng µL-1 per sample and transcribed into cDNA using a Transcriptor High Fidelity cDNA Synthesis Kit according to the manufacturer’s instructions (Roche Diagnostics GmbH, Germany). RT-PCR (Roche Light Cycler 2.0) was performed with LightCycler Faststart DNA Master SYBR Green1 (Roche Diagnostics GmbH) in a total volume of 20 µL. Primer sequences for the housekeeping gene 16sRNA and gelE were obtained from the literature and are listed in Table 1.17 The 16sRNA gene was used to normalize the expression level of gelE. Melt curve analysis was carried out to assess the specificity of each primer pair. The comparative CT method for relative quantification (∆∆CT method) was performed to analyze the data.23
Table 1. The primers for quantifying the genes of E. faecalis by RT-qPCR.

Statistical analysis
The independent samples t-test was used to compare biofilm cell CFU counts between the two groups (with/without S. aureus biofilms). One-way ANOVA was used to evaluate CFU differences within a group. The CFU counts were log-transformed before the statistical tests. A p value <0.05 was considered significant. For gene expression, the results were analyzed by t-tests and only differences of more than twofold up- or down- regulation and with a p-value <0.05 were considered significant.
RESULTS
Detection of biofilm production by E. faecalis isolates
In total 20 E. faecalis isolates were analyzed to determine the ability of biofilm formation. All of the isolates were found to be biofilm-positive by plate counting and crystal violet staining (Figures 1 and 2). In terms of biofilm-forming ability no statistically significant difference was determined between the isolates from catheters and not from catheters (Figure 2).
Figure 1.
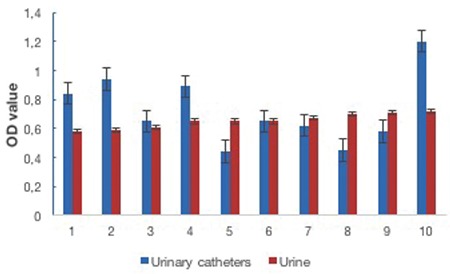
Biofilm forming ability of the Enterococcus faecalis isolates from the urine and urinary catheter samples of hospitalized patients by crystal violet staining assay
aBiofilm formation degrees of the isolates were determined by crystal violet staining assay. Results are means of at least three different experiments, OD: Optical density
Figure 2.
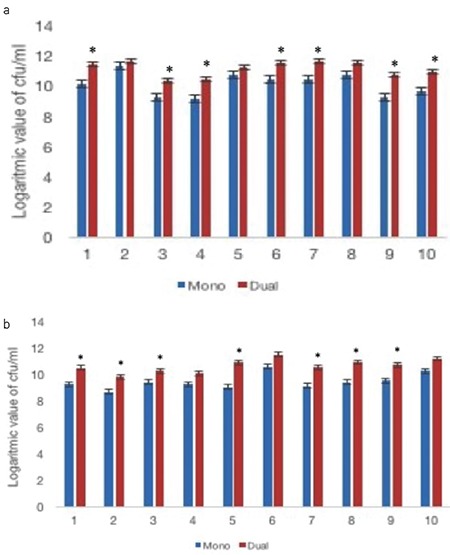
Biofilm forming ability of the Enterococcus faecalis isolates in mono and dual biofilms. Numbers of the sessile cells of the E. faecalis isolates in mono and dual species biofilms were determined by plate counting assay. (a) The results of isolates from the urinary catheter samples, (b) The results of isolates from the urine samples. Results are means of at least three different experiments. Mono: only E. faecalis biofilms, Dual: E. faecalis and S. aureus biofilms, CFU: Colony forming unit,
*: Statistically significant
In the co-culture of E. faecalis with S. aureus, the cell counts of E. faecalis were significantly higher than those in their mono-species biofilm (Figure 2). Our results showed that S. aureus contributed to the growth of E. faecalis biofilm cells by an unknown mechanism.
