Abstract
Human papillomavirus (HPV) is the main cervical cancer‐promoting element that is transmitted through sexual routes. Anal, head, and throat cancers are also reported to be accompanied by HPV infection. E6 is one of the HPV nonstructural proteins, which is responsible for cell differentiation by targeting tumor suppressor genes, p105Rb and p53. E6 was reported to be stabilized by two chaperone proteins; glucose‐regulated protein 78 (GRP78) and heat shock protein 90. GRP78 is responsible for the unfolded protein response of the cells, and it was reported to be upregulated in many cancers, including cervical cancer. It was reported that knocking out GRP78 destabilizes E6 leading to faster degradation of E6 in vivo. The current work predicts the possible binding mode between E6 and GRP78 based on sequence and structural similarities.
Keywords: BiP, GRP78, HPV E6, protein‐protein docking, structural bioinformatics
Research Highlights
Understanding the binding behavior of glucose‐regulated protein 78 (GRP78) can pave the way for drugs that can prevent this interaction between the host cell proteins and the viral protein E6 that is crucial for cancer propagation.
Once GRP78 binding is inhibited, E6 will be destabilized and prone to a faster degradation rate by the host cell degradation machinery.
1. INTRODUCTION
Human papillomavirus (HPV) is one of the papillomavirus (PVs) families that is reported to infect skin and mucosa of mammals, aves, and reptiles. 1 , 2 , 3 , 4 The cancer causative agent, which was reported in most cervical cancer cases worldwide and the majority of anal and head and throat cancer cases, is HPV. 3 , 5 It is the largest family of PVs that includes more than 150 different strains, of which HPV16 is responsible for half the cervical cancer cases worldwide. 2 , 3 There are 15 high‐risk HPV strains that are reported in cancer cases, which include HPV16, HPV18, HPV31, HPV33, HPV35, HPV39, HPV51, HPV52, HPV56, HPV58, HPV59, HPV68, HPV69, HPV73, and HPV82. 4 , 6
HPV is a nonenveloped double‐stranded DNA virus that has two structural capsid proteins, L1 (major capsid) and L2 (minor capsid), responsible for viral entry into the host cell. 4 , 7 HPV target the heparin sulfate proteoglycans (HSPGs) of the basal cell epithelium. 7 , 8 The L1 capsid protein of HPV is the primary element that recognizes HSPGs. 7 , 8 After recognition, conformational changes in L1 mediated by different host cell receptors and chaperones leads to endocytosis of the virion. 1 , 6 , 7 HPV travels to the nucleus to replicate its DNA through localization in separate host cell compartments, including the endosome, Golgi apparatus, and endoplasmic reticulum (ER). 7 , 9 E6 is one of the nonstructural proteins of HPV that was reported to be the causative element of cancer cell proliferation. 10 It binds to oncoproteins crucial for cancer development like p53 and p105Rb oncoproteins, leading to its degradation. 1 , 7 , 10 , 11 Different viral (E6 associated protein [E6‐AP] and E6^E7 splice isoform) and host‐cell (HSP90 and GRP78 chaperones) proteins are reported to bind to and stabilize the E6 protein of HPV16, increasing its presence times in vivo. 1
Glucose regulating protein 78 (GRP78), is the master player of the unfolded protein response mechanism in the ER. 12 It is a heat shock protein 70 family member, a chaperone protein, that regulates cell response under unfolded protein load stress. 13 , 14 , 15 , 16 GRP78 is localized in the lumen of the ER bound to and inactivating other enzymes responsible for cell response under accumulated unfolded proteins that include activating transcription factor 6 (ATF6), protein kinase RNA‐like endoplasmic reticulum kinase (PERK), and inositol‐requiring enzyme 1 (IRE1). 17 Upon cell stress, like a viral infection or cancer, the GRP78 releases ATF6, PERK, and IRE1 due to the accumulation of unfolded proteins leading to inhibition of protein synthesis and enhancement of refolding. 17 , 18 Besides this, cell stress leads to the upregulation of GRP78 and subsequently its escape from ER retention, translocating to the cell surface (cell‐surface GRP78). Different viral and fungal pathogens are reported to bind cell‐surface GRP78. 17 , 19 , 20 A 13 residues cyclic peptide CTVALPGGYVRVC (Pep42) is said to target, selectively, the cell‐surface GRP78 over cancer cells. 21 Pep42 is used to deliver doxorubicin to cancer cells that upregulate the cell‐surface GRP78. 17 , 22
In the current study, the binding site between GRP78 and viral E6 is predicted based on sequence and structural similarities between the Pep42 and HPV E6. Molecular docking is employed to further explore such binding using the protein‐protein docking technique that uses molecular dynamics simulation in refining the interacting residues in the binding site.
