Abstract
Canine coronavirus (CCoV) strains with the ability to spread to internal organs, also known as pantropic CCoVs (pCCoVs), have been detected in domestic dogs and wild carnivores. Our study focused on the detection and molecular characterization of pCCoV strains circulating in Italy during the period 2014–2017 in autochthonous dogs, in dogs imported from eastern Europe or illegally imported from an unknown country. Samples from the gut and internal organs of 352 dogs were screened for CCoV; putative pCCoV strains, belonging to subtype CCoV‐IIa, were identified in the internal organs of 35 of the examined dogs. Fifteen pCCoV strains were subjected to sequence and phylogenetic analyses, showing that three strains (98960‐1/2016, 98960‐3/2016, 98960‐4/2016) did not cluster either with Italian or European CCoVs, being more closely related to alphacoronaviruses circulating in Asia with which they displayed a 94%–96% nucleotide identity in partial spike protein gene sequences. The pCCoV‐positive samples were also tested for other canine viruses, showing co‐infections mainly with canine parvovirus.
Keywords: animal importation, dog, pantropic canine coronavirus, viral co‐infections
1. INTRODUCTION
Coronaviruses (CoVs) are viruses that infect a variety of mammals, including humans, and birds. Coronaviruses (CoVs, order Nidovirales, family Coronaviridae) are enveloped, single‐stranded, positive‐sense RNA viruses, commonly associated with mild enteritis and/or respiratory signs (Decaro et al., 2013). The family Coronaviridae is organized into two subfamilies, Orthocoronavirinae and Letovirinae, with the former including four different genera, Alphacoronavirus, Betacoronavirus, Gammacoronavirus and Deltacoronavirus. Canine coronavirus (CCoV) belongs to the genus Alphacoronavirus and forms a unique species, Alphacoronavirus‐1, along with feline coronaviruses (FCoVs), transmissible gastroenteritis virus of swine (TGEV), and its derivative, porcine respiratory coronavirus (PRCoV) (Decaro & Buonavoglia, 2008, 2011).
Canine coronavirus infection is restricted to the enteric tract and causes mild or asymptomatic forms of enteritis. Canine coronavirus includes two genotypes: CCoV‐I and CCoV‐II, which share up to 96% of nucleotide (nt) sequence identity in the viral genome, but are highly divergent in the spike (S) protein gene. CCoV‐II is classified into two subtypes: the classical CCoV‐IIa and the recombinant CCoV‐IIb (Decaro et al., 2009).
A hypervirulent CCoV‐IIa strain, referred to as pantropic CCoV (pCCoV), was isolated in Italy in 2005 (Buonavoglia et al., 2006) and is able to spread to extraintestinal tissues. This variant was associated with a fatal disease of dogs, characterized by leukopenia, gastroenteritis and severe lesions in the major organs (Buonavoglia et al., 2006; Chen et al., 2019; Decaro et al., 2008; Pinto et al., 2014), and subsequent studies have proved its impact on canine immune response (Marinaro et al., 2010).
Currently, there are no diagnostic tools capable to differentiate the pantropic strains from the enteric CCoVs, since they are strictly related at the genetic and antigenic level and no specific marker of pathogenicity has been detected so far (Decaro et al., 2013). Thus, the identification of pCCoV relies on the detection of CCoV‐IIa in extraintestinal tissues. To date, pCCoV infection has been reported mainly in dogs (Chen et al., 2019; Decaro et al., 2012, 2013; Ntafis et al., 2012; Pinto et al., 2014; Zicola et al., 2012), although a hypervirulent CCoV‐IIa strain was also reported in a wolf (Alfano et al., 2019). In most cases pCCoV was found in association with co‐pathogens, including canine parvovirus (CPV) (Alfano et al., 2019; Decaro et al., 2012, 2013; Ntafis et al., 2012; Pinto et al., 2014; Zicola et al., 2012) and canine adenovirus type 1 (CAdV‐1) (Alfano et al., 2019).
Taking into account the scarce information existing about the actual circulation of pCCoV in the dog population, the aims of our study were the following: (a) to conduct an epidemiological survey for this virus in autochthonous and imported dogs in Italy during years 2014–2017; (b) to investigate the genetic relatedness of the detected pCCoV strains to extant coronaviruses; and (c) to evaluate the presence of co‐infections in pCCoV positive and negative samples.
2. MATERIALS AND METHODS
2.1. Samples collection
During the period 2014–2017, 2,112 necropsy samples collected from different tissues (brain, heart, intestine, liver, spleen, lungs, kidney) of 352 dogs were submitted to molecular analysis to investigate possible viral causes of disease. The sampled animals included 141 client‐owned, 151 stray and 60 imported dogs. Two hundred ninety‐two of these dogs were from Italy, additional 56 animals had been imported by Hungary, while 4 dogs had been illegally imported from an unknown country. None of these dogs had undergone euthanasia, since their death was caused by illness or accident, but clinical signs occurring intravitam were not reported. At post‐mortem examination, the analysed dogs showed catarrhal or hemorrhagic enteritis (n = 137), enlargement of the mesenteric lymph nodes (n = 79), pneumonia or other pulmonary lesions (n = 70), and meningeal and/or encephalic hyperaemia (n = 49). For 55 animals, post‐mortem findings were not reported or were not fitting with those observed in pCCoV‐infected dogs.
2.2. Nucleic acid extraction
Samples collected for molecular investigations were homogenized with phosphate‐buffered saline (PBS), and subsequently, RNA/DNA extraction was performed using the automated extractor QIAsymphony (Qiagen) and the QIAsymphony DSP Virus/Pathogen Kit (Qiagen), following the manufacturer's instructions.