GelE gene expression in planktonic and biofilm cells of E. faecalis clinical isolates
We used qRT-PCR to compare the expression levels of gelE in planktonic and biofilm cells of 21 E. faecalis isolates (including a positive control) from urine and urinary catheter samples from hospitalized patients and E. faecalis ATCC 29212. According to the results obtained from the mRNA levels of gelE in planktonic and biofilm cells of E. faecalis, 12 of the 21 E. faecalis strains (including the positive control) exhibited increased gelE gene expression; however, only ten of them (including the positive control) were statistically significant (p<0.05). Eight isolates showed significantly decreased expression levels (p<0.05) (Figure 3).
Figure 3.
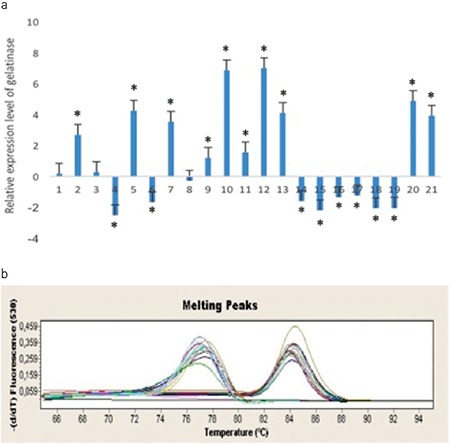
Gelatinase gene relative expression ratios of Enterococcus faecalis ATCC 29212 and clinical isolates in planktonic and biofilm cells.
*: Statistically significant (p value<0.05), (a, b) Melting curves of gelE and 16srRNA (housekeeping gene) of the bacteria, respectively
DISCUSSION
Biofilm-related urinary tract infections represent the main cause of nosocomial infections. Enterococci (especially E. faecalis) and S. aureus are a major challenging problem for treatment of urinary tract infections.24 It is widely known that the presence of bacterial biofilms on the inner or outer surface of the catheter leads to catheter-associated urinary tract infections (CAUTIs).15 The occurrence of CAUTIs, as the most common hospital-acquired infection, has an important economic and clinical impact and is directly related to the majority of uropathogens such as E. faecalis and S. aureus that may form biofilms. In the current study, we assessed the ability of biofilm formation of clinical E. faecalis isolates in alone and co-culture with S. aureus in vitro. Our results indicated that all isolates from inpatients with and without urinary catheters were biofilm positive with regard to the plate counting and crystal violet staining methods (Figures 1 and 2). The starting bacteria concentration was normalized as 6 log10 (106 CFU/mL). However, after the incubation period, the lowest bacteria number in the well plates was found to be 8.7 log10. This result showed that all the mono-species biofilm isolates of E. faecalis attached and grew on the walls of the wells in microtiter plates (Figure 2).
Interspecies interactions in polyspecies biofilm usually provide various advantages for the inhabitant species such as increased tolerance against several antimicrobials and increased virulence in infections.25 Pastar et al.26 showed that the presence of Pseudomonas inhibited the growth of S. aureus in vitro and induced expression of S. aureus virulence factors in polymicrobial wound infection. In another study, the effect of Streptococcus mutans on E. faecalis biofilm formation was investigated and an increase in biofilm formation of E. faecalis by S. mutans was obtained.27 It has been previously shown that the combined effect of C. albicans and E. faecalis in a mouse model resulted in increased growth of enterococci in the animals when C. albicans had been introduced.28 In a P. aeruginosa and C. albicans dual biofilm model, it was observed that P. aeruginosa formed biofilms on the fungal filaments of C. albicans and this close contact caused the killing of the fungal filaments.29 In our study, the number of sessile cells of E. faecalis in dual-species biofilms with S. aureus was significantly higher than in their mono-species biofilm. We concluded that the growth and biofilm formation of the E. faecalis isolates were increased by S. aureus sessile cells. According to the biofilm cell counts between the urine and urinary catheter samples, the counts of E. faecalis isolates from the urinary catheters were greater than those of the isolates from urine.
