Initial MRIs from 50 subjects with diffuse intrinsic pontine gliomas recruited for a prospective clinical trial before treatment were analyzed. Retrospective imaging analyses included FLAIR/T2 tumor volume, tumor volume enhancing, the presence of cyst and/or necrosis, median, mean, mode, skewness, kurtosis of ADC tumor volume based on FLAIR, and enhancement at baseline. Molecular subgroups based on EGFR and MGMT mutations were established. Histone mutations were also determined (H3F3A, HIST1H3B, HIST1H3C). Enhancing tumor volume was near-significantly different across molecular subgroups, after accounting for the false discovery rate. Tumor volume enhancing, median, mode, skewness, and kurtosis ADC T2-FLAIR/T2 were significantly different between patients with H3F3A and HIST1H3B/C mutations.
Abstract
BACKGROUND AND PURPOSE:
Recent advances in molecular techniques have characterized distinct subtypes of diffuse intrinsic pontine gliomas. Our aim was the identification of MR imaging correlates of these subtypes.
MATERIALS AND METHODS:
Initial MRIs from subjects with diffuse intrinsic pontine gliomas recruited for a prospective clinical trial before treatment were analyzed. Retrospective imaging analyses included FLAIR/T2 tumor volume, tumor volume enhancing, the presence of cyst and/or necrosis, median, mean, mode, skewness, kurtosis of ADC tumor volume based on FLAIR, and enhancement at baseline. Molecular subgroups based on EGFR and MGMT mutations were established. Histone mutations were also determined (H3F3A, HIST1H3B, HIST1H3C). Univariate Cox proportional hazards regression was used to test the association of imaging predictors with overall and progression-free survival. Wilcoxon rank sum, Kruskal-Wallis, and Fisher exact tests were used to compare imaging measures among groups.
RESULTS:
Fifty patients had biopsy and MR imaging. The median age at trial registration was 6 years (range, 3.3–17.5 years); 52% were female. On the basis of immunohistochemical results, 48 patients were assigned to 1 of 4 subgroups: 28 in MGMT–/epidermal growth factor receptor (EGFR)–, 14 in MGMT–/EGFR+, 3 in MGMT+/EGFR–, and 3 in MGMT+/EGFR+. Twenty-three patients had histone mutations in H3F3A, 8 in HIST1H3B, and 3 in HIST1H3C. Enhancing tumor volume was near-significantly different across molecular subgroups (P = .04), after accounting for the false discovery rate. Tumor volume enhancing, median, mode, skewness, and kurtosis ADC T2-FLAIR/T2 were significantly different (P ≤ .048) between patients with H3F3A and HIST1H3B/C mutations.
CONCLUSIONS:
MR imaging features including enhancement and ADC histogram parameters are correlated with molecular subgroups and mutations in children with diffuse intrinsic pontine gliomas.
Diffuse intrinsic pontine glioma (DIPG) is a malignant brain tumor that accounts for 75%–80% of brain stem tumors in children.1 Despite medical therapy, these tumors have a poor prognosis, with a median survival at 1, 2, and 3 years of 41%, 15.3%, and 7.2%, respectively.2 The 2016 World Health Organization classification of brain tumors introduced the term “diffuse midline glioma, H3 K27M mutant” to describe these neoplasms due to the high prevalence of unique histone protein mutations that differ from those of supratentorial (hemispheric) pediatric high-grade gliomas and adult high-grade gliomas. While H3 K27M is the most common variant detected, more recently, other distinct histone mutations have been identified in DIPG, which appear to correspond to subgroups with different prognosis and pathologic phenotypes.3 Identification of these subgroups of tumors is important to appropriately counsel families and tailor treatment strategies.
Radiogenomic techniques are emerging as valuable tools to noninvasively characterize brain tumors and are increasingly important, given the evolving molecular landscape in pediatric neuro-oncology. Although most data available to date pertain to adult high-grade gliomas,4,5 radiogenomic techniques have been successfully implemented to characterize atypical teratoid/rhabdoid tumors6 and medulloblastomas.7 A challenge to molecular analysis of DIPGs is that historically, patients with these tumors did not routinely undergo biopsy due to a perceived high likelihood of morbidity associated with the procedure; consequently, most imaging-based studies do not have pathologic or molecular correlates.8 Despite this limitation, important imaging-based prognostic factors have been described for DIPG. Jansen et al9 and Hoffman et al10 reported that ring enhancement was associated with shorter survival. Chen et al11 reported worse outcome in DIPGs with lower ADC values at baseline. Poussaint et al12 reported that lower diffusion values on ADC histogram analysis, high skewness, and enhancement were associated with shorter survival. The association among these MR imaging features, the molecular subgroups, and various histone mutations has not been elucidated.
The purpose of this study was to investigate the relationship between the molecular subgroup and histone mutations of DIPGs and MR imaging features on anatomic sequences, and ADC histogram analysis, using the baseline MR imaging from a prospective clinical trial of children with newly diagnosed DIPG who underwent biopsy.13
MATERIALS AND METHODS
Subjects
Patients for this study were recruited as part of an institutional review board (Dana Farber Cancer Institute)–approved, Health Insurance Portability and Accountability Act–compliant, multicenter clinical trial: Molecularly Determined Treatment of Diffuse Intrinsic Pontine Gliomas (DIPG) (NCT01182350). Prospective subjects had MR imaging that showed classic clinical-imaging criteria of nonmetastatic DIPG, including an expansile and infiltrative T1-isointense or -hypointense and T2-FLAIR and T2-hyperintense mass centered within the pons, encompassing at least 50% of the pons with no or little contrast enhancement, which could vary.14 Findings on clinical examination included multiple cranial nerve neuropathies, long tract signs, and ataxia.14 Detailed criteria for inclusion and exclusion are summarized in On-line Table 1. The patients enrolled in the trial underwent a biopsy before initiation of therapy; the biopsy target and approach were at the discretion of the neurosurgeon, with the goal of minimizing operative risk.13 All subjects received local radiation therapy (59.4 Gy) with adjuvant bevacizumab, as per the standard of care. Subsequently, patients were stratified into different treatment arms with erlotinib and/or temozolomide at the start of radiation therapy based on MGMT promoter methylation status and epidermal growth factor receptor (EGFR) expression in tumor tissue obtained from surgical biopsies.
