Abstract
In late December 2019, a cluster of unexplained pneumonia cases has been reported in Wuhan, China. A few days later, the causative agent of this mysterious pneumonia was identified as a novel coronavirus. This causative virus has been temporarily named as severe acute respiratory syndrome coronavirus 2 and the relevant infected disease has been named as coronavirus disease 2019 (COVID‐19) by the World Health Organization, respectively. The COVID‐19 epidemic is spreading in China and all over the world now. The purpose of this review is primarily to review the pathogen, clinical features, diagnosis, and treatment of COVID‐19, but also to comment briefly on the epidemiology and pathology based on the current evidence.
Keywords: coronavirus, COVID‐19, SARS‐CoV‐2
Highlights
This review is primarily to review the pathogen, clinical features, diagnosis, and treatment of COVID‐19, but also to comment briefly on the epidemiology and pathology based on the current evidence.
We strongly recommend that the criteria of clinical diagnosed cases based on the symptoms, exposure history, and typical manifestations on chest CT imaging should be used in COVID‐19 affected areas that are in shortage of RT‐PCR testing kits to control the COVID‐19 epidemic.
1. INTRODUCTION
In late December 2019, an outbreak of an unknown disease called pneumonia of unknown cause occurred in Wuhan, Hubei province, China. 1 The outbreak has spread substantial to infect 9720 people in China with 213 deaths and to infect 106 people in 19 other countries up to 31 January 2020 (https://www.who.int/docs/default‐source/coronaviruse/situation‐reports/20200131‐sitrep‐11‐ncov.pdf). A few days later, the causative agent of this mysterious pneumonia was identified as a novel coronavirus (nCoV) by several independent laboratories. 2 , 3 , 4 The causative virus has been temporarily named as severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) and the relevant infected disease has been named as coronavirus disease 2019 (COVID‐19) by the World Health Organization, respectively. According to the daily report of the World Health Organization, the epidemic of SARS‐CoV‐2 so far registered 78 630 cases and 2747 deaths in China, spread to 46 other countries that reported a total of 3664 cases by 27 February 2020 (https://www.who.int/docs/default‐source/coronaviruse/situation‐reports/20200227‐sitrep‐38‐covid‐19.pdf). COVID‐19 epidemic has become a global health threat.
Coronaviruses (CoVs) are a group of highly diverse, enveloped, positive‐sense, and single‐stranded RNA viruses. 5 They cause several diseases involving respiratory, enteric, hepatic, and neurological systems with vary severity among humans and animals. 5 , 6 Human CoV infections have traditionally caused a low percentage of annual respiratory infections. There are HCoV‐OC43, HCoV‐229E, HCoV‐NL63, and HCoV‐HKU1, which cause mild respiratory illness. 5 , 7 Over the past 2 decades, two novel CoVs, severe acute respiratory syndrome CoV (SARS‐CoV) and Middle East respiratory syndrome CoV (MERS‐CoV), have emerged and cause severe human diseases. 8 , 9 During the epidemic, SARS‐CoV infect more than 8000 people worldwide with nearly 800 fatalities, representing its mortality rate around 10%. Whereas MERS‐CoV infected over 857 official cases and 334 deaths, making its mortality rate approximately 35%. 10 , 11 , 12 So far, SARS‐CoV‐2 is the seventh member of the family of CoVs that infects humans. The main symptoms of COVID‐19 included fever, fatigue, and cough, which are similar to that of SARS‐CoV and MERS‐CoV infected cases. There are some overlapping and discrete aspects of the pathology and pathogenesis of these CoVs which cause severe diseases in humans. 13
Many literature reported the clinical features, virology, pathology, and radiology of COVID‐19, but the comprehensive review is few. The purpose of this review is primarily to review the pathogen, clinical features, diagnosis, and treatment of COVID‐19, but also to comment briefly on the epidemiology and pathology based on the current evidence.
