Abstract
Background
Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) is an emerging disease with fatal outcomes. In this study, a fundamental knowledge gap question is to be resolved by evaluating the differences in biological and pathogenic aspects of SARS‐CoV‐2 and the changes in SARS‐CoV‐2 in comparison with the two prior major COV epidemics, SARS and Middle East respiratory syndrome (MERS) coronaviruses.
Methods
The genome composition, nucleotide analysis, codon usage indices, relative synonymous codons usage, and effective number of codons (ENc) were analyzed in the four structural genes; Spike (S), Envelope (E), membrane (M), and Nucleocapsid (N) genes, and two of the most important nonstructural genes comprising RNA‐dependent RNA polymerase and main protease (Mpro) of SARS‐CoV‐2, Beta‐CoV from pangolins, bat SARS, MERS, and SARS CoVs.
Results
SARS‐CoV‐2 prefers pyrimidine rich codons to purines. Most high‐frequency codons were ending with A or T, while the low frequency and rare codons were ending with G or C. SARS‐CoV‐2 structural proteins showed 5 to 20 lower ENc values, compared with SARS, bat SARS, and MERS CoVs. This implies higher codon bias and higher gene expression efficiency of SARS‐CoV‐2 structural proteins. SARS‐CoV‐2 encoded the highest number of over‐biased and negatively biased codons. Pangolin Beta‐CoV showed little differences with SARS‐CoV‐2 ENc values, compared with SARS, bat SARS, and MERS CoV.
Conclusion
Extreme bias and lower ENc values of SARS‐CoV‐2, especially in Spike, Envelope, and Mpro genes, are suggestive for higher gene expression efficiency, compared with SARS, bat SARS, and MERS CoVs.
Keywords: codon bias, COVID‐19, MERS CoV, nonstructural protein, preferred codons, SARS‐CoV‐2
1. INTRODUCTION
Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) is a new emerging fatal disease emerged in Wuhan, China in December 2019. 1 , 2 This is the third CoV epidemic after SARS and Middle East respiratory syndrome (MERS) CoV outbreaks. Initial phylogenetic analysis indicates the formation of a common cluster with bat SARS‐like CoV isolated in 2015. 3 Structural studies showed the close relation of the receptor‐binding domain of SARS‐CoV‐2 with SARS CoV. 4 Three major CoV epidemics were evolved during the past few decades. The first epidemic was SARS CoV in 2003. 5 Evidence was provided that SARS CoV was raised from an animal source including an intermediate host animal, which transmitted the virus from a bat carrier to humans. 6 About a decade later, MERS CoV was first identified in the Arabian Peninsula. 7 Lastly, in December 2019, the third epidemic emerged in Wuhan, China.
CoVs are enveloped, positive‐stranded RNA viruses possessing a comparatively large genome approaching 30 kb and comprising four structural proteins, namely, spike (S), nucleocapsid (N) envelope (E), and membrane (M). 8 The S protein is responsible for virus attachment to the receptor and fusion with cell membrane. 9 , 10 The N protein interacts with the viral RNA to form the ribonucleoprotein. 11 The E protein helps in virions assembly and comprises ion channel actions 12 ; the M protein shares in the assembly of new virus particles. 13 CoV genome is organized in 10 open‐reading frames (ORFs). 14 The 5′ encodes ORF1a and ORF1b, which are translated to give large polyproteins 1a and 1b (polyprotein AB in MERS CoV), which encodes a set of nonstructural protein. The CoV polyprotein is processed by the main protease and papain‐like protease to yield the nonstructural proteins.
Analysis of genome structure and composition is a part of studies of understanding virus evolution and adaptation to host. 15 , 16 Some amino acids are encoded by one codon, while others are encoded by several alternative codons knows as synonymous codons. 17 Codon bias means a preference for one codon over another during protein translation and affects translation efficiency, which differs from one organism to another. 18 , 19
In this study, the newly emerged fatal SARS‐CoV‐2, the four structural genes, and two most important nonstructural genes were evaluated for their nucleotide composition, preferred codons, relative synonymous codons usage (RSCU), and positively and negatively biased codons. This investigation will cover a knowledge gap in our understanding of mechanisms of viral genome evolution, the underlying codon composition, and codon usage preferences in comparison with the most recent CoV epidemic viral infections. The structural genes comprises the S, E, M, and N genes, while the nonstructural genes include the RNA‐dependent RNA polymerase (RdRP) and main protease (Mpro) genes.