Of 53 patients enrolled, 50 underwent biopsy and were included in the analytic cohort of this study. Subjects without biopsy were excluded from this analysis. We performed a retrospective review that included biopsy data (histone mutations, molecular groups), MR imaging features on baseline MR imaging, and clinical outcomes. The molecular analysis determined the MGMT methylation status and expression of EGFR; 4 categories were derived from molecular analysis: MGMT–/EGFR–, MGMT–/EGFR+, MGMT+/EGFR–, MGMT+/EGFR+. Three histone mutations were identified through whole-genome RNA sequencing: H3F3A, HIST1H3B, and HISTH3C.3 Clinical end points of the study were determined with longitudinal follow-up. Overall survival (OS) was measured from the time of registration to death or to last follow-up if censored. Progression-free survival (PFS) was measured from the time of registration to progression or death (whichever was first) or to the last follow-up if censored.
Image Analysis
The MR imaging of the brain used for analysis corresponded to the baseline examination obtained before treatment and biopsy. Images were obtained using 3T (n = 17) and 1.5T (n = 33) scanners. A standard clinical protocol was used, which included the following: sagittal T1, axial T2, precontrast axial T2-FLAIR, axial DTI (TR = 6500 ms, TE = 88 ms, section thickness = 2 mm, b-values = 0 and 1000 s/mm2, 30 gradient directions), and postgadolinium (PG) 3D T1.
Volumetric data of various components of the tumor were obtained by generating 3D ROIs on a Vitrea workstation (Vital Images) on the anatomic sequences. Specifically, T2-FLAIR/T2 volume represents the volume of tissue (milliliter) with abnormal T2 hyperintense signal on a 2D TSE T2-FLAIR sequence; for cases in which T2-FLAIR was unavailable, this parameter was estimated using a 2D T2 TSE sequence. Tumor volume enhancing represents the volume (milliliter) of enhancement within the tumor estimated on the PG 3D T1 sequence.
To perform ADC histogram analysis, we registered the ADC maps to the anatomic sequence of interest (T2-FLAIR/T2 and PG T1) using tools from the FSL library (http://www.fmrib.ox.ac.uk/fsl),15 as described by Poussaint et al.12 Briefly, we transformed the b = 0 (and subsequently the ADC map) into the space of the individual anatomic sequences. Subsequently, 3D ROIs were created using the thresholding feature in Fiji (http://fiji.sc/),16 an Open Source (https://opensource.org/) distribution of Java modules along with ImageJ software (National Institutes of Health); we then extracted the values from every pixel from the ADC map within the corresponding ROI.
We applied a uniform threshold to the ADC maps, ranging from 600 to 2600 × 10−6 mm2/s to differentiate solid tumor from cyst/necrosis. ADC histograms were then generated using a uniform bin width of 10 mm2/s. Histogram metrics used for statistical analysis were mean, median, mode, skewness, and kurtosis of these histograms.
Statistical Analysis
Clinical and demographic variables were summarized using descriptive statistics. Continuous measures were summarized using medians and ranges, and categoric measures were summarized using frequencies and proportions. Kaplan-Meier curves were used to summarize OS and PFS for all patients. Univariate Cox proportional hazards regression was used to test the association of imaging predictors with OS and PFS. The Wilcoxon rank sum and Kruskal-Wallis tests were used to compare continuous imaging features among groups. The Fisher exact test was used to compare categoric imaging features among groups. When applicable, the presence of enhancement (defined as patients with tumor volume enhancing >0 mL) was recorded as a binary measure and used for subanalyses. Due to the low number of H3.1 mutations (HIST1H3B = 8, HIST1H3C = 3), we grouped them into a single category and compared them with H3F3A mutations. For each outcome tested, we calculated the false discovery rate (FDR) using the Benjamani-Hochberg method to ac-count for multiple hypothesis tests. Tests with a calculated FDR of ≤ 10% were considered statistically significant after FDR adjustment.17 R version 3.5.0 (http://www.r-project.org) and SAS version 9.4 (SAS Institute) were used for statistical analyses.
RESULTS
Patients
Of the 50 patients who underwent biopsy, all had baseline MR imaging studies. The median age at time of enrollment was 6 years (range, 3.3–17.5 years). Twenty-six (52%) subjects were female, 48 (96%) were successfully assigned to a molecular subgroup (2 unassigned), and 34 (68%) patients had (single) identifiable histone mutations (Table 1). One subject was excluded from subsequent analysis due to the concurrent presence of 2 histone mutations.
Table 1:
Baseline patient characteristics (n = 50)
| Patient Characteristic | No. (%) or Median (Range) |
|---|---|
| No. of MR imaging studies | 50 (100) |
| Sex | |
| Male | 24 (48) |
| Female | 26 (52) |
| Median age at registration (range) (yr) | 6 (3.3–17.5) |
| Molecular subgroups | |
| MGMT–/EGFR– | 28/48 (58) |
| MGMT–/EGFR+ | 14/48 (29) |
| MGMT +/EGFR– | 3/48 (6) |
| MGMT +/EGFR+ | 3/48 (6) |
| Unassigned | 2 |
| Mutational statusa | |
| Any histone mutation | 34/49 (69) |
| Wild-type | 15/49 (31) |
| Histone mutation (among patients with any histone mutations)a | |
| H3F3A | 23/34 (68) |
| HIST1H3B | 8/34 (24) |
| HIST1H3C | 3/34 (9) |
| Median follow-up time (range) (mo) | 10.9 (0.4–33.4) |
One patient with both H3F3A and HIST1H3B histone mutations was excluded.
Tumor Enhancement and Survival
The presence of enhancement (hazard ratio = 3.2; 95% CI , 1.5–7.0; P = .003) was associated with decreased OS after FDR adjustment (On-line Fig A and On-line Table 2). OS was not associated (P ≥ .07) with T2-FLAIR/T2 volume, tumor volume cyst/necrosis, the presence of necrosis, tumor vol-ume enhancing (subset of patients with enhancing tumor of >0 mL), ADC_FLAIR histogram metrics, or PG ADC enhancement histogram metrics after FDR adjustment.