2. MATERIALS AND METHODS
The X‐ray crystallography solved‐structure protein data bank (PDB) file in addition to the protein sequence (FASTA) file (released in the year 2016 with a resolution of 2.25 Å) for HPV E6 protein was downloaded from the PDB database (PDB ID: 4XR8). 23 The removal of other protein chains, water molecules, and ions from the PDB file is performed using PyMOL software. 24 The only full‐length wildtype GRP78 structure (released in the year 2016 with a resolution of 2.99 Å) in its open configuration (PDB ID: 5E84) is downloaded and prepared for the docking experiment using PyMOL. 25 , 26 The Clustal Omega web server is used to perform sequence alignment between the HPV E6 protein and the peptide Pep42, 27 while ESPript 3 software was used to represent the alignment. 28 The part of the HPV E6 protein that well represents the Pep42 (38.5% sequence identity) is further analyzed by the ProtParam web server of the ExPASy bioinformatics resource portal. 29 The Clustal Omega web server is also used to perform multiple sequence alignment (MSA) of all the present HPV E6 sequences in the National Center for Biotechnology Information (NCBI) protein database of the National Institute of Health. 30
I‐TASSER web server was used to build a model for Pep42 peptide. 31 The HpepDock web server 32 was used to dock Pep42 peptide and HPV E6 (C51:C63 region) peptide into the binding site of GRP78. Molecular docking is performed using the HADDOCK web server 33 to test the binding mode of the full‐length HPV E6 to GRP78 substrate‐binding domain β (SBDβ). The active amino acids involved in the interactions with substrates are retrieved from the literature for GRP78 (I426, T428, V429, V432, T434, F451, S452, V457, and I459), while for HPV E6, the active residues are selected based on the similarity with Pep42 peptide. 25 In contrast, the amino acid residues surrounding the active residues are chosen to be the passive amino acids in HADDOCK. The interactions that occurred between the two proteins are determined by using the Protein‐Ligand Interaction Profiler (PLIP) web server. 34
3. RESULTS AND DISCUSSION
3.1. Sequence and structural alignment
Figure 1A shows the sequence alignment between the solved structure of HPV E6 protein (PDB ID: 4XR8, F and H chains) and the sequence of the peptide Pep42. Five (red‐highlighted residues) out of 13 residues are identical (38.5% sequence identity). These identical residues are C51, V53, G57, V62, and C63 (4XR8 numbering scheme). The hydrophobicity (Kyte and Doolittle hydropathy parameters) 35 of the Pep42 and the part of HPV E6, which bear high sequence identity to Pep42, are shown in the bar graph of Figure 1B. It is clear that the high hydrophobic character is in the terminals of both Pep42 and HPV E6 C51:C63 region (around the disulfide bond) while in the central residues, the hydrophilic character is dominant. Figure S1 shows the MSA performed using the Clustal Omega web server. The retrieved 330 sequences of HPV E6 were represented with the aid of the ESPript 3.0 web server 36 after alignment with Clustal Omega. As shown in Figure S1, three CxxC motifs are very well conserved among all the HPV E6 sequences. These motifs represent the Zn finger domains present in HPV E6 crucial for its stabilization (homodimerization) and function. 1 , 37 Besides this, the motif LxxVxxxDxxxFAxC shows high sequence conservation among the aligned sequences and is surface accessible. This motif lies in the β‐hairpin fold (PDB ID: 4XR8), like the Pep42 model, and is suggested to be the binding site to GRP78.
