Abstract
Objectives:
Inflammatory response and cytokine activation are markedly stimulated in skeletal muscle during various conditions. Interleukin-6 (IL-6), a pro-inflammatory cytokine, has pleiotropic effects on skeletal muscle. Adenosine, released by all cell types, binds to a class of G protein-coupled receptors to induce various skeletal muscle effects. The aim of this work was to investigate whether activation of adenosine receptors, particularly adenosine A2B receptors, could stimulate IL-6 gene expression in rat L6 skeletal muscle cells.
Materials and Methods:
The rat L6 skeletal muscle cells were cultured in 25 cm2 flasks. These differentiated cells were treated and then quantitative reverse transcription-polymerase chain reaction (Probe-based) was used to analyze IL-6 gene expression level among different treatment conditions.
Results:
Adenosine-5′-N-ethyluronamide (NECA), a stable adenosine analogue, concentration- and time-dependently stimulates IL-6 gene expression in skeletal muscle cells. The effect of NECA is inhibited by a selective adenosine A2B receptor antagonist, PSB 603. By using cyclic adenosine monophosphate (cAMP)-arising reagent forskolin, cAMP is found to be involved in the up-regulation of IL-6 induction.
Conclusion:
Here, a novel relationship between adenosine and IL-6 up-regulation has been demonstrated for the first time; IL-6 up-regulation induced by NECA is mediated by adenosine A2B receptor activation in skeletal muscle and is dependent on mainly a cAMP pathway. Adenosine A2B receptors are, thus, potentially important pharmacological targets in treating inflammation and related diseases in skeletal muscle tissues.
Keywords: Adenosine A2B receptors, skeletal muscle, interleukin 6, cAMP, inflammation
INTRODUCTION
Adenosine is a key endogenous signalling molecule produced by all types of cells, and documented as a major local regulator of tissue function. Adenosine can modulate cellular functions via binding to the four members that belong to the cell surface G protein-coupled receptor superfamily (P1 receptors), including adenosine A1, A2A, A2B and A3 receptor subtypes.1 The adenosine A2A and A2B receptors share a relatively high homology and are coupled to Gs,2 leading to increased levels of cyclic adenosine monophosphate (cAMP). In addition, the adenosine A2B receptors has been shown to couple to Gq,3 thereby regulating intracellular calcium levels. In general, the adenosine A2B receptors have a lower affinity for adenosine.4 Among adenosine receptors, adenosine A2B receptor requires higher concentrations of adenosine in many different cellular types for activation than the adenosine A1, A2, and A3 receptors subtypes.1 Thus, adenosine A2B receptor can mostly be activated when interstitial adenosine concentrations are increased as a result of tissue hypoxia, injury, inflammation and cell stress,5 even though adenosine A2B receptors are likely to remain silent under normal physiological conditions.5
Within the skeletal muscle tissue, adenosine potentially plays important roles in a large number of physiological processes (such as glucose homeostasis and insulin sensitivity).6,7,8 Adenosine A2B receptor, in particular, has recently been proposed to act as a potentially functional adenosine receptor in skeletal muscle,9 however, its pharmacology and biological function(s) remain largely unexplored. Recently, evidence has been accumulated, suggesting that adenosine is a significant modulator of inflammation in response to various stimuli.10 There is growing evidence that the adenosine system plays an important role in regulating inflammation. Indeed, specific targeting of its components such as the adenosine A2B receptor continues to provide avenues towards the development of potential treatments for at least inflammatory diseases and related disorders, including insulin resistance and type 2 diabetes.
Inflammation is an important contributor to the pathophysiology of diseases related to skeletal muscle dysfunction.11 Pro-inflammatory cytokines are important contributors to chronic inflammation found in many diseases.12 One of these inflammatory cytokines is interleukin-6 (IL-6). IL-6, a mediator of inflammation, is a pleiotropic cytokine that has been proposed to be involved in both immune- and nonimmuno-regulation in most cell types and tissues outside the immune system, including skeletal muscle tissue.13 Indeed, IL-6 is a biologically active cytokine which has a broad range of activities regulating not only inflammatory responses but also in cell proliferation, differentiation, growth and metabolism in skeletal muscle cells.14,15,16
Elevated levels of cytokines, such as IL-6 also seems to be the main pro-inflammatory cytokine involved in the pathophysiology of insulin resistance and type 2 diabetes and obesity.17,18,19,20 Elevated circulating IL-6 levels have been observed in obese individuals and type 2 diabetic patients.21 Several association studies and numerous clinical studies have suggested that IL-6 plasma concentration are associated with insulin resistance and increased with weight gain.20 Moreover, some reports provide evidences for high circulating IL-6 levels, as a risk factor for the manifestation of type 2 diabetes.22 Hence, high circulating IL-6 levels might contribute to the progression of skeletal muscle damage and dysfunction in chronic diseases and might exert pathogenic effects in these diseases. Recent evidence has demonstrated that significant levels of IL-6 is produced in and released from skeletal muscle cells per se in the absence of inflammation and stimulated by complex signalling cascades.23,24 IL-6 is therefore considered as a myokine and its IL-6 signalling in skeletal muscle tissue has been suggested to be associated with skeletal muscle growth, myogenesis, and regulation of energy metabolism.13
Adenosine modulates the functions of many inflammatory cells such as macrophage.25,26 Moreover, increasing evidence suggests that adenosine signalling plays a role in regulating cytokine network processes. In addition, adenosine increases the release of IL-6 from various cells.27 In previous studies, it has been reported that skeletal muscle tissue is a source of IL-6 production.23 As discussed above, increasing evidence indicates that skeletal muscle plays an active role in the inflammatory process by secreting cytokines. However, the effect of adenosine or adenosine analogue on inflammatory cytokine expression by skeletal muscle cells has not been determined. At the same time, adenosine-based approaches are currently being developed for the treatment of various diseases where inflammatory modulation is a key component.28 Generally, adenosine receptors, in particular adenosine A2B receptors, are increasingly recognized as important orchestrators of inflammation. In fact, adenosine A2B receptor activation enhances the inflammatory responses of mast cells, fibroblasts, epithelial cells and smooth muscle cells.29 However, the role of this receptor in skeletal muscle cells is yet unexplored.