Higher tumor volume enhancing (P = .02) and the presence of enhancement (P = .06) showed a strong trend with decreased PFS but were not significant after FDR adjustment (On-line Fig B and On-line Table 2). T2-FLAIR/T2 tumor volume (P = .07) also showed a trend toward a significant association with decreased PFS.
Molecular Subgroup Analysis
There were differences in tumor volume enhancing across the 4 molecular subgroups within the subset of patients with tumor volume enhancing of >0 mL, but the test was not significant after FDR adjustment (P = .04; n = 30) (On-line Table 3). All MGMT+ tumors showed enhancement, most MGMT–/EGFR+ tumors (77%) showed enhancement, and 54% of MGMT–/EGFR– tumors showed enhancement (On-line Table 3). The 3 MGMT+/EGFR– tumors had the highest tumor volume enhancing values, occasionally an order of magnitude higher than those of other molecular groups, including the MGMT+/EGFR+ tumors. No other factor analyzed showed a significant association with molecular groups, including ADC histogram metrics.
Histone Mutation Analysis
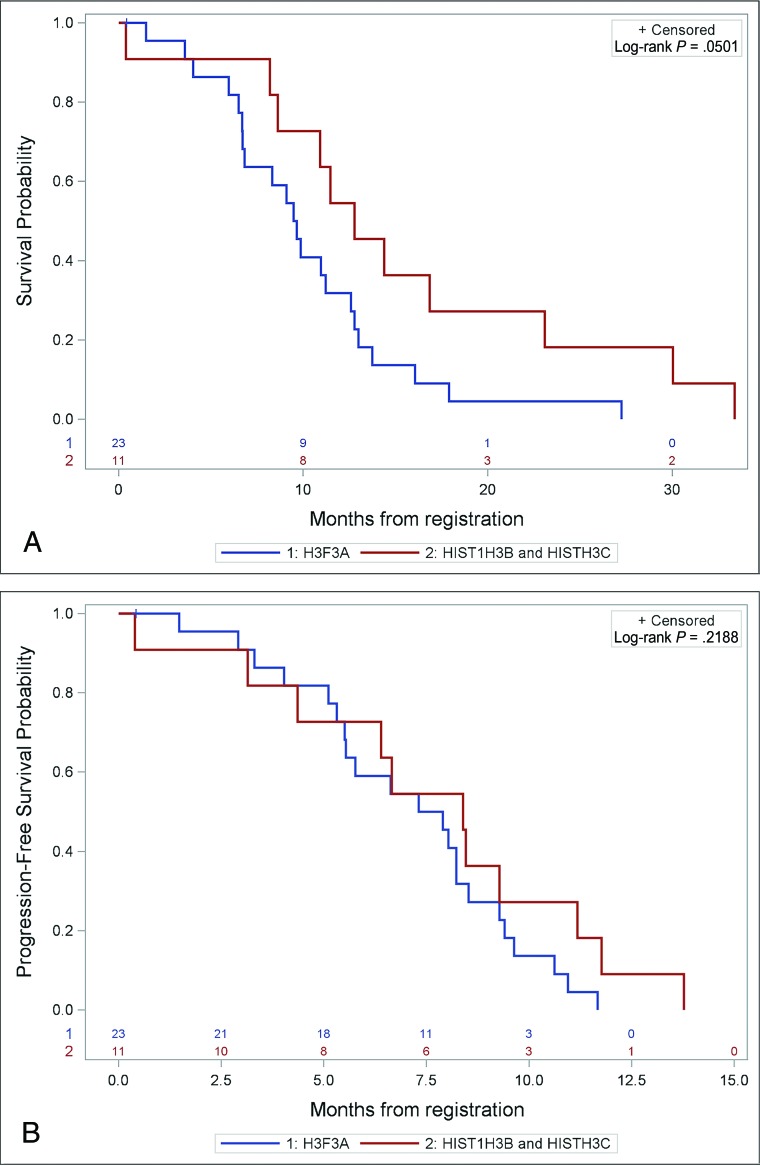
Subjects with HIST1H3B/C tumors showed a trend toward improved OS that bordered on significance (P = .0501) compared with subjects with H3FA3 tumors (Fig 1). There was no difference in PFS (P = .22) between subjects with H3FA3 and HIST1H3B/C tumors.
Fig 1.
Prognostic differences in subjects with H3F3A (n = 23) versus HIST1H3B/C (n = 11) tumors. A, Kaplan-Meier curve shows a trend that approaches significance in OS between H3F3A versus HIST1H3B/C tumors. B, Kaplan-Meier curve shows no significant difference in PFS between H3F3A versus HIST1H3B/C tumors.
Tumor volume enhancing (in a subset of patients with enhancing tumor volumes of >0 mL, P = .048), median ADC_FLAIR (P = .03), and mode ADC_FLAIR (P = .02) were significantly higher in tumors with HIST1H3B/C mutations. Skewness ADC_FLAIR (P = .009) and kurtosis ADC_FLAIR (P = .03) were significantly higher in tumors with H3F3A mutations. H3F3A mutations had lower ADC values compared with HIST1H3B tumors (Fig 2 and Table 2).