2. THE PATHOGEN
The pathogen that causes COVID‐19 is a nCoV that was first identified in the late January 2020, named SARS‐CoV‐2 (also known as 2019‐nCoV). 2 , 3 , 4
SARS‐CoV‐2 is a novel member of CoVs, which are a large group of highly diverse, enveloped, positive‐sense, and single‐stranded RNA viruses. 5 Recent research reported that SARS‐CoV‐2 likely originated in bats, based on the similarity of its genetic sequence to that of other CoVs. 14 The intermediate animal host of SARS‐CoV‐2 between a probable bat reservoir and humans is still unknown. 15 Although this nCoV has genetic features that are compatible with the family of CoV, nevertheless it has distinct gene sequences that are significantly different from previously sequenced CoVs (Table 1). The analysis of samples from seven SARS‐CoV‐2 infected patients suggested that SARS‐CoV‐2 shares 79.5% sequence identity to SARS‐CoV. 3 Simplot analysis showed that SARS‐CoV‐2 share 96.2% overall genome sequence identity to RaTG13, which is a short RdRp region from a bat CoV. 3 Phylogenetic analysis revealed that SARS‐CoV‐2 falls into the subgenus Sarbecovirus of the genus Betacoronavirus and is distinct from SARS‐CoV. 2 , 4
Table 1.
Zoonotic coronaviruses that causes serious disease in human
| Coronavirus | Affected host | Intermediate host | Potential reservoir host | Disease | Cell receptor | Reference |
|---|---|---|---|---|---|---|
| SARS‐CoV | Humans | Himalayan palm civet/raccoon | Bat | SARS | ACE2 | Li et al 16 |
| MERS‐CoV | Humans | Dromedary camels | Bat | MERS | DPP4 | Wang et al 17 |
| SARS‐CoV‐2 | Humans | NR | NR | COVID‐19 | ACE2 | Wrap et al 18 |
Abbreviations: ACE2, angiotensin‐converting enzyme 2; COVID‐19, coronavirus disease 2019; DPP4, dipeptidyl peptidase 4; MERS‐CoV, Middle East respiratory syndrome‐coronavirus; NR, no report; SARS‐CoV, severe acute respiratory syndrome‐coronavirus.
The envelope spike (S) protein is important for CoV. 19 The S protein mediates receptor binding and membrane fusion and is crucial for determining host tropism and transmission capacity. 17 , 20 , 21 Generally, the S protein is functionally divided into the S1 domain, responsible for receptor binding, and S2 domain, responsible for cell membrane fusion. 22 Structure analysis suggested that receptor‐binding domain was composed of a core and an external subdomain. 19 Angiotensin‐converting enzyme 2 (ACE2) was known as cell receptor for SARS‐CoV. 16 , 23 , 24 Similar to SARS‐CoV, SARS‐CoV‐2 also use ACE2 as an entry receptor in the ACE2‐expressing cells, 3 indicating SARS‐CoV‐2 may share the same life cycle with SARS‐CoV (Figure 1).
Figure 1.
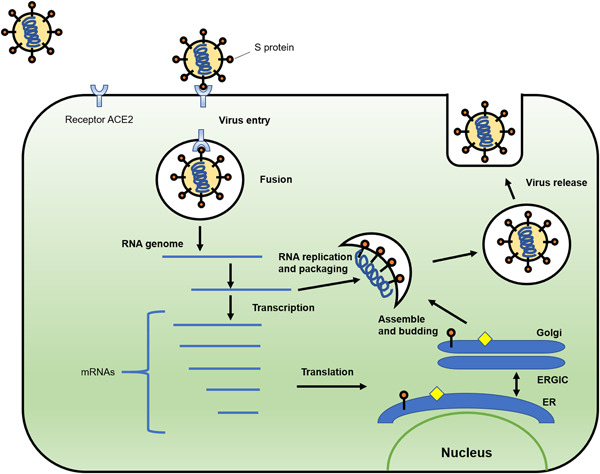
Schematic model of SARS‐CoV‐2 life cycle. S protein binds to the cellular receptor ACE2 to facilitate the entry of the virus. After the fusion of viral and plasma membranes, virus RNA undergoes replication and transcription. The proteins are synthesized. Viral proteins and new RNA genome are subsequently assembled in the ER and Golgi, followed by budding into the lumen of the ERGIC. New virions are released through vesicles. ACE2, angiotensin‐converting enzyme 2; ER, endoplasmic reticulum; ERGIC, endoplasmic reticulum‐Golgi intermediate compartment
The biophysical and structural analysis indicated that S protein of SARS‐CoV‐2 binds ACE2 with approximately10‐ to 20‐fold higher affinity than S protein of SARS‐CoV. 18 The high affinity of S protein for human ACE2 may facilitate the spread of SARS‐CoV‐2 in human populations. Meanwhile, SARS‐CoV‐2 does not use other CoV receptors, such as aminopeptidase N and dipeptidyl peptidase 4 to enter cells. 3
3. EPIDEMIOLOGY
Briefly, cases tend to be in clusters which arrive in waves, and develop into larger outbreaks all over the world. The first documented outbreak occurred primarily in Wuhan. 1 According to the daily report of the World Health Organization, the epidemic of SARS‐CoV‐2 so far registered 78 630 cases and 2747 deaths in China, spread to 46 other countries that reported a total of 3664 cases by 27 February 2020.
