Abstract
One of the most important branches of genetic engineering is the expression of recombinant proteins using biological expression systems. Nowadays, different expression systems are used for the production of recombinant proteins including bacteria, yeasts, molds, mammals, plants, and insects. Yeast expression systems such as Saccharomyces cerevisiae (S. cerevisiae) and Pichia pastoris (P. pastoris) are more popular. P. pastoris expression system is one of the most popular and standard tools for the production of recombinant protein in molecular biology. Overall, the benefits of protein production by P. pastoris system include appropriate folding (in the endoplasmic reticulum) and secretion (by Kex2 as signal peptidase) of recombinant proteins to the external environment of the cell. Moreover, in the P. pastoris expression system due to its limited production of endogenous secretory proteins, the purification of recombinant protein is easy. It is also considered a unique host for the expression of subunit vaccines which could significantly affect the growing market of medical biotechnology. Although P. pastoris expression systems are impressive and easy to use with well‐defined process protocols, some degree of process optimization is required to achieve maximum production of the target proteins. Methanol and sorbitol concentration, Mut forms, temperature and incubation time have to be adjusted to obtain optimal conditions, which might vary among different strains and externally expressed protein. Eventually, optimal conditions for the production of a recombinant protein in P. pastoris expression system differ according to the target protein.
Keywords: expression system, optimization, Pichia pastoris, recombinant proteins, subunit vaccines
The Pichia pastoris (P. pastoris) has also been established as a versatile cell factory for the production of thousands of biomolecules both on a laboratory and industrial scale. Optimal conditions for the production of a recombinant protein in P. pastoris expression system differ according to the target protein.
1. INTRODUCTION
Nowadays, biological expression systems are used for the production of heterologous proteins in industrial and medical fields. These proteins can consist of recombinant vaccines, drugs, and agricultural products (Gomes, Byregowda, Veeregowda, & Balamurugan, 2016). One of the obstacles in this area is the production of a large number of recombinant proteins in both the medical field and research. Therefore, researchers apply both prokaryotic and eukaryotic cells to overcome the difficulties associated with the production of recombinants proteins (Balamurugan, Sen, Saravanan, & Singh, 2006). Already used expression systems include: bacteria, yeasts, molds, mammals, plants, and insects. Prokaryotic cells such as Gram‐negative bacteria are among the first cells used in engineering genetic technology. One of the most important cells is Escherichia coli, that has been widely used for cloning recombinant DNA and subsequently, for the production of heterologous proteins (Baneyx, 1999). Bacterial expression system has several advantages including rapid multiplication, simple and inexpensive nutritional requirements, high‐level expression, and fast and easy transformation process. However, this cell factory has some limitations such as intracellular aggregation and misfolding of heterologous proteins, production of lipopolysaccharide, lack of posttranslational modification, and protein degradation due to proteases (Rosano & Ceccarelli, 2014). Another part of expression systems is the eukaryotic cells which include mammalian and yeast cells. The most common mammalian cell lines are Chinese hamster ovary (CHO) cells. Currently, CHO cells are used to produce biopharmaceutical compounds, monoclonal antibodies, and Fc‐fusion proteins. Apart from this, baby hamster kidney, human embryonic kidney 293 and NS0, SP2/0 (mouse‐derived myeloma) cell lines have also received legal permissions (Picanço‐Castro, Cristina Correa de Freitas, Bomfim, & Maria de Sousa Russo, 2014). Significant advantages of this system include proper protein folding, posttranslational modifications, and glycosylation of recombinant proteins in the correct sites which is important for protein stability (Khan, 2013). Besides, mammalian expression systems grow slowly and the relevant nutrient requirement is costly. On the other hand, potential contamination of culture medium with some viruses has limited its use in large‐scale production (Yin, Li, Ren, & Herrler, 2007). Yeasts are other eukaryotic cells that are widely used for the expression of several proteins in vaccine and pharmaceutical production. The mechanism of protein expression in these microorganisms is close to the ones in mammalian cells. Compared with bacteria, yeast cells have significant advantages including growth speed, posttranslational modification, secretory expression, and easy genetic manipulation. Furthermore, linearized foreign DNA can be inserted in a chromosome in high efficiency via cross recombination phenomena to generate stable cell lines (Daly & Hearn, 2005). Among yeast cells, Saccharomyces cerevisiae is used in the manufacture of hepatitis B and human papillomavirus vaccines, both of which produce a protective immune response against wild‐type viruses (Bill, 2015). The expression proteins in S. cerevisiae are often N and O‐hyperglycosylated, which may affect protein immunogenicity (Rasala & Mayfield, 2015). In recent years, to solve the problem of protein expression, methylotrophic yeasts such as Hansenulla polymorpha and Pichia pastoris (P. pastoris; syn. Komagataella phaffii) have been developed. Among these, P. pastoris has become the most popular for its cost and expression host system. This microorganism can produce high yields of recombinant proteins with the high similarity of glycosylation to the mammalian cells (Balamurugan, Reddy, & Suryanarayana, 2007). Overall, the benefits of protein production by P. pastoris system include appropriate folding (in the endoplasmic reticulum [ER]) and secretion (by Kex2 as signal peptidase) of recombinant proteins to the external environment of the cell (S. Yang et al., 2013). Given the fact that some proteins produced by their original host are secreted out of the cell; P. pastoris is suitable for the production of recombinant proteins since it is equipped with a secretion system (Ahmad, Hirz, Pichler & Schwab, 2014). The basic characteristics of different host systems for the expression of recombinant proteins are summarized in Table 1.
Table 1.
Basic characteristics of different host systems for the expression of recombinant proteins
| Characteristics | Escherichia coli | Pichia pastoris | CHO cell |
|---|---|---|---|
| Doubling time | 30 min | 60–120 min | 24 hr |
| Cost of growth medium | Low | Low | High |
| Complexity of growth medium | Minimum | Minimum | Complex |
| Expression level | High | Low to high | Low to moderate |
| Extracellular expression | Secretion to periplasm | Secretion to medium | Secretion to medium |
| Protein folding | Refolding usually required | Refolding may be required | Proper folding |
| N‐linked glycosylation | None | High mannose | Complex |
| O‐linked glycosylation | No | Yes | Yes |
| Phosphorylation & acetylation | No | Yes | Yes |
| Drawback | Accumulation of LPS | Codon bias | Contamination with animal viruses |
Abbreviations: CHO, Chinese hamster ovary; LPS, lipopolysaccharide.
2. BACKGROUND OF P. pastoris AS A EUKARYOTIC EXPRESSION SYSTEM
Historically, P. pastoris yeast was first isolated from the exudates of a chestnut tree in France and was named Zygosaccharomyces pastoris by Guilliermond (Zahrl, Peña, Mattanovich, & Gasser, 2017). Yamada et al. then categorized the organism to a novel genus, Komagataella or Pichia (Naumov, Naumova, & Boundy‐Mills, 2018). It has been proven that P. pastoris, as an engineered methylotrophic microorganism, can use methanol as sole carbon and energy source (Cereghino, Cereghino, Ilgen & Cregg, 2002). Unlike, Y‐11430 strain (wild‐type), which is not used for protein expression, GS115 is one of the most popular strains used as an important expression system particularly in industry and medicine fields (Julien, 2006). P. pastoris GS115 strain has two encoding genes (AOX1 and AOX2) of alcohol oxidase (AOX) enzyme. In the presence of methanol, the transcription of these genes is induced and finally produces a high amount of AOX enzyme (Vanz et al., 2012). Although both genes are used for the production of an AOX enzyme, AOX1 produces more enzyme. Therefore, by knocking out the AOX1 gene, the growth on methanol is slowed down drastically. This phenotype is called methanol utilization slow (MutS). A knockout of the AOX2 gene will not decelerate growth on methanol and the growth rates are comparable to methanol utilizing plus (Mut+) phenotype (wild‐type). However, by knocking out both genes, the strains are unable to grow on methanol (methanol utilizing minus [Mut−]; Cámara et al., 2017). In the KM71 strain, as a derivative of GS115, aox1 gene has been deleted; therefore, this strain is known as MutS strain (Charoenrat et al., 2013). Older strains such as KM7121, MC100‐3, and MC101‐1, cannot use methanol as a food source (Mut−), because no AOX genes were detected in these strains and therefore they were unable to grow in the presence of methanol (Cregg, Madden, Barringer, Thill, & Stillman, 1989).
3. THE CHARACTERISTIC FEATURES OF P. pastoris EXPRESSION SYSTEM
3.1. Advantages and drawbacks of the P. pastoris expression system
Pichia expression system has advantages for the expression of different recombinant proteins. As noted, P. pastoris is a methylotrophic yeast which is known as a recombinant expression host system. One of the advantages of the Pichia system is its high similarity with advanced eukaryotic expression systems such as CHO cell lines. This yeast system is inexpensive, it also has relatively rapid expression times, cotranslational and posttranslational processing. By the use of industrial bioreactors, proteins of interest can be produced on a large scale from small culture volumes. Recently, studies have shown that the Pichia expression system is unique in the production of membrane proteins including calcium and potassium channels, nitrate and phosphate transporter, and histamine H1 receptor (Byrne, 2015). Furthermore, P. pastoris is a suitable microorganism in the secretory production of recombinant proteins directly into the supernatant of the culture medium. In the P. pastoris expression system due to its limited production of endogenous secretory proteins, the purification of recombinant protein is easy (Tachioka et al., 2016).