Extensive in vitro and in vivo studies have identified potent pro-inflammatory or/and anti-inflammatory functions for all adenosine processes. Recent interest in the endocrine skeletal muscle has potentially revealed the presence of a functional adenosine system in skeletal muscle, however, the effects of adenosine and adenosine receptors modulation on downstream inflammatory signalling, in particular IL-6, still remains unclear. In the current study, an adenosine receptors, in particular adenosine A2B receptors, agonist and antagonist were evaluated for their effects on IL-6 messenger RNA (mRNA) expression level.
MATERIALS AND METHODS
Materials
N-ethylcarboxamidoadenosine (NECA), forskolin, 8-[4-[4-(4-chlorophenzyl)piperazide-1-sulfonyl) phenyl]]-1-propylxanthine (PSB 603) and 2-(4-[2-carboxyethyl]-phenethylamino) adenosine-52 -nethyluronamide (CGS21680) were obtained from Tocris Bioscience, UK; dimethyl sulphoxide reagent was sourced from Santa Cruz, USA; and Trizol and charcoal stripped serum were brought from Applied Biosystem, USA. Maxima Probe quantitative polymerase chain reaction (qPCR) Master Mix (2X) and Thermo Scientific RevertAid First Strand complementary DNA (cDNA) Synthesis were obtained from Thermo Scientific Company, USA. RNeasy Mini Total RNA Purification kits and RNase-Free DNase Set were brought from Qiagen, Germany. Fetal bovine serum (FBS) was supplied by Capricorn Scientific, USA. Horse serum was from Sigma company, Germany. Ham-F 10 was sourced from PAA Company, USA. Dulbecco’s modified essential medium (DMEM) was from Caisson, USA.
Cell culture
Rat L6 skeletal muscle cell line and myoblast cell line were originally obtained from the American Type Culture Collection (USA). Cells were maintained as an attached monolayer culture in DMEM with high glucose (4500 mg/L) and L-glutamate supplemented with 10% (v/v) heat-inactivated FBS and 100 µg/mL penicillin-streptomycin. Cells were incubated at 37°C in a 90% humidified atmosphere of 5% CO2. The cells were passaged upon reaching a state of approximately 60%-70% confluency, and the medium was changed three times per week (Figure 1).
Figure 1.
Representative myoblasts derived from passage number 7 myoblasts taken after 1 day seeding into 25 cm2 (10X)
Confluent cells in 25 cm2 flasks were cultured for a further 14 days (to allow myotube formation), according to the protocol mentioned in30 with slight modifications (Figure 2). 70%-90% confluent myotubes (approximately 2 weeks in culture) were serum-starved (incubated in Ham-F 10 medium alone) for 19 hours or 7 days. Then, cells (Figure 2) were treated for 1 hour with vehicle (0.1% dimethyl sulphoxide), NECA 100 nM and 10 µM, PSB 603 100 nM, 1 µM and 10 µM, forskolin 100 nM, NECA and PSB 603 (cells were pre-treated with PSB 603 for 10 min prior to the addition of NECA). Following treatment, cells were washed with ice cold PBS, then lysed with TRIzol (Invitrogen product name) (2 mL per flask).
Figure 2.
Representative myoblasts/myotubes derived from passage number 7 (a) myoblasts taken at (Ham-F10, 10% fetal bovine serum, 1% P/S) during 3-4 days of tissue culture (10X) (b) myoblasts taken at (Ham-F10, 6% hoarse serum, 1% P/S) during 4-5 days of tissue culture (10X) (c) myoblasts taken at (Ham-F10, 2% hoarse serum, 1% P/S) during 6-7 days of tissue culture (10X) (d) myotubes taken at (Ham-F10, 2% charcoal serum, 1% P/S) during 11-12 days of tissue culture (10X) (e) myotubes taken at (Ham-F10, 1% P/S) after 16 hours starvation (10X)
RNA extraction and cDNA synthesis
Rat L6 skeletal muscle cells (in 25 cm2 flasks) were scraped in 2 mL of ice cold TRIzol (Applied Biosystems, USA) and RNA was then isolated according to the manufacturer’s instructions. Total RNA clean-up and on-column DNAse digestion was performed using RNeasy purification columns (Qiagen, Germany). RNA concentration and purity was determined using a spectrophotometer (JENWAY Genova Nano). For cDNA synthesis, 500 ng of total RNA was reverse-transcribed using RevertAid First Strand cDNA Synthesis (Thermo Scientific, USA) in a total volume of 20 µL for 5 min at 25°C, followed by 1 hour at 42°C, and the reaction was terminated at 70°C for 5 min.
Taqman quantitative real-time polymerase chain reaction
The relative standard curve method based on Taqman quantitative real-time PCR (qRT-PCR) was used to quantify gene expression. Samples were prepared in a total reaction volume of 25 µL [13 µL Maxima Probe qPCR Master Mix 2X reagent, 1.5 µL forward primer (10 µM), 1.5 µL reverse primer (10 mM), 2.5 µL Probe (2 µM), 5 µL water, and 5 µL cDNA]. The qRT-PCR analysis was performed using a RT-PCR system (Applied Biosystems, USA). Gene expression was determined relative to reference gene, TATA. Primers and probes for all genes (Table 1) were designed using Primer Express software (Applied Biosystems, USA) and synthesised by Integrated DNA Technologies, Inc., USA. The standard curve method was used, with a slope between -3.2 and -3.6 and R2 values of more than 99%, indicating that amplification efficiency was nearly 100%.
Table 1. List of gene primer and probe sequences.