There are evidence suggest that transmission mode is human to human. 25 , 26 The major route of transmission of COVID‐19 is droplet and close contact. 26 Whether infection can occur through the oral or conjunctival routes is unknown, but SARS‐CoV‐2 has been detected in tears, 27 which is resemble to SARS‐CoV. 28 Reproductive number (R 0) was estimated by some studies. On the basis of clinical data of patients in COVID‐19 early outbreak, the mean R 0 was ranging from 2.20 to 3.58, meaning that each patient has been spreading infection to two or three other people. 25 , 29 It is still too early to develop an accurate R 0 estimate or to assess the dynamics of transmission. More research is needed in the future.
The mean incubation period is about 5 days, ranging from 1 to 14 days and 95% of patients are likely to experience symptoms within 12.5 days of contact. 25 , 30 These data suggest a 14‐day medical observation period or quarantine for exposed and close contact persons. However, an asymptomatic carrier was reported and the incubation period was 19 days, suggesting the complicated challenge to contain the outbreak. 31
4. CLINICAL FEATURES
Most case patients were 30 to 79 years of age. 32 The median age is ranging from 49 to 59 years. 25 , 26 , 33 , 34 There were few cases in children below 15 years of age. More than half the patients were male. Nearly half the cases had one or more coexisting medical conditions, such as hypertension, diabetes, and cardiovascular disease. 25 , 26 , 33 , 34 A large cases study indicated that the case‐fatality rate was elevated among those patients with coexisting medical conditions. 32
The spectrum of clinical presentations of COVID‐19 has been reported ranging from asymptomatic infection to severe respiratory failure. 25 , 26 , 30 , 32 , 33 , 34 The main symptoms include a self‐reported fever, fatigue, dry cough, myalgia, and dyspnea. The uncommon symptoms include sputum production, headache, hemoptysis, and diarrhea. 25 , 26 , 30 , 32 , 33 , 34 Although pneumonia is present in most SARS‐CoV‐2 infected patients, few cases complained of pleuritic chest pain. 26 , 33
According to the severity of symptoms, patients can be classified as mild, severe, and critical types 32 (Table 2). Mild patients had nonpneumonia or mild pneumonia. Severe patients had several clinical findings, including dyspnea, respiratory frequency ≥ 30/min, blood oxygen saturation ≤ 93%, partial pressure of arterial oxygen to fraction of inspired oxygen ratio less than 300, and/or lung infiltrates greater than 50% within 24 to 48 hours. Critical patients had severe conditions, such as respiratory failure, septic shock, and/or multiple organ dysfunction or failure. 32 If the disease progressed, the median duration period from illness onset to dyspnea was 8.0 days, and to mechanical ventilation was 10.5 days. 34
Table 2.
Clinical symptoms associated with COVID‐19
| Clinical types | Symptoms |
|---|---|
| Mild type | Nonpneumonia or mild pneumonia |
| Severe type | Dyspnea, respiratory frequency ≥ 30/min, blood oxygen saturation ≤ 93%, partial pressure of arterial oxygen to fraction of inspired oxygen ratio < 300, and/or lung infiltrates > 50% within 24/48 h |
| Critical type | Respiratory failure, septic shock, and/or multiple organ dysfunction or failure |
Abbreviation: COVID‐19, coronavirus disease 2019.
