Abstract
The emergence and spread of 2019 novel coronavirus–infected pneumonia (COVID‐19) from Wuhan, China, it has spread globally. We extracted the data on 14 patients with laboratory‐confirmed COVID‐19 from Jinhua Municipal Central hospital through 27 January 2020. We found that compared to pharyngeal swab specimens, nucleic acid detection of COVID‐19 in fecal specimens was equally accurate. And we found that patients with a positive stool test did not experience gastrointestinal symptoms and had nothing to do with the severity of the lung infection. These results may help to understand the clinical diagnosis and the changes in clinical parameters of COVID‐19.
Keywords: coronavirus, dissemination, nervous system, pathogenesis, virus classification
Highlights
To determine the accuracy of COVID‐19 diagnosis in stool samples, we analyzed 14 laboratory‐diagnosed patients with COVID‐19 pneumonia.
The results showed that fecal specimens were as accurate aspharyngeal specimens.
COVID‐19 nucleic acid positive in fecal specimens was not related to the severity of pneumonia and gastrointestinal symptoms.
1. INTRODUCTION
In December 2019, a new coronavirus (COVID‐19) pneumonia outbreak occurred in Wuhan, China. 1 Since the virus was detected in December 2019, more than 77 000 people have been infected in China, causing more than 2600 deaths. 2 Globally, cases have been reported in 33 countries and five continents. 3 Patients' clinical manifestations included fever, cough, dyspnea, myalgia, fatigue, and chest computed tomography were ground‐glass opacity. 4 A novel coronavirus currently named 2019 novel COVID‐19, real‐time reverse‐transcriptase polymerase‐chain‐reaction (RT‐PCR) method can be used for the detection of COVID‐19 in oral swabs. 5 In SARS‐CoV and MERS‐CoV infected patients, intestinal infections are very common. 6 , 7 Results have now confirmed the presence of the live virus in stool samples from patients with COVID‐19, 8 which means that stool samples may contaminate hands, food, and water, and may cause infection by invading the mouth, respiratory mucosa, and conjunctiva. We report the detection method of 14 cases with confirmed COVID‐19 infection admitted to hospitals in Jinhua, aiming to investigate the other possible transmission route of this virus.
2. METHODS
We performed a retrospective analysis of 14 laboratory‐confirmed cases with COVID‐19 pneumonia. Data were obtained from the Department of Infectious Diseases Jinhua Hospital of Zhejiang University. Patients were hospitalized from 27 January 2020 to 10 February 2020, with final follow‐up for this report on 9 February 2020. Early we are confirmed cases with COVID‐19 pneumonia were a RT‐PCR assay for pharyngeal swab specimens, and that all oropharyngeal swabs specimens were collected by a senior infectious physician with more than 10 years of experience. Starting from 4 February 2020, we have added stool samples to all cases.
3. RESULTS
The median age of the patients was 41 years (18–87 years), and 7 (50%) were female. The most common symptoms include fever (92.8%) and cough (71.4%), no fever is an 87‐year‐old female patient, all patients had no vomiting and diarrhea. Four patients (28.5%) returned from Wuhan, the remaining 10 (71.4%) had a history of contact with COVID‐19 pneumonia patients.
Since 4 February, we have collected patient samples intensively as the supply of PCR kits has increased. It was found that five of the 14 confirmed patients (35.7%) had a positive stool sample COVID‐19 nucleic acid, and patients with positive stool samples were also positive for oropharyngeal swabs specimens at least the day before (Figure 1); The same trend is that patients with negative stool samples are also negative for oropharyngeal swabs for at least the first 2 days (Figure 1). This may indicate that the accuracy of fecal specimen detection and oropharyngeal swabs is the same.
Figure 1.
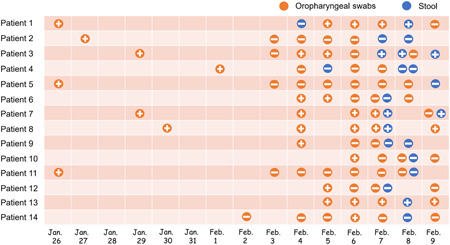
Viral nucleic acid test results in 14 patients with COVID‐19. COVID‐19, coronavirus
All 14 patients had computed tomography on admission, 92.8% manifested as pneumonia. One patient (patient 3) chest computed tomography images did not demonstrate any consolidation or scarring (Figure 2), however, this patient was positive for multiple stools and oropharyngeal swabs specimens (Figure 1). Among the remaining 13 patients, ground‐glass opacity was observed in both lungs. Two patients (patient 6 and patient 14) severely yielded more prominent abnormalities on chest computed tomography than nonsevere cases (Figure 3), the nucleic acid test was negative for stool samples in these two patients, this indicates that fecal specimens are not related to nucleic acid positivity and severity. As of 9 February 2020, three patients recovered, but 11 were still being treated in the hospital.
Figure 2.