2.3. Detection of viral pathogens
Tissues samples of dogs were submitted to molecular detection of the main viral pathogens of dogs, including CCoV (Decaro et al., 2013), CPV (Decaro, Elia, et al., 2005), CAdV‐1 and CAdV‐2, (Dowgier et al., 2016), canine distemper virus (CDV) (Elia et al., 2006), canid alphaherpesvirus type 1 (CaHV‐1) (Decaro et al., 2008) and rotaviruses (Zeng et al., 2008).
2.4. CCoV genotyping and subtyping
The detected CCoV strains were characterized by means of two distinct real‐time RT‐PCR assays, specific for the genotypes CCoV‐I and CCoV‐II, targeting a fragment of the S gene (Decaro et al., 2013).
Samples that tested positive for CCoV‐II were subjected to subtype‐specific CCoV‐IIa and CCoV‐IIb gel‐based RT‐PCR assays targeting the S gene (Table 1) (Decaro et al., 2013). The PCR products were detected using the TapeStation 2,200 (Agilent Technologies) according to the manufacturer's protocol.
TABLE 1.
Oligonucleotides used for detection and characterization of CCoV strains
| Test | Primer/probe | Reference | Sequence (5′−3′) | Sense | Position | Amplicon size | Specificity |
|---|---|---|---|---|---|---|---|
| Real‐time RT‐PCR | CCoV‐F | Decaro et al., 2004 | TTGATCGTTTTTATAACGGTTCTACAA | + | 6585−6611 a | 99 bp | CCoV‐I/II |
| CCoV‐R | AATGGGCCATAATAGCCACATAAT | − | 6660−6683 a | ||||
| CCoV‐Pb | FAM‐ACCTCAATTTAGCTGGTTCGTGTATGGCATT‐TAMRA | + | 6620−6650 a | ||||
| Real‐time RT‐PCR | CCoVI‐F | Decaro, Martella, et al., 2005 | CGTTAGTGCACTTGGAAGAAGCT | + | 478−499 b | 111 bp | CCoV‐I |
| CCoVI‐R | ACCAGCCATTTTAAATCCTTCA | − | 567−588 b | ||||
| CCoVI‐Pb | FAM ‐CCTCTTGAAGGTACACCAA‐TAMRA | + | 508−526 b | ||||
| Real‐time RT‐PCR | CCoVII‐F | Decaro, Martella, et al., 2005 | TAGTGCATTAGGAAGAAGCT | + | 6878−6897 a | 105 bp | CCoV‐II |
| CCoVII‐R | AGCAATTTTGAACCCTTC | − | 6966−6982 a | ||||
| CCoVII‐Pb | FAM ‐CCTCTTGAAGGTGTGCC‐TAMRA | + | 6906−6922 a | ||||
| RT‐PCR | 20,179 | Decaro et al., 2010 | GGCTCTATCACATAACTCAGTCCTAG | + |
12531−12556 c |
758 bp (CCoV‐IIa) |
CCoV‐I/II |
| INS‐R | GCTGTAACATAGTCATCATTCCAC | − | 1054−1077 b |
499 bp (CCoV‐IIb) |
CCoV‐IIa | ||
| 174–268 | CAACATGTAACCTTTGTCTGTGATCTGC | − | 13002−13029 c | CCoV‐IIb |
Abbreviations: FAM, 6‐carboxyfluorescein; TAMRA, 6‐carboxytetramethylrhodamine.
Oligonucleotide position is referred to the sequence of CCoV‐I strain 259/01 (GenBank accession number AF502583).
Oligonucleotide position is referred to the sequence of CCoV‐IIa strain Insavc‐1 (GenBank accession number D13096).
Oligonucleotide position is referred to the sequence of CCoV‐IIb strain 174/06 (GenBank accession number EU856362).
2.5. Molecular characterization of pCCoV and CPV
Lung samples from the pCCoV‐infected dogs were used for the molecular characterization of pCCoV. The spike protein gene (ORF2) of the putative pCCoV strains was sequenced and analysed using the protocol reported by Alfano et al. (2019). The sequences were analysed using BioEdit software package and the NCBI and EMBL analysis tools.
Samples that tested positive for CPV were further characterized by type‐specific minor groove binder probe assays (Decaro, Elia, et al., 2006; Decaro, Elia, et al., 2005; Decaro et al., 2007; Decaro, Martella, et al., 2006) and sequence analysis of partial VP2 gene (Buonavoglia et al., 2001).
2.6. Sequence analysis and phylogeny
CCoV sequences were manually edited and analysed using the Geneious platform (version 10.1.3) (Biomatters Ltd.). Nucleotide similarity with sequences deposited in the GenBank database was assessed using the BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and FASTA (http://www.ebi.ac.uk/fasta33) tools with default values used to find homologous hits.
For the construction of phylogenetic trees, a multiple alignment of all target sequences was performed, using MAFTT Multiple Sequence Alignment software version 7 (Katoh & Standley, 2013) and Geneious software, using the neighbour‐joining method, with the p‐distance model, 1,000 bootstrap replicates, and, otherwise, the default parameters in Geneious (version 10.1.3).
2.7. Nucleotide sequence accession number
The nucleotide sequences of the analysed pCCoV strains were deposited in GenBank under the following accession numbers: MN086803 (98960‐1/2016); MN086804 (98960‐3/2016); MN086805 (98960‐4/2016); MN086806 (32712/1/2016); MN086807 (78870/2016); MN086808 (103480/2014); MN086809 (31975/2015); MN086810 (43891‐1/2016); MN086811 (43891‐2/2016); MN086812 (91507/2017); MN086813 (91510/2017); MN086814 (34879/2017); MN086815 (43022/2017); MN086816 (91529/2017); and MN086817 (34865/2017).