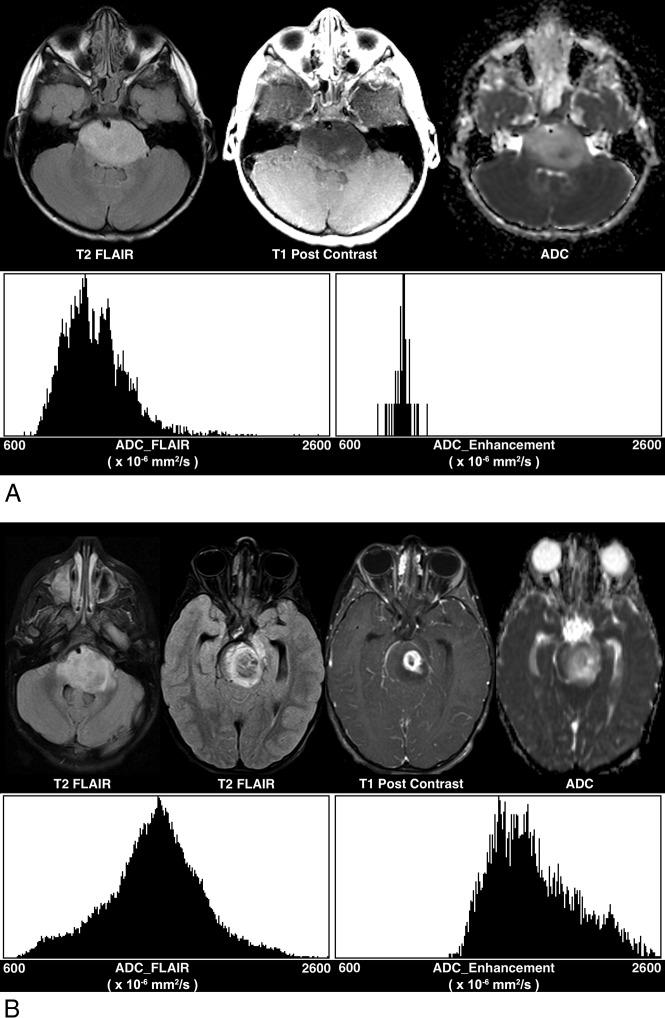
Fig 2.
Differences in tumor volume enhancing and ADC histogram parameters between histone mutations. A, H3F3A-mutated tumor shows lower tumor volume enhancing (0.08 mL), lower ADC_FLAIR mode (1099 × 10−6 mm2/s), higher ADC_FLAIR skewness (1.41), and higher ADC_FLAIR kurtosis (4.39), relative to (B) a HIST1H3B-mutated tumor (tumor volume enhancing: 3.61 mL; FLAIR_ADC mode: 1558 × 10−6 mm2/s; FLAIR_ADC skewness: −0.039; and FLAIR_ADC kurtosis: 0.33).
Table 2:
Association of imaging predictors and variant histone mutation (H3F3A versus HIST1H3B/C) (n = 34)
| Imaging Parameters (Median) (Range) | No. | Variant Histone Mutation |
Wilcoxon P | |
|---|---|---|---|---|
| H3F3A (n = 23) | HIST1H3B and HIST1H3C (n = 11) | |||
| FLAIR/T2 tumor volume | 33 | 35.1 (10.7–70.9) | 40.3 (11.6–63) | .4 |
| Tumor volume cyst/necrosis (mL) in patients with cyst/necrosis | 16 | 0.8 (0.07–8.1) | 2.1 (0.09–4.1) | .8 |
| Presence of necrosis (yes vs no) in patients with enhancing tumor volume >0 | 22 | 11/16 (69%) | 5/6 (83%) | .6a |
| Tumor volume enhancing (mL) in patients with enhancing tumor volume >0 | 22 | 2.5 (0.08–12.4) | 6.3 (3.6–15.2) | .04b |
| Presence of enhancement (yes vs no) | 31 | 16/21 (76%) | 6/10 (60%) | .4a |
| Mean ADC_FLAIR × 10−6 mm2/s | 31 | 1306 (893.9–1915.2) | 1411 (1168.8–2098.3) | .06 |
| Median ADC_FLAIR × 10−6 mm2/s | 31 | 1260 (906.7–1913) | 1370 (1189.4–2146.0) | .03b |
| Mode ADC_FLAIR × 10−6 mm2/s | 31 | 1257 (895.5–1886.4) | 1396 (1150.8–2214.4) | .02b |
| Skewness ADC_FLAIR | 29 | 1 (−0.6–2.0) | 0.03 (−1.3–0.9) | .009b |
| Kurtosis ADC_FLAIR | 29 | 1.8 (−0.3–8.2) | 0.3 (−1.1–2.4) | .03a |
Fisher exact test P value.
Test is significant after FDR adjustment <.1.
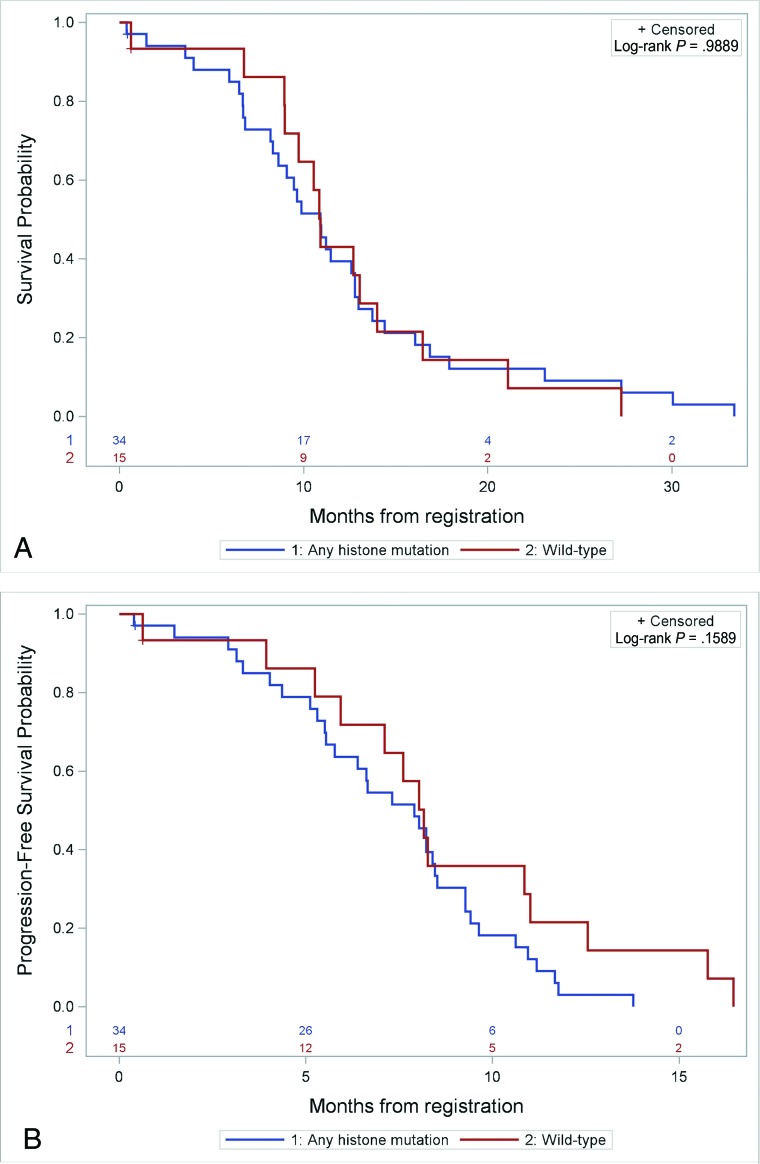
A comparison of ADC T2-FLAIR/T2 mean, median, and mode; skewness; and kurtosis did not show differences between wild-type tumors and grouped histone mutation (H3F3A and HIST1H3B/C) tumors (Table 3). Kaplan-Meier analysis also failed to demonstrate significant differences in OS or PFS between wild-type and grouped histone-mutated tumors (Fig 3).