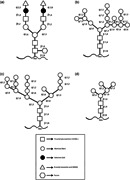
Another advantage of P. pastoris as a protein production host is its ability to perform posttranslational modifications such as O‐ and N‐linked glycosylation and disulfide bond formation. Many therapeutic proteins are glycoproteins and require the attachment of carbohydrate structures to the protein backbone (glycosylation) to allow for correct folding, solubility, stability, and proper biological activity (Cereghino et al., 2002). There are two main types of glycosylation in yeast cells (N‐linked and O‐linked glycosylation) that takes place in the ER or Golgi apparatus. In yeast, the structure of N‐linked glycans is typical of the hypermannose type, whereas, in humans, complex and hybrid structures are of the predominant type. For providing N‐linked glycosylation, oligosaccharides are attached to the amide nitrogen of asparagine (Asn) residue through an N‐glycosyl linkage within the consensus sequence Asn‐X‐Ser/Thr (where X is any amino acid except proline). For yielding the O‐linked type of glycosylation, oligosaccharides are attached to serine or threonine residues through a glycosidic linkage. The O‐linked saccharides are typically much smaller than N‐linked saccharides (<5 residues). N‐glycosylation in S. cerevisiae is characterized by hypermannosylation with α‐1,2‐, α‐1,6‐, and α‐1,3‐mannosyltransferases (Figure 1b). In comparison to S. cerevisiae, P. pastoris may have an advantage in the glycosylation of secreted proteins because it may not hyperglycosylated. In P. pastoris N‐glycans (Man8‐14GlcNAc2) are frequently shorter than the long oligosaccharide chains (Man > 50GlcNAc2) found in the S. cerevisiae. Besides, S. cerevisiae core oligosaccharides have terminal α‐1,3 glycan linkages whereas P. pastoris does not. Unlike S. cerevisiae, P. pastoris does not hyperglycosylated therapeutic proteins and does not contain potentially immunogenic terminal α‐1,3‐linked mannoses (Figure 1c). It is believed that the α‐1,3 glycan linkages in glycosylated proteins produced from S. cerevisiae are primarily responsible for the hyper‐antigenic nature of these proteins making them particularly unsuitable for therapeutic use. Moreover, very little O‐linked glycosylation has been observed in P. pastoris (Bretthauer & Castellino, 1999). Due to its attractive characteristics for heterologous protein production (low incidence of hyperglycosylation), P. pastoris is an interesting organism for the production of therapeutic glycoproteins. The N‐glycosylation plays an important role in achieving complete biological activities of therapeutic proteins such as interferon, erythropoietin, and monoclonal antibodies. The N‐glycosylation pathway in mammalian cells consists of the addition of one or more N‐acetylglucosamine (GlcNAc) residues followed by the sequential addition of galactose (Gal) and sialic acid which creates a complex type of N‐glycans (Figure 1a). While, hypermannosylation of recombinant protein in the P. pastoris expression system can lead to immunologic reaction and decreased serum half‐life (Laukens, De Wachter, & Callewaert, 2015). Recently, new strategies have been designed to engineer the P. pastoris N‐glycosylation pathway. With Pichia GlycoSwitch, the yeast's own hyperglycosyl N‐glycans are switched to the more human biantennary complex‐type N‐glycans. In glycoengineering strategy by disruption of an endogenous glycosyltransferase gene (OCH1) and introducing heterologous enzyme activities, Pichia has been engineered to produce human‐like glycoproteins (Jacobs, Geysens, Vervecken, Contreras, & Callewaert, 2009). Indeed, the first step of humanizing Pichia glycosylation or GlycoSwitch® strategy is the knockout of the gene coding for α‐1,6‐mannosyltransferase (OCH1) which is responsible for the initiation of hypermannosylation. The next step in this process is the co‐overexpression of several glycosyltransferases or glycosidase to produce human‐like glycoproteins. Commonly employed Pichia GlycoSwitch® strains (BioGrammatics, Carlsbad, CA) are SuperMan5, SuperMan5HIS−, SuperMan5pep4–, SuperMan5(aox1–, Muts), SuperMan5(pep4–, prb1–) and SuperMan5(pep4–, sub2–). These strains, OCH1 inactivated strains that express an ER‐targeted α‐1,2‐mannosidase, express target protein with a mannose‐5 structure at N‐linked site (Ahmad et al., 2014; Figure 1d). The SuperMan5 strain is commonly used to express of vaccine antigens. Moreover, by introducing heterologous enzyme activities and adding of N‐acetyl glucose amine (GN) or Gal, the SuperMan5 strain has been engineered to produce human‐like glycoproteins (M5GN and M5GNGal strains). M5GN and M5GNGal strains (BioGrammatics) are commonly used to express vaccine antigens, cytokines, and antibodies.
Figure 1.
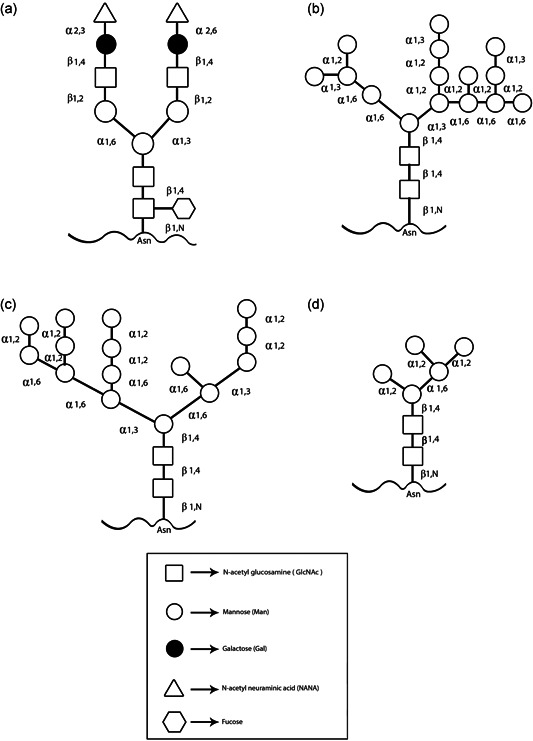
Schematic diagram of N‐linked glycan structure in a mammalian cell, Saccharomyces cerevisiae and, Pichia pastoris. (a) N‐linked glycan structure in mammalian cells commonly generates complex terminally sialylated structures. (b) In S. cerevisiae, the N‐linked glycan structure is typically hypermannosylated (Man > 50GlcNAc2). Moreover, S. cerevisiae core oligosaccharides have terminal α‐1,3 glycan linkages. (c) N‐linked glycan structure in P. pastoris typically is of the Man8‐14GlcNAc2 type with a triantennary‐branched structure. Unlike S. cerevisiae, P. pastoris does not contain potentially immunogenic terminal α‐1,3‐linked mannoses. (d) In Pichia GlycoSwitch® strains (SuperMan5) N‐linked glycan structure is typically hypomannosylated (with a mannose‐5 structure)
However, like other expression systems, this eukaryotic system has some disadvantages. In the transformation stage, unlike the bacterial system, competent cells require large (µg‐level) amounts of the plasmid. The number of E. coli transformants (108–1011) is higher than P. pastoris transformants (103–104) per µg of DNA (S. Wu & Letchworth, 2004). The production of recombinant protein in this system is regulated through two promoters: Promoter of glyceraldehyde‐3‐phosphate dehydrogenase (PGAP) and promoter of AOX (PAOX1). Despite several advantages, both promoters have no tenability (Rajamanickam, Metzger, Schmid, & Spadiut, 2017). Protein production in yeast systems is dependent on the consumption of methanol. Based on EasySelect™ Pichia Expression Kit (Catalog no. K1750‐01, Invitrogen, Carlsbad, CA), at least 0.5% concentration of methanol is required for the expression of recombinant proteins. The production could reach to the maximum level of 2–2.5% (wt/vol) of methanol (Z. Wang et al., 2010). Normally, the concentration of methanol up to 5% is tolerable for the organisms, but high levels of methanol concentrations (above 5%) are very toxic for cell viability and can stop the production process (Santoso, Herawati, & Rubiana, 2012). Another limitation in the Pichia system is the presence of a few selectable markers for P. pastoris transformation. Selectable marker genes include his4, arg4, and Shble (needed to resist against Zeocin antibiotic) (Cereghino & Cregg, 1999). A common occurrence in this system is the contamination of expressive broth culture with saprophytic bacteria and fungi. The secreted proteases of these microorganisms potentially hydrolyze the secreted proteins to the supernatant (Stewart, 2015). Indeed, one of the problems in the P. pastoris expression system is the destruction of proteins produced by proteases. New P. pastoris strains such as SMD1163 (his4 pep4 prb1), SMD1165 (his4 pep4), SMD1168 (his4 pep4), BG21, and Pichia pink have no protease; therefore, the degradation of a secreted protein is prevented. In these strains, to achieve high product yields and the quality of recombinant proteins, the genes encoding of proteinase A (pep4) and proteinase B (prb1) have been disrupted (Safder, Khan, Islam, & Kazim, 2018).
3.2. General topics for cloning and expression in P. pastoris
The expression of any recombinant gene in P. pastoris has three phase: (a) Cloning of a new gene into a suitable expression vector, (b) insertion of the cloned vector into P. pastoris host genome; and (c) trial of the potential different strains for the expression of the recombinant integrated gene (Macauley‐Patrick, Fazenda, McNeil, & Harvey, 2005). Based on EasySelect™ Pichia Expression Kit (Invitrogen), to increase the efficacy of external DNA integration into the Pichia genome, first, the cloned vector should be linear with restriction enzymes such as Sac I, PmeI, and BstX I. Then, linear DNA should be inserted into the competent cells by electroporation. The entered gene is integrated into the cell genome via the crossover recombination phenomenon (Figure 2), and consequently, recombinant cells are formed. In most cases, a single crossover occurs in the genome, but multiple insertions occur in 1–10% of the cases (Kit).
Figure 2.
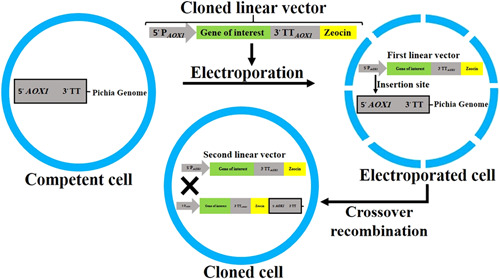
Crossover recombination phenomenon in the Pichia pastoris genome. Following the electroporation process of competent yeast cells, cloned linear vectors are inserted into the electroporated cells. Crossover recombination occurs between 5′ promoter (5′ PAOX1) of vector and AOX1 region of P. pastoris genome. Consequently, cloned cells with a recombinant genome are formed. AOX1, alcohol oxidase 1; TT, transcription termination region
The expression vectors in the P. pastoris expression system are one of the major components of this system. These vectors are composed of three sequences: Promoter sequence (most often AOX1) in 5′ region; transcriptional termination sequence in 3′ region which is essential in the processing and polyadenylation of messenger RNAs; and one sequence that contains single or multiple cloning sites, necessary for the insertion of the gene of interest. The episomal vectors can replicate either autonomously in the cytoplasm or as part of a chromosome. But the vectors used in P. pastoris have no stable episomal status; therefore, they should first be linearized with enzymes and then be integrated into the P. pastoris chromosome (Li et al., 2007). Like E. coli, expression vectors in P. pastoris are shuttle vectors that is they can propagate in two different host species. The vectors also contain one drug resistance genes such as Kan, Shble, Bsd, Amp, or FLD1, which are resistant to geneticin, zeocin, blasticidin, ampicillin, and formaldehyde, respectively (Ilgen, Lin‐Cereghino, & Cregg, 2005). Common plasmids used in the P. pastoris expression system to produce extracellular and intracellular proteins are listed in Tables 2 and 3, respectively.
Table 2.
Common Pichia pastoris expression vectors for the production of secretory proteins
| Vector name | Marker gene | Used strain | Recombinant protein | Reference |
|---|---|---|---|---|
| pPIC9K | His4, Kan, Amp | GS115 | Xylanase | Fu, Zhao, Xiong, Tian, and Peng (2011) |
| GS115 | Porcine circovirus type 2 | Tu et al. (2013) | ||
| GS115 | Endo‐1,3(4)‐b‐d‐glucanase | X. Chen et al. (2012) | ||
| GS115 | Staphylokinase | Apte‐Deshpnade, Mandal, Soorapaneni, Prasad, Kumar, and Padmanabhan (2009) | ||
| pPICZα | Shble | SMD1168 | Human chitinase | Goodrick et al. (2001) |
| GS115 | Human topoisomerase I | Chan et al. (2018) | ||
| GS115 | Human interferon gamma | Prabhu, Veeranki, and Dsilva (2016) | ||
| X‐33 | C‐reactive protein | J. Li et al. (2017) | ||
| SuperMan5 | Insulin | Baeshen et al. (2016) | ||
| X‐33 | Human RNase4 | Bardiya and Chang (2017) | ||
| pHIL‐S1 | His4, Amp | GS115 | Rabies virus glycoprotein | Ben Azoun, Belhaj, Göngrich, Gasser, and Kallel (2016) |
| GS115 | Rhizopus oryzae Lipase | Satomura, Kuroda, and Ueda (2015) | ||
| KM71 | Camel lactoferricin | Chahardooli, Niazi, Aram, and Sohrabi (2016) | ||
| pGAPZα | Shble | GS115 | Acyl homoserine lactonase | J. Wu et al. (2016) |
| SMD1168 | Variable lymphocyte receptor B | J. S. Lee et al. (2018) | ||
| X‐33 | Human gastric lipase | Sams et al. (2017) | ||
| pJL‐SX | FLD1, Amp | MS105 | Formaldehyde dehydrogenase | Sunga and Cregg (2004) |
| pBLHIS‐SX | His4, Amp | JC100 | Leukocyte protease inhibitor | Li et al. (2010) |
Table 3.