2.8. Data analysis
The comparison between positive and negative pCCoV dogs was carried out by examining the data with a chi‐squared test, considering the statistically significant values p < .05, using the IBM SPSS Statistics 25 software. The confidence interval (CI 95%) was calculated using the prevalence parameter estimate with the Excel software.
3. RESULTS
3.1. Detection of CCoV and pCCoV in autochthonous and imported dogs
CCoV RNA was detected in the gut of 76 (21.59%) out of 352 tested dogs, while 35 animals (9.94%), about half of the positive dogs, tested positive for CCoV‐IIa in internal organs by means of typing and subtyping molecular assays (Decaro et al., 2013), thus showing the presence of putative pCCoV strains in the infected dogs (Table 2). Of these 35 dogs, 6 lived in Italy and 29 had been imported from other countries (25 from Hungary and 4 illegally imported from an unknown country). The proportion of pCCoV‐infected dogs varies markedly between regions: we found 6 pCCoV‐infected dogs from Italy (6/292, 2%), 25 infected animals from Hungary (25/56, 45%) and 4 animals of unknown origin (4/4, 100%). At necropsy, apart from acute, sometime hemorrhagic gastroenteritis, most of the pCCoV positive dogs had displayed lesions suggestive of systemic involvement, including pneumonia and encephalitis (Table 2).
TABLE 2.
Signalment of dogs tested positive to pantropic CCoV and detection of viral co‐infections
| Pantropic CCoV‐positive dogs | Year | Prot. no. | Breed | Age | Country of origin | Life conditions | Gross lesions | Extraintestinal tissues positive to pCCoV | Co‐infecting viruses |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2014 | 103480 | Pomeranian | Y | Italy | Client‐owned | UKN | Heart, liver, spleen, lungs | CPV (2a), CDV |
| 2 | 2015 | 31975 | Mixed breed | Y | Italy | Stray dog | UKN | Heart, liver, spleen, lungs | CPV (2b) |
| 3 | 2015 | 15910‐3 |
Cavalier king charles spaniel |
Y | Hungary | Imported | Enteritis, splenomegaly, pneumonia, meningeal and encephalic hyperaemia | Brain, heart, liver, spleen, lungs, kidney | CPV (2a), CDV |
| 4 | 2016 | 32712‐1 | Mixed breed | Y | Italy | Stray dog | UKN | Brain, heart, liver, spleen, lungs, kidney | CPV (2b), CAdV‐1 |
| 5 | 2016 | 43891/1 | Mixed breed | J | Italy | Stray dog | Enteritis, meningeal and encephalic hyperaemia | Brain, heart, liver, spleen, lungs | None |
| 6 | 2016 | 43891‐2 | Mixed breed | J | Italy | Stray dog | Enteritis, meningeal and encephalic hyperaemia | Brain, heart, liver, spleen, lungs | None |
| 7 | 2016 | 78870 | Chihuahua | Y | Italy | Client‐owned | UKN | Brain, heart, liver, spleen, lungs, kidney | CPV (2b), CAdV‐2, CDV |
| 8 | 2016 | 98960‐1 | English Bulldog | Y | UKN | Illegally imported | Enteritis, enlargement of mesenteric lymph nodes, pulmonary atelectasis and emphysema, meningeal hyperaemia | Brain, heart, liver, spleen, lungs | CPV (2a), CAdV‐2 |
| 9 | 2016 | 98960‐2 | Pug | Y | UKN | Illegally imported | Enteritis, enlargement of mesenteric lymph nodes, pneumonia, meningeal and encephalic hyperaemia | Brain, heart, liver, spleen, lungs | CPV (2a), CAdV‐2 |
| 10 | 2016 | 98960‐3 | English bulldog | Y | UKN | Illegally imported | Enteritis, enlargement of mesenteric lymph nodes, pneumonia and pulmonary oedema | Brain, heart, liver, spleen, lungs | CPV (2a), CAdV‐2 |
| 11 | 2016 | 98960‐4 | Dogue de bordeaux | Y | UKN | Illegally imported | Enteritis, enlargement of mesenteric lymph nodes, pneumonia and pulmonary oedema, meningeal and encephalic hyperaemia | Brain, heart, liver, spleen, lungs | CPV (2a), CAdV‐2 |
| 12 | 2017 | 34865 | Husky | Y | Hungary | Imported | Haemorrhagic enteritis, enlargement of mesenteric lymph nodes, meningeal and encephalic hyperaemia | Brain, heart, liver, spleen, lungs | None |
| 13 | 2017 | 34873 | Spitz | Y | Hungary | Imported | Haemorrhagic enteritis, enlargement of mesenteric lymph nodes, meningeal and encephalic hyperaemia | Brain, heart, liver, spleen, lungs | None |
| 14 | 2017 | 34875 | Spitz | Y | Hungary | Imported | Enteritis, enlargement of mesenteric lymph nodes, pneumonia | Brain, heart, liver, spleen, lungs | CPV (2a) |
| 15 | 2017 | 34876 | Spitz | Y | Hungary | Imported | Haemorrhagic enteritis, enlargement of mesenteric lymph nodes, pulmonary oedema | Brain, heart, liver, spleen, lungs | None |
| 16 | 2017 | 34879 | Shiba inu | Y | Hungary | Imported | Enlargement of mesenteric lymph nodes, meningeal hyperaemia | Brain, heart, liver, spleen, lungs | None |