Figure 1.

A, Sequence alignment of the solved structures for HPV E6 oncoprotein (PDB ID: 4XR8) and the sequence of Pep42. The alignment is made using the Clustal Omega web server and represented by ESPript 3 software. The red‐highlighted residues are the identical residues found in both proteins. B, Hydrophobicity plot (Kyte and Doolittle) for both Pep42 and HPV E6 C51:C63 peptides. HPV, human papillomavirus; PDB, Protein Data Bank
Figure 2A shows the structure of modeled Pep42 (using the I‐TASSER web server) and the HPV E6 C51:C63 region (PDB ID: 4XR8). Figure 2B shows the superposition between the two peptides with a root mean square deviation (RMSD) value of 1.67 Å (66 atoms included in the fitting). Identical amino acids are represented in red cartoons in both peptides, while the remaining residues are represented in colored cartoons (green for Pep42 and cyan for HPV E6 C51:C63). The structural alignment shows that the two peptides share the same structural folding (β‐hairpin structures).
Figure 2.
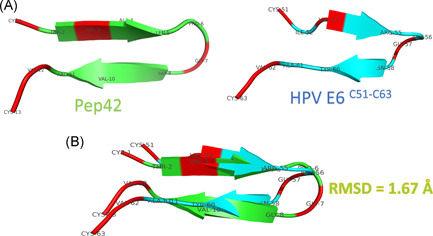
A, Structure of the Pep42 model (built by I‐TASSER web server) and the HPV E6 C51:C63 (PDB ID: 4XR8) peptides represented in green and cyan cartoon, respectively. Residues are labeled in both peptides using the three‐letter naming scheme. B, Structural alignment of the two peptides using the align option of PyMOL software. RMSD of the superposition is 1.67 Å. HPV, human papillomavirus; PDB, Protein Data Bank; RMSD, root mean square deviation
3.2. Peptide/protein docking
Before docking experiments, both proteins and peptides are prepared by PyMOL software. Missing H‐atoms are added while water molecules and ions are removed from the PDB files. A redocking test is performed to ensure the suitability of the docking software on the protein system under the study. The RMSD of the docked structures shows good agreement with the experimentally solved structures for both software used for the docking (RMSDs are 1.97 and 1.51 Å for HpepDock and HADDOCK softwares, respectively).
HpepDock web server is used to dock the peptides (both cyclic Pep42 and HPV E6 C51:C63) into the protein (GRP78 SBDβ). The docking is accomplished by defining the binding site of GRP78 (I426, T428, V429, V432, T434, F451, S452, V457, and I459) while the peptides (Pep42 and HPV E6 C51:C63) are treated as rigid structures. After docking, the PLIP web server is used to quantify the interactions of the resulting docking complexes. We compared the interactions settled down upon binding the peptides into GRP78 SBDβ. There are four H‐bonds, and four hydrophobic interactions are established between the Pep42 peptide and GRP78 SBDβ, while four H‐bonds and five hydrophobic interactions are settled in the case of HPV E6 C51:C63 peptide. Pep42 interacts by six residues (C1, T2, V3, A4, L5, and Y9) while HPV E6 C51:C63 interacts through five residues (C51, I52, Y54, D56, and G57). The HpepDock docking score for Pep42 is slightly more negative (−72.4) (means has better binding) compared with HPV E6 C51:C63 (−62.18). Figure 3 summarizes the interactions established after docking for Pep42 (A) and HPV E6 C51:C63 (B) peptides. GRP78 is represented in Figure 3 as a green cartoon with its domains marked on the figure; nucleotide binding domain (NBD) and substrate‐binding domains α and β (SBDα and SBDβ). The interactions with the peptides are maintained by the H‐bonding (blue‐colored residues) and hydrophobic interactions (magenta‐colored residues) with the peptides (red cartoons). The enlarged panels in Figure 3A,B show the interacted amino acids from both GRP78 SBDβ and the peptides.