Table 3:
Association of imaging predictors with mutational status (any histone mutation versus wild-type) (n = 49)
| Imaging Parameters (Median) (Range) | No. | Any Histone Mutation (n = 34) | Wild-Type (n = 15) | Wilcoxon P |
|---|---|---|---|---|
| Mean ADC_T2-FLAIR/T2 × 10−6 mm2/s | 45 | 1360 (893.9–2098.3) | 1327 (1040.4–1838.5) | .9 |
| Median ADC_T2-FLAIR/T2 × 10−6 mm2/s | 45 | 1309 (906.7–2146.0) | 1316 (952.0–1925.7) | .9 |
| Mode ADC_T2-FLAIR/T2 × 10−6 mm2/s | 45 | 1311 (895.5–2214.4) | 1342 (893.8–1965.4) | .99 |
| Skewness ADC_T2-FLAIR/T2 | 42 | 0.5 (−1.3–2.0) | 0.8 (−0.9–2.8) | .4 |
| Kurtosis ADC_T2-FLAIR/T2 | 42 | 1.1 (−1.1–8.2) | 2.0 (−0.1–9.0) | .1 |
Fig 3.
Prognostic differences in subjects with any histone mutation (n = 34) versus wild-type (n = 15) tumors. A, Kaplan-Meier curve shows no significant difference in OS between mutant versus wild-type tumors. B, Kaplan-Meier curve shows no significant difference in PFS between mutant versus wild-type tumors.
DISCUSSION
Even though MR imaging has traditionally played a central role in the evaluation and monitoring of patients with DIPG, little is known regarding the biologic and molecular correlates of imaging features such as the presence of enhancement, low diffusivity, and findings on ADC histogram analysis.12,18 Recently, more centers are performing biopsies for DIPG, which has permitted identification of molecular subgroups with distinct clinical characteristics.3,19-21 This work demonstrates the feasibility of a radiogenomic approach to investigate differences among various tumor groups. Specifically, we identified significant differences in volume of enhancing tumor between molecular subgroups and differences in volume of enhancing tumor and ADC histogram parameters across tumors with different histone mutations. We followed a previously validated approach for patients with DIPG, thus, minimizing the risk of introducing methodologic bias in our analysis.12
Molecular characteristics of brain tumors are now widely accepted as important diagnostic criteria for many entities as well as independent prognostic markers.22,23 Castel et al3 investigated the molecular profile of DIPG in 91 children and identified 2 distinct subgroups with mutations in H3.3 (H3F3A) and H3.1 (HIST1H3B/C) histone variants. Tumors with H3F3A mutations had a worse prognosis, with a poor response to radiation therapy and earlier relapse compared with HIST1H3B/C mutated tumors.20 Similar to the observations by Castel et al, we found a strong trend toward shorter OS in patients with H3F3A mutations relative to HIST1H3B/C, though this did not reach statistical significance, likely due to the smaller sample size in our study.
ADC histogram analysis revealed differences across histone mutations, with H3F3A-mutated tumors having lower ADC T2-FLAIR/T2, higher skewness, and higher kurtosis than HIST1H3B/C-mutated tumors. This finding suggests that histogram metrics may help differentiate tumors that have the H3F3A mutation, which carries a worse prognosis, from those with HIST1H3B/C mutations.3,20,24,25 Poussaint et al12 found that tumors with lower preradiotherapy mean, median, and mode ADC FLAIR (analogous to T2-FLAIR/T2 in our study) had shorter PFS and that tumors with higher skewness and kurtosis had shorter OS and PFS. Although that study did not include pathologic or molecular analysis of the tumors, the findings consistently indicated that ADC histogram metrics can identify tumors with more aggressive biology; it is therefore possible that some of the tumors with these ADC histogram features in the study by Poussaint et al may have had H3F3A mutations.
The differences in ADC and ADC histogram parameters among histone-mutated tumors is likely multifactorial. As postulated by Poussaint et al,12 Lober et al,26 Zukotynski et al,27 Chen et al,11 and Lobel et al28 on prior studies of DIPG, and by Nowosielski et al29 on studies of adult high-grade gliomas, the lower diffusivity and increased kurtosis possibly reflect higher cellularity, which could be related to the more aggressive biology of H3F3A-mutated tumors. Additionally, Castel et al3 reported that the H3.1 tumors had an enrichment in genes associated with glioblastoma with edema, which correlated with exuberant extracellular edema in their histologic analysis; these factors may also contribute to overall higher ADC and lower skewness/kurtosis in H3.1-mutated tumors. A difference between the study by Poussaint et al12 and the current study is that we did not find an association of ADC histogram metrics with PFS or OS, which could be related to a larger sample size in the former study, which analyzed approximately 3 times as many patients. However, the significant trends we observed showed an association of other adverse prognostic factors with the more aggressive (H3F3A) tumors.