Data analysis
Data are expressed as means ± standard error of mean of triplicate or quadruplicate wells generated from at least three independent experimental group. All mRNA data were analysed using one-way ANOVA with a Tukey test. Analysis was performed using GraphPad Prism, version 5.03 (GraphPad Software Inc). The level of statistical significance was set at p<0.05.
RESULTS
N-ethylcarboxamidoadenosine stimulates IL-6 mRNA gene expression in skeletal muscle cells
To assess whether stimulation of adenosine A2 receptors could induce IL-6 mRNA gene expression in rat L6 skeletal muscle cells, the effects of NECA were relatively quantified using a non-selective adenosine receptor agonist on IL-6 mRNA gene expression by qRT-PCR (probe-based). Starved skeletal muscle cells were incubated with NECA (10 µM) for one hour, 3 hours and 24 hours, and mRNA gene expression of IL-6 was subsequently quantified.
Incubation of one week starved L6 skeletal muscle cells with 10 µM of the non-selective adenosine analogue NECA for one hour increases significantly (p<0.001) mRNA gene expression of IL-6 (an average around 2.3-fold change compared to vehicle) (Figure 3). Moreover, incubation of 19 hours starved L6 skeletal muscle cells with 10 µM (but, not 100 nM) of the non-selective adenosine analogue NECA for 3 hours (but, not for either one hour or 24 hours) increases significantly (p<0.05) mRNA gene expression of IL-6 (an average around 2.2-fold change compared to vehicle) (Figure 4).
Figure 3.
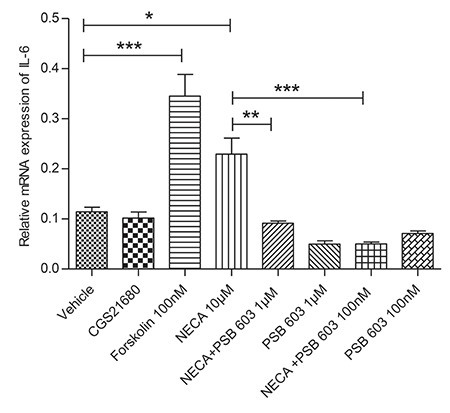
Effects of 5’-N-ethylcarboxamidoadenosine (NECA), 2-(4-[2-carboxyethyl]-phenethylamino) adenosine-52-nethyluronamide (CGS21680), and 8-[4-[4-(4-chlorophenzyl) piperazide-1-sulfonyl) phenyl]]-1-propylxanthine (PSB 603) on interleukin 6 (IL-6) messenger RNA (mRNA) gene expression in rat L6 skeletal muscle myotubes using charcoal serum rat L6 skeletal muscle myotubes (70-90% confluent) were serum starved for 7 days and then stimulated for 1 hour IL-6 mRNA levels were measured relative to TATA-BOX using real-time quantitative polymerase chain reaction; stimulation was performed with vehicle (0.1% dimethyl sulphoxide), NECA (10 μM), PSB 603 (100 nM and 1 μM), CGS21680 (100 nM), and forskolin (100 nM), data were represented as means ± standard error of mean of at least three independent experimental groups *denotes p<0.05, **denotes p<0.01 and ***denotes p<0.001, data were analysed using a one-way ANOVA test followed by a Tukey test
Figure 4.
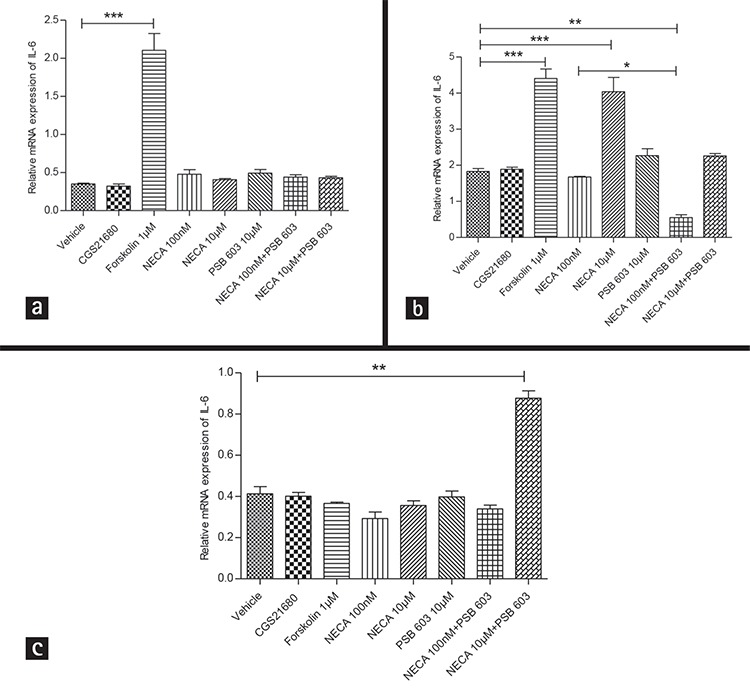
Effects of 5’-N-ethylcarboxamidoadenosine (NECA), 2-(4-[2-carboxyethyl]-phenethylamino) adenosine-52-nethyluronamide (CGS21680) and 8-[4-[4-(4-chlorophenzyl) piperazide-1-sulfonyl) phenyl]]-1-propylxanthine (PSB 603) on interleukin 6 (IL-6) messenger RNA (mRNA) gene expression in rat L6 skeletal muscle myotubes using charcoal serum, rat L6 skeletal muscle myotubes were stimulated for the indicated time from 1 hour to 24 hours and interleukin-6 mRNA levels, relative to TATA-BOX, was measured by quantitative real time polymerase chain reaction; stimulation was performed with vehicle (0.1% dimethyl sulphoxide), NECA (100 nM and 10 μM), PSB 603 (10 μM), CGS21680 (100 nM) and forskolin (1 μM) (a) Stimulation was performed up to 1 hour (b) Stimulation was performed for up to 3 hours (c) Stimulation was performed for up to 24 hours, data were represented as means ± standard error of mean of three independent experimental group (n=3; *denotes p<0.05, **denotes p<0.01 and ***denotes p<0.001), data were analyzed using one way ANOVA test followed by Tukey test
Adenosine A2B receptors mediates N-ethylcarboxamidoa-denosine-induced IL-6 mRNA gene expression in skeletal muscle cells
To determine which subtype of adenosine A2 receptors mediate the increase IL-6 mRNA gene expression level, the adenosine receptor agonist CGS21680 (subtype A2A selective) was used. The concentration applied for CGS21680 could selectively activate the indicated subtype (Ki=27 nM).31
Furthermore, since no selective agonist exists for adenosine A2B receptors, the effect of a selective antagonist to the adenosine A2B receptors (PSB 603) was investigated. PSB 603 exhibits a strong affinity to adenosine A2B receptors and very weak affinity to three other adenosine receptors subtypes. Adenosine A2B receptors display >17000-fold selectivity over other adenosine receptors (Ki values: 0.553, >10000, >10000, and >10000 nM for A2B, A1, A2A, and A3 receptors, respectively).32
As shown in Figure 3 and Figure 4a, 4b, NECA (10 µM) increases the mRNA gene expression level of IL-6 significantly. In contrast, the adenosine A2A receptor selective agonist CGS21680 (100 nM) does not cause a significant increase in the mRNA gene expression level of IL-6.