Common Pichia pastoris expression vectors for the production of intracellular proteins
| Vector name | Marker gene | Used strain | Recombinant protein | Reference |
|---|---|---|---|---|
| pPIC3.5K | His4, Kan, Amp | KM71 | Maltooligosyltrehalose synthase | Han, Su, Hong, Wu, and Wu (2017) |
| SMD1168 | Camellia sinensis heat shock protein | Wang, Zou et al. (2017) | ||
| GS115 | Pleurotus ostreatus laccases | Zhuo et al. (2018) | ||
| GS115 | Rhizopus oryzae Lipase | Jiao, Zhou, Su, Xu, and Yan (2018) | ||
| GS115 | HSA/GH fusion protein | M. Wu et al. (2014) | ||
| pPICZ | Shble | X‐33 | Aquaporin | Nordén et al. (2011) |
| KM71 | Membrane protein | J. Y. Lee, Chen, Liu, Alba, and Lim (2017) | ||
| KM71 | Dengue virus envelope glycoprotein | Khetarpal et al. (2017) | ||
| pHIL‐D2 | His4, Amp | GS115 | Prostaglandin H synthase‐2 | Kukk and Samel (2016) |
| GS115 | CatA1 and SODC | Mina et al. (2017) | ||
| KM71 | Rhodococcus nitrile hydratase | Pratush, Seth, and Bhalla (2017) | ||
| GS115 | Feline serum albumin | Yokomaku, Akiyama, Morita, Kihira, and Komatsu (2018) | ||
| pGAPZ | GS115 | GTPase RabA4c | Glöckner and Voigt (2015) | |
| GS115 | Xylose isomerase | Li, Sun, Chen, Li, and Zhu (2015) | ||
| GS115 | β‐Galactosidase | H. Sun et al. (2017) | ||
| pJL‐IX | FLD1, Amp | MS105 | Formaldehyde dehydrogenase | Sunga and Cregg (2004) |
| pBLHIS‐IX | His4, Amp | KM71 | L1‐L2 proteins of HPV virus type 16 | Bredell, Smith, Görgens, and van Zyl (2018) |
Abbreviation: HPV, human papillomavirus.
3.3. Subunit vaccines expressed in Pichia pastoris
Recombinant protein therapeutics is a growing market within the human medical biotechnology industry. The majority of all approved biopharmaceuticals are protein‐based and include blood factors, anticoagulants, hormones, hematopoietic growth factors, interferons, interleukins, vaccines, and monoclonal antibodies. Vaccine production is one of the major protective strategies against infectious diseases. In general, vaccines used for all pathogens are divided into three groups: live attenuated, inactivated/killed, and subunit vaccines (Schiller & Lowy, 2014). After the injection of live attenuated vaccines, microorganisms start to replicate in the injection area and could induce strong immune responses. However, reversion of attenuated pathogen to its wild‐type strain especially in immunocompromised individuals has restricted the application of these vaccines (Minor, 2015). Although, inactivated/killed vaccines are safe, they are relatively less effective in the induction of strong immune responses. Therefore, these vaccines should be injected in multiple doses along with suitable adjuvants (S. Lee & Nguyen, 2015). Subunit vaccines consist of one or more immunodominant antigens of pathogens that can be produced in eukaryotic and prokaryotic systems. Unlike live attenuated and killed/inactivated vaccines, subunit vaccines are completely safe and cost‐effective. Recently, subunit vaccines have been replaced with other forms of vaccines. Also, for the production of recombinant subunit vaccines, P. pastoris is more famous than other expression systems (Wang, Jiang, & Wang, 2016). Subunit vaccines often suffer from poor immunogenicity and require certain helper molecules known as adjuvants to induce or enhance an appropriate immune response to the antigen. T‐cell activation is crucial in inducing protective immune responses (Todryk, 2018). Antigens mannosylated by P. pastoris have shown to have enhanced antigen presentation and T‐cell activation properties compared with their nonglycosylated counterparts. Therefore, glycoproteins derived from P. pastoris have the potential to function as adjuvants. The increased immunogenicity of mannosylated glycoproteins is thought to be linked to certain mannose‐binding receptors carried by professional antigen‐presenting cells, like dendritic cells and macrophages (Luong, Lam, Chen, & Levitz, 2007). Some of the new recombinant subunit vaccines that have been expressed in P. pastoris system are listed in Table 4.
Table 4.
Recombinant subunit vaccine expressed in Pichia pastoris
| Construct name | Used strain | Used vector | Targeted disease | Reference |
|---|---|---|---|---|
| PIMP‐V1 and PIMP‐V2 | KM71 | pPICZαA | Malaria | Spiegel et al. (2015) |
| P1‐3CD | PichiaPink | pPink‐HC | Hand–foot–mouth disease | C. Zhang et al. (2015) |
| DENV‐3 E | KM71 | pPICZ‐A | Dengue | Tripathi et al. (2015) |
| CHIKV‐C‐E3‐E2‐6K‐E1 | GS115 | pPIC9K | Chikungunya | Saraswat et al. (2016) |
| Gp350 | GS115 | pPICZαA | EBV infection | Wang et al. (2016) |
| RBD219‐N1 | X‐33 | pPICZαA | SARS | W.‐H. Chen et al. (2017) |
| VP2–VP5–Fc | GS115 | pPIC9K | Infectious bursal | H. Wang et al. (2017) |
| F protein | GS115 | pPICZαA | Newcastle | Kang et al. (2016) |
| OmpA | GS115 | pPIC9K | P. mirabilis infection | Y. Zhang et al. (2015) |
| BoNT Hc | X‐33 | pPICZ‐A | Botulism | Webb et al. (2017) |
| Tc52 | GS115 | pPICZαA | Chagas | Matos, Alberti, Morales, Cazorla, and Malchiodi (2016) |
| Apa | GS115 | pPIC9K | Tuberculosis | S. Wang, Wang, Wang, Chen, and Kong (2018) |
| HBHA | GS115 | pPIC9K | Tuberculosis | Teng, Chen, Zhu, and Xu (2018) |
| CFP10‐Fcγ2a | GS115 | pPICZαA | Tuberculosis | Baghani et al. (2017) |
| ESAT6‐CFP10‐Fcγ2a | GS115 | pPICZαA | Tuberculosis | Farsiani et al. (2016) |
| ESAT6‐Fcγ2a | GS115 | pPICZαA | Tuberculosis | Kebriaei et al. (2016) |
| CFP10‐HspX‐Fcγ2a | GS115 | pPICZαA | Tuberculosis | Mosavat et al. (2016) |
| ESAT6‐HspX‐Fcγ2a | GS115 | pPICZαA | Tuberculosis | Soleimanpour et al. (2015) |
| Glycoprotein D | GS115 | pPIC9K | HSV‐2 infection | Wang, Jiang et al. (2017) |
| OmpA‐Fc | GS115 | pPIC9K | Bordetellosis | Dong et al. (2016) |
Abbreviations: EBV, Epstein‐Barr virus; SARS, severe acute respiratory syndrome.
4. STRATEGIES TO IMPROVE PROTEIN EXPRESSION IN P. pastoris
Methanol and sorbitol concentration, Mut forms, temperature and incubation time have to be adjusted to obtain optimal conditions, which might vary among different strains and externally expressed protein.
4.1. The impact of Mut forms and methanol on protein concentration
Methanol is the principal carbon source and gene expression induction agent in most P. pastoris fermentation strategies. Monitoring methanol levels during fermentation enables cell growth and optimizes productivity, whilst methanol toxicity is avoided. Methanol is used for both biomass growth and the production of protein in P. pastoris. Based on the utilization of methanol, P. pastoris strains are divided into three phenotypes: Mut+ strains with both AOX1 and AOX2 genes in their chromosomes; MutS strains with only AOX2 gene; and Mut− mutant strains without any AOX genes (Anggiani, Helianti, & Abinawanto, 2018). Therefore, the growth rate of different strains depends on their Mut forms. Mut+ strains can grow in a wide range of micron from 0.028 to 0.154 per hour, while MutS strains at a narrow range of 0.011–0.035 per hour (Looser et al., 2015). As noted above, based on EasySelect™ Pichia Expression Kit, at least 0.5% concentration of methanol is necessary for the production of recombinant protein, and to fully express the protein, the concentration must be at most 2–2.5% (wt/vol). Besides, high levels of methanol (concentrations above 5%) are toxic to the cells, leading to the accumulation of formaldehyde and hydrogen peroxide, and consequently the death of the cells. On the other hand, low levels of methanol trigger proteolytic degradation of heterologous proteins, which results in lower productivity (W. Zhang, Bevins, Plantz, Smith, & Meagher, 2000). The optimum methanol concentration used in some research articles is listed in Table 5.
Table 5.
Optimum concentration of methanol for the expression of recombinant protein
| Used strain | Mut form | Optimum methanol concentration (%) | Reference |
|---|---|---|---|
| GS115 | Mut+ | 2 | Z. Wang et al. (2010) |
| X‐33 | Mut+ | 2.5 | Santoso et al. (2012) |
| GS115 | Mut+ | 3 | Anggiani et al. (2018) |
| GS115 | Mut+ | 2 | Farsiani et al. (2016) |
| GS115 | Mut+ | 2.5 | Mosavat et al. (2016) |
| GS115 | Mut+ | 2 | Soleimanpour et al. (2015) |
| X‐33 | Mut+ | 1 | Tyagi et al. (2016) |
| X‐33 | Mut+ | 0.5 | T. Zhao et al. (2018) |
| GS115 | Mut+ | 2 | Cunha, Gama, Cintra, Bataus, and Ulhoa (2018) |
| GS115 | Mut+ | 1 | Camattari et al. (2016) |
| GS115 | Mut+ | 0.5 | J. Wang et al. (2017) |
| KM71 | MutS | 1 | Han et al. (2017) |
| GS115 | Mut+ | 0.5 | Apte‐Deshpnade et al. (2009) |
| GS115 | Mut+ | 1 | Dehnavi, Siadat, Roudsari, and Khajeh (2016) |
| X‐33 | Mut+ | 0.5 | Jain, Kumar, Bhardwaj, and Kuhad (2018) |
| GS115 | Mut+ | 2 | Farsiani et al. (2016) |
| GS115 | Mut+ | 2.5 | Soleimanpour et al. (2015) |
| GS115 | Mut+ | 2 | Mosavat et al. (2016) |
Abbreviation: Mut+, methanol utilizing plus.