We did not identify differences in mean, median, mode ADC histogram parameters, or survival between wild-type histone tumors and H3K27M-mutated tumors (collectively), consistent with the results from a study published by Aboian et al.30 Similarly, the study by Castel et al3 did not identify differences in survival between wild-type tumors and histone-mutated tumors (collectively). Chen et al31 found lower ADC in the tumor and peritumoral regions of patients with H3K27M mutations compared with wild-type tumors; however, the patient population analyzed in that study is vastly different, possibly accounting for the discrepancy. For instance, the study by Chen et al included H3K27M tumors outside of the brain stem (spinal cord) and included substantially older subjects (most of subjects were older than 18 years of age).31
In our cohort, higher tumor volume enhancing had a significant association with shorter OS and PFS, as has been shown in multiple prior studies.12,18 We also observed significant differences across molecular subgroups in tumor volume enhancing. Specifically, MGMT+ tumors (EGFR– and EGFR+) showed enhancement regardless of their EGFR status, most MGMT–/EGFR+ tumors showed enhancement, and slightly more than 50% of MGMT–/EGFR– tumors showed enhancement. These findings suggest that a radiogenomic approach could contribute to identifying specific mutations in tumors and could help individualize therapy to target the oncogenic pathways involved.19,21,32 Differences in tumor volume enhancing were also observed between tumors with H3F3A and HIST1H3B/C mutations, a finding that has not been previously reported in the literature.
Most interesting, higher tumor volume enhancing, which is thought to have a negative prognostic association, was higher in HIST1H3B/C tumors, which have a better prognosis compared with H3F3A tumors.3 We believe that this observation is likely related to the higher expression of genes responsible for angiogenesis in H3.1-mutated tumors, and these findings correlate with preliminary observations described by Castel et al3 in a subanalysis of their radiologic data. Our sample size was too small to perform a subgroup analysis between enhancing and nonenhancing tumors for each histone mutation; additional work is needed to further elucidate the implications of differences in enhancement in tumors with different histone mutations.24 This complex landscape suggests that a multiparametric approach may be useful when evaluating DIPG, because multiple genetic pathways (including MGMT, EGFR, histone) probably account for the pleomorphic MR imaging appearance and clinical course of these tumors, and these factors likely interact with each other to determine the clinical course.33
There are several limitations to this study. First, we conducted our analysis on a small sample size from a single prospective clinical trial. Consequently, our results are preliminary observations that need to be corroborated in a larger separate cohort. Due to the small sample size, we were unable to explore the prognostic implications of differences in enhancement and ADC histogram metrics within individual molecular subgroups and histone variants. Although we identified group differences in diffusion metrics, ADC histogram analysis, and enhancement, the exact biologic substrates for these were not explored. More detailed genetic analyses being performed on this cohort of patients may uncover biologic explanations for these differences. Finally, we analyzed only the baseline MR imaging and did not analyze the changes in any of the baseline parameters with treatment on subsequent MRIs. ADC histogram metrics and tumor enhancing volume could potentially serve as valuable markers of treatment response.
CONCLUSIONS
MR imaging features including enhancement and ADC histogram metrics are correlated with molecular subgroups and histone mutations in children with DIPG. Noninvasive markers that allow identification of molecular subgroups of DIPG could help prognosticate survival and guide therapy. Future studies in a larger cohort are required to verify these findings.
ABBREVIATIONS:
- DIPG
diffuse intrinsic pontine glioma
- EGFR
epidermal growth factor receptor
- FDR
false discovery rate
- PFS
progression-free survival
- PG
postgadolinium
- OS
overall survival
Footnotes
Disclosures: Sridhar Vajapeyam—RELATED: Consulting Fee or Honorarium: Dana Farber Cancer Institute, Comments: salary support for MRI data-processing and analysis.* Nalin Gupta—UNRELATED: Grants/Grants Pending National Institutes of Health/National Institute of Neurological Disorders and Stroke, Chad Moser Foundation, Comments: Research grants funded by named entities*; Royalties: Millipore Corporation, Comments: Cell lines were submitted with named entity.* Liliana Goumnerova—UNRELATED: Payment for Lectures Including Service on Speakers Bureaus: Stanford University, Department of Neurosurgery, Comments: invited as a visiting professor in October 2019 and received an honorarium ($500); Travel/Accommodations/Meeting Expenses Unrelated to Activities Listed: Stanford University, Department of Pediatric Neurosurgery, Comments: visiting professor (invited by them) October 2019, received transportation/accommodation and an honorarium. Pratiti Bandopahayay—RELATED: Grant: Novartis Institute of Biomedical Research, Comments: My lab receives grant funding for an unrelated project.