To investigate the effect of a selective adenosine A2B receptor antagonist, PSB 603, rat L6 skeletal muscle cells were incubated with 10 µM NECA in combination with PSB 603 (which was added to cells 10 min prior to adding NECA) in concentrations of 100 nM, 1 µM or 10 µM, which blocks IL-6 mRNA gene expression significantly (p<0.01, p<0.05 and p<0.05, respectively) (Figure 3, 4a, 4b).
It is interesting to note that treatment the 19 hours starved cells for 24 hours with PSB 603 and NECA up-regulates IL-6 mRNA expression level. However, treatment the 19 hours starved cells for 24 hours with either PSB 603 or NECA alone does not modulate the IL-6 mRNA expression level.
Collectively, these results indicate that adenosine A2B receptors are the functionally expressed receptors of adenosine A2 receptors in skeletal muscle, whereas no functional expression of the adenosine A2A receptors was detected using mRNA gene expression levels for IL-6 as a functional readout.
It is worth to mention that the adenosine A2B receptors antagonists/inverse agonists, PSB 603 (at 100 nM, 1 µM and 10 µM), does not mediate a significant change in baseline IL-6 mRNA gene expression levels in skeletal muscle cells (Figure 3, 4). However, even though treatment the 19 hours starved skeletal muscle cells with 100 nM NECA does not up-regulates the IL-6 mRNA expression, incubation that cells with 100 nM NECA in combination with PSB 603 (which was added to cells 10 min prior to adding NECA) decrease baseline IL-6 mRNA gene expression level significantly (p<0.01) (Figure 4a, 4b).
Role of cyclic adenosine monophosphate pathway in the mRNA gene expression of IL-6
In previous studies, activation of adenosine A2B receptors in skeletal muscle by NECA increased cAMP accumulation,33,34 and the NR4A mRNA gene expression.9
Experiments were conducted to investigate if the adenylyl cyclase pathway is involved in the activation of IL-6 transcription profile in skeletal muscle cells. In rat skeletal muscle cells, transcripts of adenosine A1, A2, and A3 receptors were detected.33,34 In addition to gene expression, using cAMP accumulation as a functional readout, it has been confirmed the presence of functional adenosine A2B receptors in skeletal muscle cells,33,34 whereas the presence of functional adenosine A1, A2A, and A3 receptors were not detected. Furthermore, using NR4A expression as a functional downstream signalling readout, it has been confirmed the presence of functional adenosine A2B receptors in skeletal muscle cells,9 whereas the presence of functional adenosine A2A receptors were not detected.
Many physiologic roles of adenosine are mediated through cell surface adenosine receptors. In this present study, i provided evidence that the adenosine A2B receptors subtype mediates the effect of the adenosine analogue NECA on IL-6 expression. Our results show the following:
(1) The nonselective agonist NECA increases the expression of IL-6, whereas selective agonist for adenosine A2A receptors CGS21680 had no effect. This agonist is very potent and, at this concentration (100 nM), it fully activates their cognate receptors without significant activation of the adenosine A2B receptors.31 For this purpose, the adenylyl cyclase activator forskolin was used in the present study to understand the potential role of the cAMP pathway in NECA induces IL-6 mRNA gene expression in skeletal muscle.
Forskolin (100 nM and 1 µM) increases the expression of IL-6 significantly (an average around 3.5 and 2.4 fold change compared to vehicle, respectively) (Figure 3, 4a, 4b), a result similar to that of NECA. These findings support the idea that the cAMP pathway plays an important role in NECA-induces IL-6 mRNA gene expression. However, treatment the 19 hours starved skeletal muscle cells for 24 hours with forskolin (1 µM) does not induce the expression of IL-6.
DISCUSSION
The novel findings of this present study are that stable adenosine analogue NECA increases the expression of IL-6 by skeletal muscle cells, and that this effect of NECA is mediated by the adenosine A2B receptor subtype. To our knowledge, this is the first paper on the effect of adenosine analogue and its receptor subtype on inflammatory cytokine expression by skeletal muscle cells, and it may represent a novel mechanism for the role of adenosine analogue in skeletal muscle cytokine network. Several reports demonstrated the presence of adenosine receptors in skeletal muscle cells from different species.35 This is the rationale for determining the effect of this agonist at a concentration of 100 nM.
