Abstract
By 27 February 2020, the outbreak of coronavirus disease 2019 (COVID‐19) caused 82 623 confirmed cases and 2858 deaths globally, more than severe acute respiratory syndrome (SARS) (8273 cases, 775 deaths) and Middle East respiratory syndrome (MERS) (1139 cases, 431 deaths) caused in 2003 and 2013, respectively. COVID‐19 has spread to 46 countries internationally. Total fatality rate of COVID‐19 is estimated at 3.46% by far based on published data from the Chinese Center for Disease Control and Prevention (China CDC). Average incubation period of COVID‐19 is around 6.4 days, ranges from 0 to 24 days. The basic reproductive number (R0 ) of COVID‐19 ranges from 2 to 3.5 at the early phase regardless of different prediction models, which is higher than SARS and MERS. A study from China CDC showed majority of patients (80.9%) were considered asymptomatic or mild pneumonia but released large amounts of viruses at the early phase of infection, which posed enormous challenges for containing the spread of COVID‐19. Nosocomial transmission was another severe problem. A total of 3019 health workers were infected by 12 February 2020, which accounted for 3.83% of total number of infections, and extremely burdened the health system, especially in Wuhan. Limited epidemiological and clinical data suggest that the disease spectrum of COVID‐19 may differ from SARS or MERS. We summarize latest literatures on genetic, epidemiological, and clinical features of COVID‐19 in comparison to SARS and MERS and emphasize special measures on diagnosis and potential interventions. This review will improve our understanding of the unique features of COVID‐19 and enhance our control measures in the future.
Keywords: COVID‐19, diagnosis and interventions, features
1. INTRODUCTION
In December 2019, a cluster of patients with pneumonia of unknown cause was observed in Wuhan, China. A novel coronavirus was identified as the causative pathogen, 1 , 2 , 3 , 4 , 5 , 6 provisionally named as 2019 novel coronavirus (2019‐nCoV) by the World Health Organization (WHO). On 11 February 2020, WHO named this novel coronavirus pneumonia as “COVID‐19” (coronavirus disease 2019). On the basis of phylogeny, taxonomy, and established practice, the Coronavirus Study Group of the International Committee on Taxonomy of Viruses formally recognizes this virus as a sister to severe acute respiratory syndrome coronavirus (SARS‐CoV) and renamed it as SARS‐CoV‐2. 7 SARS‐CoV‐2 belongs to species of severe acute respiratory syndrome‐related coronavirus (SARSr‐CoV) and genus Betacoronavirus. 2 COVID‐19 rapidly triggered a global health emergency alert and spread to 46 countries by 27 February 2020. SARS‐CoV‐2 is the seventh member of the family of coronaviruses that infects humans. Like SARS‐CoV and Middle East respiratory syndrome coronavirus (MERS‐CoV), SARS‐CoV‐2 is responsible for lower respiratory infection and can cause acute respiratory distress syndromes (ARDS). Other human coronaviruses (HCoV 229E, NL63, OC43, and HKU1) are responsible for upper respiratory infections and common cold. 8
By 27 February 2020, according to open data from China CDC as shown in Table 1 and Figure 1, COVID‐19 has caused 82 623 confirmed cases and 2858 deaths globally. The total case‐fatality rate is 3.46% as shown in Table 1. Because COVID‐19 started from Wuhan, the capital city of Hubei province with a large population of nearly 14 million people, 58.3% cases are in Wuhan. A total of 1932 health workers have been infected in Wuhan alone, 9 which overwhelmed the local health system and resulted in the highest case‐fatality rate (4.42%). Excluding Hubei province, the rest of China has 13 045 cases, 109 fatalities (0.84%). Outside of China, COVID‐19 has spread to 46 countries and has caused 3664 infections and 67 fatalities (1.83%). Overall, the case‐fatality rate of COVID‐19 so far is much lower than either SARS (9.6%) or MERS (34.5%). 10 Here, we summarized common and discrete features of SARS‐CoV‐2 in comparison to its two predecessors (SARS‐CoV and MERS‐CoV) in genetics, epidemiology, clinical features, and further discussed challenges for diagnosis and special control measures for COVID‐19.
Table 1.
The global distribution of mortality of COVID‐19 (by 27 February 2020)
| Number of confirmed cases | Percentage of total cases | Number of deaths | Fatality rate | |
|---|---|---|---|---|
| Wuhan | 48 137 | 58.3% | 2132 | 4.42% |
| Rest of Hubei | 17 777 | 21.5% | 550 | 3.09% |
| Hubei | 65 914 | 79.8% | 2682 | 4.07% |
| Rest of China | 13 045 | 15.8% | 109 | 0.84% |
| China | 78 959 | 95.6% | 2791 | 3.53% |
| International (46 countries) | 3664 | 4.43% | 67 | 1.83% |
| Total | 82 623 | 100% | 2858 | 3.46% |
Abbreviation: COVID‐19, coronavirus disease 2019.
Figure 1.