* Rameen Beroukhim—UNRELATED: Grant: Novartis, Board Membership: Ampressa Therapeutics, Comments: member of scientific advisory board; Stock/Stock Options: Ampressa Therapeutics. Keith L. Ligon—UNRELATED: Consultancy: Bristol Myers Squibb for diffuse intrinsic pontine glioma trial design consulting. Susan Chi—RELATED: Grant: philanthropic and foundation, Comments: Zach Carson Fund, Ellie Kavalieros Diffuse Intrinsic Pontine Glioma Fund, Ryan Harvey Fund, Mikey Czech Diffuse Intrinsic Pontine Glioma Foundation, Caroline Cronk Fund, Prayers from Maria, Guglietti Diffuse Intrinsic Pontine Glioma Fund, Markoff Art in Giving Foundation, Brock Fleming Fund, Stop and Shop Pediatric Brain Tumor Program, the Diffuse Intrinsic Pontine Glioma Collaborative (which includes the Hope for Caroline Foundation, Reflections of Grace Foundation, Cure Starts Now Foundation, Soar with Grace Foundation, Abbie’s Army Charity Trust, Julian Boivin Courage for Cures Foundation, Smiles for Sophie Forever, Caroline’s Miracle Foundation, Love, Chloe Foundation, Benny’s World Foundation, Pray Hope Believe Foundation, Jeffrey Thomas Hayden Foundation) and 5P30 CA006516*; UNRELATED: Consultancy: Epizyme Inc. Mark W. Kieran—UNRELATED: Consultancy: Novartis, Boehringer Ingelheim, Eli Lilly, Sanofi, Merck, Bristol Myers Squibb, Bayer Pharmaceutics Company (Bayer), Takeda Pharmaceutical Company, Comments: Advisory Board; Travel/Accommodations/Meeting Expenses Unrelated to Activities Listed: Novartis, Comments: travel to Advisory Board. Karen D. Wright—RELATED: Grant: multiple grant funders, Comments: Zach Carson Fund, Ellie Kavalieros Diffuse Intrinsic Pontine Glioma Fund, Ryan Harvey Fund, Mikey Czech Diffuse Intrinsic Pontine Glioma Foundation, Caroline Cronk Fund, Prayers from Maria, Guglietti Diffuse Intrinsic Pontine Glioma Fund, Markoff Art in Giving Foundation, Brock Fleming Fund, Stop and Shop Pediatric Brain Tumor Program, the Diffuse Intrinsic Pontine Glioma Collaborative (which includes the Hope for Caroline Foundation, Reflections of Grace Foundation, Cure Starts Now Foundation, Soar with Grace Foundation, Abbie’s Army Charity Trust, Julian Boivin Courage for Cures Foundation, Smiles for Sophie Forever, Caroline’s Miracle Foundation, Love, Chloe Foundation, Benny’s World Foundation, Pray Hope Believe Foundation, Jeffrey Thomas Hayden Foundation) and 5P30 CA006516*; Support for Travel to Meetings for the Study or Other Purposes: Diffuse Intrinsic Pontine Glioma Collaborative, Comments: fund travel to/from diffuse intrinsic pontine glioma meeting; specifically, Diffuse Intrinsic Pontine Glioma Collaborative paid for flights for attendees to a meeting in Sydney, Australia, August 2–4, 2019. This meeting, however, was not related to this work, only diffuse intrinsic pontine glioma in general and plans for future studies; UNRELATED: Board Membership: Canines-N-Kids, Comments: Scientific advisory board; Consultancy: Grand Rounds through association with Boston Children’s Hospital, Boehringer Ingelheim, listed as consultant but have not done anything as of yet to warrant payment; Guidepoint Global Advisors, on list but have not participated in paid consult; Infinite MD, paid consultant for patients; Takeda Pharmaceutical Company, previously consulting because I was Principal Investigator/author for the trial they support with the drug TAK580, which has since been sold to Day One Biopharma; Grants/Grants Pending: 6314201, PLGA, $300,000, 9619394; Team Jack Foundation gift funding, to be split between TAK580 and MEK162, $600,000 (includes recent $100,000 donation that is not intended for TAK580 but was deposited here as a placeholder); 6310103, Team Jack Foundation, $300,000; Specialized Programs of Research Excellence in Human Cancers (SPORE) award fund 122249*; Patents (Planned, Pending or Issued): Dana Farber Cancer Institute (DFCI) case No. C2843 (participant is Karen Wright): Clinical Use of Day101 (formerly TAK580/MLN2840) for the treatment of low-grade glioma; and DFCI case No. C2844 (Participants are from DFCI, Day One Biopharmaceuticals, and Imaging Endpoints Core Lab): use of DWI/ADC as a biomarker of response specific to DAY101 (formerly TAK580/MLN2840). Tina Young Poussaint—UNRELATED: Grants/Grants Pending: Pediatric Brain Tumor Consortium neuroimaging center grant, National Institutes of Health*; Royalties: Springer. Frank Dubois—RELATED: Grant: German Research Foundation. *Money paid the institution.
Paper previously presented at: Annual Meeting of the American Society of Neuroradiology, May 18–23, 2019, Boston, Massachusetts.
C. Jaimes was partially supported by the Schlaeger Fellowship for Neuroscience Research.
References
- 1.Ostrom QT, Gittleman H, Truitt G, et al. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2011–2015. Neuro Oncol 2018;20:iv1–86 10.1093/neuonc/noy131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hassan H, Pinches A, Picton SV, et al. Survival rates and prognostic predictors of high-grade brain stem gliomas in childhood: a systematic review and meta-analysis. J Neurooncol 2017;135:13–20 10.1007/s11060-017-2546-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Castel D, Philippe C, Calmon R, et al. Histone H3F3A and HIST1H3B K27M mutations define two subgroups of diffuse intrinsic pontine gliomas with different prognosis and phenotypes. Acta Neuropathol 2015;130:815–27 10.1007/s00401-015-1478-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pope WB, Chen JH, Dong J, et al. Relationship between gene expression and enhancement in glioblastoma multiforme: exploratory DNA microarray analysis. Radiology 2008;249:268–77 10.1148/radiol.2491072000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gutman DA, Cooper LA, Hwang SN, et al. MR imaging predictors of molecular profile and survival: multi-institutional study of the TCGA glioblastoma data set. Radiology 2013;267:560–69 10.1148/radiol.13120118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nowak J, Nemes K, Hohm A, et al. Magnetic resonance imaging surrogates of molecular subgroups in atypical teratoid/rhabdoid tumor. Neuro Oncol 2018;20:1672–79 10.1093/neuonc/noy111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perreault S, Ramaswamy V, Achrol AS, et al. MRI surrogates for molecular subgroups of medulloblastoma. AJNR Am J Neuroradiol 2014;35:1263–69 10.3174/ajnr.A3990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kieran MW, Goumnerova LC, Prados M, et al. Biopsy for diffuse intrinsic pontine glioma: a reappraisal. J Neurosurg Pediatr 2016;18:390–91 10.3171/2015.6.PEDS15374 [DOI] [PubMed] [Google Scholar]