Common clinical laboratory findings include leucopenia and lymphopenia. 25 , 30 , 33 , 34 Lymphopenia is a cardinal feature of COVID‐19. Lactate dehydrogenase and creatinine kinase are all elevated. Half of patients had abnormal liver function, with elevated alanine aminotransferase or aspartate aminotransferase. Most patients had abnormal myocardial zymogram, which showed the elevation of creatine kinase and lactate dehydrogenase. Most patients showed normal serum levels of procalcitonin, but the C‐reactive protein was above the normal range. One‐third of patients had the elevation of D‐dimer. 25 , 30 , 33 , 34
One study investigated the changes of several cytokines in serum in the COVID‐19 patients. 34 Initial plasma IL1B, interleukin‐1 receptor antagonist (IL1RA), IL7, IL8, IL9, IL10, basic fibroblast growth factor, granulocyte colony‐stimulating factor (GCSF), granulocyte‐macrophage colony‐stimulating factor, interferon γ, IP10, MCP1, MIP1A, MIP1B, platelet‐derived growth factor, tumor necrosis factor (TNF‐α), and vascular endothelial growth factor concentrations were higher in patients than in healthy adults. Plasma levels of IL5, IL12p70, IL15, eotaxin, and RANTES were similar between patients and healthy adults. Further comparison between intensive care unit (ICU) and non‐ICU patients showed that plasma concentrations of IL2, IL7, IL10, GCSF, IP10, MCP1, MIP1A, and TNF‐α were higher in ICU patients than non‐ICU patients. 34 These findings suggested that the initiation of the immune response result in the production of chemokines and cytokines, which damage normal host lung.
The radiologic manifestations of SARS‐CoV‐2 infected patients are diverse and progressing rapidly. 35 , 36 , 37 , 38 Two‐third of patients had at least two affected lobes, nearly half of patients had five affected lobes. 37 , 38 The most common manifestations are patchy ground‐glass opacities (GGO) and patchy consolidation which were mainly distributed in the middle and outer zone of the lung 37 , 38 (Figure 2). A little fibrous stripe may appear if the condition was improved. 37 One report suggested that there are four stages defined on CT scan. 36 In early stage, GGO was the main radiological demonstration distributed in the lower lobes unilaterally or bilaterally. In progressive stage, diffuse and bilateral GGO and consolidation in more than two lobes became the main manifestation. In peak stage, the diffuse GGO and dense consolidation became more prevalent. In absorption stage, extensive GGO could be observed and the consolidation was gradually absorbed.
Figure 2.
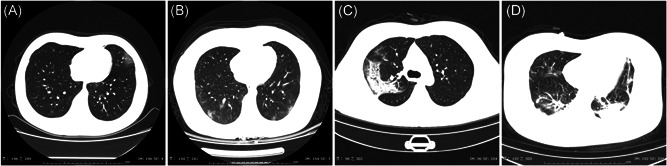
Chest CT Manifestations of COVID‐19. A, Single GGO; B, diffuse GGO; C, consolidation; D, both GGO and consolidation. COVID‐19, coronavirus disease 2019; CT, computed tomography; GGO, ground‐glass opacities
5. PATHOLOGY
The pathological findings of human SARS‐CoV‐2 infection have been limited due to the rare number of biopsies or autopsies. In a case reported by Xu et al, 39 a 50‐year‐old man died 14 days after admission due to respiratory failure and cardiac arrest. 39 The primary finding of biopsy at autopsy was bilateral diffuse alveolar damage with cellular fibromyxoid exudates and interstitial mononuclear inflammatory infiltrates dominated by lymphocytes. Multinucleated syncytial cells with atypical enlarged pneumocytes characterized by large nuclei, amphophilic granular cytoplasm, and prominent nucleoli were identified in the intraalveolar spaces, showing viral cytopathic‐like changes. No obvious intranuclear or intracytoplasmic viral inclusions were identified. These pathological features show great similarities to SARS‐CoV and MERS‐CoV infection. 40 , 41 , 42 In addition, liver and heart were studied. There is moderate microvascular steatosis and mild lobular and portal activity in the liver tissue and a few interstitial mononuclear inflammatory infiltrates in the heart tissue.