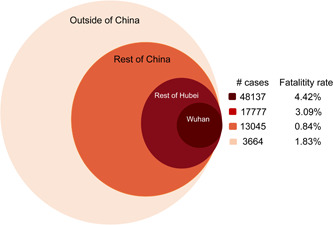
Case numbers and fatality rates of COVID‐19 in Wuhan and other areas (by 27 February 2020). COVID‐19, coronavirus disease 2019
2. GENETIC BIOLOGY OF SARS‐COV‐2
Full‐length genome sequences of SARS‐CoV‐2 were obtained from early infected individuals related to a wild animal market in Wuhan by different research groups through next‐generation sequencing. 1 , 2 , 11 Full genomic length of this novel coronavirus ranges from 29 891 to 29 903 nucleotides (nt). 2 , 4 All viral genome sequences obtained are extremely similar, showing more than 99.98% sequence identity. SARS‐CoV‐2 is 96.2% identical at the whole‐genome level to a bat coronavirus isolate RaTG13 (Global initiative on sharing all influenza data [GISAID] accession no. EPI_ISL_402131) collected from Yunnan province, China, and is 88% identical to two bat‐derived SARS‐like coronaviruses, bat‐SL‐CoVZC45 and bat‐SL‐CoVZXC21, collected in 2018 in Zhoushan, Eastern China. 1 , 2 , 11 The close phylogenetic relationship to RaTG13 suggests bats are probably natural hosts for SARS‐CoV‐2. 2 Human SARS‐CoV‐2 have a unique RRAR motif in the spike protein which is not found in coronaviruses isolated from pangolins, suggesting SARS‐CoV‐2 may not come directly from pangolins. 12 An evolutionary study 13 based on 86 genomic sequences from GISAID (https://www.gisaid.org/) showed three deletions were found in isolates from Japan, USA, and Australia, 93 mutations found over the entire genomes. Of note, eight mutations were found in the spike surface glycoprotein, especially three mutations (D354, Y364, and F367) located in the spike surface glycoprotein receptor‐binding domain (RBD), which suggested SARS‐CoV‐2 may rapidly evolve to evade immune response and adapt to other hosts in the future.
SARS‐CoV‐2 share 79% nt sequence identity to SARS‐CoV and around 50% to MERS‐CoV. 2 However, the seven conserved replicase domains in ORF1ab (used for CoV species classification) of SARS‐CoV‐2 are 94.6% identical to SARS‐CoV, implying the two belong to same species. 1 , 2 The receptor‐binding protein spike (S) gene of SARS‐CoV‐2 is highly divergent to all previously described SARSr‐CoVs with less than 75% nt sequence identity to except a 93.1% nt identity to RaTG13. Homology modeling revealed SARS‐CoV‐2 had a similar RBD structure to that of SARS‐CoV. 11 Further study showed SARS‐CoV‐2 uses the same cell entry receptor, ACE2, as SARS‐CoV, not CD26 as MERS‐CoV. 2 Structural analysis by cryo‐electron microscopy revealed SARS‐CoVs protein binds ACE2 with 10 to 20 folds higher affinity than SARS‐CoV, 14 which suggests that SARS‐CoV‐2 may be more infectious to human than SARS‐CoV.
3. EPIDEMIOLOGY OF COVID‐19
Transmission of infectious diseases must rely on three conditions: sources of infection, routes of transmission, and susceptible hosts. As the COVID‐19 continues spreading, more epidemiologic features of SARS‐CoV‐2 have been revealed. On the basis of recently published literatures, we compared transmission features of SARS‐CoV‐2 with SARS‐CoV and MERS‐CoV in Table 2. All three epidemics caused by these three coronaviruses are linked to wild animal markets. SARS and MERS are defined as zoonotic disease, and transmitted by intermediated hosts (palm civets and dromedary camels respectively). 11 Recent studies showed pangolins 15 and snakes 16 at wild animal markets were likely to be intermediate hosts of SARS‐CoV‐2.
Table 2.
Epidemiological characteristics of SARS‐CoV, MERS‐CoV, and SARS‐CoV‐2
| SARS‐CoV | MERS‐CoV | SARS‐CoV‐2 | |
|---|---|---|---|
| Estimated R0 | 2‐5 63 , 64 | <1 63 | 2.68 (95% CI, 2.48‐2.86) 19 |
| Host of virus | Natural host: Chinese horseshoe bats, 65 intermediate host: masked palm civet, 11 and terminal host: humans. 11 | Natural host: bats, 11 intermediate host: dromedary camels, 11 and terminal hosts: humans. 11 | Natural host: bats, 11 intermediate host: pangolins, 15 and terminal hosts: humans. 11 |
| Virus transmission mode | Person‐to‐person transmission through droplets, 66 opportunistic airborne transmission, 67 nosocomial transmission, 68 sporadic zoonotic transmission, aerosol transmission, 69 and fecal‐oral transmission. 65 | Respiratory transmission, 70 sporadic zoonotic transmission, 67 nosocomial transmission, 68 via aerosols, 69 and limited human‐to‐human transmission. 71 | Person‐to‐person transmission through respiratory droplets, contact and fomites, 63 zoonotic transmission, 72 nosocomial transmission, 9 fecal‐oral transmission, and aerosol transmission is highly possible. 34 |
| Median incubation period | 4.6 d (95% CI, 3.8‐5.8 d). 67 | 5.2 d (95% CI, 1.9‐14.7 d). 67 | 6.4 d (range, 0‐24.0 d). 73 |
| Case‐fatality rate | Worldwide (WHO): 9.6%, mainland China: 6.4%, and Hong Kong: 17%. 19 | Worldwide (WHO): 34.5% and South Korea: 20.4%. 19 | Wuhan, China: 3%. 63 |
Abbreviations: CI, confidence interval; MERS‐CoV, Middle East respiratory syndrome coronavirus; SARS‐CoV, severe acute respiratory syndrome coronavirus; WHO, World Health Organization.