- 9.Jansen MH, Veldhuijzen van Zanten SE, Sanchez Aliaga E, et al. Survival prediction model of children with diffuse intrinsic pontine glioma based on clinical and radiological criteria. Neuro Oncol 2015;17:160–66 10.1093/neuonc/nou104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoffman LM, Veldhuijzen van Zanten SE, Colditz N, et al. Clinical, radiologic, pathologic, and molecular characteristics of long-term survivors of diffuse intrinsic pontine glioma (DIPG): A Collaborative Report from the International and European Society for Pediatric Oncology DIPG Registries. J Clin Oncol 2018;36:1963–72 10.1200/JCO.2017.75.9308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen HJ, Panigrahy A, Dhall G, et al. Apparent diffusion and fractional anisotropy of diffuse intrinsic brain stem gliomas. AJNR Am J Neuroradiol 2010;31:1879–85 10.3174/ajnr.A2179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poussaint TY, Vajapeyam S, Ricci KI, et al. Apparent diffusion coefficient histogram metrics correlate with survival in diffuse intrinsic pontine glioma: a report from the Pediatric Brain Tumor Consortium. Neuro Oncol 2016;18:725–34 10.1093/neuonc/nov256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gupta N, Goumnerova LC, Manley P, et al. Prospective feasibility and safety assessment of surgical biopsy for patients with newly diagnosed diffuse intrinsic pontine glioma. Neuro Oncol 2018;20:1547–55 10.1093/neuonc/noy070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fischbein NJ, Prados MD, Wara W, et al. Radiologic classification of brain stem tumors: correlation of magnetic resonance imaging appearance with clinical outcome. Pediatr Neurosurg 1996;24:9–23 10.1159/000121010 [DOI] [PubMed] [Google Scholar]
- 15.Jenkinson M, Beckmann CF, Behrens TE, et al. FSL. Neuroimage 2012;62:782–90 10.1016/j.neuroimage.2011.09.015 [DOI] [PubMed] [Google Scholar]
- 16.Schindelin J, Arganda-Carreras I, Frise E, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods 2012;9:676–82 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society, Series B (Methodological) 1995;57:289–300 10.1111/j.2517-6161.1995.tb02031.x [DOI] [Google Scholar]
- 18.Poussaint TY, Kocak M, Vajapeyam S, et al. MRI as a central component of clinical trials analysis in brainstem glioma: a report from the Pediatric Brain Tumor Consortium (PBTC). Neuro Oncol 2011;13:417–27 10.1093/neuonc/noq200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Infinger LK, Stevenson CB. Re-examining the need for tissue diagnosis in pediatric diffuse intrinsic pontine gliomas: a review. Curr Neuropharmacol 2017;15:129–33 10.2174/1570159x14666160425114024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khuong-Quang DA, Buczkowicz P, Rakopoulos P, et al. K27M mutation in histone H3.3 defines clinically and biologically distinct subgroups of pediatric diffuse intrinsic pontine gliomas. Acta Neuropathol 2012;124:439–47 10.1007/s00401-012-0998-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buczkowicz P, Bartels U, Bouffet E, et al. Histopathological spectrum of paediatric diffuse intrinsic pontine glioma: diagnostic and therapeutic implications. Acta Neuropathol 2014;128:573–81 10.1007/s00401-014-1319-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol 2016;131:803–20 10.1007/s00401-016-1545-1 [DOI] [PubMed] [Google Scholar]
- 23.Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med 2009;360:765–73 10.1056/NEJMoa0808710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goya-Outi JC, Orlhac R, Philippe F, et al. Can structural MRI radiomics predict DIPG histone H3 mutation and patient overall survival at diagnosis time? In: Proceedings of the Institute of Electrical and Electronics Engineers Engineering in Medicine and Biology Society International Conference on Biomedical & Health Informatics (BHI’19), Chicago, Illinois: May 19–22, 2019: 1–4 [Google Scholar]
- 25.Goya-Outi J, Orlhac F, Calmon R, et al. Computation of reliable textural indices from multimodal brain MRI: suggestions based on a study of patients with diffuse intrinsic pontine glioma. Phys Med Biol 2018;63:105003 10.1088/1361-6560/aabd21 [DOI] [PubMed] [Google Scholar]
- 26.Lober RM, Cho YJ, Tang Y, et al. Diffusion-weighted MRI derived apparent diffusion coefficient identifies prognostically distinct subgroups of pediatric diffuse intrinsic pontine glioma. J Neurooncol 2014;117:175–82 10.1007/s11060-014-1375-8 [DOI] [PubMed] [Google Scholar]
- 27.Zukotynski KA, Vajapeyam S, Fahey FH, et al. Correlation of (18)F-FDG PET and MRI apparent diffusion coefficient histogram metrics with survival in diffuse intrinsic pontine glioma: a report from the Pediatric Brain Tumor Consortium. J Nucl Med 2017;58:1264–69 10.2967/jnumed.116.185389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lobel U, Sedlacik J, Reddick WE, et al. Quantitative diffusion-weighted and dynamic susceptibility-weighted contrast-enhanced perfusion MR imaging analysis of T2 hypointense lesion components in pediatric diffuse intrinsic pontine glioma. AJNR Am J Neuroradiol 2011;32:315–22 10.3174/ajnr.A2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nowosielski M, Recheis W, Goebel G, et al. ADC histograms predict response to anti-angiogenic therapy in patients with recurrent high-grade glioma. Neuroradiology 2011;53:291–302 10.1007/s00234-010-0808-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aboian MS, Tong E, Solomon DA, et al. Diffusion characteristics of pediatric diffuse midline gliomas with histone H3-K27M mutation using apparent diffusion coefficient histogram analysis. AJNR Am J Neuroradiol 2019;40:1804–10 10.3174/ajnr.A6302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen H, Hu W, He H, et al. Noninvasive assessment of H3 K27M mutational status in diffuse midline gliomas by using apparent diffusion coefficient measurements. Eur J Radiol 2019;114:152–59 10.1016/j.ejrad.2019.03.006 [DOI] [PubMed] [Google Scholar]
- 32.Bailey S, Howman A, Wheatley K, et al. Diffuse intrinsic pontine glioma treated with prolonged temozolomide and radiotherapy: results of a United Kingdom phase II trial (CNS 2007 04). Eur J Cancer 2013;49:3856–62 10.1016/j.ejca.2013.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aboian MS, Solomon DA, Felton E, et al. Imaging characteristics of pediatric diffuse midline gliomas with histone H3 K27M mutation. AJNR Am J Neuroradiol 2017;38:795–800 10.3174/ajnr.A5076 [DOI] [PMC free article] [PubMed] [Google Scholar]