4.2. The impact of sorbitol and temperature on protein concentration
In the P. pastoris expression system, one of the most well‐known carbon sources which can be used with methanol is sorbitol. Sorbitol does not induce or repress AOX promoters, hence using sorbitol instead of glycerol in mixed substrate methods could reduce cell growth rate, and increase specific product formation rates. The use of mixed substrates increases productivity and cell density and reduces induction time (Orman, Calik, & Ozdamar, 2009). Moreover, cofeeding of sorbitol with methanol reduces the toxic effects of intermediate metabolites and oxygen consumption. However, sorbitol has a negative impact on the specific activity of the AOX1 promoter (Çelik, Çalık, & Oliver, 2009). Several studies revealed that methanol/sorbitol cofeeding increases the expression of the recombinant proteins (Gao et al., 2012; Z. Wang et al., 2010; T. Zhu et al., 2011). The results obtained by Azadi, Mahboubi, Naghdi, Solaimanian, and Mortazavi (2017) showed that sorbitol at a concentration of 50 g/L (5%) could significantly increase the expression of recombinant protein. Moreover, according to the results of other studies, 2.5% methanol with 1% sorbitol (Mosavat et al., 2016), 2% methanol with 0.5% sorbitol (Farsiani et al., 2016), and 2% methanol with 1% sorbitol (Soleimanpour et al., 2015) were considered optimal for the highest recombinant protein production in shake flask culture. Therefore, the presence of other carbon sources such as glycerol and sorbitol can be beneficial to increase protein production. Since the AOX1 promoter is repressed severely by glycerol, sorbitol is considered an appropriate replacement (M. Gao & Shi, 2013).
The required growth temperature for P. pastoris is 28–30°C. Temperatures above 32°C could be detrimental to protein expression induction and can even cause cell death. Several studies revealed that by lowering the cultivation temperature from 30°C to 20°C, protein production can be improved according to higher yeast cell viability, decreased folding stress and lower proteolytic activity against the target protein (Dragosits et al., 2009; Gao et al., 2015; Gasser et al., 2007; Li et al., 2001; Zhong et al., 2014). The decreased synthetic rate of target protein in low‐temperature cultivation causes ER stress reduction, preserves the folding capacity of the ER and enhances cell viability (Zhong et al., 2014).
4.3. The impact of expression time on protein concentration
Incubation time is one of the most critical factors for acquiring the highest protein expression level in the P. pastoris expression system. In the P. pastoris expression system, production time is relatively long (about 100 hr). The incubation time is related to the number of yeast cells and the degree of target protein degradation. Santoso et al. (2012) showed that the highest growth of P. pastoris cells was at 96 hr incubation time, whereas the highest protein expression occurred at 48 hr. This suggests that it is very possible that longer incubation time may cause more proteolytic digestion of the expressed protein. Moreover, other studies discussed that optimal protein expression occurred at 72 to 96 hr incubation time (Farsiani et al., 2016; Soleimanpour et al., 2015).
5. OTHER BIOMOLECULES PRODUCED BY P. pastoris
The P. pastoris has also been established as a versatile cell factory for the production of thousands of biomolecules both on a laboratory and industrial scale. Some of the new recombinant biological molecules that have been expressed in the P. pastoris system are listed in Table 6.
Table 6.
Pichia pastoris as a suitable host for the production of recombinant biological molecules
| Product | Used strain | Used vector | Usage | Reference |
|---|---|---|---|---|
| Lycopene and β‐carotene | X‐33 | pGAPZA | Feed supplements | Araya‐Garay, Feijoo‐Siota, Rosa‐dos‐Santos, Veiga‐Crespo, and Villa (2012) |
| Plectasin | X‐33 | pPICZαA | Antibacterial peptide | J. Zhang et al. (2011) |
| Bovine lactoferrin | KM71H | pJ902 | Transferrin and antibacterial protein | Iglesias‐Figueroa et al. (2016) |
| Bovine IFN‐α | GS115 | pPIC9K | Prevention and therapy of viral diseases | Tu et al. (2016) |
| Apidaecin | SMD1168 | pPIC9K | Antibacterial peptide | X. Chen et al. (2017) |
| hPAB‐β | GS115 | pPIC9K | Antibacterial peptide | Z. Chen et al. (2011) |
| Tachyplesin I | GS115 | pGAPZαB | Antibacterial peptide | H. Li et al. (2019) |
| Snakin‐1 | GS11 | pPIC9 | Antimicrobial peptide | Kuddus et al. (2016) |
| PAF102 | X‐33 | pGAPZA | Antifungal peptide | Popa, Shi, Ruiz, Ferrer, and Coca (2019) |
| Pisum sativum defensin 1 | GS115 | pPIC9K | Antifungal peptide | Cabral, Almeida, Valente, Almeida, and Kurtenbach (2003) |
| Class I chitinase | KM71H | pPICZαA | Antifungal peptide | Landim et al. (2017) |
| Ch‐penaeidin | KM71H | pPIC9K | Antimicrobial peptide | L. Li et al. (2005) |
| Hispidalin | GS115 | pPICZαA | Antimicrobial peptide | Meng et al. (2019) |
| Fowlicidin‐2 | X‐33 | pPICZαA | Antimicrobial peptide | Xing et al. (2016) |
| Parasin I | X‐33 | pPICZαA | Antimicrobial peptide | H. Zhao et al. (2015) |
| CecropinA‐thanatin | X‐33 | pPICZαA | Antimicrobial peptide | Z. Liu et al. (2018) |
| Type I collagen | Connective tissue | Nokelainen et al. (2001) | ||
| Human serum albumin | GS115 | pPIC9K | Maintaining osmolarity and carrier in blood | W. Zhu et al. (2018) |
| Legumain | X‐33 | pPICZαA | Lysosomal protease | T. Zhao et al. (2018) |
| Goat chymosin | X‐33 | pPICZαA | Hydrolysis of κ‐casein | Tyagi et al. (2016) |
| Carrot antifreeze protein | GS115 | pPIC9K | Inhibition of gluten deterioration | M. Liu et al. (2018) |
| Proinsulin | SuperMan5 | pPICZαA | Treatment of diabetes mellitus | Baeshen et al. (2016) |
| hIFN‐γ | X‐33, GS115, KM71H, CBS7435 | pPICZα, pPIC9, pPpT4aS | Critical cytokine for innate and adaptive immunity | Razaghi et al. (2017) |
| IL‐1β | GS115, SMD1168, X‐33 | pPICZ‐A | Proinflammatory cytokine | Li et al. (2016) |
| IL‐3 | X‐33 | pPICZαA | Multipotent hematopoietic cytokine | Dagar and Khasa (2018) |
| IL‐11 | GS115 | pPINKαHC | Thrombopoietic growth factor | Yu et al. (2018) |
| IL‐15 | X‐33 | pPICZαA | Differentiation and proliferation of T, B, and NK cells | W. Sun et al. (2016) |
| Cyanate hydratase | GS115 | pPICZαA | Detoxification of cyanate and cyanide | Ranjan, Pillai, Permaul, and Singh (2017) |
| Human antiplatelet scFv antibody | X‐33 | pPICZαA | Treatment of atherosclerosis | Vallet‐Courbin et al. (2017) |
| α‐Amylase | X‐33 | pPICZαA | Starch saccharification | Parashar and Satyanarayana (2017) |
| Human epidermal growth factor | GS115 | pPIC9K | Generation of new epithelial and endothelial cells | Eissazadeh et al. (2017) |
| Bromelain | KM71H | pPICZαA | Oedematous swellings | Luniak, Meiser, Burkart, and Müller (2017) |
| Keratinocyte growth factor | X‐33 | pPICZαA | Epithelialization‐phase of wound healing | Kalhor (2016) |
| DM64 | X‐33 | pPICZαA | Anti‐myotoxic | Vieira, da Rocha, da Costa Neves‐Ferreira, Almeida, and Perales (2017) |
| Trypsin | GS115 | pPIC9K | Hydrolysis of proteins in the digestive system | Y. Zhang et al. (2018) |
| Human sialyltransferase | KM71H | pPICZαB | Pharmacological uses | Luley‐Goedl et al. (2016) |
| Transglutaminase | GS115 | pPIC9K | Restructured meat products | X. Yang and Zhang (2019) |
| Streptokinase | X‐33 | pPICZαA | Thrombolytic medication | Dagar, Devi, and Khasa (2016) |
| Staphylokinase | GS115, KM71H | pPICZαA | Thrombolytic medication | Faraji et al. (2017) |
| TFPR1 | X‐33 | pPICZαA | Adjuvant | Ning et al. (2016) |
Abbreviations: hIFN‐γ, human interferon γ; IL, interleukin; NK, natural killer.