Human‐to‐human transmission was considered as a major transmission mode. According to the sixth version of the guidance for diagnosis and treatments for COVID‐19 issued by the National Health Commission of China, SARS‐CoV‐2 was transmitted through respiratory aspirates, droplets, contacts, and feces, and aerosols transmission is highly possible. Chan et al 6 reported that six familial members got COVID‐19, but none of them had contacts with Wuhan markets or animals, although two had visited a Wuhan hospital. On the bais of data from China CDC, 58.3% of COVID‐19 cases are in Wuhan, while the rest of cases are imported from Wuhan. Wuhan is the capital of China's Hubei province, with over 14 million inhabitants, and is a major transportation hub, which increases person‐to‐person contacts and adds to the possibility of exporting cases to other locations. The early outbreak data largely followed the exponential growth before the implementation of quarantine strategies by governments on 24 January 2020. The basic reproductive values (R0 ) of COVID‐19 at the early stage were calculated between 2 and 3.5, indicating that one patient could transmit the disease to two to three other people, 17 , 18 , 19 , 20 which was higher than SARS and MERS. Phylodynamic analysis based on 52 genomic sequences of SARS‐CoV‐2 strains sampled in different countries publicly available at GISAID showed the estimated mean evolutionary rate was 7.8 × 10−4 subs/site/year (range 1.1 × 10−4 to 15 × 10−4), which was in line with that of SARS and MERS, and the mean time of the most recent common ancestor was 73 days. 21 With enforced implementation of isolation strategies, R0 was expected to decline in coming days. The mean incubation period was around 6.4 days (ranges from 0 to 24 days). 19 , 22
Similar to SARS and MERS, nosocomial transmission was a severe problem to COVID‐19, and even worse. A recent retrospective study 9 indicated that a total of 1716 health workers were infected, accounting for 3.84% of total cases. Nosocomial infections extremely burdened the health system and hindered early infected individuals from getting immediate medical supports, therefore resulting in high case‐fatality rate in Wuhan as shown in Table 1. In Wuhan alone, 1080 health workers were infected, in return case‐fatality rate of Wuhan is the highest. Wang et al 23 reported that among 138 hospitalized patients with COVID‐19, 41% of patients were suspected to be infected via hospital‐related transmission, 26% of patients received intensive care unit (ICU) care, and mortality was 4.3%. A lot of respiratory treatments for critically ill patients are deemed as high‐risk factors for nosocomial transmission, such as intubation, manual ventilation by resuscitator, noninvasive ventilation, high‐flow nasal cannula, bronchoscopy examination, suction and patient transportation. 24 Unexpectedly, a large portion of nosocomial transmissions occurred through contacts between clinicians and visitors with no or mild symptoms of COVID‐19 at the early phase of this outbreak. Similarly, presymptomatic transmission occurred through familial 25 , 26 , 27 and social gatherings, 27 such as banquets, church activities, sports, cruise traveling.
Vertical transmission was sporadically reported in some media but not yet proved. Chen et al 28 investigated nine pregnant women with COVID‐19 in their third trimester who underwent cesarean section. SARS‐CoV‐2 was tested in the amniotic fluid, cord blood, neonatal throat swab, and breast milk samples from six pregnant women with COVID‐19 pneumonia, and got all negative results. None of the neonates has clinical signs of infection. This result suggested no intrauterine fetal infections occurred as a result of COVID‐19 infection during the late stage of pregnancy. Previous studies also showed no evidence of perinatal infection of SARS‐CoV or MERS‐CoV during pregnancy. 29 However, a neonate born to a pregnant woman with COVID‐19 pneumonia tested positive for SARS‐CoV‐2 infection 36 hours after birth at Wuhan Tongji Hospital. 28 It is reasonable to assume that a newborn could be infected, either in utero or perinatally, and, thus, newborns should be placed in isolation to avoid exposure to any source of infection.
In terms of susceptible populations, all groups were generally susceptible to COVID‐19 regardless of age or sex. 9 Patients aged from 30 to 79 accounted for 86.6% of all cases. 9 The median age of the patients was 47 years. 30
Unlike SARS and MERS, patients diagnosed as COVID‐19 have presented with high viral loads even when those have no fever or mild symptoms. 31 High titers of SARS‐CoV‐2 were detected in travelers who recently visited Wuhan and have no fever or mild symptoms in the United States 31 and Germany 32 and other places. 33 A study showed high viral loads were detected in upper respiratory specimens of patients with COVID‐19, and viral shedding pattern of patients resembles that of patients with influenza. 33 This suggests SARS‐CoV‐2 may stay around for some time like influenza viruses.
4. CLINICAL FEATURES OF COVID‐19
The full spectrum of disease severity as shown in the guidelines for diagnosis and treatments for COVID‐19 34 issued by the National Health Commission of China had been updated for six times by 19 February 2020. COVID‐19 is now classified as four levels based on the severity of symptoms: mild, moderate, severe, and critical. Mild patients only present mild symptoms without radiographic features. Moderate patients present with fever, respiratory symptoms, and radiographic features. Severe patients meet one of three criteria: (a) dyspnea, RR greater than 30 times/min, (b) oxygen saturation less than 93% in ambient air, and (c) PaO2/FiO2 less than 300 mm Hg. Critical patients meet one of three criteria: (a) respiratory failure, (b) septic shock, and (c) multiple organ failure. The largest epidemiology study done by China CDC 9 showed among 44 672 confirmed cases, 86.6% of confirmed patients were aged 30 to 79 years, 80.9% were considered mild/common pneumonia, 13.8% were severe cases, and 4.7% were critical cases. Case‐fatality rate for critical patients was 49%. Patients with comorbidities (cardiovascular disease, diabetes, chronic respiratory disease, hypertension, and cancers) had higher case‐fatality rates (10.5%, 7.3%, 6.5%, 6.0%, and 5.6%, respectively) than those without comorbidities (0.9%). This indicated comorbidities were high‐risk factors for patients with COVID‐19.