6. DIAGNOSIS
Although a good contact history, systemic symptoms, and radiographic changes of pneumonia make the diagnosis likely, the laboratory diagnosis is more reliable. Real time‐polymerase chain reaction (RT‐PCR) is routinely used to detect causative viruses from respiratory secretions. 43 , 44 During COVID‐19 transmission events, RT‐PCR has served as the primary clinical laboratory diagnostic test. 25 , 26 , 30 , 33 Success of these tests are very important to understand the viral kinetics and tissue tropism found in COVID‐19 cases. Several specific and sensitive assays targeting RdRP, N, and E genes of the SARS‐CoV‐2 genome were designed to detect viral RNA in clinical specimens. 44 Lower respiratory tract samples provide the higher viral loads. 45 The sampling source or operation may affect RT‐PCR testing results. 43
The positive rate of RT‐PCR for throat swab samples was reported to be about 60% in early stage of COVID‐19. 46 These findings suggested that the result of RT‐PCR should be interpret with caution. One study investigated the diagnostic value and consistency of chest computed tomography (CT) compared with RT‐PCR test in 1014 patients with suspected SARS‐CoV‐2 infection. The results suggest that the sensitivity of chest CT in suspected patients was 97% based on positive RT‐PCR result and 75% based on negative RT‐PCR results. These findings indicated that chest CT is a sensitive modality to detect SARS‐CoV‐2 infection.
During the COVID‐19 epidemic in China, 10 567 patients were diagnosed as clinical diagnosed cases. This designation is being used in Hubei province, where is the worst affected area in China. In these cases, no RT‐PCR test was performed but diagnosis was made based on typical symptoms, exposure history, and chest CT manifestations consistent with COVID‐19 pneumonia. Under this criteria, 10 567 cases were diagnosed and isolated. This strategy quarantined a large number of suspected people and protected the healthy people to the most extent. On the basis of experiences above, we strongly recommend that the criteria of clinical diagnosed cases based on the symptoms, exposure history, and typical manifestations on chest CT imaging should be used in COVID‐19 affected areas that are in shortage of RT‐PCR testing kits to control the COVID‐19 epidemic.
7. TREATMENT
Until the diagnosis is confirmed, SARS‐CoV‐2 infected patients are treated in single rooms. 25 , 30 As SARS‐CoV‐2 is an emerging virus, an effective antiviral treatment has not been identified. The main treatment of COVID‐19 is symptomatic treatment. The antiviral drugs, including oseltamivir, ribavirin, ganciclovir, lopinavir, and ritonavir have been used in attempts to reduce viral load and to prevent the likelihood of respiratory complications in several studies. 25 , 26 , 30 , 33 , 34 Remdesivir was reported in the treatment of a patient with COVID‐19 in the United States and got an effective result. 47 However, the efficacy of these antiviral drugs for COVID‐19 need to be verified by randomized‐controlled clinical trials.
The antibiotics used generally covered common pathogens and some atypical pathogens. When secondary bacterial infection occurred, medication was administered according to the results of bacterial culture and drug sensitivity. 33 Current evidence in patients with SARS and MERS suggests that receiving corticosteroids did not have a survival benefit, but rather delayed viral clearance. 48 , 49 , 50 Therefore, routine corticosteroids should be avoided unless they are indicated for other reason. Arbidol is used empirically in China because of its direct antiviral effect on SARS‐CoV in cell culture. 51 Chinese herbal medicine formulae are used to prevent SARS‐CoV‐2 infection in 23 provinces in China. 52
Noninvasive or mechanical ventilation should be considered in patients with hypoxia despite oxygen supplement and worsening shortness of breath. Extracorporeal membrane oxygenation is used as a last resort. 30 , 33 , 34
8. PROGNOSIS
As of 27 February 2020, a total of 2747 deaths in China and 57 deaths outside of China have been reported. The number of laboratory‐confirmed cases and deaths continues to rise. The current reported mortality for COVID‐19 is approximately 3.41% compared to 10% for SARS and 35% for MERS. 10 , 11 , 12 , 53 The mortality rate was higher than 3.41% in Iran and France, lower in Italy, Japan, Republic of Korea, and United States (Figure 3). Considering the quick spread of COVID‐19, it is still too early to assess the mortality. All countries in the world should respond to the epidemic effectively. Approximately, 20% to 25% SARS‐CoV‐2‐infected patients developed acute respiratory distress syndrome and required ICU care. 30 , 33 , 34 Current evidence indicated that older age and comorbidity may be risk factors for poor outcome. 30
Figure 3.