6. CONCLUSION AND PERSPECTIVES
For the production of recombinant proteins with medical and industrial purposes, researchers should apply biological expression systems. Yeast expression systems such as S. cerevisiae and P. pastoris are more popular. P. pastoris expression system is one of the most popular and standard tools for the production of recombinant protein in molecular biology. It is also considered a unique host for the expression of subunit vaccines which could significantly affect the growing market of medical biotechnology. Although P. pastoris expression systems are impressive and easy to use with well‐defined process protocols, some degree of process optimization is required to achieve maximum production of the target proteins. Eventually, optimal conditions for the production of a recombinant protein in P. pastoris expression system differ according to the target protein.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
Karbalaei M, Rezaee SA, Farsiani H. Pichia pastoris: A highly successful expression system for optimal synthesis of heterologous proteins. J Cell Physiol. 2020;235:5867–5881. 10.1002/jcp.29583
DATA AVAILABILITY STATEMENT
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
REFERENCES
- Ahmad, M. , Hirz, M. , Pichler, H. , & Schwab, H. (2014). Protein expression in Pichia pastoris: Recent achievements and perspectives for heterologous protein production. Applied Microbiology and Biotechnology, 98(12), 5301–5317. 10.1007/s00253-014-5732-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anggiani, M. , Helianti, I. , & Abinawanto, A. (2018). Optimization of methanol induction for expression of synthetic gene Thermomyces lanuginosus lipase in Pichia pastoris . Paper presented at the AIP Conference Proceedings. [Google Scholar]
- Apte‐Deshpnade, A. , Mandal, G. , Soorapaneni, S. , Prasad, B. , Kumar, J. , & Padmanabhan, S. (2009). High‐level expression of non‐glycosylated and active staphylokinase from Pichia pastoris . Biotechnology Letters, 31(6), 811–817. [DOI] [PubMed] [Google Scholar]
- Araya‐Garay, J. M. , Feijoo‐Siota, L. , Rosa‐dos‐Santos, F. , Veiga‐Crespo, P. , & Villa, T. (2012). Construction of new Pichia pastoris X‐33 strains for production of lycopene and β‐carotene. Applied Microbiology and Biotechnology, 93(6), 2483–2492. [DOI] [PubMed] [Google Scholar]
- Azadi, S. , Mahboubi, A. , Naghdi, N. , Solaimanian, R. , & Mortazavi, S. A. (2017). Evaluation of sorbitol‐methanol co‐feeding strategy on production of recombinant human growth hormone in Pichia pastoris . Iranian Journal of Pharmaceutical Research, 16(4), 1555–1564. [PMC free article] [PubMed] [Google Scholar]
- Ben Azoun, S. , Belhaj, A. E. , Göngrich, R. , Gasser, B. , & Kallel, H. (2016). Molecular optimization of rabies virus glycoprotein expression in Pichia pastoris . Microbial Biotechnology, 9(3), 355–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeshen, M. N. , Bouback, T. A. , Alzubaidi, M. A. , Bora, R. S. , Alotaibi, M. A. , Alabbas, O. T. , … Al‐Hejin, A. (2016). Expression and purification of C‐peptide containing insulin using Pichia pastoris expression system. BioMed Research International, 2016, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baghani, A. A. , Soleimanpour, S. , Farsiani, H. , Mosavat, A. , Yousefi, M. , Meshkat, Z. , … Sadeghian, H. (2017). CFP10: MFcγ2 as a novel tuberculosis vaccine candidate increases immune response in mouse. Iranian Journal of Basic Medical Sciences, 20(2), 122–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balamurugan, V. , Reddy, G. , & Suryanarayana, V. (2007). Pichia pastoris: A notable heterologous expression system for the production of foreign proteins—vaccines. Indian Journal of Biotechnology, 6(2), 175–186. [Google Scholar]
- Balamurugan, V. , Sen, A. , Saravanan, P. , & Singh, R. (2006). Biotechnology in the production of recombinant vaccine or antigen for animal health. Journal of Animal and Veterinary Advances, 5(6), 487–495. [Google Scholar]
- Baneyx, F. (1999). Recombinant protein expression in Escherichia coli . Current Opinion in Biotechnology, 10(5), 411–421. [DOI] [PubMed] [Google Scholar]
- Bardiya, N. , & Chang, S. I. (2017). Cloning and expression of human ribonuclease 4 in methylotrophic yeast Pichia pastoris . Journal of Biotechnology Science Research, 4(2). [Google Scholar]
- Bill, R. M. (2015). Recombinant protein subunit vaccine synthesis in microbes: A role for yeast? Journal of Pharmacy and Pharmacology, 67(3), 319–328. [DOI] [PubMed] [Google Scholar]
- Bredell, H. , Smith, J. J. , Görgens, J. F. , & van Zyl, W. H. (2018). Expression of unique chimeric human papilloma virus type 16 (HPV‐16) L1‐L2 proteins in Pichia pastoris and Hansenula polymorpha . Yeast, 35, 519–529. [DOI] [PubMed] [Google Scholar]
- Bretthauer, R. K. , & Castellino, F. J. (1999). Glycosylation of Pichia pastoris‐derived proteins. Biotechnology and Applied Biochemistry, 30(Pt 3), 193–200. [PubMed] [Google Scholar]
- Byrne, B. (2015). Pichia pastoris as an expression host for membrane protein structural biology. Current Opinion in Structural Biology, 32, 9–17. [DOI] [PubMed] [Google Scholar]
- Cabral, K. M. , Almeida, M. S. , Valente, A. P. , Almeida, F. C. , & Kurtenbach, E. (2003). Production of the active antifungal Pisum sativum defensin 1 (Psd1) in Pichia pastoris: Overcoming the inefficiency of the STE13 protease. Protein Expression and Purification, 31(1), 115–122. [DOI] [PubMed] [Google Scholar]
- Cámara, E. , Landes, N. , Albiol, J. , Gasser, B. , Mattanovich, D. , & Ferrer, P. (2017). Increased dosage of AOX1 promoter‐regulated expression cassettes leads to transcription attenuation of the methanol metabolism in Pichia pastoris . Scientific Reports, 7, 44302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camattari, A. , Goh, A. , Yip, L. Y. , Tan, A. H. M. , Ng, S. W. , Tran, A. , … Rancati, G. (2016). Characterization of a panARS‐based episomal vector in the methylotrophic yeast Pichia pastoris for recombinant protein production and synthetic biology applications. Microbial Cell Factories, 15(1), 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Çelik, E. , Çalık, P. , & Oliver, S. G. (2009). Fed‐batch methanol feeding strategy for recombinant protein production by Pichia pastoris in the presence of co‐substrate sorbitol. Yeast, 26(9), 473–484. [DOI] [PubMed] [Google Scholar]
- Cereghino, G. P. L. , Cereghino, J. L. , Ilgen, C. , & Cregg, J. M. (2002). Production of recombinant proteins in fermenter cultures of the yeast Pichia pastoris . Current Opinion in Biotechnology, 13(4), 329–332. [DOI] [PubMed] [Google Scholar]
- Cereghino, G. P. L. , & Cregg, J. M. (1999). Applications of yeast in biotechnology: Protein production and genetic analysis. Current Opinion in Biotechnology, 10(5), 422–427. [DOI] [PubMed] [Google Scholar]
- Chahardooli, M. , Niazi, A. , Aram, F. , & Sohrabi, S. M. (2016). Expression of recombinant Arabian camel lactoferricin‐related peptide in Pichia pastoris and its antimicrobial identification. Journal of the Science of Food and Agriculture, 96(2), 569–575. [DOI] [PubMed] [Google Scholar]
- Chan, M. K. , Lim, S. K. , Miswan, N. , Chew, A. L. , Noordin, R. , & Khoo, B. Y. (2018). Expression of stable and active human DNA topoisomerase I in Pichia pastoris . Protein Expression and Purification, 141, 52–62. [DOI] [PubMed] [Google Scholar]
- Charoenrat, T. , Khumruaengsri, N. , Promdonkoy, P. , Rattanaphan, N. , Eurwilaichitr, L. , Tanapongpipat, S. , & Roongsawang, N. (2013). Improvement of recombinant endoglucanase produced in Pichia pastoris KM71 through the use of synthetic medium for inoculum and pH control of proteolysis. Journal of Bioscience and Bioengineering, 116(2), 193–198. [DOI] [PubMed] [Google Scholar]
- Chen, W.‐H. , Chag, S. M. , Poongavanam, M. V. , Biter, A. B. , Ewere, E. A. , Rezende, W. , … McAtee, C. P. (2017). Optimization of the production process and characterization of the yeast‐expressed SARS‐CoV recombinant receptor‐binding domain (RBD219‐N1), a SARS vaccine candidate. Journal of Pharmaceutical Sciences, 106(8), 1961–1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X. , Li, J. , Sun, H. , Li, S. , Chen, T. , Liu, G. , & Dyson, P. (2017). High‐level heterologous production and functional secretion by recombinant Pichia pastoris of the shortest proline‐rich antibacterial honeybee peptide Apidaecin. Scientific Reports, 7(1), 14543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X. , Meng, K. , Shi, P. , Bai, Y. , Luo, H. , Huang, H. , … Yao, B. (2012). High‐level expression of a novel Penicillium endo‐1, 3 (4)‐β‐D‐glucanase with high specific activity in Pichia pastoris . Journal of industrial microbiology & biotechnology, 39(6), 869–876. [DOI] [PubMed] [Google Scholar]
- Chen, Z. , Wang, D. , Cong, Y. , Wang, J. , Zhu, J. , Yang, J. , … Hu, F. (2011). Recombinant antimicrobial peptide hPAB‐β expressed in Pichia pastoris, a potential agent active against methicillin‐resistant Staphylococcus aureus . Applied Microbiology and Biotechnology, 89(2), 281–291. [DOI] [PubMed] [Google Scholar]
- Cregg, J. M. , Madden, K. , Barringer, K. , Thill, G. , & Stillman, C. (1989). Functional characterization of the two alcohol oxidase genes from the yeast Pichia pastoris . Molecular and Cellular Biology, 9(3), 1316–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha, C. C. d Q. B. , Gama, A. R. , Cintra, L. C. , Bataus, L. A. M. , & Ulhoa, C. J. (2018). Improvement of bread making quality by supplementation with a recombinant xylanase produced by Pichia pastoris . PLOS One, 13(2), 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagar, V. K. , Devi, N. , & Khasa, Y. P. (2016). High level production of active streptokinase in Pichia pastoris fed‐batch culture. International Journal of Biological Macromolecules, 83, 50–60. [DOI] [PubMed] [Google Scholar]
- Dagar, V. K. , & Khasa, Y. P. (2018). Combined effect of gene dosage and process optimization strategies on high‐level production of recombinant human interleukin‐3 (hIL‐3) in Pichia pastoris fed‐batch culture. International Journal of Biological Macromolecules, 108, 999–1009. [DOI] [PubMed] [Google Scholar]
- Daly, R. , & Hearn, M. T. (2005). Expression of heterologous proteins in Pichia pastoris: A useful experimental tool in protein engineering and production. Journal of Molecular Recognition: An Interdisciplinary Journal, 18(2), 119–138. [DOI] [PubMed] [Google Scholar]
- Dehnavi, E. , Siadat, S. O. R. , Roudsari, M. F. , & Khajeh, K. (2016). Cloning and high‐level expression of β‐xylosidase from Selenomonas ruminantium in Pichia pastoris by optimizing of pH, methanol concentration and temperature conditions. Protein Expression and Purification, 124, 55–61. [DOI] [PubMed] [Google Scholar]