Figure 3.
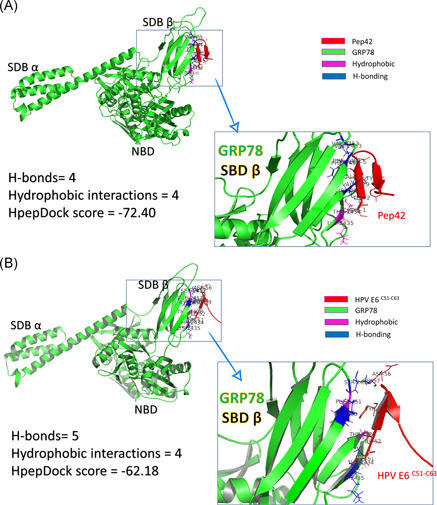
Peptide/protein docking experiment using HpepDock. GRP78 solved structure (5E84) represented in the green‐colored cartoon is docked with (A) Pep42 and (B) HPV E6 C51:C63 peptides (red‐colored cartoons). Residues from GRP78 SBDβ that are involved in H‐bond formation with the peptides are represented in blue‐colored cartoons. The docking scores and number of interactions (H‐bonds and hydrophobic interactions) are listed in the left panel of the figure. HPV, human papillomavirus; GRP, glucose‐regulated protein; SBD, substrate‐binding domain
3.3. GRP78/HPV E6 full‐length protein docking
After predicting the binding site using sequence and structural alignments, testing the binding mode and binding energy is the next step. Haddock web server (easy interface) is used to test the binding mode, and the potency of the interactions settled between GRP78 SBDβ (PDB ID: 5E84, chain A) to the full‐length HPV E6 protein (PDB ID: 4XR8, chain H). The active residues are defined for GRP78 (I426, T428, V429, V432, T434, F451, S452, V457, and I459) while for the HPV E6, it is the C51:C63 region (C51, I52, V53, Y54, R55, D56, G57, N58, P59, Y60, A61, V62, and C63). Passive residues are selected to be the residues surrounding the active residues for both proteins.
A total of 153 docked structures are clustered into 10 different clusters according to HADDOCK scores. The number of structures in every cluster varies from 6 (clusters 8, 9, and 10) up to 43 (cluster 1). The HADDOCK scores for each cluster varies from −68.8 ± 3.8 (the best‐scored cluster 8) to −26.8 ± 3.2 (the worst scored cluster 7). The top‐ranked cluster (cluster 8) is used to follow the docking pose with the aid of PLIP. Table 1 summarizes the interactions established in the first four, top‐ranked, docking structures of cluster 8 (top‐ranked cluster). The most stable interactions are the three H‐bonds that are created between the R55 of HPV E6 and both T456 and T458 of GRP78. The Y60 of HPV E6 forms H‐bonds to G430 of GRP78 in two (out of four) docking complexes, while at least two H‐bonds are formed between D56 of HPV E6 and S452 of GRP78 in two complexes (out of four). Some hydrophobic interactions are established between the two proteins upon dockings, such as between V429, F451, and V453 of GRP78 and Y54, D56, and Y60 of HPV E6.
Table 1.