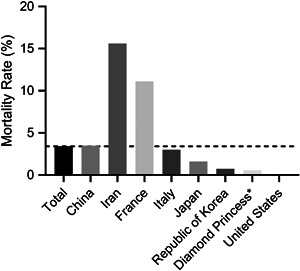
Mortality rates of different countries or regions, 27 February 2020. *A cruise ship currently in Japanese territorial waters
9. SUMMARY AND OUTLOOK
This review summarizes the current findings of SARS‐CoV‐2 along with the treatment for this severe CoV infection. The most common symptoms were addressed. Due to the only biopsy report, the pathological findings associated with SARS‐CoV‐2 infection have been limited. Autopsy is warranted and valuable for future research.
The WHO issued a public health emergency of international concern on 30 January 2020. SARS‐CoV‐2 epidemic is becoming a global concern. At the moment, there is no vaccine and no specific treatment for COVID‐19. The best strategy to deal with SARS‐CoV‐2 epidemic includes controlling the sources of infection, protecting the susceptible people, and cutting off the transmission. The infected patients should be identified early by rapid and robust detection technologies, provided with optimized treatment in isolation timely. The close contact people should be quarantined with follow‐up. The healthy people should be aware of the severity of COVID‐19 and take measures to protect themselves, such as staying at home, limiting social contacts, and wearing protective mask in public. The authorities should encourage people to stay at home; discourage mass gathering; postpone or cancel public events; and close public institutions. These control measures will help COVID‐19 infected countries to prevent the epidemic effectively. Future research will focus on improving the accuracy of early diagnostic tests, developing the vaccine and identifying effective drugs. Therefore, elucidating the pathogenesis of SARS‐CoV‐2 infection is imperative for achieving such goals.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
ACKNOWLEDGMENT
The authors would like to thank all the doctors and nurses who fight the virus during the COVID‐19 epidemic bravely.
He F, Deng Y, Li W. Coronavirus disease 2019: What we know? J Med Virol. 2020;92:719–725. 10.1002/jmv.25766
REFERENCES
- 1. Wuhan Municipal Health Commission . Report of Clustering Pneumonia of Unknown Etiology in Wuhan City. 2019. http://wjw.wuhan.gov.cn/front/web/showDetail/2019123108989. Accessed December 31, 2019.
- 2. Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565‐574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270‐273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727‐733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zumla A, Chan JF, Azhar EI, Hui DS, Yuen KY. Coronaviruses—drug discovery and therapeutic options. Nat Rev Drug Discov. 2016;15(5):327‐347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chan JF, Lau SK, Woo PC. The emerging novel Middle East respiratory syndrome coronavirus: the “knowns” and “unknowns”. J Formos Med Assoc. 2013;112(7):372‐381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Channappanavar R, Zhao J, Perlman S. T cell‐mediated immune response to respiratory coronaviruses. Immunol Res. 2014;59(1‐3):118‐128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cheng VC, Lau SK, Woo PC, Yuen KY. Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clin Microbiol Rev. 2007;20(4):660‐694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chan JF, Lau SK, To KK, Cheng VC, Woo PC, Yuen KY. Middle East respiratory syndrome coronavirus: another zoonotic betacoronavirus causing SARS‐like disease. Clin Microbiol Rev. 2015;28(2):465‐522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gretebeck LM, Subbarao K. Animal models for SARS and MERS coronaviruses. Curr Opin Virol. 2015;13:123‐129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gralinski LE, Baric RS. Molecular pathology of emerging coronavirus infections. J Pathol. 2015;235(2):185‐195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Drosten C, Günther S, Preiser W, et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348(20):1967‐1976. [DOI] [PubMed] [Google Scholar]
- 13. Liu J, Zheng X, Tong Q, et al. Overlapping and discrete aspects of the pathology and pathogenesis of the emerging human pathogenic coronaviruses SARS‐CoV, MERS‐CoV, and 2019‐nCoV. J Med Virol. 2020;92(5):491‐494. 10.1002/jmv.25709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhou P, Yang X‐L, Wang X‐G, et al. Discovery of a novel coronavirus associated with the recent pneumonia outbreak in humans and its potential bat origin. bioRxiv. 2020. 10.1101/2020.01.22.914952. [published online ahead of print January 2020]. [DOI] [Google Scholar]