- Dong, W. , Zhang, H. , Huang, H. , Zhou, J. , Hu, L. , Lian, A. , … Wei, K. (2016). Chicken IgY Fc linked to Bordetella avium ompA and Taishan Pinus massoniana pollen polysaccharide adjuvant enhances macrophage function and specific immune responses. Frontiers in Microbiology, 7, 1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragosits, M. , Stadlmann, J. , Albiol, J. , Baumann, K. , Maurer, M. , Gasser, B. , … Mattanovich, D. (2009). The effect of temperature on the proteome of recombinant Pichia pastoris . Journal of Proteome Research, 8(3), 1380–1392. 10.1021/pr8007623 [DOI] [PubMed] [Google Scholar]
- Eissazadeh, S. , Moeini, H. , Dezfouli, M. G. , Heidary, S. , Nelofer, R. , & Abdullah, M. P. (2017). Production of recombinant human epidermal growth factor in Pichia pastoris . Brazilian Journal of Microbiology, 48(2), 286–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faraji, H. , Ramezani, M. , Sadeghnia, H. R. , Abnous, K. , Soltani, F. , & Mashkani, B. (2017). High‐level expression of a biologically active staphylokinase in Pichia pastoris . Preparative Biochemistry and Biotechnology, 47(4), 379–387. [DOI] [PubMed] [Google Scholar]
- Farsiani, H. , Mosavat, A. , Soleimanpour, S. , Sadeghian, H. , Eydgahi, M. R. A. , Ghazvini, K. , … Rezaee, S. A. (2016). Fc‐based delivery system enhances immunogenicity of a tuberculosis subunit vaccine candidate consisting of the ESAT‐6: CFP‐10 complex. Molecular BioSystems, 12(7), 2189–2201. [DOI] [PubMed] [Google Scholar]
- Fu, X.‐Y. , Zhao, W. , Xiong, A.‐S. , Tian, Y.‐S. , & Peng, R.‐H. (2011). High expression of recombinant Streptomyces sp. S38 xylanase in Pichia pastoris by codon optimization and analysis of its biochemical properties. Molecular Biology Reports, 38(8), 4991–4997. [DOI] [PubMed] [Google Scholar]
- Gao, M. J. , Li, Z. , Yu, R. S. , Wu, J. R. , Zheng, Z. Y. , Shi, Z. P. , … Lin, C. C. (2012). Methanol/sorbitol co‐feeding induction enhanced porcine interferon‐alpha production by P. pastoris associated with energy metabolism shift. Bioprocess and Biosystems Engineering, 35(7), 1125–1136. 10.1007/s00449-012-0697-1 [DOI] [PubMed] [Google Scholar]
- Gao, M. , & Shi, Z. (2013). Process control and optimization for heterologous protein production by methylotrophic Pichia pastoris . Chinese Journal of Chemical Engineering, 21(2), 216–226. [Google Scholar]
- Gao, M.‐J. , Zhan, X.‐B. , Gao, P. , Zhang, X. , Dong, S.‐J. , Li, Z. , … Lin, C.‐C. (2015). Improving performance and operational stability of porcine interferon‐α production by Pichia pastoris with combinational induction strategy of low temperature and methanol/sorbitol co‐feeding. Applied Biochemistry and Biotechnology, 176(2), 493–504. [DOI] [PubMed] [Google Scholar]
- Gasser, B. , Maurer, M. , Rautio, J. , Sauer, M. , Bhattacharyya, A. , Saloheimo, M. , … Mattanovich, D. (2007). Monitoring of transcriptional regulation in Pichia pastoris under protein production conditions. BMC Genomics, 8, 179. 10.1186/1471-2164-8-179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glöckner, A. , & Voigt, C. A. (2015). Expression, purification and in vitro enzyme activity assay of plant derived GTPase. BIO‐PROTOCOL, 5(22), 1–6. [Google Scholar]
- Gomes, A. R. , Byregowda, S. M. , Veeregowda, B. M. , & Balamurugan, V. (2016). An overview of heterologous expression host systems for the production of recombinant proteins. Advances in Animal Veterinary Sciences, 4(7), 346–356. [Google Scholar]
- Goodrick, J. , Xu, M. , Finnegan, R. , Schilling, B. , Schiavi, S. , Hoppe, H. , & Wan, N. (2001). High‐level expression and stabilization of recombinant human chitinase produced in a continuous constitutive Pichia pastoris expression system. Biotechnology and Bioengineering, 74(6), 492–497. [DOI] [PubMed] [Google Scholar]
- Han, C. , Su, L. , Hong, R. , Wu, S. , & Wu, J. (2017). A comparative study of maltooligosyltrehalose synthase from Sulfolobus acidocaldarius expressed in Pichia pastoris and Escherichia coli . Process Biochemistry, 60, 35–41. [Google Scholar]
- Iglesias‐Figueroa, B. , Valdiviezo‐Godina, N. , Siqueiros‐Cendón, T. , Sinagawa‐García, S. , Arévalo‐Gallegos, S. , & Rascón‐Cruz, Q. (2016). High‐level expression of recombinant bovine lactoferrin in Pichia pastoris with antimicrobial activity. International Journal of Molecular Sciences, 17(6), 902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilgen, C. , Lin‐Cereghino, J. , & Cregg, J. M. (2005). Pichia pastoris, Production of recombinant proteins–Novel microbial and eukaryotic expression systems (pp. 143–162). Colorado State University, CO: John Wiley & Sons. [Google Scholar]
- Jacobs, P. P. , Geysens, S. , Vervecken, W. , Contreras, R. , & Callewaert, N. (2009). Engineering complex‐type N‐glycosylation in Pichia pastoris using GlycoSwitch technology. Nature Protocols, 4(1), 58–70. 10.1038/nprot.2008.213 [DOI] [PubMed] [Google Scholar]
- Jain, K. K. , Kumar, S. , Bhardwaj, K. N. , & Kuhad, R. C. (2018). Functional expression of a thermostable Endoglucanase from Thermoascus aurantiacus RCKK in Pichia pastoris X‐33 and its characterization. Molecular Biotechnology, 60(10), 736–748. [DOI] [PubMed] [Google Scholar]
- Jiao, L. , Zhou, Q. , Su, Z. , Xu, L. , & Yan, Y. (2018). High‐level extracellular production of Rhizopus oryzae lipase in Pichia pastoris via a strategy combining optimization of gene‐copy number with co‐expression of ERAD‐related proteins. Protein Expression and Purification, 147, 1–12. [DOI] [PubMed] [Google Scholar]
- Julien, C. (2006). Production of humanlike recombinant proteins in Pichia pastoris . BioProcess International, 22–30. [Google Scholar]
- Kalhor, H. R. (2016). Expression of the full‐length human recombinant keratinocyte growth factor in Pichia pastoris . Journal of Cell and Molecular Research, 8(1), 1–7. [Google Scholar]
- Kang, X. , Wang, J. , Jiao, Y. , Tang, P. , Song, L. , Xiong, D. , … Jiao, X. (2016). Expression of recombinant Newcastle disease virus F protein in Pichia pastoris and its immunogenicity using flagellin as the adjuvant. Protein Expression and Purification, 128, 73–80. [DOI] [PubMed] [Google Scholar]
- Kebriaei, A. , Derakhshan, M. , Meshkat, Z. , Eidgahi, M. R. A. , Rezaee, S. A. , Farsiani, H. , … Ghazvini, K. (2016). Construction and immunogenicity of a new Fc‐based subunit vaccine candidate against Mycobacterium tuberculosis . Molecular Biology Reports, 43(9), 911–922. [DOI] [PubMed] [Google Scholar]
- Khan, K. H. (2013). Gene expression in mammalian cells and its applications. Advanced Pharmaceutical Bulletin, 3(2), 257–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khetarpal, N. , Shukla, R. , Rajpoot, R. K. , Poddar, A. , Pal, M. , Swaminathan, S. , … Khanna, N. (2017). Recombinant dengue virus 4 envelope glycoprotein virus‐like particles derived from Pichia pastoris are capable of eliciting homotypic domain III‐directed neutralizing antibodies. The American Journal of Tropical Medicine and Hygiene, 96(1), 126–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuddus, M. R. , Rumi, F. , Tsutsumi, M. , Takahashi, R. , Yamano, M. , Kamiya, M. , … Aizawa, T. (2016). Expression, purification and characterization of the recombinant cysteine‐rich antimicrobial peptide snakin‐1 in Pichia pastoris . Protein Expression and Purification, 122, 15–22. [DOI] [PubMed] [Google Scholar]
- Kukk, K. , & Samel, N. (2016). Enhanced expression of human prostaglandin H synthase‐2 in the yeast Pichia pastoris and removal of the C‐terminal tag with bovine carboxypeptidase A. Journal of Biotechnology, 231, 224–231. [DOI] [PubMed] [Google Scholar]
- Landim, P. G. C. , Correia, T. O. , Silva, F. D. , Nepomuceno, D. R. , Costa, H. P. , Pereira, H. M. , … Medeiros, S. C. (2017). Production in Pichia pastoris, antifungal activity and crystal structure of a class I chitinase from cowpea (Vigna unguiculata): Insights into sugar binding mode and hydrolytic action. Biochimie, 135, 89–103. [DOI] [PubMed] [Google Scholar]
- Laukens, B. , De Wachter, C. , & Callewaert, N. (2015). Engineering the Pichia pastoris N‐glycosylation pathway using the GlycoSwitch Technology. Methods in Molecular Biology, 1321, 103–122. 10.1007/978-1-4939-2760-9_8 [DOI] [PubMed] [Google Scholar]
- Lee, J. Y. , Chen, H. , Liu, A. , Alba, B. M. , & Lim, A. C. (2017). Auto‐induction of Pichia pastoris AOX1 promoter for membrane protein expression. Protein Expression and Purification, 137, 7–12. [DOI] [PubMed] [Google Scholar]
- Lee, J. S. , Kim, J. , Im, S. P. , Kim, S. W. , Jung, J. W. , Lazarte, J. M. S. , … Jung, T. S. (2018). Expression and characterization of monomeric variable lymphocyte receptor B specific to the glycoprotein of viral hemorrhagic septicemia virus (VHSV). Journal of Immunological Methods, 462, 48–53. [DOI] [PubMed] [Google Scholar]
- Lee, S. , & Nguyen, M. T. (2015). Recent advances of vaccine adjuvants for infectious diseases. Immune Network, 15(2), 51–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. , Ali, Z. , Liu, X. , Jiang, L. , Tang, Y. , & Dai, J. (2019). Expression of recombinant tachyplesin I in Pichia pastoris . Protein Expression and Purification, 157, 50–56. [DOI] [PubMed] [Google Scholar]
- Li, P. , Anumanthan, A. , Gao, X.‐G. , Ilangovan, K. , Suzara, V. V. , Düzgüneş, N. , & Renugopalakrishnan, V. (2007). Expression of recombinant proteins in Pichia pastoris . Applied Biochemistry and Biotechnology, 142(2), 105–124. [DOI] [PubMed] [Google Scholar]
- Li, Z. , Moy, A. , Gomez, S. R. , Franz, A. H. , Lin‐Cereghino, J. , & Lin‐Cereghino, G. P. (2010). An improved method for enhanced production and biological activity of human secretory leukocyte protease inhibitor (SLPI) in Pichia pastoris . Biochemical and Biophysical Research Communications, 402(3), 519–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, P. , Sun, H. , Chen, Z. , Li, Y. , & Zhu, T. (2015). Construction of efficient xylose utilizing Pichia pastoris for industrial enzyme production. Microbial Cell Factories, 14(1), 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. , Sun, C. , Chen, L. , Sun, L. , Duan, L. , Zheng, Q. , & Hu, X. (2017). Optimization of the secretory expression of recombinant human C‐reactive protein in Pichia pastoris . 3 Biotech, 7(5), 291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L. , Wang, J.‐X. , Zhao, X.‐F. , Kang, C.‐J. , Liu, N. , Xiang, J.‐H. , … Kondo, H. (2005). High level expression, purification, and characterization of the shrimp antimicrobial peptide, Ch‐penaeidin, in Pichia pastoris . Protein Expression and Purification, 39(2), 144–151. [DOI] [PubMed] [Google Scholar]
- Li, Z. , Xiong, F. , Lin, Q. , d'Anjou, M. , Daugulis, A. J. , Yang, D. S. , & Hew, C. L. (2001). Low‐temperature increases the yield of biologically active herring antifreeze protein in Pichia pastoris . Protein Expression and Purification, 21(3), 438–445. [DOI] [PubMed] [Google Scholar]
- Li, P. , Yang, G. , Geng, X. , Shi, J. , Li, B. , Wang, Z. , … Xu, C. (2016). High‐level secretory expression and purification of recombinant human interleukin 1 beta in Pichia pastoris . Protein and Peptide Letters, 23(8), 763–769. [DOI] [PubMed] [Google Scholar]