The interactions formed between HPV E6 and GRP78 SBDβ upon docking
| H‐bonding | Hydrophobic interaction | ||||||
|---|---|---|---|---|---|---|---|
| HADDOCK cluster 8 | HADDOCK score | Number | Amino acids involved from HPV E6 | Amino acids involved from GRP78 | Number | Amino acids involved from HPV E6 | Amino acids involved from GRP78 |
| First model | −68.8 ± 3.8 | 4 | R55 | T456 | 1 | Y54 | V453 |
| R55 | T458 | ||||||
| R55 | T458 | ||||||
| Y60 | G430 | ||||||
| Second model | 3 | R55 | T456 | 0 | |||
| R55 | T458 | ||||||
| R55 | T458 | ||||||
| Third model | 7 | R55 | T456 | 1 | Y60 | V429 | |
| R55 | T456 | ||||||
| R55 | T458 | ||||||
| D56 | S452 | ||||||
| D56 | S452 | ||||||
| D56 | V453 | ||||||
| Y60 | G430 | ||||||
| Fourth model | 8 | R55 | T458 | 1 | D56 | F451 | |
| R55 | T458 | ||||||
| R55 | T458 | ||||||
| D56 | S452 | ||||||
| D56 | S452 | ||||||
| D56 | S452 | ||||||
| D56 | S452 | ||||||
| G57 | S452 | ||||||
Note: Four different docked complexes are analyzed here, representing the best cluster of HADDOCK.
Abbreviations: HPV, human papillomavirus; GRP, glucose‐regulated protein; SBD, substrate‐binding domain.
Figure 4 shows one of the docking complexes of cluster 8. GRP78 is represented in a green cartoon with its domain marked in Figure 4A (NBD, SBDα, and SBDβ). HPV E6 is represented in the yellow cartoon with its C51:C63 region in red. H‐bonds that formed between the two interacted proteins are expressed in blue dashed lines. As shown in the left panel of Figure 4A, the number of H‐bonds between GRP78 and HPV E6 is four (R55::T456, R55::T458(2), and Y60::G430), while only one hydrophobic interaction (Y54::V453) is established upon binding with HADDOCK cluster score of −68.8 ± 3.8. Figure 4B shows how the HPV E6 C51:C63 region (red cartoon) fits inside the SBDβ of GRP78 (molecular surface) between its loops.
Figure 4.
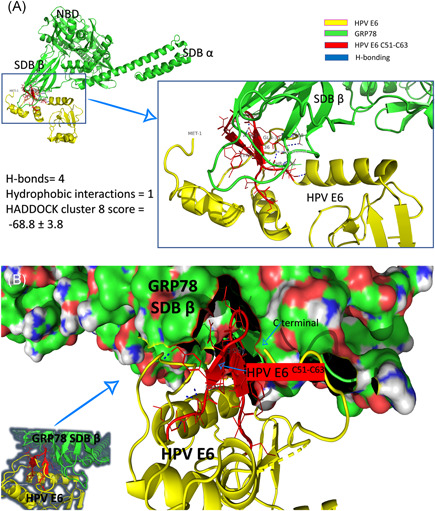
Protein/protein docking experiment using HADDOCK. A, GRP78 solved structure (5E84) represented in green‐colored cartoon labeled with the full‐length HPV E6 solved structure (4XR8) (yellow‐colored cartoons). The C51:C63 region of HPV E6 is represented in the red‐colored cartoon. Residues from both GRP78 and HPV E6 that involved in the interactions are labeled and represented in lines, while the H‐bonds are represented in blue dashed lines. B, The same docking complex but showing the molecular structure of GRP78 to show how HPV E6 C51:C63 fits inside the SBDβ of GRP78. HPV, human papillomavirus; GRP, glucose‐regulated protein; SBD, substrate‐binding domain
The docking study illustrates how the binding is established between HPV E6 and the chaperone, GRP78, protein. Interestingly, the N‐terminal of the HPV E6 lies near the suggested binding site of HPV E6 (Figure 4B). Further molecular dynamics study is needed to check the contribution of this N‐terminal region in GRP78 binding.