| 17 | 2017 | 42211 | Shitzu | Y | Hungary | Imported | Enteritis, enlargement of mesenteric lymph nodes | Brain, heart, liver, spleen, lungs | CPV (2a) |
| 18 | 2017 | 43020 | Maltese | Y | Hungary | Imported | Enteritis, enlargement of mesenteric lymph nodes, pneumonia | Brain, heart, liver, spleen, lungs | CPV (2a) |
| 19 | 2017 | 43021 | Mixed breed | Y | Hungary | Imported | Enteritis, enlargement of mesenteric lymph nodes | Brain, heart, liver, spleen, lungs | CPV (2a, 2b) |
| 20 | 2017 | 43022 | Mixed breed | Y | Hungary | Imported | Enteritis, enlargement of mesenteric lymph nodes, pneumonia | Brain, heart, liver, spleen, lungs | CPV (2b), CaHV‐1 |
| 21 | 2017 | 91490 | Spitz | Y | Hungary | Imported | Enteritis, enlargement of mesenteric lymph nodes, pneumonia, meningeal and encephalic hyperaemia | Brain, heart, liver, spleen, lungs | CPV (2a) |
| 22 | 2017 | 91494 | Spitz | Y | Hungary | Imported | Enteritis, enlargement of mesenteric lymph nodes, pneumonia, meningeal and encephalic hyperaemia | Brain, heart, liver, spleen, lungs | CPV (2a), CAdV2 |
| 23 | 2017 | 91498 | Maltese | Y | Hungary | Imported | Enlargement of mesenteric lymph nodes | Brain, heart, liver, spleen, lungs | None |
| 24 | 2017 | 91501 | French Bulldog | Y | Hungary | Imported | Enteritis, enlargement of mesenteric lymph nodes | Brain, heart, liver, spleen, lungs | CPV (2a), CAdV2 |
| 25 | 2017 | 91504 | Spitz | Y | Hungary | Imported | Enteritis, enlargement of mesenteric lymph nodes | Brain, heart, liver, spleen, lungs | CPV (2a), CAdV2 |
| 26 | 2017 | 91507 | Spitz | Y | Hungary | Imported | Enteritis, enlargement of mesenteric lymph nodes, pneumonia, meningeal and encephalic hyperaemia | Brain, heart, intestine, liver, spleen, lungs | CPV (2a), CAdV2 |
| 27 | 2017 | 91510 | Maltese | Y | Hungary | Imported | Enteritis, enlargement of mesenteric lymph nodes, pneumonia | Brain, heart, intestine, liver, spleen, lungs | CPV (2a), CAdV2 |
| 28 | 2017 | 91513 | Yorkshire | Y | Hungary | Imported | Enteritis, enlargement of mesenteric lymph nodes, pneumonia, meningeal hyperaemia | Brain, heart, intestine, liver, spleen, lungs | CPV (2a), CAdV2 |
| 29 | 2017 | 91517 | Maltese | Y | Hungary | Imported | Enteritis, enlargement of mesenteric lymph nodes, pneumonia | Brain, heart, intestine, liver, spleen, lungs | CPV (2a), CAdV1, CAdV2 |
| 30 | 2017 | 91519 | Maltese | Y | Hungary | Imported | Haemorrhagic enteritis, enlargement of mesenteric lymph nodes, pneumonia | Brain, heart, intestine, liver, spleen, lungs | CPV (2a), CAdV2 |
| 31 | 2017 | 91529 | Poodle | Y | Hungary | Imported | Enteritis, enlargement of mesenteric lymph nodes, pulmonary oedema | Brain, heart, intestine, liver, spleen, lungs | CPV (2a), CAdV2 |
| 32 | 2017 | 91531 | Yorkshire | Y | Hungary | Imported | Haemorrhagic enteritis, enlargement of mesenteric lymph nodes, pneumonia | Brain, heart, intestine, liver, spleen, lungs | CPV (2a), CAdV2 |
| 33 | 2017 | 91538 | Maltese | Y | Hungary | Imported | Enteritis, enlargement of mesenteric lymph nodes | Brain, heart, intestine, liver, spleen, lungs | CPV (2a), CAdV2 |
| 34 | 2017 | 91542 | Maltese | Y | Hungary | Imported | Enteritis, enlargement of mesenteric lymph nodes | Brain, heart, intestine, liver, spleen, lungs | CPV (2a, 2b), CAdV2 |
| 35 | 2017 | 94583 | Maltese | Y | Hungary | Imported | Enteritis, pneumonia | Brain, heart, intestine, liver, spleen, lungs | CPV (2a), CAdV2 |
Abbreviations: J, juvenile (6‐ to 12‐month‐old); UKN, unknown; Y, young (0‐ to 6‐month‐old).
3.2. Molecular characterization of putative pCCoV strains
Fifteen pCCoV strains were sequenced: 6 were from dogs of Italy and 9 from animals imported from other countries (6 from Hungary and 3 from an unknown country). The sequenced pCCoV strains presented neither the deletion in the genes of the accessory 3abc proteins nor the D125N mutation that had been suggested as potential markers for the pantropic behaviour (Decaro et al., 2008, 2013).
The phylogenetic tree, generated from partial ORF2 gene sequences, based on neighbour‐joining method (Figure 1) showed 5 different clustering groups. Strain 103480 felt in a cluster quite distant from all the others; 8 strains (32712, 78870, 43891‐1, 43891‐2, 34879, 43022, 91529, 34865) clustered with the Italian pCCoV strain 120/10; 2 strains (91507, 91510) were intermingled with enteric CCoVs; 3 other strains (98960‐1, 98960‐3, 98960‐4) clustered with strain pCCoV/wolf/2016/IT and 3 Asian enteric CCoVs. Another strain (31975) clustered with pCCoV strains identified in Italy and Greece (Ntafis et al., 2012).