Many virulence factors have significant roles in the pathogenesis of enterococcal infections such as adhesion, colonization, and invasion. Although it has been indicated that some of the major virulence genes were related to biofilm formation on abiotic surfaces in hospital environments, research on the virulence mechanism and related genes in biofilm formation is still needed.30,31,32 A high amount of gelE gene expression in E. faecalis biofilm cells was shown in some studies, whereas others were in contradiction with this finding.33,34,35 Arciola et al.15 showed the importance of gelE in biofilm formation in implant infections. In a recent study, the prevalence of the gelE gene was determined as 64.3% among 510 clinical Enterococcus spp. isolates from UTI and wound infections.30 However, Kafil and Mobarez24 did not find a significant effect of the presence or absence of gelE on biofilm production by Enterococcus species. We examined the gelE mRNA levels of both planktonic and sessile cells of E. faecalis ATCC 29212 and 20 E. faecalis isolates by RT-qPCR. Our results showed that the gelE expression levels of ten isolates were significantly enhanced, but eight of the isolates were significantly decreased in biofilms when compared to their planktonic forms (p≤0.05) (Figure 3). Based on this result, we concluded that gelE expression had no effect on biofilm formation of the isolates collected from urinary tract infections (p>0.05) (Figure 3). The comparison of gelE mRNA levels of the isolates from the two different samples showed no significant difference either.
CONCLUSIONS
There was no statistically significance between the isolates from catheters and not from catheters in terms of biofilm-forming capability. E. faecalis sessile cell counts were increased in the presence of S. aureus. Expression of the gelE gene was not affected by E. faecalis biofilm formation of the isolates collected from the patients with urinary tract infections.
Acknowledgments
We gratefully acknowledge Prof. Dr. Gülşen Hasçelik and Dr. Dolunay Gülmez from Hacettepe University, Faculty of Medicine, Department of Medical Microbiology, for providing the E. faecalis clinical strains.
Footnotes
Conflict of Interest: No conflict of interest was declared by the authors.
References
- 1.Van Acker H, Van Dijck P, Coenye T. Molecular mechanisms of antimicrobial tolerance and resistance in bacterial and fungal biofilms. Trends Microbiol. 2014;22:326–333. doi: 10.1016/j.tim.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 2.Bjarnsholt T, Alhede M, Alhede M, Eickhardt-Sørensen SR, Moser C, Kühl M, Jensen PØ, Høiby N. The in vivo biofilm. Trends Microbiol. 2013;21:466–474. doi: 10.1016/j.tim.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 3.Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol. 2004;2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 4.Tay WH, Chong KK, Kline KA. Polymicrobial-Host Interactions during Infection. J Mol Biol. 2016;428:3355–3371. doi: 10.1016/j.jmb.2016.05.006. [DOI] [PubMed] [Google Scholar]
- 5.Roberts FA, Darveau RP. Microbial protection and virulence in periodontal tissue as a function of polymicrobial communities: symbiosis and dysbiosis. Periodontol. 2015;69:18–27. doi: 10.1111/prd.12087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hajishengallis G. Periodontitis: from microbial immune subversion to systemic inflammation. Nat Rev Immunol. 2015;15:30–44. doi: 10.1038/nri3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Darveau RP. Periodontitis: a polymicrobial disruption of host homeostasis. Nat Rev Microbiol. 2010;8:481–490. doi: 10.1038/nrmicro2337. [DOI] [PubMed] [Google Scholar]