2. MATERIALS AND METHODS
2.1. Gene data collection and analytical programs
SARS‐CoV‐2 and Pangolins Beta‐CoV complete genomes sequences were downloaded either from the NCBI GenBank or GISAID (https://www.gisaid.org/). The sequences of SARS CoV, bat SARS CoV, MERS CoV were retrieved from the gene databases at NCBI.
The CLC Genomics Workbench 12.0 (QIAGEN, Aarhus, Denmark) was used to handle the sequences. 20 The patterns of codon usage were assessed using CodonW 1.4.2. 21
2.2. Nucleotide composition and codon usage parameters
The nucleotide composition of the four structure genes, S, E, M, and N, were analyzed to reveal the nucleotides (A, T, G, and C) percentages. A/T, G/C percentage, the percentage of G or C nucleotides at the first position of codons (GC1), the percentage of each nucleotide at the third position of codons, the percentage of G or C nucleotides at the third position of codons (GC3) were calculated.
2.3. Relative synonymous codons usage
RSCU is calculated by dividing the expected frequencies of synonymous codon against their observed frequencies, according to Equation (1) 22 :
| (1) |
where X ij implies the observed number of codons used and ni stands for the sum of synonymous codons. The raw data for RSCU are provided in the Supporting Information Material.
2.4. The effective number of codons
ENc values range from 20 to 61. The obtained ENc value can be used to conclude the codon usage bias. An ENc value of <35 indicates strong codon usage bias due to lower number of codons used in protein translation. Higher ENc value indicates low codon usage bias. The raw data for ENc are provided in the Supporting Information Material.
3. RESULTS
3.1. Nucleotide compositions of the non structural proteins of SARS‐CoV‐2, SARS CoV, bat CoV, and MERS CoV
In S gene, T nucleotides were the most predominant (32%‐34%) followed by A (25%‐29%). SARS‐CoV‐2 showed the lowest GC% (37%) and the highest AT% (63%), compared with bat SARS, MERS, and SARS CoV. In addition, SARS‐CoV‐2 has the lowest percentage of G/C nucleotides at the third position of codons followed by pangolins Beta‐CoV, which showed the highest A3s. In all of the examined CoVs, the nucleotide percentages in the S gene were in the following order: T>A>C>G. In NT3s, the T3s and G3s were the most and the least frequent nucleotides, respectively (Table 1).
Table 1.
The codon usage indices of S, E, M, and N genes SARS‐CoV‐2, pangolins Beta‐CoV, SARS CoV, Bat CoV, and MERS CoV
| Virus | T3s | C3s | A3s | G3s | Nc | GC3s | GC | Gravy | Aromo | |
|---|---|---|---|---|---|---|---|---|---|---|
| Spike | SARS‐CoV‐2 | 0.55 | 0.19 | 0.38 | 0.13 | 44 | 0.25 | 0.37 | −0.08 | 0.11 |
| Beta‐CoV pangolin | 0.51 | 0.21 | 0.40 | 0.13 | 45 | 0.27 | 0.38 | −0.04 | 0.11 | |
| SARS CoV | 0.54 | 0.22 | 0.34 | 0.15 | 46 | 0.29 | 0.39 | −0.05 | 0.12 | |
| Bat SARS CoV | 0.52 | 0.23 | 0.34 | 0.15 | 48 | 0.30 | 0.39 | −0.05 | 0.11 | |
| MERS CoV | 0.53 | 0.23 | 0.28 | 0.19 | 48 | 0.33 | 0.41 | 0.05 | 0.11 | |
| Envelope | SARS‐CoV‐2 | 0.49 | 0.20 | 0.27 | 0.21 | 42 | 0.34 | 0.39 | 1.13 | 0.12 |
| Beta‐CoV pangolin | 0.45 | 0.23 | 0.28 | 0.21 | 46 | 0.37 | 0.39 | 1.13 | 0.12 | |