FIGURE 1.
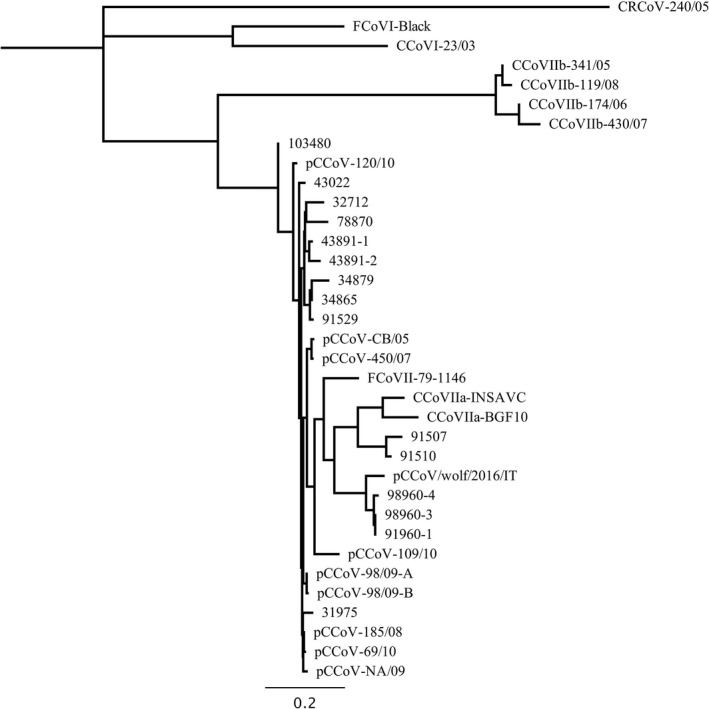
Phylogenetic tree generated with the neighbour‐joining method from partial spike protein gene (ORF2) sequences of the putative pantropic canine coronavirus strains and reference carnivore alphacoronaviruses
3.3. Detection of viral co‐infections
Only 7 dogs (two adults and five pups) showed a single pCCoV infection, while the other 28 pCCoV‐infected animals displayed co‐infections caused by CPV, CDV, CAdV‐1, CAdV‐2, and CHV‐1 (Table 2). Pantropic CCoV/CPV infections were found in 6 animals, with 4 and 1 dogs being infected by CPV‐2a and CPV‐2b, respectively, whereas one animal was co‐infected by both CPV variants. Twenty dogs had triple infections caused by pCCoV, CPV and CAdV‐2 (n = 16); pCCoV, CPV and CAdV‐1 (n = 1); pCCoV, CPV and CDV (n = 2); and pCCoV, CPV and CHV‐1 (n = 1), and the remaining 2 dogs displayed quadruple infections caused by CCoV, CPV‐2, CAdV‐1 and CAdV‐2 (n = 1), or pCCoV, CPV‐2, CDV and CAdV‐2 (n = 1). The statistical association of co‐pathogens to pCCoV is shown in Table 3, which reports the detection rates of selected viral agents in pCCoV positive and negative samples. Both CPV and CAdV‐2 were significantly associated to pCCoV infection (p < .00001), whereas CAdV‐1 and CDV were not (p = .50 and p = .62, respectively).
TABLE 3.
Prevalence of viral co‐pathogens in dogs with and without pCCoV infection
| Virus | pCCoV positive (n = 35) | pCCoV negative (n = 317) | p‐value | ||
|---|---|---|---|---|---|
| Positive (%) | 95% CI | Positive (%) | 95% CI | ||
| CPV | 28 (80.0%) | (0.693984–0.906016) | 94 (30.0%) | (0.281621–0.311439) | <.00001 |
| CAdV‐1 | 2 (5.7%) | (0.052749–0.061537) | 11 (3.5%) | (0.034001–0.035399) | .504 |
| CAdV‐2 | 18 (51.4%) | (0.429129–0.599442) | 24 (7.6%) | (0.073505–0.077915) | <.00001 |
| CDV | 3 (8.6%) | (0.077765–0.093664) | 36 (11.0%) | (0.109598–0.117531) | .618 |
Bold numbers indicate statistically significant p values (p < .05).
4. DISCUSSION
To date, pCCoV has been detected only sporadically in Italy and other countries (Buonavoglia et al., 2006; Chen et al., 2019; Decaro et al., 2012, 2013; Ntafis et al., 2012; Pinto et al., 2014; Zicola et al., 2012). In this study, we examined 352 dogs and identified 35 putative pCCoV‐infected animals, thus showing a wide distribution of this emerging virus. Twenty‐eight of the putative pCCoV‐infected dogs displayed double, triple or quadruple co‐infections with other viral pathogens. Therefore, in most cases, the identification of other pathogens in the same animals does not allow a clear association between the detected pCCoV strains and the death of the animals. Even for 7 dogs (two juvenile dogs and five pups), in which no associations with other viral pathogens were demonstrated, there is no definitive evidence that pCCoV was the cause of their death. In fact, previous studies have demonstrated that this virus is frequently associated to subclinical infections and impairment of the lymphocyte counts, rather than to severe clinical signs and death of the infected dogs (Marinaro et al., 2010). Most of the pCCoV‐positive animals also displayed post‐mortem findings accounting for a systemic involvement, but at which extent those lesions were induced by pCCoV or by other co‐pathogens, including the highly pathogenic CPV, CAdV‐1 and CDV, infecting the same dogs could not be assessed.