- Liu, M. , Liang, Y. , Zhang, H. , Wu, G. , Wang, L. , Qian, H. , & Qi, X. (2018). Production of a recombinant carrot antifreeze protein by Pichia pastoris GS115 and its cryoprotective effects on frozen dough properties and bread quality. LWT‐Food Science and Technology, 96, 543–550. [Google Scholar]
- Liu, Z. , Zhu, M. , Chen, X. , Yang, G. , Yang, T. , Yu, L. , … Wang, X. (2018). Expression and antibacterial activity of hybrid antimicrobial peptide cecropinA‐thanatin in Pichia pastoris . Frontiers in Laboratory Medicine, 2(1), 23–29. [Google Scholar]
- Looser, V. , Bruhlmann, B. , Bumbak, F. , Stenger, C. , Costa, M. , Camattari, A. , … Kovar, K. (2015). Cultivation strategies to enhance productivity of Pichia pastoris: A review. Biotechnology Advances, 33(6), 1177–1193. [DOI] [PubMed] [Google Scholar]
- Luley‐Goedl, C. , Czabany, T. , Longus, K. , Schmölzer, K. , Zitzenbacher, S. , Ribitsch, D. , … Nidetzky, B. (2016). Combining expression and process engineering for high‐quality production of human sialyltransferase in Pichia pastoris . Journal of Biotechnology, 235, 54–60. [DOI] [PubMed] [Google Scholar]
- Luniak, N. , Meiser, P. , Burkart, S. , & Müller, R. (2017). Heterologous expression of the plant cysteine protease bromelain and its inhibitor in Pichia pastoris . Biotechnology Progress, 33(1), 54–65. [DOI] [PubMed] [Google Scholar]
- Luong, M. , Lam, J. S. , Chen, J. , & Levitz, S. M. (2007). Effects of fungal N‐ and O‐linked mannosylation on the immunogenicity of model vaccines. Vaccine, 25(22), 4340–4344. 10.1016/j.vaccine.2007.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macauley‐Patrick, S. , Fazenda, M. L. , McNeil, B. , & Harvey, L. M. (2005). Heterologous protein production using the Pichia pastoris expression system. Yeast, 22(4), 249–270. [DOI] [PubMed] [Google Scholar]
- Matos, M. N. , Alberti, A. S. , Morales, C. , Cazorla, S. I. , & Malchiodi, E. L. (2016). A prime‐boost immunization with Tc52 N‐terminal domain DNA and the recombinant protein expressed in Pichia pastoris protects against Trypanosoma cruzi infection. Vaccine, 34(28), 3243–3251. [DOI] [PubMed] [Google Scholar]
- Meng, D.‐M. , Li, W.‐J. , Shi, L.‐Y. , Lv, Y.‐J. , Sun, X.‐Q. , Hu, J.‐C. , & Fan, Z.‐C. (2019). Expression, purification and characterization of a recombinant antimicrobial peptide Hispidalin in Pichia pastoris . Protein Expression and Purification, 160, 19–27. [DOI] [PubMed] [Google Scholar]
- Mina, S. , Staerck, C. , Marot, A. , Godon, C. , Calenda, A. , Bouchara, J.‐P. , & Fleury, M. J. (2017). Scedosporium boydii CatA1 and SODC recombinant proteins, new tools for serodiagnosis of Scedosporium infection of patients with cystic fibrosis. Diagnostic Microbiology and Infectious Disease, 89(4), 282–287. [DOI] [PubMed] [Google Scholar]
- Minor, P. D. (2015). Live attenuated vaccines: Historical successes and current challenges. Virology, 479, 379–392. [DOI] [PubMed] [Google Scholar]
- Mosavat, A. , Soleimanpour, S. , Farsiani, H. , Sadeghian, H. , Ghazvini, K. , Sankian, M. , … Rezaee, S. A. (2016). Fused Mycobacterium tuberculosis multi‐stage immunogens with an Fc‐delivery system as a promising approach for the development of a tuberculosis vaccine. Infection, Genetics and Evolution, 39, 163–172. [DOI] [PubMed] [Google Scholar]
- Naumov, G. I. , Naumova, E. S. , & Boundy‐Mills, K. L. (2018). Description of Komagataella mondaviorum sp. nov., a new sibling species of Komagataella (Pichia) pastoris. Antonie Van Leeuwenhoek, 111, 1–11. [DOI] [PubMed] [Google Scholar]
- Ning, X. , Kou, Z. , Sun, W. , Zhu, Q. , Yang, Y. , Qiu, H. , … Zhou, Y. (2016). Expression of a novel adjuvant TFPR1 in Pichia pastoris and its identification. Chinese Journal of Microbiology and Immunology, 36(4), 294–299. [Google Scholar]
- Nokelainen, M. , Tu, H. , Vuorela, A. , Notbohm, H. , Kivirikko, K. I. , & Myllyharju, J. (2001). High‐level production of human type I collagen in the yeast Pichia pastoris . Yeast, 18(9), 797–806. [DOI] [PubMed] [Google Scholar]
- Nordén, K. , Agemark, M. , Danielson, J. Å. , Alexandersson, E. , Kjellbom, P. , & Johanson, U. (2011). Increasing gene dosage greatly enhances recombinant expression of aquaporins in Pichia pastoris . BMC Biotechnology, 11(1), 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orman, M. A. , Calik, P. , & Ozdamar, T. H. (2009). The influence of carbon sources on recombinant‐human‐ growth‐hormone production by Pichia pastoris is dependent on phenotype: A comparison of Muts and Mut + strains. Biotechnology and Applied Biochemistry, 52(Pt 3), 245–255. 10.1042/ba20080057 [DOI] [PubMed] [Google Scholar]
- Parashar, D. , & Satyanarayana, T. (2017). Production of chimeric acidic α‐amylase by the recombinant Pichia pastoris and its applications. Frontiers in Microbiology, 8, 493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picanço‐Castro, V. , Cristina Correa de Freitas, M. , Bomfim, A. , & Maria de Sousa Russo, E. (2014). Patents in therapeutic recombinant protein production using mammalian cells. Recent Patents on Biotechnology, 8(2), 165–171. [DOI] [PubMed] [Google Scholar]
- Popa, C. , Shi, X. , Ruiz, T. , Ferrer, P. , & Coca, M. (2019). Biotechnological production of the cell penetrating antifungal PAF102 peptide in Pichia pastoris . Frontiers in Microbiology, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhu, A. A. , Veeranki, V. D. , & Dsilva, S. J. (2016). Improving the production of human interferon gamma (hIFN‐γ) in Pichia pastoris cell factory: An approach of cell level. Process Biochemistry, 51(6), 709–718. [Google Scholar]
- Pratush, A. , Seth, A. , & Bhalla, T. C. (2017). Expression of nitrile hydratase gene of mutant 4D strain of Rhodococcus rhodochrous PA 34 in Pichia pastoris . Biocatalysis and Biotransformation, 35(1), 19–26. [Google Scholar]
- Rajamanickam, V. , Metzger, K. , Schmid, C. , & Spadiut, O. (2017). A novel bi‐directional promoter system allows tunable recombinant protein production in Pichia pastoris . Microbial Cell Factories, 16(1), 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjan, B. , Pillai, S. , Permaul, K. , & Singh, S. (2017). Expression of a novel recombinant cyanate hydratase (rTl‐Cyn) in Pichia pastoris, characteristics and applicability in the detoxification of cyanate. Bioresource Technology, 238, 582–588. [DOI] [PubMed] [Google Scholar]
- Rasala, B. A. , & Mayfield, S. P. (2015). Photosynthetic biomanufacturing in green algae; production of recombinant proteins for industrial, nutritional, and medical uses. Photosynthesis Research, 123(3), 227–239. [DOI] [PubMed] [Google Scholar]
- Razaghi, A. , Tan, E. , Lua, L. H. , Owens, L. , Karthikeyan, O. , & Heimann, K. (2017). Is Pichia pastoris a realistic platform for industrial production of recombinant human interferon gamma? Biologicals, 45, 52–60. [DOI] [PubMed] [Google Scholar]
- Rosano, G. L. , & Ceccarelli, E. A. (2014). Recombinant protein expression in Escherichia coli: Advances and challenges. Frontiers in Microbiology, 5, 172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safder, I. , Khan, S. , Islam, I. , & Kazim, M. (2018). Pichia pastoris expression system: A potential candidate to express protein in industrial and biopharmaceutical domains. Biomedical Letters, 4(1), 1–13. [Google Scholar]
- Sams, L. , Amara, S. , Chakroun, A. , Coudre, S. , Paume, J. , Giallo, J. , & Carrière, F. (2017). Constitutive expression of human gastric lipase in Pichia pastoris and site‐directed mutagenesis of key lid‐stabilizing residues. Biochimica et Biophysica Acta (BBA)‐Molecular and Cell Biology of Lipids, 1862(10), 1025–1034. [DOI] [PubMed] [Google Scholar]
- Santoso, A. , Herawati, N. , & Rubiana, Y. (2012). Effect of methanol induction and incubation time on expression of human erythropoietin in methylotropic yeast Pichia pastoris . Makara Journal of Technology, 16(1), 29–34. [Google Scholar]
- Saraswat, S. , Athmaram, T. , Parida, M. , Agarwal, A. , Saha, A. , & Dash, P. K. (2016). Expression and characterization of yeast derived Chikungunya virus like particles (CHIK‐VLPs) and its evaluation as a potential vaccine candidate. PLOS Neglected Tropical Diseases, 10(7), 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satomura, A. , Kuroda, K. , & Ueda, M. (2015). Generation of a functionally distinct Rhizopus oryzae lipase through protein folding memory. PLOS One, 10(5), 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller, J. T. , & Lowy, D. R. (2014). Raising expectations for subunit vaccine. The Journal of Infectious Diseases, 211(9), 1373–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soleimanpour, S. , Farsiani, H. , Mosavat, A. , Ghazvini, K. , Eydgahi, M. R. A. , Sankian, M. , … Rezaee, S. A. (2015). APC targeting enhances immunogenicity of a novel multistage Fc‐fusion tuberculosis vaccine in mice. Applied Microbiology and Biotechnology, 99(24), 10467–10480. [DOI] [PubMed] [Google Scholar]
- Spiegel, H. , Schinkel, H. , Kastilan, R. , Dahm, P. , Boes, A. , Scheuermayer, M. , … Schillberg, S. (2015). Optimization of a multi‐stage, multi‐subunit malaria vaccine candidate for the production in Pichia pastoris by the identification and removal of protease cleavage sites. Biotechnology and Bioengineering, 112(4), 659–667. [DOI] [PubMed] [Google Scholar]
- Stewart, G. (2015). Yeast quality assessment, management and culture maintenance, Brewing Microbiology (pp. 11–29). Edinburgh, Scotland, UK: Elsevier. [Google Scholar]
- Sun, H. , Bankefa, O. E. , Ijeoma, I. O. , Miao, L. , Zhu, T. , & Li, Y. (2017). Systematic assessment of Pichia pastoris system for optimized β‐galactosidase production. Synthetic and Systems Biotechnology, 2(2), 113–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, W. , Lai, Y. , Li, H. , Nie, T. , Kuang, Y. , Tang, X. , … Li, P. (2016). High level expression and purification of active recombinant human interleukin‐15 in Pichia pastoris . Journal of Immunological Methods, 428, 50–57. [DOI] [PubMed] [Google Scholar]
- Sunga, A. J. , & Cregg, J. M. (2004). The Pichia pastoris formaldehyde dehydrogenase gene (FLD1) as a marker for selection of multicopy expression strains of P. pastoris . Gene, 330, 39–47. [DOI] [PubMed] [Google Scholar]