Clinical symptoms of severe and critical patients with COVID‐19 resembled most of SARS and MERS as listed in Table 3, including fever, dry cough, myalgia, fatigue, dyspnea, anorexia, diarrhea, ARDS, arrhythmia, acute kidney injury, various degrees of liver damage, and septic shock. Common symptoms of hospitalized patients with COVID‐19 included fever (98.6%), fatigue (69.6%), dry cough, 23 , 35 and diarrhea. 36 Less common symptoms included muscle ache, confusion, headache, sore throat, rhinorrhoea, chest pain, sputum production, 36 and nausea and vomiting. 35 Severe complications included ARDS, RNAaemia, acute cardiac injury, and multiple organ failure. 36 The median time from first symptom to dyspnea was 5.0 days, to hospital admission was 7.0 days, and to ARDS was 8.0 days. 23 A few patients had symptoms, such as nasal congestion, runny nose, sore throat, myalgia, and diarrhea. 34 Most patients had a good prognosis according to the guidelines for diagnosis and treatments for COVID‐19. 34 Wang et al 23 reported that age and comorbidity may be risk factors. A recent study showed that renal damage was caused by virus and antiviral drugs. 37 Meanwhile, SARS‐CoV‐2 might cause various degrees of liver damage 38 and damages in testicular tissue. 37 Mild patients showed only low fever, mild fatigue, and no pneumonia. Severe patients usually had dyspnea/hypoxemia 1 week after the onset. Critical patients could quickly progress to ARDS, shock, metabolic acidosis, coagulation dysfunction, and multiple organ functional failure. 34 Dyspnea, abdominal pain, and anorexia were also more common in critically ill patients. 23 It had to be noted that severe and critically ill patients might present moderate to low fever, even without obvious fever. 34
Table 3.
Clinical, laboratory, and radiologic characteristics of SARS‐CoV, MERS‐CoV, and SARS‐CoV‐2
| SARS‐CoV | MERS‐CoV | SARS‐CoV‐2 | |
|---|---|---|---|
| Clinical features |
|
|
|
| Laboratory features |
|
|
|
| Radiologic features |
Abbreviations: ALT, alanine aminotransferase; APTT, activated partial thromboplastin time; ARDS, acute respiratory distress syndrome; AST, aspartate aminotransferase; CK, creatine kinase. CRP, C reactive protein; DIC, disseminated intravascular coagulation; LDH, lactate dehydrogenase; MERS‐CoV, Middle East respiratory syndrome coronavirus; PT, prothrombin time; SARS‐CoV, severe acute respiratory syndrome‐coronavirus.
Laboratory features of COVID‐19 included Lymphopenia with depletion of CD4 and CD8 lymphocytes, prolonged prothrombin time, elevated lactate dehydrogenase, 23 elevated D‐Dimer, elevated alanine transaminase, C‐reactive protein, and creatinine kinase. 36 ICU patients had higher plasma levels of IL2, IL7, IL10, GSCF, IP10, MCP1, MIP1A, and tumor necrosis factor‐α, compared with non‐ICU patients. 36 Patients who received ICU care had numerous laboratory abnormalities that suggested COVID‐19 might be associated with cellular immune deficiency, coagulation activation, myocardia injury, hepatic injury, and kidney injury as showed in Table 3. Laboratory abnormalities were similar to those previously observed in patients with MERS‐CoV and SARS‐CoV infection. 23 , 39 Most patients had elevated C reactive protein, erythrocyte sedimentation rate, and normal procalcitonin. In severe cases, D‐dimer increased and peripheral blood lymphocytes progressively decreased. Severe and critically ill patients had elevated inflammatory factors. 34 In the nonsurvivors, the neutrophil count, D‐dimer, blood urea, and creatinine levels continued to increase. 23
X‐ray and chest computed tomographic scans showed bilateral patchy shadows or ground‐glass opacity in the lungs of moderate and severe patients. 35 It had to be noted that majority of COVID‐19 patients with mild/moderate symptoms can quickly transit into severe or critical statuses if without immediate care. 40 These virus‐ carriers with no or mild symptoms can fool health workers and could be huge challenges for controlling this epidemic.
5. CHALLENGES FOR DIAGNOSIS OF COVID‐19
Diagnosis of COVID‐19 has been facing difficulties because laboratory detections and radiographic images are not always in agreement with clinical features and contact histories of patients. 41 Laboratory detections included genomic sequencing, reverse‐transcription polymerase chain reaction (RT‐PCR), and serological methods (such as enzyme‐linked immunoassay [ELISA]). In addition, because manifestations of the novel coronavirus pneumonia were diverse and changed rapidly, judging by radiographic images for early detection and evaluation of disease severity and follow‐up of patients were heavily depended on experience. 40 As a result, clinically suspected patients, with a history of exposure, fever, and positive findings on chest CT, had to receive rapid diagnosis with molecular technologies. 42
Genomic sequencing was a way for identifying disease‐associated pathogens 2 , 11 at the beginning of the outbreak of COVID‐19. But it was too complicated and expensive for a large scale of detections. RT‐PCR methods based on spike gene and N gene developed by several companies and China CDC were widely used for detecting viral RNA, and were considered a gold standard. 2 , 11 , 34 , 43 However, this method had its limitations, such as short detection window from nasopharyngeal swabs, false sampling, cross‐contamination of samples, and inconsistence of sample collections and preparations.
RT‐PCR methods generated false‐positive or false‐negative results, 42 , 44 which caused troubles for isolating sources of infections and determining hospitalization days. According to current guidelines for diagnosis and treatments for COVID‐19, if one is tested by RT‐PCR negative for twice, he/she is considered the cured and should be discharged. However, some of cured and discharged patients later have been tested positive by RT‐PCR. 45 Presumably, many factors mentioned above could lead to “false negative” in these cases. On the other hand, a proportion of patients with fever or pneumonia were wrongly isolated together with other confirmed patients with COVID‐19 in general medical wards because RT‐PCR could produce false‐positive results due to sample contaminations or other reasons. These patients turned out to be infected by influenza or other pneumonia associated pathogens. A recent large diagnostic study 42 showed 316 patients were confirmed infected with multiple respiratory pathogens including common HCoV (5 cases), influenza A virus (2 cases), rhinovirus (12 cases), and influenza A H3N2 (12 cases), respiratory syncytial virus (7 cases), influenza B virus (6 cases), and metapneumovirus (4 cases). In addition, RT‐PCR methods could generate inconsistent results. A fluorescence‐based quantitative PCR kit urgently distributed by the China CDC was designed to detect NP and ORF1ab regions on the SARS‐CoV‐2 genome. Sometimes, the results from the two pairs of primers did not agree with each other. 42 Besides technical difficulties, deletions and mutations in genome of SARS‐CoV‐2 occurred during viral evolution may also contribute to false results generated by RT‐PCR.