- 15. Li X, Zai J, Zhao Q, et al. Evolutionary history, potential intermediate animal host, and cross‐species analyses of SARS‐CoV‐2. J Med Virol. 2020. 10.1002/jmv.25731. [published online ahead of print February 27, 2020]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li W, Moore MJ, Vasilieva N, et al. Angiotensin‐converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450‐454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang Q, Wong G, Lu G, Yan J, Gao GF. MERS‐CoV spike protein: targets for vaccines and therapeutics. Antiviral Res. 2016;133:165‐177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wrapp D, Wang N, Corbett KS, et al. Cryo‐EM structure of the 2019‐nCoV spike in the prefusion conformation. Science. 2020;367:1260‐1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Du L, He Y, Zhou Y, Liu S, Zheng BJ, Jiang S. The spike protein of SARS‐CoV–a target for vaccine and therapeutic development. Nat Rev Microbiol. 2009;7(3):226‐236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lu G, Wang Q, Gao GF. Bat‐to‐human: spike features determining 'host jump' of coronaviruses SARS‐CoV, MERS‐CoV, and beyond. Trends Microbiol. 2015;23(8):468‐478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li F. Structure, function, and evolution of coronavirus spike. Proteins. Annu Rev Virol. 2016;3(1):237‐261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. He Y, Zhou Y, Liu S, et al. Receptor‐binding domain of SARS‐CoV spike protein induces highly potent neutralizing antibodies: implication for developing subunit vaccine. Biochem Biophys Res Commun. 2004;324(2):773‐781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Oudit GY, Kassiri Z, Jiang C, et al. SARS‐coronavirus modulation of myocardial ACE2 expression and inflammation in patients with SARS. Eur J Clin Invest. 2009;39(7):618‐625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kuba K, Imai Y, Rao S, et al. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus‐induced lung injury. Nat Med. 2005;11(8):875‐879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li Q, Guan X, Wu P, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus‐infected pneumonia. N Engl J Med. 2020. 10.1056/NEJMoa2001316. [published online ahead of print January 29, 2020]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chan JFW, Yuan S, Kok KH, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person‐to‐person transmission: a study of a family cluster. Lancet. 2020;395(10223):514‐523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xia J, Tong J, Liu M, Shen Y, Guo D. Evaluation of coronavirus in tears and conjunctival secretions of patients with SARS‐CoV‐2 infection. J Med Virol. 2020. 10.1002/jmv.25725. [published online ahead of print February 26, 2020]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Loon SC. The severe acute respiratory syndrome coronavirus in tears. Br J Ophthalmol. 2004;88(7):861‐863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhao S, Lin Q, Ran J, et al. Preliminary estimation of the basic reproduction number of novel coronavirus (2019‐nCoV) in China, from 2019 to 2020: a data‐driven analysis in the early phase of the outbreak. Int J Infect Dis. 2020;92:214‐217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang D, Hu B, Hu C, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus‐infected pneumonia in Wuhan, China. JAMA. 2020. 10.1001/jama.2020.1585. [published online ahead of print February 7, 2020]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bai Y, Yao L, Wei T, et al. Presumed asymptomatic carrier transmission of COVID‐19. JAMA. 2020. 10.1001/jama.2020.2565. [published online ahead of print February 21, 2020]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wu Z, McGoogan JM. Characteristics of and important lessons from the coronavirus disease 2019 (COVID‐19) outbreak in China: summary of a report of 72314 cases from the Chinese center for disease control and prevention. JAMA. 2020. 10.1001/jama.2020.2648. [published online ahead of print February 24, 2020]. [DOI] [PubMed] [Google Scholar]
- 33. Chen N, Zhou M, Dong X, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507‐513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497‐506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ai T, Yang Z, Hou H, et al. Correlation of chest CT and RT‐PCR testing in coronavirus disease 2019 (COVID‐19) in China: a report of 1014 Cases. Radiology. 2020. 10.1148/radiol.2020200642. [published online ahead of print February 26, 2020]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pan F, Ye T, Sun P, et al. Time course of lung changes on chest ct during recovery from 2019 novel coronavirus (COVID‐19) pneumonia. Radiology. 2020. 10.1148/radiol.2020200370. [published online ahead of print February 13, 2020]. [DOI] [Google Scholar]