- Tachioka, M. , Sugimoto, N. , Nakamura, A. , Sunagawa, N. , Ishida, T. , Uchiyama, T. , … Samejima, M. (2016). Development of simple random mutagenesis protocol for the protein expression system in Pichia pastoris . Biotechnology for Biofuels, 9(1), 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng, X. , Chen, X. , Zhu, K. , & Xu, H. (2018). Immunogenicity of heparin‐binding hemagglutinin expressed by Pichia pastoris GS115 strain. Iranian Journal of Basic Medical Sciences, 21(2), 219–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todryk, S. M. (2018). T cell memory to vaccination. Vaccines, 6(4), 84. 10.3390/vaccines6040084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi, L. , Mani, S. , Raut, R. , Poddar, A. , Tyagi, P. , Arora, U. , … Khanna, N. (2015). Pichia pastoris‐expressed dengue 3 envelope‐based virus‐like particles elicit predominantly domain III‐focused high titer neutralizing antibodies. Frontiers in Microbiology, 6, 1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu, Y. , Wang, G. , Wang, Y. , Chen, W. , Zhang, L. , Liu, Y. , … Cai, X. (2016). Extracellular expression and antiviral activity of a bovine interferon‐alpha through codon optimization in Pichia pastoris . Microbiological Research, 191, 12–18. [DOI] [PubMed] [Google Scholar]
- Tu, Y. , Wang, Y. , Wang, G. , Wu, J. , Liu, Y. , Wang, S. , … Cai, X. (2013). High‐level expression and immunogenicity of a porcine circovirus type 2 capsid protein through codon optimization in Pichia pastoris . Applied Microbiology and Biotechnology, 97(7), 2867–2875. [DOI] [PubMed] [Google Scholar]
- Tyagi, A. , Kumar, A. , Yadav, A. K. , Saklani, A. C. , Grover, S. , & Batish, V. K. (2016). Functional expression of recombinant goat chymosin in Pichia pastoris bioreactor cultures: A commercially viable alternate. LWT‐Food Science and Technology, 69, 217–224. [Google Scholar]
- Vallet‐Courbin, A. , Larivière, M. , Hocquellet, A. , Hemadou, A. , Parimala, S.‐N. , Laroche‐Traineau, J. , … Noubhani, A. (2017). A recombinant human anti‐platelet SCFV antibody produced in Pichia pastoris for atheroma targeting. PLOS One, 12(1), 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanz, A. , Lünsdorf, H. , Adnan, A. , Nimtz, M. , Gurramkonda, C. , Khanna, N. , & Rinas, U. (2012). Physiological response of Pichia pastoris GS115 to methanol‐induced high level production of the Hepatitis B surface antigen: Catabolic adaptation, stress responses, and autophagic processes. Microbial Cell Factories, 11(1), 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira, S. M. , da Rocha, S. L. G. , da Costa Neves‐Ferreira, A. G. , Almeida, R. V. , & Perales, J. (2017). Heterologous expression of the antimyotoxic protein DM64 in Pichia pastoris . PLOS Neglected Tropical Diseases, 11(7), 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, M. , Jiang, S. , Han, Z. , Zhao, B. , Zhou, Z. , & Wang, Y. (2016). Expression and immunogenic characterization of recombinant gp350 for developing a subunit vaccine against Epstein‐Barr virus. Applied Microbiology and Biotechnology, 100(3), 1221–1230. [DOI] [PubMed] [Google Scholar]
- Wang, M. , Jiang, S. , & Wang, Y. (2016). Recent advances in the production of recombinant subunit vaccines in Pichia pastoris . Bioengineered, 7(3), 155–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, M. , Jiang, S. , Zhou, L. , Wang, C. , Mao, R. , & Ponnusamy, M. (2017). Efficient production of recombinant glycoprotein D of herpes simplex virus type 2 in Pichia pastoris and its protective efficacy against viral challenge in mice. Archives of Virology, 162(3), 701–711. [DOI] [PubMed] [Google Scholar]
- Wang, H. , Shan, S. , Wang, S. , Zhang, H. , Ma, L. , Hu, L. , … Zhu, R. (2017). Fused IgY Fc and polysaccharide adjuvant enhanced the immune effect of the recombinant VP2 and VP5 subunits—A prospect for improvement of infectious bursal disease virus subunit vaccine. Frontiers in Microbiology, 8, 2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J. , Wang, X. , Shi, L. , Qi, F. , Zhang, P. , Zhang, Y. , … Cai, M. (2017). Methanol‐independent protein expression by AOX1 promoter with trans‐acting elements engineering and glucose‐glycerol‐shift induction in Pichia pastoris . Scientific Reports, 7, 41850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S. , Wang, Y. , Wang, P. G. , Chen, M. , & Kong, Y. (2018). High level expression and glycosylation of recombinant Mycobacterium tuberculosis Ala‐Pro‐rich antigen in Pichia pastoris . Protein Expression and Purification, 150, 67–71. [DOI] [PubMed] [Google Scholar]
- Wang, Z. , Wang, Y. , Zhang, D. , Li, J. , Hua, Z. , Du, G. , & Chen, J. (2010). Enhancement of cell viability and alkaline polygalacturonate lyase production by sorbitol co‐feeding with methanol in Pichia pastoris fermentation. Bioresource Technology, 101(4), 1318–1323. [DOI] [PubMed] [Google Scholar]
- Wang, M. , Zou, Z. , Li, Q. , Xin, H. , Zhu, X. , Chen, X. , & Li, X. (2017). Heterologous expression of three Camellia sinensis small heat shock protein genes confers temperature stress tolerance in yeast and Arabidopsis thaliana . Plant Cell Reports, 36(7), 1125–1135. [DOI] [PubMed] [Google Scholar]
- Webb, R. P. , Smith, T. J. , Smith, L. A. , Wright, P. M. , Guernieri, R. L. , Brown, J. L. , & Skerry, J. C. (2017). Recombinant botulinum neurotoxin Hc subunit (BoNT Hc) and catalytically inactive Clostridium botulinum holoproteins (ciBoNT HPs) as vaccine candidates for the prevention of botulism. Toxins, 9(9), 269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J. , Jiao, Z. , Guo, F. , Chen, L. , Ding, Z. , & Qiu, Z. (2016). Constitutive and secretory expression of the AiiA in Pichia pastoris inhibits Amorphophallus konjac soft rot disease. American Journal of Molecular Biology, 6(2), 79–87. [Google Scholar]
- Wu, S. , & Letchworth, G. J. (2004). High efficiency transformation by electroporation of Pichia pastoris pretreated with lithium acetate and dithiothreitol. Biotechniques, 36(1), 152–154. [DOI] [PubMed] [Google Scholar]
- Wu, M. , Liu, W. , Yang, G. , Yu, D. , Lin, D. , Sun, H. , & Chen, S. (2014). Engineering of a Pichia pastoris expression system for high‐level secretion of HSA/GH fusion protein. Applied Biochemistry and Biotechnology, 172(5), 2400–2411. [DOI] [PubMed] [Google Scholar]
- Xing, L. W. , Tian, S. X. , Gao, W. , Yang, N. , Qu, P. , Liu, D. , … Feng, X. J. (2016). Recombinant expression and biological characterization of the antimicrobial peptide fowlicidin‐2 in Pichia pastoris . Experimental and Therapeutic Medicine, 12(4), 2324–2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, S. , Kuang, Y. , Li, H. , Liu, Y. , Hui, X. , Li, P. , … Xu, A. (2013). Enhanced production of recombinant secretory proteins in Pichia pastoris by optimizing Kex2 P1'site. PLOS One, 8(9):e75347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X. , & Zhang, Y. (2019). Expression of recombinant transglutaminase gene in Pichia pastoris and its uses in restructured meat products. Food Chemistry, 291, 245–252. [DOI] [PubMed] [Google Scholar]
- Yin, J. , Li, G. , Ren, X. , & Herrler, G. (2007). Select what you need: A comparative evaluation of the advantages and limitations of frequently used expression systems for foreign genes. Journal of Biotechnology, 127(3), 335–347. [DOI] [PubMed] [Google Scholar]
- Yokomaku, K. , Akiyama, M. , Morita, Y. , Kihira, K. , & Komatsu, T. (2018). Core–shell protein clusters comprising haemoglobin and recombinant feline serum albumin as an artificial O2 carrier for cats. Journal of Materials Chemistry B, 6(16), 2417–2425. [DOI] [PubMed] [Google Scholar]
- Yu, K.‐M. , Lau, J. Y.‐N. , Fok, M. , Yeung, Y.‐K. , Fok, S.‐P. , Shek, F. , … Choo, Q.‐L. (2018). Efficient expression and isolation of recombinant human interleukin‐11 (rhIL‐11) in Pichia pastoris . Protein Expression and Purification, 146, 69–77. [DOI] [PubMed] [Google Scholar]
- Zahrl, R. J. , Peña, D. A. , Mattanovich, D. , & Gasser, B. (2017). Systems biotechnology for protein production in Pichia pastoris . FEMS Yeast Research, 17(7). [DOI] [PubMed] [Google Scholar]
- Zhang, W. , Bevins, M. A. , Plantz, B. A. , Smith, L. A. , & Meagher, M. M. (2000). Modeling Pichia pastoris growth on methanol and optimizing the production of a recombinant protein, the heavy‐chain fragment C of botulinum neurotoxin, serotype A. Biotechnology and Bioengineering, 70(1), 1–8. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Huang, H. , Yao, X. , Du, G. , Chen, J. , & Kang, Z. (2018). High‐yield secretory production of stable, active trypsin through engineering of the N‐terminal peptide and self‐degradation sites in Pichia pastoris . Bioresource Technology, 247, 81–87. [DOI] [PubMed] [Google Scholar]
- Zhang, C. , Ku, Z. , Liu, Q. , Wang, X. , Chen, T. , Ye, X. , … Huang, Z. (2015). High‐yield production of recombinant virus‐like particles of enterovirus 71 in Pichia pastoris and their protective efficacy against oral viral challenge in mice. Vaccine, 33(20), 2335–2341. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Yang, S. , Dai, X. , Liu, L. , Jiang, X. , Shao, M. , … Wei, K. (2015). Protective immunity induced by the vaccination of recombinant Proteus mirabilis OmpA expressed in Pichia pastoris . Protein Expression and Purification, 105, 33–38. [DOI] [PubMed] [Google Scholar]
- Zhang, J. , Yang, Y. , Teng, D. , Tian, Z. , Wang, S. , & Wang, J. (2011). Expression of plectasin in Pichia pastoris and its characterization as a new antimicrobial peptide against Staphyloccocus and Streptococcus . Protein Expression and Purification, 78(2), 189–196. [DOI] [PubMed] [Google Scholar]
- Zhao, T. , Li, Z. , Guo, Z. , Wang, A. , Liu, Z. , Zhao, Q. , … Diao, A. (2018). Functional recombinant human Legumain protein expression in Pichia pastoris to enable screening for Legumain small molecule inhibitors. Protein Expression and Purification, 150, 12–16. [DOI] [PubMed] [Google Scholar]
- Zhao, H. , Tang, J. , Cao, L. , Jia, G. , Long, D. , Liu, G. , … Shang, H. (2015). Characterization of bioactive recombinant antimicrobial peptide parasin I fused with human lysozyme expressed in the yeast Pichia pastoris system. Enzyme and Microbial Technology, 77, 61–67. [DOI] [PubMed] [Google Scholar]
- Zhong, Y. , Yang, L. , Guo, Y. , Fang, F. , Wang, D. , Li, R. , … Xiao, W. (2014). High‐temperature cultivation of recombinant Pichia pastoris increases endoplasmic reticulum stress and decreases production of human interleukin‐10. Microbial Cell Factories, 13, 163. 10.1186/s12934-014-0163-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, W. , Gong, G. , Pan, J. , Han, S. , Zhang, W. , Hu, Y. , & Xie, L. (2018). High level expression and purification of recombinant human serum albumin in Pichia pastoris . Protein Expression and Purification, 147, 61–68. [DOI] [PubMed] [Google Scholar]
- Zhu, T. , You, L. , Gong, F. , Xie, M. , Xue, Y. , Li, Y. , & Ma, Y. (2011). Combinatorial strategy of sorbitol feeding and low‐temperature induction leads to high‐level production of alkaline beta‐mannanase in Pichia pastoris . Enzyme and Microbial Technology, 49(4), 407–412. 10.1016/j.enzmictec.2011.06.022 [DOI] [PubMed] [Google Scholar]
- Zhuo, R. , Yu, H. , Yuan, P. , Fan, J. , Chen, L. , Li, Y. , … Zhang, X. (2018). Heterologous expression and characterization of three laccases obtained from Pleurotus ostreatus HAUCC 162 for removal of environmental pollutants. Journal of Hazardous Materials, 344, 499–510. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.