Remarkably, most of the pCCoV‐infected dogs had been recently imported from Hungary, which may account for a wider circulation of this virus in eastern Europe. This finding is in contrast with those of a previous study aiming to assess the pCCoV circulation in Europe, which reported similar prevalences in Italy and Hungary, with detection rates of 8.69% and 9.33%, respectively (Decaro et al., 2013).
Currently, no test is available to differentiate the pantropic from the enteric CCoV strains, since no specific genetic markers have been identified in pCCoV so far. As a consequence, only the detection of a CCoV‐IIa strain in extraintestinal tissues accounts for a possible pCCoV infection in dogs. This situation is similar to that observed in calicivirus infections in cats, where markers of pathogenicity have been not yet detected in highly virulent strains, so that diagnosis of systemic calicivirosis is obtained when the virus is detected in extrarespiratory tissues (Caringella et al., 2019).
Different from coronavirus infections in dogs, in cats potential genetic signatures were recently detected, which are able to discriminate between feline infectious peritonitis and feline enteric coronavirus strains (Felten et al., 2017).
According to previous observations (Decaro et al., 2013), two of the putative pCCoV strains identified in this study (91507 and 91510) fell in the same cluster with enteric CCoV strains, while other strains (103480, 32712, 78870, 43891‐1, 43891‐2, 34879, 43022, 91529, 34865, 91507, 91510, 31975) were more closely related to pCCoVs reference strains. Surprisingly, 3 pCCoVs (98960‐1, 98960‐3, 98960‐4) clustered with viruses detected in wildlife or domestic cats in Asia (Wang, Ma, Lu, & Wen, 2006). These strains were from dogs illegally imported from an unknown country, which highlights the role of illegal trade of dogs in the introduction of pathogens into Italy (Decaro et al., 2007; Mira et al., 2018).
An additional finding of the present study is the high frequency of co‐infections with pCCoV and other viruses. Enteric CCoV infections in dogs are very frequent (Decaro & Buonavoglia, 2008; Decaro et al., 2008; Pratelli et al., 2003; Priestnall, Pratelli, Brownlie, & Erles, 2007). Co‐infection by CPV and CCoV in dogs is known to enhance the severity of clinical signs (Decaro, Elia, et al., 2006; Evermann, Abbott, & Han, 2005; Pratelli, 2006), with fatal outcomes being frequently reported in pups (Decaro, Elia, et al., 2006). A high frequency of co‐infections with pCCoV and other pathogens has been previously reported (Alfano et al., 2019; Decaro et al., 2013; Ntafis et al., 2012; Pinto et al., 2014; Zicola et al., 2012). In the present study, we found a significant association of pCCoV with CPV and CAdV‐2 infections. Pantropic CCoV is able to affect lymphocyte counts, thus causing a prolonged lymphopenia, so that it has been postulated that pCCoV infection may predispose to the increase in virulence of other pathogens by inducing immunosuppression in the infected dogs (Marinaro et al., 2010).
The large population of unvaccinated free‐ranging dogs present in Italy (Corrain et al., 2007; Verardi, Lucchini, & Randi, 2006) considerably increases the density of susceptible hosts, and may thus importantly impact the spread and maintenance of canine pathogens in the environment. Therefore, it is strongly recommended to vaccinate not only private‐owned dogs but also stray dogs whenever possible. In addition, the epidemiological risk related to the legal and illegal trade of carnivores from Asian countries must be taken into account, since this trade may represent a source of several emerging pathogens in domestic and wild canids (Mira et al., 2018, 2019).
5. CONCLUSION
The present study demonstrates an increasing circulation of pCCoV in Italy, which reinforces the need for intensive and continuous surveillance on the importation and illegal trade in animals and the need for increased controls on both autochthonous and imported dogs.
ETHICAL APPROVAL
Ethical statement is not applicable since sample collection was obtained from dead animals that were submitted to diagnostic investigations upon request of the owners or public authorities.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
ACKNOWLEDGEMENTS
We thank Dr Gianvito Lanave for his help in the deposit of the sequences in the GenBank database, Dr Lorena Cardillo, Dr Lucia Vangone and Dr Antonella De Angelis for their assistance with part of the experimental work. We are also grateful to Dr Loredana Baldi for her excellent support in the epidemiological and statistical investigation.
Alfano F, Fusco G, Mari V, et al. Circulation of pantropic canine coronavirus in autochthonous and imported dogs, Italy. Transbound Emerg Dis. 2020;67:1991–1999. 10.1111/tbed.13542
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are openly available in the GenBank database at https://www.ncbi.nlm.nih.gov/nucleotide/ under accession numbers MN086803‐MN086817.