- 8.Marom T, Nokso-Koivisto J, Chonmaitree T. Viral-bacterial interactions in acute otitis media. Curr Allergy Asthma Rep. 2012;12:551–558. doi: 10.1007/s11882-012-0303-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wolcott R, Costerton JW, Raoult D, Cutler SJ. The polymicrobial nature of biofilm infection. Clin Microbiol Infect. 2013;19:107–112. doi: 10.1111/j.1469-0691.2012.04001.x. [DOI] [PubMed] [Google Scholar]
- 10.Weimer KE, Juneau RA, Murrah KA, Pang B, Armbruster CE, Richardson SH, Swords WE. Divergent mechanisms for passive pneumococcal resistance to beta-lactam antibiotics in the presence of Haemophilus influenzae. J Infect Dis. 2011;203:549–555. doi: 10.1093/infdis/jiq087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elias S, Banin E. Multi-species biofilms: living with friendly neighbors. FEMS Microbiol Rev. 2012;36:990–1004. doi: 10.1111/j.1574-6976.2012.00325.x. [DOI] [PubMed] [Google Scholar]
- 12.Madsen JS, Burmolle M, Sorensen SJ. The interconnection between biofilm formation and horizontal gene transfer. FEMS Immunol Med Microbiol. 2012;65:183–195. doi: 10.1111/j.1574-695X.2012.00960.x. [DOI] [PubMed] [Google Scholar]
- 13.Ruoff KL, De La Maza L, Murtagh Mj, Spargo JD, Ferraro MJ. Species identities of Enterococci isolated from clinical specimen. J Clin Microbiol. 1990;28:435–437. doi: 10.1128/jcm.28.3.435-437.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bulacio Mde L, Galván LR, Gaudioso C, Cangemi R, Erimbaue MI. Enterococcus faecalis biofilm formation and development in vitro observed by scanning electron microscopy. Acta Odontol Latinoam. 2015;28:210–214. [PubMed] [Google Scholar]
- 15.Arciola CR, Baldassarri L, Campoccia D, Creti R, Prini V, Huebner J, Montanaro L. Strong biofilm production, antibiotic multi-resistance and high gelE expression in epidemic clones of Enterococcus faecalis from orthopaedic implant infections. Biomaterials. 2008;29:580–586. doi: 10.1016/j.biomaterials.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 16.Pérez-Laguna V, García-Luque I, Ballesta S, Pérez-Artiaga L, Lampaya-Pérez V, Samper S, Soria-Lozano P, Rezusta A, Gilaberte Y. Antimicrobial photodynamic activity of Rose Bengal, alone or in combination with Gentamicin, against planktonic and biofilm Staphylococcus aureus. Photodiagnosis Photodyn Ther. 2018;21:211–216. doi: 10.1016/j.pdpdt.2017.11.012. [DOI] [PubMed] [Google Scholar]
- 17.Otto M. Staphylococcal Biofilms. Curr Top Microbiol Immunol. 2008;322:207–228. doi: 10.1007/978-3-540-75418-3_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Archer NK, Mazaitis MJ, Costerton JW, Leid JG, Powers ME, Shirtliff ME. Staphylococcus aureus biofilms: properties, regulation and roles in human disease. Virulence. 2011;2:445–459. doi: 10.4161/viru.2.5.17724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stickler DJ. Bacterial biofilms in patients with indwelling urinary catheters. Nat Clin Pract Urol. 2008;5:598–608. doi: 10.1038/ncpuro1231. [DOI] [PubMed] [Google Scholar]
- 20.Crémet L, Corvec S, Batard E, Auger M, Lopez I, Pagniez F, Dauvergne S, Caroff N. Comparison of three methods to study biofilm formation by clinical strains of Escherichia coli. Diagn Microbiol Infect Dis. 2013;75:252–255. doi: 10.1016/j.diagmicrobio.2012.11.019. [DOI] [PubMed] [Google Scholar]