(2) The effects of NECA on cytokine expression are dependently blocked by selective antagonist of the adenosine A2B receptors subtype, PSB 603. Collectively, these findings provide strong evidence for the role of the adenosine A2B receptors in up-regulating the expression of IL-6 caused by NECA.
These results are in agreement with those obtained in various cell types of different origins, including intestinal36 and airway epithelial cells,37 macrophages,38,39 pulmonary fibroblasts,40 bronchial smooth muscle cells,41 astrocytoma cells,42 astroglioma cells,43 astrocytes44 and cardiomyocytes,45 osteoblasts,46 and pituitary folliculostellate cells,47 that all show NECA-induced IL-6 release was via the adenosine A2B receptors. Moreover, numerous in vivo studies have also demonstrated that adenosine A2B receptors activation can stimulate the release of IL-6, an important pro-inflammatory cytokine.36,48 Accordingly, adenosine A2B receptors have been suggested to mediate the pro-inflammatory actions of adenosine. Yet, those results in this current study and above studies contradict a gene-knockout study in which it was reported that adenosine A2B receptors knockout mice show evidence of increased inflammation at baseline, in that levels of cytokines such as IL-6 were elevated in naive adenosine A2B receptors knockout mice.39,49
In this present study, cAMP elevation in skeletal muscle cells induces IL-6 expression in a similar to that effect of NECA is expected, because it is well documented that the Gs protein-coupled adenosine A2B receptor increases the formation of cAMP. In fact, several studies have supported the role of cAMP pathway. Indeed, some researchers have also shown that cAMP elevation induced IL-6 release in various cells.50,51,52 In addition, recently, it has been demonstrated that NECA increased cAMP concentration in rat skeletal muscle cells.33,34 These data suggest that adenosine A2B receptors mediate IL-6 expression through mainly a cAMP-dependent mechanism in rat skeletal muscle cells, although future studies are recommended to validate and investigate the exact signaling mechanism.
It is interesting to note that PSB 603 might act as a positive allosteric modulator for adenosine A2B receptors in case of treatment the 19 hours starved skeletal muscle cells with PSB 603 and NECA (10 µM but not 100 nM) (cells were pre-treated with PSB 603 for 10 min prior to the addition of NECA) for 24 hours (but not 1 or 3 hours). In this situation, it seems that cAMP is not involved in the (NECA 10 µM and PSB 603) induces IL-6 expression as forskolin does not induce that. However, whether or not Gq-signalling or other downstream targets is involved in the regulation of (NECA 10 µM and PSB 603) induces IL-6 expression in skeletal muscle cells needs to be investigated further. In the literature, the adenosine A2B receptors driven production of these pro-inflammatory molecules has been attributed to both Gs and Gq pathways.36,53
It is reported in the current study that the NECA effect in Figure 3 is made on skeletal muscle cells that are starved for one week, and this is not physiologically relevant. However, the idea behind experimenting such a condition is that starvation might change the rate of RNA synthesis for IL-6 and/or the expression level of adenosine receptors subtypes.54,55
It should be highlighted that the 1 hour and 24 hours NECA stimulations did not work, but the 3 hours did on 19 hour-starved cells (Figure 4). The reason behind that that adenosine A2B receptors, as a G-protein coupled receptor, might need enough time to couple to G-protein and, then induce downstream signaling pathway to target IL-6 mRNA gene expression. While the cells concentrations of NECA do not explain the disparate effects of 3 hours and 24 hours treatment, duration of exposure may be a pivotal factor. The demonstration of adenosine A2B receptors expression level/density in skeletal muscle cells might fulfill a necessary condition for the specific action of a adenosine A2B receptors activator to alter expression of adenosine A2B receptor-responsive genes.
It is difficult to determine what a 2.5-fold increase in IL-6 transcripts at a single time point translates into IL-6 production. However, it is important to suggest that the relationship between the protein synthesis and RNA content in skeletal muscle cells might not be in a definite linear correlation.56 One may argue that the changes in expression of IL-6 involved in inflammation observed in the present study may be transient and not reflected by changes in protein expression and therefore have little significance for inflammation developing in response to stable adenosine analogue NECA. The main reason total protein measurements were not included in the present study is the short duration of the treatment (i.e. 1 hour, 3 hours and up to 24 hours), which in line with my main aim was employed to identify the transcriptional events that precede the induction of inflammation under the activation of adenosine A2B receptors. It has shown previously57 that most of the inflammatory transcripts studied (such as IL-6) in skeletal muscle cells are not translated into protein within that time frame. Therefore my future studies using longer durations of NECA (after 24 hours) will be recommended to investigate the translational changes in response to treatment. The use of primary skeletal muscle cells in future studies may also provide a better opportunity to investigate both transcriptional and translational changes over the time course of prolonged durations of NECA (up to four days).
In this regard, it is clear that adenosine receptors, in particular adenosine A2B receptors, are important molecular targets for adenosine-based therapeutics for inflammation. Approaches utilizing adenosine receptor-based therapeutics will be dependent on further investigation of signalling mechanism for adenosine A2B receptors in skeletal muscle, timing and duration of treatment, and monitoring of beneficial and adverse effects.
CONCLUSION
I showed that the activation of adenosine A2B receptors increases the expression of IL-6 in time- and concentration-dependent manners in rat skeletal muscle cells. These findings provide a novel mechanism whereby adenosine analogue acts as a pro-inflammatory mediator in the skeletal muscle tissue. Furthermore, these findings suggest that the adenosine A2B receptor antagonist at acute early states might have a potential therapeutic utility for the treatment of inflammatory-related skeletal muscle dysfunction.
Acknowledgments
I would like to thank Abdul Hameed Shoman Foundation for supporting scientific research in Jordan and for their kind generous financial support of this project. Without this support, I could not perform this work. Indeed, this project was supported by grants from mainly Abdul Hameed Shoman Foundation (Grant number 12/2015) and Philadelphia University.
Footnotes
Conflict of Interest: No conflict of interest was declared by the author.
