Abstract
The emergence of the novel betacoronavirus, recently renamed as severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) has raised serious concerns due to the virus's rapid dissemination worldwide. Nevertheless, there is limited information about the genomic epidemiology of SARS‐CoV‐2 circulating in Italy from surveillance studies. The shortage of complete genomic sequences available impairs our understanding of the SARS‐CoV‐2 introduction and establishment in the country. To better understand its dynamics in Italy, we analyzed complete genomes of SARS‐CoV‐2 isolates, obtained directly from clinical samples. Our phylogenetic reconstructions suggest possible multiple introduction of SARS‐CoV‐2. Continued genomic surveillance strategies are needed to improve monitoring and understanding of the current SARS‐CoV‐2 epidemics, which might help to attenuate public health impact of infectious diseases.
Keywords: COVID‐19, epidemic, Italian outbreak, phylogenetic inference
Research Highlights
COVId‐19 epidemic spread in Italy.
Italian circulation started form mutiple introduction.
Genomic data availabilty can help in epidemiological dynamic understanding improvement.
Italian epidemic was probably caused by two independent introductions mediated by China and Germany.
1. INTRODUCTION
Coronaviruses (CoVs) are enveloped, positive‐sensed, single‐stranded RNA viruses belonging to the Coronaviridae family. 1 They have been categorized based on antigenic properties and phylogenetic analyses in three main groups 2 : (a) alpha‐CoVs, mainly responsible for gastrointestinal disorders; (b) beta‐CoVs, that include: (i) bat coronavirus, (ii) the human severe acute respiratory syndrome (SARS) virus, (iii) the Middle Eastern respiratory syndrome (MERS) virus; (c) gamma‐CoVs, that mainly infect avian species. Some variants of CoVs are associated with outbreaks, some others are continuously circulating and cause mostly mild respiratory infections (eg, the common cold). The most well known of these CoVs is the SARS‐CoV, which between 2002 and 2003 was responsible to cause an outbreak that spread around the world and resulted in over 8000 cases and 774 deaths, with a case fatality rate of around 9% to 11%. 3 In 2012, a novel CoV, MERS‐CoV, causing severe respiratory symptoms was identified. 4 Very recently, in December 2019, a novel beta‐CoV, was responsible of a new illness, first detected in Wuhan, China. 5 We now know this to be another outbreak of CoV in humans (the 7th), that was recently renamed as SARS‐CoV‐2. The origin of the virus is still unclear, however, genomic analysis suggests nCoV is most closely related to viruses previously identified in bats. 6
In the first week of March 2020, over 98.192 confirmed cases and 3,380 deaths have been reported globally. 7 The case counts are dramatically rising in part due to increased surveillance and testing. While the outbreak seems to be centered in Wuhan, which is now under quarantine, the virus has spread throughout China and abroad, including Western and South‐Eastern Pacific regions, European regions, Eastern Mediterranean regions as well as many states in the United States of America. 8 It comes amid signs of the virus's shift away from its origins in China, with Italy, Iran, and South Korea emerging as the other countries hit hardest by the deadly disease. Italy is the worst affected country in Europe, with more than 4636 cases, a rise of 778 in a day, and a death toll of 197, an increase of 49. 9 Nevertheless, there is still limited information about the genomic epidemiology of the SARS‐CoV‐2 circulating in Italy from genomic surveillance studies that impairs our understanding of the virus introduction, establishment, and dissemination in the country. Thus, to better understand the emergence of the recent SARS‐CoV‐2 epidemic in Italy, we analyzed a larger and updated dataset of recently released data of the COVID‐2019 epidemic to improve more effective intervention strategies.
2. MATERIALS AND METHODS
2.1. Phylogenetic analysis
The dataset comprised (n = 141) complete genomes sequences from the current (2019‐2020) SARS‐CoV‐2 epidemic retrieved from GISAID (https://www.gisaid.org/) database. The alignment was performed using MAFFT online program. 10 The complete dataset was assessed for the presence of phylogenetic signal by applying the likelihood mapping analysis implemented in the IQ‐TREE 1.6.8 software (http://www.iqtree.org). 11 A maximum likelihood (ML) phylogeny was reconstructed using IQ‐TREE 1.6.8 software under the general time‐reversible nucleotide substitution model with a proportion of invariant sites (GTR+I) which was inferred in jModelTest (https://github.com/ddarriba/jmodeltest2) as the best fitting model. 12
3. RESULTS
To better understand the establishment and transmission of the novel SARS‐CoV‐2 in Italy, the recent Italian genomes available on the GISAID database were added to a dataset of 138 SARS‐CoV‐2 genomes from the global epidemic. An ML phylogenetic tree (Figure 1) was built to explore the recent Italian genomes relationship with previous worldwide isolates. ML phylogenies suggested that the three Italian strains, grouped into two different well‐supported phylogenetic clades (bootstrap score [BS] 0.80, 0.99, respectively). We found that two isolates from Italy (EPI_ISL_410546 a Chinese tourist from Hubei Province, and EPI_ISL_412974 the first Italian case isolate in Rome Italy) fall within a clade (BS = 94%) (Figure 1A clade 1) including sequences from several countries as China, Japan, Singapore, England, Australia, and Canada. However, it is important to note that sequences from China are positioned basally to this first clade, reinforcing the idea that the first introduction event could be mediated, as suspected by China.
Figure 1.