ELISA was highly recommended and expected to improve detection rate for COVID‐19 42 because sampling blood was much less stringent than sampling nasal or oral swabs for detecting viruses, and antibodies allows much longer detection window than viruses. Furthermore, ELISA had a quick turnaround time and relatively low costs. The strength of ELISA methods could make up of the shortages of RT‐PCR. 44 ELISA method based on SARSr‐CoV Rp3 nucleocapsid protein was successfully developed to detect immunoglobulin M and immunoglobulin G against SARS‐CoV‐2 in early COVID‐19 cases. 2 A caveat is that this ELISA method may generate false‐positive results as N protein is the most conservative viral protein among human β‐coronavirus genus. 46 Antigens used in ELISA may react with antibodies against four other HCoV that occurred in common colds. S protein is the most diverse protein and may be good candidate for ELISA development. 2
Differential diagnosis is also critical for confirming cases of COVID‐19. Winter usually has higher prevalence of flu and other pathogens associated pneumonia. In all, diagnosis of COVID‐19 has to be based on comprehensive understanding of epidemic history, clinical features, radiographic features, and laboratory detection. 34
6. POTENTIAL INTERVENTIONS
Isolation is still the most effective means of containing COVID‐19. 47 Effective surveillance is the prerequisite for blocking the source of infections. Many methods are applied to recognize source of infections (laboratory confirmed patients, suspected infected persons, and closely contacted persons), including community registration, tracing suspected carrier by cell phones. Those evaluated at moderate/high risk of exposure are encouraged to report conditions daily. General medical wards were used to collectively monitor and treat mild patients. 42 , 48
Therapeutics of COVID‐19 primarily include symptomatic treatments and antiviral therapies. Early supportive interventions are critical for treating mild patients including nutrient supplements, oxygen therapy, Chinese herbal medicine, and antibacterial therapy. Patients infected with COVID‐19 are mostly middle aged and elderly generally with low resistance to infection. 42 Supportive treatments are necessary for patients with mild symptoms at the early stage of infection. For critically ill patients, high‐flow oxygen therapy, extracorporeal membrane oxygenation, glucocorticoid therapy, and administration of convalescent plasma are applied. 34 , 49 There are several suggested antiviral treatments: lopinavir/ritonavir, ribavirin, interferon‐α, chloroquine phosphate, and Abidor. It is not recommended to use three or more antiviral drugs above simultaneously. 34 , 50 , 51 Combinational use of lopinavir, ritonavir, and ribavirin in the treatment of SARS was reported to be associated with better outcome. 52 It was previously reported that chloroquine could inhibit SARS‐CoV 53 and was tested for treating COVID‐19 in clinical trials in China. Recently, several studies suggested remdesivir effectively inhibit RNA viruses (including SARS/MERS‐CoV/2019‐nCoV) infection. 51 , 54 , 55 Remdesivir was used on the first patient in the United States and showed promising results. 31 Currently, three clinical trials were registered for testing remdesivir on the treatment of COVID‐19 (http://clinicaltrials.gov). In addition, a pan‐coronavirus fusion inhibitor EK1 targeting the HR1 domain of HCoV spike was reported to have the potential to treat COVID‐19. 56 Convalescent sera were used for treating critically ill patients and show some effects. 57
Recent report indicated that transmission may occur from individuals with no symptoms or from convalescents. 42 , 58 It is likely that SARS‐CoV‐2 will stay around for some time and even coevolve with its hosts. Due to uncertainty of clinical spectrum of virus carriers, vaccination is highly recommended for susceptible populations, especially for those with comorbidities (cardiovascular disease, diabetes, chronic respiratory disease, hypertension, and cancers). Vaccines based on S protein, T cell epitopes, and RBD have been studied for SARS and MERS. 56 , 59 , 60 , 61 Recently, an oral vaccine based on yeast expressed S protein developed by a group of scientists in Tianjing University of China provoked a lot of interests after it was reported by Jingyun News. However, it needs to be further tested in clinical trials. On 24 February 2020, Moderna, a pharmaceutical company in the United States announced that its experimental messenger RNA (mRNA) COVID‐19 vaccine, known as mRNA‐1273, was ready for human testing and the initial batch of the vaccine were shipped to US government researchers from the National Institute of Allergy and Infectious Diseases. We have reasons to believe that vaccines would be a way to prevent viral infection because convalescent sera could improve conditions of critically ill patients. 57 In addition to the protective and therapeutic measures mentioned above, psychological interventions were expected to be helpful for infection control. 62
7. CONCLUSION
Latest literatures and official data from China CDC revealed the epidemic of COVID‐19 caused more infections and deaths than either SARS or MERS by far, despite the fact that its case‐fatality rate is much lower. SARS‐CoV‐2 appears to be more infectious than SARS‐CoV or MERS‐CoV based on R0 values calculated at the early stage of this outbreak. Majority of infected individuals with no or mild symptoms can release viruses and spread viruses to others, which is extremely challenging for preventing the spread of COVID‐19. Therefore, intense surveillance is vital for preventing sustained transmission. Active interventions including nutrition supplement, symptomatic treatment, and antiviral treatment are critical for mild patients as well as severe patients. Finally, prophylactic vaccination is highly demanded for future prevention of emerging coronavirus‐related epidemics or pandemics.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
QQ conceived, wrote, and revised the paper. YW, YW, and YC equally contribute to writing. All authors approved the final version of the manuscript before submission.
ACKNOWLEDGMENTS
Qingsong Qin was supported from the Department of Education of Guangdong Province of China (No. 2017KTSCX067) and the Natural Scientific Foundation of Guangdong Province of China (No.2018A030307061).