4. CONCLUSION
HPV is the primary causative agent for cervical cancer and other related diseases. The viral protein E6 is the primary oncogenic protein signal that promotes p53 degradation. Preventing such binding is crucial in fighting against, this cancer‐promoting, viral infection. GRP78 is said to cause E6 stabilization in vivo among other host cell and viral proteins. Once bound to GRP78, the HPV E6 is stabilized (saved from the degradation through host cell machinery and proteasome degradation pathway). E6 destabilization is reported to be a cancer‐preventing strategy. Inhibiting the GRP78‐E6 association would help in cancer cessation after the identification of the binding site. The present work suggests the GRP78‐E6 binding site using in silico techniques. Further dynamic simulations and experimental work are required to prove our hypothesis; besides, this study paves the way for exploring GRP78‐E6 binding inhibitors to fight against this cancer‐promoting viral infection.
CONFLICT OF INTEREST
The author declares that there is no conflict of interest.
Supporting information
Supporting information
Supporting information
ACKNOWLEDGMENT
Hazem Reda is thankful for the helpful discussions.
Elfiky AA. Human papillomavirus E6: Host cell receptor, GRP78, binding site prediction. J Med Virol. 2020;92:3759–3765. 10.1002/jmv.25737
REFERENCES
- 1. Ajiro M, Zheng Z‐M. E6^E7, a novel splice isoform protein of human papillomavirus 16, stabilizes viral E6 and E7 oncoproteins via HSP90 and GRP78. mBio. 2015;6(1):e02068‐e02014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Koutsky LA, Galloway DA, Holmes KK. Epidemiology of genital human papillomavirus infection. Epidemiol Rev. 1988;10:122‐163. [DOI] [PubMed] [Google Scholar]
- 3. Bosch FX, Manos MM, Munoz N, et al. Prevalence of human papillomavirus in cervical cancer: a worldwide perspective. J Natl Cancer Inst. 1995;87(11):796‐802. [DOI] [PubMed] [Google Scholar]
- 4. Schiffman M, Castle PE, Jeronimo J, Rodriguez AC, Wacholder S. Human papillomavirus and cervical cancer. Lancet 2007;370(9590):890‐907. [DOI] [PubMed] [Google Scholar]
- 5. Zur Hausen H. Papillomavirus infections—a major cause of human cancers. Biochim Biophys Acta. 1996;1288(2):F55‐F78. [DOI] [PubMed] [Google Scholar]
- 6. Baseman JG, Koutsky LA. The epidemiology of human papillomavirus infections. J Clin Virol. 2005;32:16‐24. [DOI] [PubMed] [Google Scholar]
- 7. Aksoy P, Gottschalk EY, Meneses PI. HPV entry into cells. Mutat Res Rev Mutat Res. 2017;772:13‐22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Stanley M. Immune responses to human papillomavirus. Vaccine. 2006;24:S16‐S22. [DOI] [PubMed] [Google Scholar]
- 9. Moody CA, Laimins LA. Human papillomavirus oncoproteins: pathways to transformation. Nat Rev Cancer. 2010;10(8):550‐560. [DOI] [PubMed] [Google Scholar]
- 10. Kaliamurthi S, Selvaraj G, Kaushik AC, Gu K‐R, Wei D‐Q. Designing of CD8+ and CD8+‐overlapped CD4+ epitope vaccine by targeting late and early proteins of human papillomavirus. Biologics. 2018;12:107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Scheffner M, Huibregtse JM, Vierstra RD, Howley PM. The HPV‐16 E6 and E6‐AP complex functions as a ubiquitin‐protein ligase in the ubiquitination of p53. Cell. 1993;75(3):495‐505. [DOI] [PubMed] [Google Scholar]
- 12. Gething M‐J, Sambrook J. Protein folding in the cell. Nature. 1992;355(6355):33‐45. [DOI] [PubMed] [Google Scholar]
- 13. Lee AS. The ER chaperone and signaling regulator GRP78/BiP as a monitor of endoplasmic reticulum stress. Methods. 2005;35(4):373‐381. [DOI] [PubMed] [Google Scholar]
- 14. Li J, Lee AS. Stress induction of GRP78/BiP and its role in cancer. Curr Mol Med. 2006;6(1):45‐54. [DOI] [PubMed] [Google Scholar]