- 21.Chavant P, Gaillard-Martinie B, Talon R, Hébraud M, Bernardi T. A new device for rapid evaluation of biofilm formation potential by bacteria. J Microbiol Methods. 2007;68:605–612. doi: 10.1016/j.mimet.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 22.Naves P, del Prado G, Huelves L, Gracia M, Ruiz V, Blanco J, Dahbi G, Blanco M, Ponte Mdel C, Soriano F. Correlation between virulence factors and in vitro biofilm formation by Escherichia coli strains. Microb Pathog. 2008;45:86–91. doi: 10.1016/j.micpath.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C (T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Kafil HS, Mobarez AM. Assessment of biofilm formation by enterococci isolates from urinary tract infections with different virulence profiles. Journal of King Saud University Science. 2015;27:312–317. [Google Scholar]
- 25.Henriette LR, Søren JS, Mette B. Studying Bacterial Multispecies Biofilms: Where to Start? Trends Microbiol. 2016;24:503–513. doi: 10.1016/j.tim.2016.02.019. [DOI] [PubMed] [Google Scholar]
- 26.Pastar I, Nusbaum AG, Gil J, Patel SB, Chen J, Valdes J, Stojadinovic O, Plano LR, Tomic-Canic M, Davis SC. Interactions of methicillin resistant Staphylococcus aureus USA300 and Pseudomonas aeruginosa in polymicrobial wound infection. PLoS ONE. 2013;8:e56846. doi: 10.1371/journal.pone.0056846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deng DM, Hoogenkamp MA, Exterkate RA, Jiang LM, van der Sluis LW, Ten Cate JM, Crielaard W. Influence of S. mutans on E. faecalis Biofilm Formation. J Endod. 2009;35:1249–1252. doi: 10.1016/j.joen.2009.05.038. [DOI] [PubMed] [Google Scholar]
- 28.Mason KL, Downward JR, Falkowski NR, Young VB, Kao JY, Huffnagle GB. Interplay between the gastric bacterial microbiota and Candida albicans during postantibiotic recolonization and gastritis. Infect Immun. 2012;80:150–158. doi: 10.1128/IAI.05162-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harriott MM, Noverr MC. Importance of Candida-bacterial polymicrobial biofilms in disease. Trends Microbiol. 2011;19:557–563. doi: 10.1016/j.tim.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stratevaa T, Atanasova D, Savov E, Petrovac G, Mitov I. Incidence of virulence determinants in clinical Enterococcus faecalis and Enterococcus faecium isolates collected in Bulgaria. Braz J Infect Dis. 2016;20:127–133. doi: 10.1016/j.bjid.2015.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oli AK, Raju S, Rajeshwari Nagaveni S, Kelmani CR. Biofilm formation by Multidrug resistant Enterococcus faecalis (MREF) originated from clinical samples. J Microbiol Biotechnol Res. 2012;2:284–288. [Google Scholar]
- 32.Biswas PP, Dey S, Adhikari L, Sen A. Virulence markers of vancomycin resistant enterococci isolated from infected and colonized patients. J Glob Infect Dis. 2014;6:157–163. doi: 10.4103/0974-777X.145242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang L, Dong M, Zheng J, Song Q, Yin W, Li J, Niu W. Relationship of biofilm formation and gelE gene expression in Enterococcus faecalis recovered from root canals in patients requiring endodontic retreatment. J Endod. 2011;37:631–636. doi: 10.1016/j.joen.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 34.Di Rosa R, Creti R, Venditti M, D’Amelio R, Arciola CR, Montanaro L, Baldassarri L. Relationship between biofilm formation, the enterococcal surface protein (Esp) and gelatinase in clinical isolates of Enterococcus faecalis and Enterococcus faecium. FEMS Microbiol Lett. 2006;256:145–150. doi: 10.1111/j.1574-6968.2006.00112.x. [DOI] [PubMed] [Google Scholar]
- 35.Seno Y, Kariyama R, Mitsuhata R, Monden K, Kumon H. Clinical implications of biofilm formation by Enterococcus faecalis in the urinary tract. Acta Med Okayama. 2005;59:7987. doi: 10.18926/AMO/31979. [DOI] [PubMed] [Google Scholar]