| SARS CoV | 0.39 | 0.24 | 0.33 | 0.21 | 61 | 0.38 | 0.41 | 1.14 | 0.11 | |
| Bat SARS CoV | 0.43 | 0.23 | 0.30 | 0.21 | 53 | 0.36 | 0.40 | 1.15 | 0.11 | |
| MERS CoV | 0.38 | 0.24 | 0.44 | 0.18 | 53 | 0.34 | 0.40 | 0.78 | 0.14 | |
| Membrane | SARS‐CoV‐2 | 0.42 | 0.26 | 0.32 | 0.17 | 54 | 0.36 | 0.43 | 0.45 | 0.12 |
| Beta‐CoV pangolin | 0.39 | 0.24 | 0.36 | 0.28 | 57 | 0.39 | 0.40 | 0.22 | 0.08 | |
| SARS CoV | 0.36 | 0.29 | 0.31 | 0.23 | 60 | 0.43 | 0.46 | 0.41 | 0.12 | |
| Bat SARS CoV | 0.39 | 0.28 | 0.30 | 0.22 | 57 | 0.41 | 0.44 | 0.43 | 0.12 | |
| MERS CoV | 0.43 | 0.27 | 0.28 | 0.19 | 60 | 0.38 | 0.43 | 0.45 | 0.12 | |
| Nucleocapsid | SARS‐CoV‐2 | 0.40 | 0.29 | 0.38 | 0.16 | 53 | 0.36 | 0.47 | −0.97 | 0.07 |
| Beta‐CoV pangolin | 0.41 | 0.29 | 0.38 | 0.15 | 54 | 0.35 | 0.47 | −0.99 | 0.07 | |
| SARS CoV | 0.38 | 0.31 | 0.39 | 0.15 | 54 | 0.37 | 0.48 | −1.02 | 0.07 | |
| Bat SARS CoV | 0.39 | 0.30 | 0.39 | 0.16 | 55 | 0.37 | 0.48 | −1.00 | 0.07 | |
| MERS CoV | 0.45 | 0.29 | 0.31 | 0.17 | 50 | 0.37 | 0.48 | −0.87 | 0.07 |
Abbreviations: CoV, coronavirus; MERS, Middle East respiratory syndrome; SARS, severe acute respiratory syndrome.
In the E gene, T nucleotides were the most predominant (34.5%‐40.4%) followed by A (21.5%‐25.7%). SARS‐CoV‐2 showed the lowest GC% (38.2%) and the highest AT% (63%), compared with bat SARS, MERS, and SARS CoV. In addition, SARS‐CoV‐2 has the lowest frequency of G/C nucleotides. In all of the examined dCoVs, the nucleotide percentages in the E gene were in the following order: T>A>C>G. In NT3s, the T3s and G3s were the most and the least frequent nucleotides, respectively.
In the N gene, A nucleotides were the most predominant (29.6%‐31.7%) followed by C (25%‐29%). There was little or no differences in GC% and AT% between the CoVs with a conserved tendency for higher AT%. The nucleotide percentages in the N gene were in the following order: A>C>G>T for SARS‐CoV‐2, SARS, and bat SARS CoVs, while MERS CoV showed a revised order of A>C>T>G. In NT3s, the T3s and G3s were the most and the least frequent nucleotides, respectively.
In the M gene, T nucleotides were the most predominant (29.9%‐31.9%) followed by A (24.4%‐25.6%). SARS‐CoV‐2 showed the lowest GC% (42.6%) and the highest AT% (57.4%), compared with bat SARS, MERS, and SARS CoV. In addition, SARS‐CoV‐2 and pangolins Beta‐CoV showed slightly lower G/C nucleotides at the third position of codons. In all of the examined CoVs, the nucleotide percentages in the M gene were in the following order: T>A>C>G. In NT3s, similar to other structural genes, the T3s and G3s were the most and the least frequent nucleotides, respectively.
In RdRP, T and A nucleotides were the most predominant nucleotides. In addition, SARS‐CoV‐2 showed the highest T3s and the lowest G3s (Table 2). In contrast, pangolins Beta‐CoV and MERS CoV showed the lowest A3s and the highest G3s frequencies. Therefore, similar to structural genes, RdRP contained pyrimidine nucleotides more frequent than purines. For Mpro, there is a conserved profile of general preference for T3s and low frequencies for G3s. Both SARS‐CoV‐2 and pangolin Beta‐CoV showed the lowest G3s frequencies.
Table 2.