Wang Y, Wang Y, Chen Y, Qin Q. Unique epidemiological and clinical features of the emerging 2019 novel coronavirus pneumonia (COVID‐19) implicate special control measures. J Med Virol. 2020;92:568–576. 10.1002/jmv.25748
REFERENCES
- 1. Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727‐733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579, 10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wu F, Zhao S, Yu B, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579, 10.1038/s41586-020-2008-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen L, Liu W, Zhang Q, et al. RNA based mNGS approach identifies a novel human coronavirus from two individual pneumonia cases in 2019 Wuhan outbreak. Emerg Microbes Infect. 2020;9(1):313‐319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ren LL, Wang YM, Wu ZQ, et al. Identification of a novel coronavirus causing severe pneumonia in human: a descriptive study. Chin Med J. 2020. 10.1097/CM9.0000000000000722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chan JFW, Yuan S, Kok KH, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person‐to‐person transmission: a study of a family cluster. Lancet. 2020;395(10223):514‐523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gorbalenya AE, Baker SC, Baric RS, et al. Severe acute respiratory syndrome‐related coronavirus: the species and its viruses—a statement of the Coronavirus Study Group. bioRxiv. 2020. [Google Scholar]
- 8. Chen Y, Liu Q, Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J Med Virol. 2020;92(4):418‐423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Novel Coronavirus Pneumonia Emergency Response Epidemiology Team . The epidemiological characteristics of an outbreak of 2019 novel coronavirus diseases (COVID‐19) in China. Zhonghua Liu Xing Bing Xue Za Zhi. 2020;41(2):145‐151.32064853 [Google Scholar]
- 10. Zumla A, Chan JF, Azhar EI, Hui DS, Yuen KY. Coronaviruses—drug discovery and therapeutic options. Nat Rev Drug Discov. 2016;15(5):327‐347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565‐574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li X, Zai J, Zhao Q, et al. Evolutionary history, potential intermediate animal host, and cross‐species analyses of SARS‐CoV‐2. J Med Virol. 2020. 10.1002/jmv.25731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Phan T. Genetic diversity and evolution of SARS‐CoV‐2. Infect Genet Evol. 2020;81:104260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wrapp D, Wang N, Corbett KS, et al. Cryo‐EM structure of the 2019‐nCoV spike in the prefusion conformation. bioRxiv. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lam TT‐Y, Shum MH‐H, Zhu H‐C, et al. Identification of 2019‐nCoV related coronaviruses in Malayan pangolins in southern China. bioRxiv. 2020. [Google Scholar]
- 16. Ji W, Wang W, Zhao X, Zai J, Li X. Homologous recombination within the spike glycoprotein of the newly identified coronavirus may boost cross‐species transmission from snake to human. J Med Virol. 2020;92:433‐440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li Q, Guan X, Wu P, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus‐infected pneumonia. N Engl J Med. 2020. 10.1056/NEJMoa2001316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Riou J, Althaus CL. Pattern of early human‐to‐human transmission of Wuhan 2019 novel coronavirus (2019‐nCoV), December 2019 to January 2020. Euro Surveill. 2020;25(4):2000058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wu JT, Leung K, Leung GM. Nowcasting and forecasting the potential domestic and international spread of the 2019‐nCoV outbreak originating in Wuhan, China: a modelling study. Lancet. 2020;395:689‐697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu Y, Gayle AA, Wilder‐Smith A, Rocklov J. The reproductive number of COVID‐19 is higher compared to SARS coronavirus. J Travel Med. 2020;27(2), 10.1093/jtm/taaa021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lai A, Bergna A, Acciarri C, Galli M, Zehender G. Early phylogenetic estimate of the effective reproduction number of SARS‐CoV‐2. J Med Virol. 2020. 10.1002/jmv.25723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Backer JA, Klinkenberg D, Wallinga J. Incubation period of 2019 novel coronavirus (2019‐nCoV) infections among travellers from Wuhan, China, 20‐28 January 2020. Euro Surveill. 2020;25(5):2000062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang D, Hu B, Hu C, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus‐infected pneumonia in Wuhan, China. JAMA. 2020. 10.1001/jama.2020.1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Respiratory Care Committee of Chinese Thoracic Society . Expert consensus on preventing nosocomial transmission during respiratory care for critically ill patients infected by 2019 novel coronavirus pneumonia. Zhonghua Jie He He Hu Xi Za Zhi. 2020;17:E020. [DOI] [PubMed] [Google Scholar]
- 25. Tong ZD, Tang A, Li KF, et al. Potential presymptomatic transmission of SARS‐CoV‐2, Zhejiang Province, China, 2020. Emerg Infect Dis. 2020;26(5), 10.3201/eid2605.200198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bai SL, Wang JY, Zhou YQ, et al. Analysis of the first cluster of cases in a family of novel coronavirus pneumonia in Gansu Province. Zhonghua Yu Fang Yi Xue Za Zhi. 2020;54:E005. [DOI] [PubMed] [Google Scholar]
- 27. Rocklov J, Sjodin H, Wilder‐Smith A. COVID‐19 outbreak on the Diamond Princess cruise ship: estimating the epidemic potential and effectiveness of public health countermeasures. J Travel Med. 2020. 10.1093/jtm/taaa030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen H, Guo J, Wang P, et al. Clinical characteristics and intrauterine vertical transmission potential of COVID‐19 infection in nine pregnant women: a retrospective review of medical records. Lancet. 2020;395:809‐815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rasmussen SA, Smulian JC, Lednicky JA, Wen TS, Jamieson DJ. Coronavirus disease 2019 (COVID‐19) and pregnancy: what obstetricians need to know. Am J Obstet Gynecol. 2020. 10.1016/j.ajog.2020.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Guan WJ, Ni ZY, Hu Y, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020. 10.1056/NEJMoa2002032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Holshue ML, DeBolt C, Lindquist S, et al. First case of 2019 novel coronavirus in the United States. N Engl J Med. 2020;382:929‐936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hoehl S, Berger A, Kortenbusch M, et al. Evidence of SARS‐CoV‐2 infection in returning travelers from Wuhan, China. N Engl J Med. 2020. 10.1056/NEJMc2001899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zou L, Ruan F, Huang M, et al. SARS‐CoV‐2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382(12). 10.1056/NEJMc2001737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.National Health Commission of China. The guidelines for diagnosis and treatment of novel coronavirus (2019‐nCoV) infected pneumonia (the sixth edition draft) issued by the National Health Commission of China. http://www.gov.cn/zhengce/zhengceku/2020‐02/19/content_5480948.htm2020. Accessed February, 2020.