- 15. Rao RV, Peel A, Logvinova A, et al. Coupling endoplasmic reticulum stress to the cell death program: role of the ER chaperone GRP78. FEBS Lett. 2002;514(2‐3):122‐128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Quinones QJ, Ridder GGd, Pizzo SV. GRP78, a chaperone with diverse roles beyond the endoplasmic reticulum. Histol Histopathol. 2008;23:1409‐1416. [DOI] [PubMed] [Google Scholar]
- 17. Ibrahim IM, Abdelmalek DH, Elfiky AA. GRP78: a cell's response to stress. Life Sci. 2019;226:156‐163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shen J, Chen X, Hendershot L, Prywes R. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev Cell. 2002;3(1):99‐111. [DOI] [PubMed] [Google Scholar]
- 19. Gebremariam T, Liu M, Luo G, et al. CotH3 mediates fungal invasion of host cells during mucormycosis. J Clin Invest. 2014;124(1):237‐250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ibrahim I, Abdelmalek D, Elshahat M, Elfiky A. COVID‐19 spike‐host cell receptor GRP78 binding site prediction. 2020. Journal of Infection , In press. [DOI] [PMC free article] [PubMed]
- 21. Kim Y, Lillo AM, Steiniger SCJ, et al. Targeting heat shock proteins on cancer cells: selection, characterization, and cell‐penetrating properties of a peptidic GRP78 ligand. Biochemistry. 2006;45(31):9434‐9444. [DOI] [PubMed] [Google Scholar]
- 22. Martin S, Hill DS, Paton JC, et al. Targeting GRP78 to enhance melanoma cell death. Pigment Cell Melanoma Res. 2010;23(5):675‐682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Berman HM. The protein data bank. Nucleic Acids Res. 2000;28(1):235‐242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. DeLano WL. The PyMOL Molecular Graphics System, Version 1.2r3pre, Schrödinger, LLC. 2002. [Google Scholar]
- 25. Yang J, Nune M, Zong Y, Zhou L, Liu Q. Close and allosteric opening of the polypeptide‐binding site in a human Hsp70 chaperone BiP. Structure. 2015;23(12):2191‐2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yang J, Zong Y, Su J, et al. Conformation transitions of the polypeptide‐binding pocket support an active substrate release from Hsp70s. Nat Commun. 2017;8(1):1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sievers F, Wilm A, Dineen D, et al. Fast, scalable generation of high‐quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7(1):539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gouet P, Courcelle E, Stuart DI, Metoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15(4):305‐308. [DOI] [PubMed] [Google Scholar]
- 29. Garg VK, Avashthi H, Tiwari A, et al. MFPPI—multi FASTA ProtParam interface. Bioinformation. 2016;12(2):74‐77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. NCBI . National Center of Biotechnology Informatics (NCBI) database. 2020. http://www.ncbi.nlm.nih.gov/
- 31. Zhang Y. I‐TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9(1):40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhou P, Jin B, Li H, Huang S‐Y. HPEPDOCK: a web server for blind peptide–protein docking based on a hierarchical algorithm. Nucleic Acids Res. 2018;46(W1):W443‐W450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. van Dijk AD, Bonvin AM. Solvated docking: introducing water into the modelling of biomolecular complexes. Bioinformatics. 2006;22(19):2340‐2347. [DOI] [PubMed] [Google Scholar]
- 34. Salentin S, Schreiber S, Haupt VJ, Adasme MF, Schroeder M. PLIP: fully automated protein–ligand interaction profiler. Nucleic Acids Res. 2015;43(W1):W443‐W447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157(1):105‐132. [DOI] [PubMed] [Google Scholar]
- 36. Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42(W1):W320‐W324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ansari T, Brimer N, Pol SBV. Peptide interactions stabilize and restructure human papillomavirus type 16 E6 to interact with p53. J Virol. 2012;86(20):11386‐11391. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information
Supporting information