References
- 1.Alexander SP, Mathie A, Peters JA. Guide to Receptors and Channels (GRAC), 5th edition. Br J Pharmacol. 2011;164 Suppl 1:S1–324. doi: 10.1111/j.1476-5381.2011.01649_1.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fredholm BB, IJzerman AP, Jacobson KA, Linden J, Müller CE. International Union of Basic and Clinical Pharmacology. LXXXI. Nomenclature and classification of adenosine receptors--an update. Pharmacol Rev. 2011;63:1–34. doi: 10.1124/pr.110.003285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feoktistov I, Goldstein AE, Ryzhov S, Zeng D, Belardinelli L, Voyno- Yasenetskaya T, Biaggioni I. Differential expression of adenosine receptors in human endothelial cells: role of A2B receptors in angiogenic factor regulation. Circ Res. 2002;90:531–538. doi: 10.1161/01.res.0000012203.21416.14. [DOI] [PubMed] [Google Scholar]
- 4.Fredholm BB1, Chern Y, Franco R, Sitkovsky M. Aspects of the general biology of adenosine A2A signaling. Prog Neurobiol. 2007;83:263–276. doi: 10.1016/j.pneurobio.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 5.Fredholm BB. Adenosine, an endogenous distress signal, modulates tissue damage and repair. Cell Death Differ. 2007;14:1315–1323. doi: 10.1038/sj.cdd.4402132. [DOI] [PubMed] [Google Scholar]
- 6.Rüsing D, Müller CE, Verspohl EJ. The impact of adenosine and A(2B) receptors on glucose homoeostasis. J Pharm Pharmacol. 2006;58:1639–1645. doi: 10.1211/jpp.58.12.0011. [DOI] [PubMed] [Google Scholar]
- 7.Johnston-Cox H, Koupenova M, Yang D, Corkey B, Gokce N, Farb MG, LeBrasseur N, Ravid K. The A2b adenosine receptor modulates glucose homeostasis and obesity. PLoS One. 2012;7:e40584. doi: 10.1371/journal.pone.0040584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnston-Cox H, Eisenstein AS, Koupenova M, Carroll S, Ravid K. The macrophage A2B adenosine receptor regulates tissue insulin sensitivity. PLoS One. 2014;9:e98775. doi: 10.1371/journal.pone.0098775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haddad M. The Impact of Adenosine A2B Receptors Modulation on Nuclear Receptors (NR4A) Gene Expression BPJ. 2016;9:177–185. [Google Scholar]
- 10.Hasko G, Cronstein B. Regulation of inflammation by adenosine. Front Immunol. 2013;4:85. doi: 10.3389/fimmu.2013.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Remels AH, Gosker HR, van der Velden J, Langen RC, Schols AM. Systemic inflammation and skeletal muscle dysfunction in chronic obstructive pulmonary disease: state of the art and novel insights in regulation of muscle plasticity. Clin Chest Med. 2007;28:537–552. doi: 10.1016/j.ccm.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 12.Gosker HR, Wouters EF, van der Vusse GJ, Schols AM. Skeletal muscle dysfunction in chronic obstructive pulmonary disease and chronic heart failure: underlying mechanisms and therapy perspectives. Am J Clin Nutr. 2000;71:1033–1047. doi: 10.1093/ajcn/71.5.1033. [DOI] [PubMed] [Google Scholar]
- 13.Munoz-Canoves P, Scheele C, Pedersen BK, Serrano AL. Interleukin-6 myokine signaling in skeletal muscle: a double-edged sword? FEBS J. 2013;280:4131–4148. doi: 10.1111/febs.12338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Londhe P, Guttridge DC. Inflammation induced loss of skeletal muscle. Bone. 2015;80:131–142. doi: 10.1016/j.bone.2015.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Begue G, Douillard A, Galbes O, Rossano B, Vernus B, Candau R, Py G. Early activation of rat skeletal muscle IL-6/STAT1/STAT3 dependent gene expression in resistance exercise linked to hypertrophy. PLoS One. 2013;8:e57141. doi: 10.1371/journal.pone.0057141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Serrano AL, Baeza-Raja B, Perdiguero E, Jardí M, Muñoz-Cánoves P. Interleukin-6 is an essential regulator of satellite cell-mediated skeletal muscle hypertrophy. Cell Metab. 2008;7:33–44. doi: 10.1016/j.cmet.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 17.Kristiansen OP, Mandrup-Poulsen T. Interleukin-6 and diabetes: the good, the bad, or the indifferent? Diabetes. 2005;54 Suppl 2:S114–124. doi: 10.2337/diabetes.54.suppl_2.s114. [DOI] [PubMed] [Google Scholar]
- 18.Illig T, Bongardt F, Schöpfer-Wendels A, Huth C, Heid I, Rathmann W, Martin S, Vollmert C, Holle R, Thorand B, Wichmann HE, Koenig W, Kolb H, Herder C; KORA Study Group. Genetics of type 2 diabetes: impact of interleukin-6 gene variants. Gesundheitswesen. 2005;67 Suppl 1:S122–126. doi: 10.1055/s-2005-858396. [DOI] [PubMed] [Google Scholar]
- 19.Araki A, Hosoi T, Orimo H, Ito H. Association of plasma homocysteine with serum interleukin-6 and C-peptide levels in patients with type 2 diabetes. Metabolism. 2005;54:809–814. doi: 10.1016/j.metabol.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 20.Tangvarasittichai S, Pongthaisong S, Tangvarasittichai O. Tumor Necrosis Factor-Alpha, Interleukin-6, C-Reactive Protein Levels and Insulin Resistance Associated with Type 2 Diabetes in Abdominal Obesity Women. Indian J Clin Biochem. 2016;31:68–74. doi: 10.1007/s12291-015-0514-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vozarova B, Weyer C, Hanson K, Tataranni PA, Bogardus C, Pratley RE. Circulating interleukin-6 in relation to adiposity, insulin action, and insulin secretion. Obes Res. 2001;9:414–417. doi: 10.1038/oby.2001.54. [DOI] [PubMed] [Google Scholar]