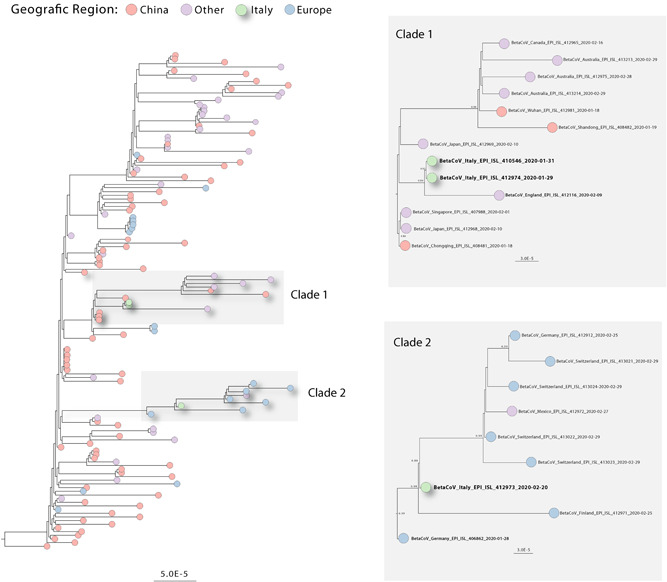
Maximum likelihood phylogeny, estimated from complete and near‐complete coronavirus (CoV) genomes using genome data available in GISAID. Colors represent different locations (panels A and B represent expansions of the clades containing the Italian CoV isolates [green]) in clade 1 closely related with sequence from England, in clade B closely related with sequences from Germany
We also identified that one Italian strain (EPI_ISL_412973 from the province of Lombardy) isolated in February 2020, grouped separately into a second clade (Figure 1B clade 2) including isolates from Germany, Finland, Switzerland, and Mexico. Phylogenetic analyses further suggest that the CoV epidemic might have been reintroduce to the country via Germany, since the basal sequence of this clade is a case that emerged in Munich in Germany in January 2020, highlighting a complex dynamic of transmission and a tight connection within European countries.
4. DISCUSSION
In this study, we investigate the transmission path of the novel SARS‐CoV‐2 in Italy. Our results indicate that the SARS‐CoV‐2 circulation in Italy likely started from different multiple introductions from China and Germany (Figure 1, clades 1 and 2) (suggesting a possible European linkage) followed by autochthonous transmission within Italy. Moreover, more genomic data will be fundamental to shed light on the epidemiological dynamics of the virus in the country helping to understand how the virus is spreading both around the world and thought time. More genomic surveillance efforts during epidemics should be undertaken to provide enough data to make reliable inferences on the molecular epidemiology of the rapidly evolving virus. Phylogenetic inference robustness will be improved if other genomic sequences will be available to build an even more informative dataset despite the low heterogeneity of the sequenced strains. Consequently, this analysis has to be considered as the first preliminary report regarding the SARS‐CoV‐2 epidemic in Italy that highlights two independent introductions probably mediated by China and Germany, respectively. This could reinforce the tight connection within samples that have been isolated all over Europe, indicating that the virus is spreading quite widely in Europe suggesting that it could be likely spreading in the general population.
In conclusion, our study shows that genomic data generated in real‐time can be employed to assist public health in monitoring and understanding the outbreak of novel emerging viral 13 pathogens.
ACKNOWLEDGMENTS
The authors would like to thank all personnel from Health Surveillance Systems who coordinated surveillance in Italy. MG is supported by Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro—FAPERJ.
Giovanetti M, Angeletti S, Benvenuto D, Ciccozzi M. A doubt of multiple introduction of SARS‐CoV‐2 in Italy: A preliminary overview. J Med Virol. 2020;92:1634–1636. 10.1002/jmv.25773
REFERENCES
- 1. Cui J, Li F, Shi Z‐L. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17(3):181‐192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Schoeman D, Fielding BC. Coronavirus envelope protein: current knowledge. Virol J. 2019;16(1):69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Peiris JS, Guan Y, Yuen KY. Severe acute respiratory syndrome. Nat Med. 2004;10(suppl 12):S88‐S97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814‐1820. [DOI] [PubMed] [Google Scholar]
- 5. Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727‐733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Benvenuto D, Giovanetti M, Ciccozzi A, Spoto S, Angeletti S, Ciccozzi M. The 2019‐new coronavirus epidemic: evidence for virus evolution. J Med Virol. 2020;92:455‐459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. World Health Organization , Coronavirus disease 2019 (COVID‐19) Situation Report – 46, March 2020. https://www.who.int/docs/default‐source/coronaviruse/situation‐reports/20200306‐sitrep‐46‐covid‐19.pdf?sfvrsn=96b04adf_2
- 8. World Health Organization , Novel Coronavirus (2019‐nCoV) Situation Report – 19, February 2020. https://www.who.int/docs/default‐source/coronaviruse/situation‐reports/20200208‐sitrep‐19‐ncov.pdf
- 9. Italian Ministry of Health , COVID‐19 ‐ Situazione in Italia, March 2020, http://www.salute.gov.it/portale/nuovocoronavirus/dettaglioContenutiNuovoCoronavirus.jsp?lingua=italiano&id=5351&area=nuovoCoronavirus&menu=vuoto
- 10. Katoh K, Rozewicki J, Yamada KD. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 2019;20(4):1160‐1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ‐TREE: a fast and effective stochastic algorithm for estimating maximum‐likelihood phylogenies. Mol Biol Evol. 2015;32(1):268‐274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 2012;9(8):772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Benvenuto D, Giovanetti M, Salemi M, et al. The global spread of 2019‐nCoV: a molecular evolutionary analysis. Pathog Glob Health. 2020;12:1‐4. [DOI] [PMC free article] [PubMed] [Google Scholar]