- 35. Chen N, Zhou M, Dong X, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507‐513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497‐506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fan C, Li K, Ding Y, Lu W, Wang J. ACE2 expression in kidney and testis may cause kidney and testis damage after 2019‐nCoV infection. medRxiv. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chai X, Hu L, Zhang Y, et al. Specific ACE2 expression in cholangiocytes may cause liver damage after 2019‐nCoV infection. bioRxiv. 2020. [Google Scholar]
- 39. Hui DS, Wong PC, Wang C. SARS: clinical features and diagnosis. Respirology. 2003;8(suppl):S20‐S24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pan Y, Guan H, Zhou S, et al. Initial CT findings and temporal changes in patients with the novel coronavirus pneumonia (2019‐nCoV): a study of 63 patients in Wuhan, China. Eur Radiol. 2020. 10.1007/s00330-020-06731-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fang Y, Zhang H, Xie J, et al. Sensitivity of chest CT for COVID‐19: comparison to RT‐PCR. Radiology. 2020. 10.1148/radiol.2020200432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang M, Wu Q, Xu W, et al. Clinical diagnosis of 8274 samples with 2019‐novel coronavirus in Wuhan. medRxiv. 2020. 10.1101/2020.02.12.20022327 [DOI] [Google Scholar]
- 43. Chu DKW, Pan Y, Cheng SMS, et al. Molecular diagnosis of a novel coronavirus (2019‐nCoV) causing an outbreak of pneumonia. Clin Chem. 2020. 10.1093/clinchem/hvaa029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Xiao SY, Wu Y, Liu H. Evolving status of the 2019 novel coronavirus infection: proposal of conventional serologic assays for disease diagnosis and infection monitoring. J Med Virol. 2020;92(5), 10.1002/jmv.25702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lan L, Xu D, Ye G, et al. Positive RT‐PCR test results in patients recovered from COVID‐19. JAMA. 2020. 10.1001/jama.2020.2783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sun ZF, Meng XJ. Antigenic cross‐reactivity between the nucleocapsid protein of severe acute respiratory syndrome (SARS) coronavirus and polyclonal antisera of antigenic group I animal coronaviruses: implication for SARS diagnosis. J Clin Microbiol. 2004;42(5):2351‐2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhou G, Chi C. A model simulation study on effects of intervention measures in Wuhan COVID‐19 epidemic. med Rxiv. 2020. 10.1101/2020.02.14.20023168 [DOI] [Google Scholar]
- 48. Bernard Stoecklin S, Rolland P, Silue Y, et al. First cases of coronavirus disease 2019 (COVID‐19) in France: surveillance, investigations and control measures, January 2020. Euro Surveill. 2020;25(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Notice on the issuance of strategic guidelines for diagnosis and treatment of novel coronavirus (2019‐nCoV) infected pneumonia (first edition draft).
- 50. Lim J, Jeon S, Shin HY, et al. Case of the index patient who caused tertiary transmission of COVID‐19 infection in Korea: the application of lopinavir/ritonavir for the treatment of COVID‐19 infected pneumonia monitored by quantitative RT‐PCR. J Korean Med Sci. 2020;35(6):e79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang M, Cao R, Zhang L, et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019‐nCoV) in vitro. Cell Res. 2020;30:269‐271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chu CM, Cheng VC, Hung IF, et al. Role of lopinavir/ritonavir in the treatment of SARS: initial virological and clinical findings. Thorax. 2004;59(3):252‐256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Vincent MJ, Bergeron E, Benjannet S, et al. Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol J. 2005;2:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sheahan TP, Sims AC, Graham RL, et al. Broad‐spectrum antiviral GS‐5734 inhibits both epidemic and zoonotic coronaviruses. Sci Transl Med. 2017;9(396), 10.1126/scitranslmed.aal3653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Warren TK, Jordan R, Lo MK, et al. Therapeutic efficacy of the small molecule GS‐5734 against Ebola virus in rhesus monkeys. Nature. 2016;531(7594):381‐385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Xia S, Yan L, Xu W, et al. A pan‐coronavirus fusion inhibitor targeting the HR1 domain of human coronavirus spike. Sci Adv. 2019;5(4), 10.1126/sciadv.aav4580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Li XY, Du B, Wang YS, et al. The keypoints in treatment of the critical coronavirus disease 2019 patient. Zhonghua Jie He He Hu Xi Za Zhi. 2020;43:E026. [DOI] [PubMed] [Google Scholar]
- 58. Bai Y, Yao L, Wei T, et al. Presumed asymptomatic carrier transmission of COVID‐19. JAMA. 2020. 10.1001/jama.2020.2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Liu WJ, Zhao M, Liu K, et al. T‐cell immunity of SARS‐CoV: implications for vaccine development against MERS‐CoV. Antiviral Res. 2017;137:82‐92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chen WH, Du L, Chag SM, et al. Yeast‐expressed recombinant protein of the receptor‐binding domain in SARS‐CoV spike protein with deglycosylated forms as a SARS vaccine candidate. Hum Vaccin Immunother. 2014;10(3):648‐658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yu F, Du L, Ojcius DM, Pan C, Jiang S. Measures for diagnosing and treating infections by a novel coronavirus responsible for a pneumonia outbreak originating in Wuhan, China. Microbes Infect. 2020;22(2), 10.1016/j.micinf.2020.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Chen Q, Liang M, Li Y, et al. Mental health care for medical staff in China during the COVID‐19 outbreak. Lancet. 2020;7(4), 10.1016/S2215-0366(20)30078-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Chen J. Pathogenicity and transmissibility of 2019‐nCoV—a quick overview and comparison with other emerging viruses. Microbes Infect. 2020;22(2), 10.1016/j.micinf.2020.