- 22.Park J, Chang JH, Park SH, Lee HJ, Lim YS, Kim TH, Kim CW, Han SW. Interleukin-6 is associated with obesity, central fat distribution, and disease severity in patients with acute pancreatitis. Pancreatology. 2015;15:59–63. doi: 10.1016/j.pan.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 23.Pal M, Febbraio MA, Whitham M. From cytokine to myokine: the emerging role of interleukin-6 in metabolic regulation. Immunol Cell Biol. 2014;92:331–339. doi: 10.1038/icb.2014.16. [DOI] [PubMed] [Google Scholar]
- 24.Febbraio MA, Pedersen BK. Contraction-induced myokine production and release: is skeletal muscle an endocrine organ? Exerc Sport Sci Rev. 2005;33:114–119. doi: 10.1097/00003677-200507000-00003. [DOI] [PubMed] [Google Scholar]
- 25.Hasko G, Pacher P. Regulation of macrophage function by adenosine. Arterioscler Thromb Vasc Biol. 2012;32:865–869. doi: 10.1161/ATVBAHA.111.226852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feng W, Song Y, Chen C, Lu ZZ, Zhang Y. Stimulation of adenosine A(2B) receptors induces interleukin-6 secretion in cardiac fibroblasts via the PKC-delta-P38 signalling pathway. Br J Pharmacol. 2010;159:1598–1607. doi: 10.1111/j.1476-5381.2009.00558.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abbracchio MP, Ceruti S. P1 receptors and cytokine secretion. Purinergic Signal. 2007;3:13–25. doi: 10.1007/s11302-006-9033-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sachdeva S, Gupta M. Adenosine and its receptors as therapeutic targets: An overview. Saudi Pharm J. 2013;21:245–253. doi: 10.1016/j.jsps.2012.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feoktistov I, Biaggioni I. Role of adenosine A(2B) receptors in inflammation. Adv Pharmacol. 2011;61:115–144. doi: 10.1016/B978-0-12-385526-8.00005-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haddad M. Do CB1 Cannabinoid Receptors Regulate Insulin Signalling in Rat Primary Skeletal Muscle Cells? J Phys Pharm Adv. 2013;3:277–291. [Google Scholar]
- 31.Ongini E, Dionisotti S, Gessi S, Irenius E, Fredholm BB. Comparison of CGS 15943, ZM 241385 and SCH 58261 as antagonists at human adenosine receptors. Naunyn Schmiedebergs Arch Pharmacol. 1999;359:7–10. doi: 10.1007/pl00005326. [DOI] [PubMed] [Google Scholar]
- 32.Borrmann T, Hinz S, Bertarelli DC, Li W, Florin NC, Scheiff AB, Müller CE. 1-alkyl-8-(piperazine-1-sulfonyl)phenylxanthines: development and characterization of adenosine A2B receptor antagonists and a new radioligand with subnanomolar affinity and subtype specificity. J Med Chem. 2009;52:3994–4006. doi: 10.1021/jm900413e. [DOI] [PubMed] [Google Scholar]
- 33.Haddad M. Adenosine Receptors Machinery and Purinergic Receptors in Rat Primary Skeletal Muscle Cells. Biomed Pharmacol J. 2014;7:383–398. [Google Scholar]
- 34.Lynge J, Schulte G, Nordsborg N, Fredholm BB, Hellsten Y. Adenosine A2B receptors modulate cAMP levels and induce CREB but not ERK1/2 and p38 phosphorylation in rat skeletal muscle cells. Biochem Biophys Res Commun. 2003;307:180–187. doi: 10.1016/s0006-291x(03)01125-2. [DOI] [PubMed] [Google Scholar]
- 35.Haddad M. mRNA expression of GPCRs in rat skeletal muscle tissues. IJBPAS. 2014;3:2506–2536. [Google Scholar]
- 36.Sitaraman SV, Merlin D, Wang L, Wong M, Gewirtz AT, Si-Tahar M, Madara JL. Neutrophil-epithelial crosstalk at the intestinal lumenal surface mediated by reciprocal secretion of adenosine and IL-6. J Clin Invest. 2001;107:861–869. doi: 10.1172/JCI11783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun Y, Wu F, Sun F, Huang P. Adenosine promotes IL-6 release in airway epithelia. J Immunol. 2008;180:4173–4181. doi: 10.4049/jimmunol.180.6.4173. [DOI] [PubMed] [Google Scholar]
- 38.Ritchie PK, Spangelo BL, Krzymowski DK, Rossiter TB, Kurth E, Judd AM. Adenosine increases interleukin 6 release and decreases tumour necrosis factor release from rat adrenal zona glomerulosa cells, ovarian cells, anterior pituitary cells, and peritoneal macrophages. Cytokine. 1997;9:187–198. doi: 10.1006/cyto.1996.0153. [DOI] [PubMed] [Google Scholar]
- 39.Ryzhov S, Zaynagetdinov R, Goldstein AE, Novitskiy SV, Blackburn MR, Biaggioni I, Feoktistov I. Effect of A2B adenosine receptor gene ablation on adenosine-dependent regulation of proinflammatory cytokines. J Pharmacol Exp Ther. 2008;324:694–700. doi: 10.1124/jpet.107.131540. [DOI] [PubMed] [Google Scholar]
- 40.Zhong H, Belardinelli L, Maa T, Zeng D. Synergy between A2B adenosine receptors and hypoxia in activating human lung fibroblasts. Am J Respir Cell Mol Biol. 2005;32:2–8. doi: 10.1165/rcmb.2004-0103OC. [DOI] [PubMed] [Google Scholar]