REFERENCES
- Alfano, F. , Dowgier, G. , Valentino, M. P. , Galiero, G. , Tinelli, A. , Nicola, D. , & Fusco, G. (2019). Identification of pantropic canine coronavirus in a wolf (Canis lupus italicus) in Italy. Journal of Wildlife Diseases, 55(2), 504–508. 10.7589/2018-07-182 [DOI] [PubMed] [Google Scholar]
- Buonavoglia, C. , Decaro, N. , Martella, V. , Elia, G. , Campolo, M. , Desario, C. , … Tempesta, M. (2006). Canine coronavirus highly pathogenic for dogs. Emerging Infectious Diseases, 12(3), 492–494. 10.3201/eid1203.050839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonavoglia, C. , Martella, V. , Pratelli, A. , Tempesta, M. , Cavalli, A. , Buonavoglia, D. , … Carmichael, L. (2001). Evidence for evolution of canine parvovirus type 2 in Italy. Journal of General Virology, 82, 3021–3025. 10.1099/0022-1317-82-12-3021 [DOI] [PubMed] [Google Scholar]
- Caringella, F. , Elia, G. , Decaro, N. , Martella, V. , Lanave, G. , Varello, K. , … Buonavoglia, C. (2019). Feline calicivirus infection in cats with virulent systemic disease, Italy. Research in Veterinary Science, 124, 46–51. 10.1016/j.rvsc.2019.02.008 [DOI] [PubMed] [Google Scholar]
- Chen, S. , Liu, D. , Tian, J. , Kang, H. , Guo, D. , Jiang, Q. , … Qu, L. (2019). Molecular characterization of HLJ‐073, a recombinant canine coronavirus strain from China with an ORF3abc deletion. Archives of Virology, 164(8), 2159–2164. 10.1007/s00705-019-04296-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrain, R. , Di Francesco, A. , Bolognini, M. , Ciucci, P. , Baldelli, R. , & Guberti, V. (2007). Serosurvey for CPV‐2, distemper virus, ehrlichiosis and leishmaniosis in free‐ranging dogs in Italy. Veterinary Record, 160(3), 91–92. 10.1136/vr.160.3.91 [DOI] [PubMed] [Google Scholar]
- Decaro, N. , & Buonavoglia, C. (2008). An update on canine coronaviruses: Viral evolution and pathobiology. Veterinary Microbiology, 132, 221–234. 10.1016/j.vetmic.2008.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro, N. , & Buonavoglia, C. (2011). Canine coronavirus: Not only an enteric pathogen. Veterinary Clinics of North America: Small Animal Practice, 41, 1121–1132. 10.1016/j.cvsm.2011.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro, N. , Campolo, M. , Lorusso, A. , Desario, C. , Mari, V. , Colaianni, M. L. , … Buonavoglia, C. (2008). Experimental infection of dogs with a novel strain of canine coronavirus causing systemic disease and lymphopenia. Veterinary Microbiology, 128, 253–260. 10.1016/j.vetmic.2007.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro, N. , Cordonnier, N. , Demeter, Z. , Egberink, H. , Elia, G. , Grellet, A. , … Buonavoglia, C. (2013). European surveillance for pantropic canine coronavirus. Journal of Clinical Microbiology, 51(1), 83–88. 10.1128/JCM.02466-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro, N. , Desario, C. , Addie, D. D. , Martella, V. , Vieira, M. J. , Elia, G. , … Buonavoglia, C. (2007). The study molecular epidemiology of canine parvovirus, Europe. Emerging Infectious Diseases, 13(8), 1222–2122. 10.3201/eid1308.070505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro, N. , Elia, G. , Martella, V. , Campolo, M. , Desario, C. , Camero, M. , … Buonavoglia, C. (2006). Characterisation of the canine parvovirus type 2 variants using minor groove binder probe technology. Journal of Virological Methods, 133(1), 92–99. 10.1016/j.jviromet.2005.10.026 [DOI] [PubMed] [Google Scholar]
- Decaro, N. , Elia, G. , Martella, V. , Desario, C. , Campolo, M. , Trani, L. D. , … Buonavoglia, C. (2005). A real‐time PCR assay for rapid detection and quantitation of canine parvovirus type 2 in the feces of dogs. Veterinary Microbiology, 105(1), 19–28. 10.1016/j.vetmic.2004.09.018 [DOI] [PubMed] [Google Scholar]
- Decaro, N. , Mari, V. , Campolo, M. , Lorusso, A. , Camero, M. , Elia, G. , … Buonavoglia, C. (2009). Recombinant canine coronaviruses related to transmissible gastroenteritis virus of Swine are circulating in dogs. Journal of Virology, 83(3), 1532–1537. 10.1128/JVI.01937-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro, N. , Mari, V. , Elia, G. , Addie, D. D. , Camero, M. , Lucente, M. S. , … Buonavoglia, C. (2010). Recombinant canine coronaviruses in dogs. Europe. Emerging Infectious Diseases, 16(1), 41–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro, N. , Mari, V. , von Reitzenstein, M. , Lucente, M. S. , Cirone, F. , Elia, G. , … Buonavoglia, C. (2012). A pantropic canine coronavirus genetically related to the prototype isolate CB/05. Veterinary Microbiology, 159(1–2), 239–244. 10.1016/j.vetmic.2012.03.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro, N. , Martella, V. , Desario, C. , Bellacicco, A. L. , Camero, M. , Manna, L. , … Buonavoglia, C. (2006). First detection of canine parvovirus type 2c in pups with haemorrhagic enteritis in Spain. Journal of Veterinary Medicine. B, Infectious Diseases and Veterinary Public Health, 53(10), 468–472. 10.1111/j.1439-0450.2006.00974.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro, N. , Martella, V. , Ricci, D. , Elia, G. , Desario, C. , Campolo, M. , … Buonavoglia, C. (2005). Genotype‐specific fluorogenic RT‐PCR assays for the detection and quantitation of canine coronavirus type I and type II RNA in faecal samples of dogs. Journal of Virological Methods, 130(1–2), 72–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro, N. , Pratelli, A. , Campolo, M. , Elia, G. , Martella, V. , Tempesta, M. , & Buonavoglia, C. (2004). Quantitation of canine coronavirus RNA in the faeces of dogs by TaqMan RT‐PCR. Journal of Virological Methods, 119(2), 145–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowgier, G. , Mari, V. , Losurdo, M. , Larocca, V. , Colaianni, M. L. , Cirone, F. , … Decaro, N. (2016). A duplex real‐time PCR assay based on TaqMan technology for simultaneous detection and differentiation of canine adenovirus types 1 and 2. Journal of Virological Methods, 234, 1–6. 10.1016/j.jviromet.2016.03.011 [DOI] [PubMed] [Google Scholar]