- 37. Pan Y, Guan H, Zhou S, et al. Initial CT findings and temporal changes in patients with the novel coronavirus pneumonia (2019‐nCoV): a study of 63 patients in Wuhan, China. Eur Radiol. 2020. 10.1007/s00330-020-06731-x. [published online ahead of print February 13, 2020]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shi H, Han X, Jiang N, et al. Radiological findings from 81 patients with COVID‐19 pneumonia in Wuhan, China: a descriptive study. Lancet Infect Dis. 2020. 10.1016/S1473-3099(20)30086-4. [published online ahead of print February 24, 2020]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Xu Z, Shi L, Wang Y, et al. Pathological findings of COVID‐19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020. 10.1016/S2213-2600(20)30076-X. [published online ahead of print February 18, 2020]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hui DSC, Zumla A. Severe acute respiratory syndrome: historical, epidemiologic, and clinical features. Infect Dis Clin North Am. 2019;33(4):869‐889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. van den Brand JM, Smits SL, Haagmans BL. Pathogenesis of Middle East respiratory syndrome coronavirus. J Pathol. 2015;235(2):175‐184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nassar MS, Bakhrebah MA, Meo SA, Alsuabeyl MS, Zaher WA. Middle east respiratory syndrome coronavirus (MERS‐CoV) infection: epidemiology, pathogenesis and clinical characteristics. Eur Rev Med Pharmacol Sci. 2018;22(15):4956‐4961. [DOI] [PubMed] [Google Scholar]
- 43. Binnicker MJ. Emergence of a novel coronavirus disease (COVID‐19) and the importance of diagnostic testing: why partnership between clinical laboratories, public health agencies, and industry is essential to control the outbreak. Clin Chem. 2020. 10.1093/clinchem/hvaa071. [published online ahead of print March 12, 2020]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Corman VM, Landt O, Kaiser M, et al. Detection of 2019 novel coronavirus (2019‐nCoV) by real‐time RT‐PCR. Euro Surveill. 2020;25(3):2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zou L, Ruan F, Huang M, et al. SARS‐CoV‐2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020. 10.1056/NEJMc2001737. [published online ahead of print March 19, 2020]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yang Y, Yang M, Shen C, et al. Evaluating the accuracy of different respiratory specimens in the laboratory diagnosis and monitoring the viral shedding of 2019‐nCoV infections. MedRxiv. 2020. 10.1101/2020.02.11.20021493. [published online ahead of print February 17, 2020]. [DOI] [Google Scholar]
- 47. Holshue ML, DeBolt C, Lindquist S, et al. First case of 2019 novel coronavirus in the United States. N Engl J Med. 2020;382:929‐936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Arabi YM, Mandourah Y, Al‐Hameed F, et al. Corticosteroid therapy for critically ill patients with Middle East respiratory syndrome. Am J Respir Crit Care Med. 2018;197(6):757‐767. [DOI] [PubMed] [Google Scholar]
- 49. Lansbury L, Rodrigo C, Leonardi‐Bee J, Nguyen‐Van‐Tam J, Lim WS. Corticosteroids as adjunctive therapy in the treatment of influenza. Cochrane Database Syst Rev. 2019;2(2):CD010406. 10.1002/14651858.CD010406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Stockman LJ, Bellamy R, Garner P. SARS: systematic review of treatment effects. PLoS Med. 2006;3(9):e343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Khamitov RA, Loginova S, Shchukina VN, Borisevich SV, Maksimov VA, Shuster AM. Antiviral activity of arbidol and its derivatives against the pathogen of severe acute respiratory syndrome in the cell cultures. Vopr Virusol. 2008;53(4):9‐13. [PubMed] [Google Scholar]
- 52. Luo H, Tang Q, Shang Y, et al. Can Chinese medicine be used for prevention of corona virus disease 2019 (COVID‐19)? a review of historical classics, research evidence and current prevention programs. Chin J Integr Med. 2020. 10.1007/s11655-020-3192-6. [published online ahead of print February 17, 2020]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wang W, Tang J, Wei F. Updated understanding of the outbreak of 2019 novel coronavirus (2019‐nCoV) in Wuhan, China. J Med Virol. 2020;92(4):441‐447. [DOI] [PMC free article] [PubMed] [Google Scholar]