- 41.Zhong H, Belardinelli L, Maa T, Feoktistov I, Biaggioni I, Zeng D. A(2B) adenosine receptors increase cytokine release by bronchial smooth muscle cells. Am J Respir Cell Mol Biol. 2004;30:118–125. doi: 10.1165/rcmb.2003-0118OC. [DOI] [PubMed] [Google Scholar]
- 42.Fiebich BL, Akundi RS, Biber K, Hamke M, Schmidt C, Butcher RD, van Calker D, Willmroth F. IL-6 expression induced by adenosine A2b receptor stimulation in U373 MG cells depends on p38 mitogen activated kinase and protein kinase C. Neurochem Int. 2005;46:501–512. doi: 10.1016/j.neuint.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 43.Fiebich BL, Biber K, Gyufko K, Berger M, Bauer J, van Calker D. Adenosine A2b receptors mediate an increase in interleukin (IL)-6 mRNA and IL-6 protein synthesis in human astroglioma cells. J Neurochem. 1996;66:1426–1431. doi: 10.1046/j.1471-4159.1996.66041426.x. [DOI] [PubMed] [Google Scholar]
- 44.Schwaninger M, Neher M, Viegas E, Schneider A, Spranger M. Stimulation of interleukin-6 secretion and gene transcription in primary astrocytes by adenosine. J Neurochem. 1997;69:1145–1150. doi: 10.1046/j.1471-4159.1997.69031145.x. [DOI] [PubMed] [Google Scholar]
- 45.Wagner DR, Kubota T, Sanders VJ, McTiernan CF, Feldman AM. Differential regulation of cardiac expression of IL-6 and TNF-alpha by A2- and A3- adenosine receptors. Am J Physiol. 1999;276:H2141–2147. doi: 10.1152/ajpheart.1999.276.6.H2141. [DOI] [PubMed] [Google Scholar]
- 46.Evans BA, Elford C, Pexa A, Francis K, Hughes AC, Deussen A, Ham J. Human osteoblast precursors produce extracellular adenosine, which modulates their secretion of IL-6 and osteoprotegerin. J Bone Miner Res. 2006;21:228–236. doi: 10.1359/JBMR.051021. [DOI] [PubMed] [Google Scholar]
- 47.Rees DA, Lewis BM, Lewis MD, Francis K, Scanlon MF, Ham J. Adenosineinduced IL-6 expression in pituitary folliculostellate cells is mediated via A2b adenosine receptors coupled to PKC and p38 MAPK. Br J Pharmacol. 2003;140:764–772. doi: 10.1038/sj.bjp.0705488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vazquez JF, Clement HW, Sommer O, Schulz E, van Calker D. Local stimulation of the adenosine A2B receptors induces an increased release of IL-6 in mouse striatum: an in vitro microdialysis study. J Neurochem. 2008;105:904–909. doi: 10.1111/j.1471-4159.2007.05191.x. [DOI] [PubMed] [Google Scholar]
- 49.Yang D, Zhang Y, Nguyen HG, Koupenova M, Chauhan AK, Makitalo M, Jones MR, St Hilaire C, Seldin DC, Toselli P, Lamperti E, Schreiber BM, Gavras H, Wagner DD, Ravid K. The A2B adenosine receptor protects against inflammation and excessive vascular adhesion. J Clin Invest. 2006;116:1913–1923. doi: 10.1172/JCI27933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kiriyama Y, Murayama T, Tokumitsu Y, Nomura Y. Protein kinase A-dependent IL-6 production induced by calcitonin in human glioblastoma A172 cells. J Neuroimmunol. 1997;76:139–144. doi: 10.1016/s0165-5728(97)00044-1. [DOI] [PubMed] [Google Scholar]
- 51.Chio CC, Chang YH, Hsu YW, Chi KH, Lin WW. PKA-dependent activation of PKC, p38 MAPK and IKK in macrophage: implication in the induction of inducible nitric oxide synthase and interleukin-6 by dibutyryl cAMP. Cell Signal. 2004;16:565–575. doi: 10.1016/j.cellsig.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 52.Carapau D, Kruhofer M, Chatalbash A, Orengo JM, Mota MM, Rodriguez A. Transcriptome profile of dendritic cells during malaria: cAMP regulation of IL-6. Cell Microbiol. 2007;9:1738–1752. doi: 10.1111/j.1462-5822.2007.00910.x. [DOI] [PubMed] [Google Scholar]
- 53.Feoktistov I, Goldstein AE, Biaggioni I. Role of p38 mitogen-activated protein kinase and extracellular signal-regulated protein kinase kinase in adenosine A2B receptor-mediated interleukin-8 production in human mast cells. Mol Pharmacol. 1999;55:726–734. [PubMed] [Google Scholar]
- 54.Fegatella F, Cavicchioli R. Physiological responses to starvation in the marine oligotrophic ultramicrobacterium Sphingomonas sp. strain RB2256. Appl Environ Microbiol. 2000;66:2037–2044. doi: 10.1128/aem.66.5.2037-2044.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ogata ES, Foung SK, Holliday MA. The effects of starvation and refeeding on muscle protein synthesis and catabolism in the young rat. J Nutr. 1978;108:759–765. doi: 10.1093/jn/108.5.759. [DOI] [PubMed] [Google Scholar]
- 56.Millward DJ, Garlick PJ, James WP, Nnanyelugo DO, Ryatt JS. Relationship between protein synthesis and RNA content in skeletal muscle. Nature. 1973;241:204–205. doi: 10.1038/241204a0. [DOI] [PubMed] [Google Scholar]
- 57.Scheler M, Irmler M, Lehr S, Hartwig S, Staiger H, Al-Hasani H, Beckers J, de Angelis MH, Häring HU, Weigert C. Cytokine response of primary human myotubes in an in vitro exercise model. Am J Physiol Cell Physiol. 2013;305:C877–886. doi: 10.1152/ajpcell.00043.2013. [DOI] [PubMed] [Google Scholar]