- Elia, G. , Decaro, N. , Martella, V. , Cirone, F. , Lucente, M. S. , Lorusso, E. , … Buonavoglia, C. (2006). Detection of canine distemper virus in dogs by real‐time RT‐PCR. Journal of Virological Methods, 136(1–2), 171–176. 10.1016/j.jviromet.2006.05.004 [DOI] [PubMed] [Google Scholar]
- Evermann, J. F. , Abbott, J. R. , & Han, S. (2005). Canine coronavirus‐associated puppy mortality without evidence of concurrent canine parvovirus infection. Journal of Veterinary Diagnostic Investigation, 17, 610–614. 10.1177/104063870501700618 [DOI] [PubMed] [Google Scholar]
- Felten, S. , Weider, K. , Doenges, S. , Gruendl, S. , Matiasek, K. , Hermanns, W. , … Hartmann, K. (2017). Detection of feline coronavirus spike gene mutations as a tool to diagnose feline infectious peritonitis. Journal of Feline Medicine and Surgery, 19(4), 321–335. 10.1177/1098612X15623824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh, K. , & Standley, D. M. (2013). MAFTT multiple sequence alignment software version 7: Improvements in performance and usability. Molecular Biology and Evolution, 30, 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinaro, M. , Mari, V. , Bellacicco, A. L. , Tarsitano, E. , Elia, G. , Losurdo, M. , … Decaro, N. (2010). Prolonged depletion of circulating CD4+ T lymphocytes and acute monocytosis after pantropic canine coronavirus infection in dogs. Virus Research., 152(1–2), 73–78. 10.1016/j.virusres.2010.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira, F. , Purpari, G. , Di Bella, S. , Colaianni, M. L. , Schirò, G. , Chiaramonte, G. , … Guercio, A. (2019). Spreading of canine parvovirus type 2c mutants of Asian origin in southern Italy. Transboundary and Emerging Diseases, 66(6), 2297–2304. 10.1111/tbed.13283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira, F. , Purpari, G. , Lorusso, E. , Di Bella, S. , Gucciardi, F. , Desario, C. , … Guercio, A. (2018). Introduction of Asian canine parvovirus in Europe through dog importation. Transboundary and Emerging Diseases, 65(1), 16–21. 10.1111/tbed.12747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntafis, V. , Xylouri, E. , Mari, V. , Papanastassopoulou, M. , Papaioannou, N. , Thomas, A. , … Decaro, N. (2012). Molecular characterization of a canine coronavirus NA/09 strain detected in a dog's organs. Archives of Virology, 157(1), 171–175. 10.1007/s00705-011-1141-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto, L. D. , Barros, I. N. , Budaszewski, R. F. , Weber, M. N. , Mata, H. , Antunes, J. R. , … Canal, C. W. (2014). Characterization of pantropic canine coronavirus from Brazil. The Veterinary Journal, 202(3), 659–662. 10.1016/j.tvjl.2014.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratelli, A. (2006). Genetic evolution of canine coronavirus and recent advances in prophylaxis. Veterinary Research, 37(2), 191–200. 10.1051/vetres:2005053 [DOI] [PubMed] [Google Scholar]
- Pratelli, A. , Martella, V. , Decaro, N. , Tinelli, A. , Camero, M. , Cirone, F. , … Buonavoglia, C. (2003). Genetic diversity of a canine coronavirus detected in pups with diarrhoea in Italy. Journal of Virological Methods, 110(1), 9–17. 10.1016/s0166-0934(03)00081-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priestnall, S. L. , Pratelli, A. , Brownlie, J. , & Erles, K. (2007). Serological prevalence of canine respiratory coronavirus in southern Italy and epidemiological relationship with canine enteric coronavirus. Journal of Veterinary Diagnostic Investigation, 19(2), 176–180. 10.1177/104063870701900206 [DOI] [PubMed] [Google Scholar]
- Verardi, A. , Lucchini, V. , & Randi, E. (2006). Detecting introgressive hybridization between free‐ranging domestic dogs and wild wolves (Canis lupus) by admixture linkage disequilibrium analysis. Molecular Ecology, 15(10), 2845–2855. 10.1111/j.1365-294X.2006.02995.x [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Ma, G. , Lu, C. , & Wen, H. (2006). Detection of canine coronaviruses genotype I and II in raised Canidae animals in China. Berliner und Münchener tierärztliche Wochenschrift, 119(1–2), 35–39. [PubMed] [Google Scholar]
- Zeng, S. Q. , Halkosalo, A. , Salminen, M. , Szakal, E. D. , Puustinen, L. , & Vesikari, T. (2008). One‐step quantitative RT‐PCR for the detection of rotavirus in acute gastroenteritis. Journal of Virological Methods, 153(2), 238–240. 10.1016/j.jviromet.2008.08.004 [DOI] [PubMed] [Google Scholar]
- Zicola, A. , Jolly, S. , Mathijs, E. , Ziant, D. , Decaro, N. , Mari, V. , & Thiry, E. (2012). Fatal outbreaks in dogs associated with pantropic canine coronavirus in France and Belgium. Journal of Small Animal Practice, 53(5), 297–300. 10.1111/j.1748-5827.2011.01178.x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are openly available in the GenBank database at https://www.ncbi.nlm.nih.gov/nucleotide/ under accession numbers MN086803‐MN086817.