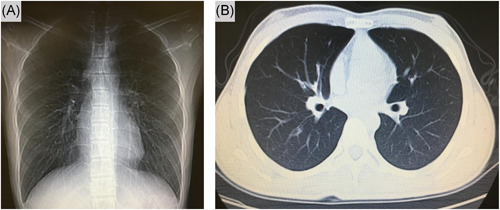
Chest computed tomographic images, 5 February 2020 (illness day 10, hospital day 6)
Figure 3.
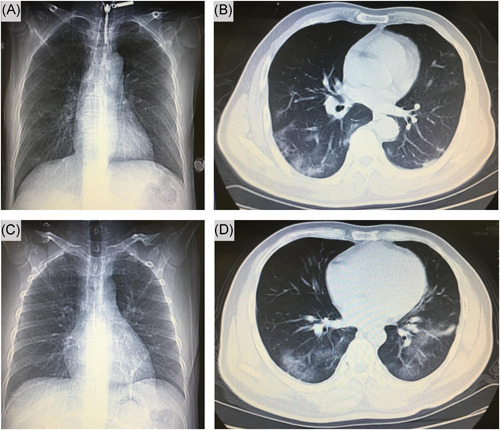
Chest computed tomographic images. Shown patient are chest images obtained at on day 5 (A, B) and another patient day 5 (C, D) after admission
4. DISCUSSION
According to the latest pilot experiment, four out of 62 stool specimens (6.5%) tested positive to COVID‐19, 9 but we found very high accuracy in detecting nucleic acids in stool samples. Recently, reports of fecal nucleic acid tests to diagnose COVID‐19 have increased. 10 , 11 In our cases, we found that patients with a positive stool test did not experience gastrointestinal symptoms and had nothing to do with the severity of the lung infection. Stool specimen collection will reduce medical staff infections compared to oropharyngeal swabs specimens, especially when protective equipment in China is inadequate. We are not sure if there is a fecal‐oral transmission, but fomite transmission might have contributed considerably to the rapid spread of COVID‐19. Finally, this report highlights the need for health protection, which should be considered for transmission through stool, these discoveries will curb the rapid spread of COVID‐19 worldwide.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
ACKNOWLEDGMENT
The author thanks Dr. Saibin Wang for sharing her personal observations.
Zhang J, Wang S, Xue Y. Fecal specimen diagnosis 2019 novel coronavirus–infected pneumonia. J Med Virol. 2020;92:680–682. 10.1002/jmv.25742
REFERENCES
- 1. Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumoniain China. N Engl J Med. 2020;382:727‐733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. National Health Commission of the People's Republic of China . http://www.nhc.gov.cn/xcs/yqfkdt/202002/945bd98a9d884aeeb54d76afa02ca813.shtml. Accessed on 25 February 2020. [DOI] [PMC free article] [PubMed]
- 3. World Health Organization . Novel coronavirus(2019‐nCoV): situation report‐36. https://www.who.int/docs/default‐source/coronaviruse/situation‐reports/20200225‐sitrep‐36‐covid‐19.pdf?sfvrsn=2791b4e0_2. Accessed 25 February 2020.
- 4. Wang D, Hu B, Hu C, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. JAMA. 2020. 10.1001/jama.2020.1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhou P, Yang XL, Wang XG, et al. Discovery of a novel coronavirus associated with the recent pneumonia outbreak in humans and its potential bat origin. Nature. 2020. 10.1038/s41586-020-2012-7 [DOI] [Google Scholar]
- 6. Shi X, Gong E, Gao D, et al. Severe acute respiratory syndrome associated coronavirus is detected in intestinal tissues of fatal cases. Am J Gastroenterol. 2005;100(1):169‐176. [DOI] [PubMed] [Google Scholar]
- 7. Ding Y, He L, Zhang Q, et al. Organ distribution of severe acute respiratory syndrome (SARS) associated coronavirus (SARS‐CoV) in SARS patients: implications for pathogenesis and virus transmission pathways. J Pathol. 2004;203(2):622‐630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang Y, Chen H, Zhu S, et al. Isolation of 2019‐nCoV from a stool specimen of a laboratory‐confirmed case of the coronavirus disease 2019 (COVID‐19). China CDC Weekly. 2020;2(8):123‐124. [PMC free article] [PubMed] [Google Scholar]
- 9. Guan W, Ni Z, Hu Y, et al. Clinical characteristics of 2019 novel coronavirus infection in China. N Engl J Med. 2020. 10.1056/NEJMoa2002032 [DOI] [Google Scholar]
- 10. Zhang H, Kang Z, Gong H, et al. The digestive system is a potential route of 2019‐nCov infection: a bioinformatics analysis based on single‐cell transcriptomes. 2020. Preprint at https://www.biorxiv.org/content/10.1101/2020.01.30.927806v1
- 11. Zhang W, Du RH, Li B, et al. Molecular and serological investigation of 2019‐nCoV infected patients: implication of multiple shedding routes. Emerging Microbes Infect. 2020;9:386‐389. [DOI] [PMC free article] [PubMed] [Google Scholar]