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zhao S, Lin Q, Ran J, et al. Preliminary estimation of the basic reproduction number of novel coronavirus (2019‐nCoV) in China, from 2019 to 2020: a data‐driven analysis in the early phase of the outbreak. Int J Infect Dis. 2020;92:214‐217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hui DSC, Zumla A. Severe acute respiratory syndrome: historical, epidemiologic, and clinical features. Infect Dis Clin North Am. 2019;33(4):869‐889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Cao C, Chen W, Zheng S, Zhao J, Wang J, Cao W. Analysis of spatiotemporal characteristics of pandemic SARS spread in Mainland China. BioMed Res Int. 2016;2016:7247983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hui DS. Epidemic and emerging coronaviruses (severe acute respiratory syndrome and Middle East respiratory syndrome). Clin Chest Med. 2017;38(1):71‐86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. de Wit E, van Doremalen N, Falzarano D, Munster VJ. SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol. 2016;14(8):523‐534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Judson SD, Munster VJ. Nosocomial transmission of emerging viruses via aerosol‐generating medical procedures. Viruses. 2019;11(10):940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Sharif‐Yakan A, Kanj SS. Emergence of MERS‐CoV in the Middle East: origins, transmission, treatment, and perspectives. PLoS Pathog. 2014;10(12):e1004457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Aevermann BD, Pickett BE, Kumar S, et al. A comprehensive collection of systems biology data characterizing the host response to viral infection. Sci Data. 2014;1:140033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Malik YS, Sircar S, Bhat S, et al. Emerging novel coronavirus (2019‐nCoV)—current scenario, evolutionary perspective based on genome analysis and recent developments. Vet Q. 2020;40:68‐76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Guan W‐J, Ni Z‐Y, Hu Y, et al. Clinical characteristics of 2019 novel coronavirus infection in China. medRxiv. 2020. [Google Scholar]
- 74. Peiris JS, Lai ST, Poon LL, et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361(9366):1319‐1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Leung WK, To KF, Chan PK, et al. Enteric involvement of severe acute respiratory syndrome‐associated coronavirus infection. Gastroenterology. 2003;125(4):1011‐1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. GENG QS. Guidelines for the prevention and treatment of SARS. Dongguan Sci Technol J. 2003;5:7. [Google Scholar]
- 77. Chau TN, Lee KC, Yao H, et al. SARS‐associated viral hepatitis caused by a novel coronavirus: report of three cases. Hepatology. 2004;39(2):302‐310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Du HWQ, Ma Y. Analysis of SARS inpatients in Beijing in 2003. Chin Gen Pract 2004;7(4):231‐232. [Google Scholar]
- 79. Assiri A, Al‐Tawfiq JA, Al‐Rabeeah AA, et al. Epidemiological, demographic, and clinical characteristics of 47 cases of Middle East respiratory syndrome coronavirus disease from Saudi Arabia: a descriptive study. Lancet Infect Dis. 2013;13(9):752‐761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Hijawi B, Abdallat M, Sayaydeh A, et al. Novel coronavirus infections in Jordan, April 2012: epidemiological findings from a retrospective investigation. East Mediterr Health J. 2013;19(suppl 1):S12‐S18. [PubMed] [Google Scholar]
- 81. Assiri AM, Mcgeer A, Perl TM, et al. Hospital outbreak of Middle East respiratory syndrome coronavirus. N Engl J Med. 2013;369:407‐416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. State of knowledge and data gaps of Middle East respiratory syndrome coronavirus (MERS‐COV) in humans. PLoS Curr. 2013;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Guery B, Poissy J, el Mansouf L, et al. Clinical features and viral diagnosis of two cases of infection with Middle East respiratory syndrome coronavirus: a report of nosocomial transmission. Lancet. 2013;381(9885):2265‐2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Memish ZA, Zumla AI, Al‐Hakeem RF, Al‐Rabeeah AA, Stephens GM. Family cluster of Middle East respiratory syndrome coronavirus infections. N Engl J Med. 2013;368(26):2487‐2494. [DOI] [PubMed] [Google Scholar]
- 85. National Health and Family Planning Commission of People's Republicao of China . Diagnosis and treatment plan for Middle East respiratory syndrome cases (2015 edition). Virologica Sinica. 2015;(5):352354. [Google Scholar]
- 86. Lee JY, Kim YJ, Chung EH, et al. The clinical and virological features of the first imported case causing MERS‐CoV outbreak in South Korea, 2015. BMC Infect Dis. 2017;17(1):498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. World Health Organization. WHO MERS global summary and assessment of risk. https://www.who.int/csr/disease/coronavirus_infections/risk‐assessment‐august‐2018.pdf. Accessed July, 2019.
- 88. Wong RS, Wu A, To KF, et al. Haematological manifestations in patients with severe acute respiratory syndrome: retrospective analysis. BMJ. 2003;326(7403):1358‐1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367(19):1814‐1820. [DOI] [PubMed] [Google Scholar]
- 90. Drosten C, Seilmaier M, Corman VM, et al. Clinical features and virological analysis of a case of Middle East respiratory syndrome coronavirus infection. Lancet Infect Dis. 2013;13(9):745‐751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Dyall J, Gross R, Kindrachuk J, et al. Middle East respiratory syndrome and severe acute respiratory syndrome: current therapeutic options and potential targets for novel therapies. Drugs. 2017;77(18):1935‐1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Chan PK, Tang JW, Hui DS. SARS: clinical presentation, transmission, pathogenesis and treatment options. Clin Sci. 2006;110(2):193‐204. [DOI] [PubMed] [Google Scholar]