The codon usage indices of RdRP and Mpro genes SARS‐CoV‐2, pangolins Beta‐CoV, SARS CoV, Bat CoV, and MERS CoV
| Virus | T3s | C3s | A3s | G3s | Nc | GC3s | GC | Gravy | Aromo | |
|---|---|---|---|---|---|---|---|---|---|---|
| RdRP | SARS‐CoV‐2 | 0.42 | 0.25 | 0.39 | 0.20 | 50.91 | 0.35 | 0.39 | 0.02 | 0.14 |
| Beta‐CoV pangolin | 0.38 | 0.23 | 0.39 | 0.27 | 51.81 | 0.39 | 0.39 | 0.24 | 0.11 | |
| SARS CoV | 0.40 | 0.25 | 0.36 | 0.24 | 53.32 | 0.38 | 0.42 | 0.10 | 0.11 | |
| Bat SARS CoV | 0.41 | 0.22 | 0.36 | 0.25 | 52.23 | 0.37 | 0.41 | 0.20 | 0.09 | |
| MERS CoV | 0.38 | 0.25 | 0.33 | 0.27 | 55.57 | 0.41 | 0.43 | 0.37 | 0.11 | |
| Mpro | SARS‐CoV‐2 | 0.52 | 0.21 | 0.42 | 0.13 | 45.68 | 0.27 | 0.37 | −0.23 | 0.13 |
| Beta‐CoV pangolin | 0.54 | 0.20 | 0.40 | 0.13 | 46.65 | 0.26 | 0.37 | −0.21 | 0.13 | |
| SARS CoV | 0.51 | 0.22 | 0.37 | 0.18 | 48.91 | 0.31 | 0.39 | −0.20 | 0.13 | |
| Bat SARS CoV | 0.49 | 0.24 | 0.37 | 0.19 | 50.23 | 0.32 | 0.40 | −0.20 | 0.13 | |
| MERS CoV | 0.54 | 0.25 | 0.28 | 0.21 | 50.95 | 0.35 | 0.40 | −0.18 | 0.14 |
Abbreviations: CoV, coronavirus; Mpro, main protease; MERS, Middle East respiratory syndrome; RdRP, RNA‐dependent RNA polymerase; SARS, severe acute respiratory syndrome.
3.2. RSCU analysis
In Tables 3, 4, and Table S1, the RSCU values for codons of CoV structural and nonstructural genes are provided, respectively. The tables are colored by a color scheme to denote the levels of codon usage bias. A value of RSCU =1 means that the observed frequency of codon is equivalent to the predictable frequency and indicating the lack of any codon usage bias. The underrepresented or negatively biased codons denote RSCU <0.6 (blue color), the overexpressed or positively biased codons with RSCU >0.6 (red color). The range between 0.6 and 1.6 conforms to the nonbiased codons.
Table 3.
RSCU values of structural genes (S and E) from SARS‐CoV‐2, pangolins Beta‐CoV, SARS CoV, Bat CoV, and MERS CoV
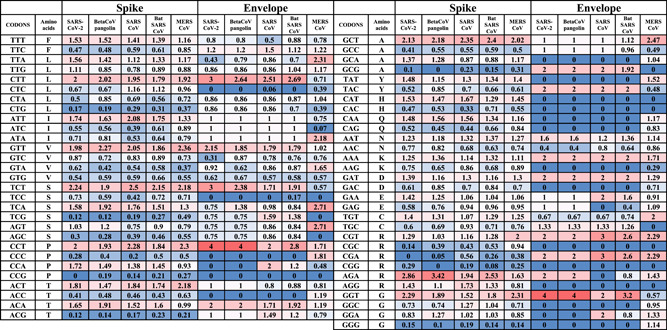
|
Abbreviations: CoV, coronavirus; MERS, Middle East respiratory syndrome; RSCU, relative synonymous codons usage; SARS, severe acute respiratory syndrome.
Table 4.
RSCU values of structural genes (M and N genes) from SARS‐CoV‐2, pangolins Beta‐CoV, SARS CoV, Bat CoV, and MERS CoV
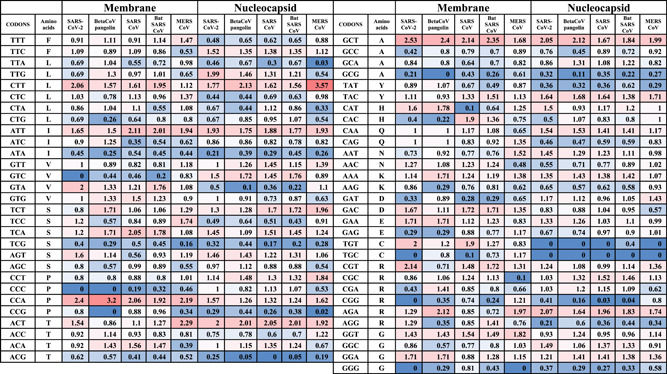
|
Abbreviations: CoV, coronavirus; MERS, Middle East respiratory syndrome; RSCU, relative synonymous codons usage; SARS, severe acute respiratory syndrome.
In the S gene, the over‐biased codons, SARS‐CoV‐2 showed the highest number of over‐biased codons (10 codons), including CTT, ATT, GTT, TCT, CCT, CCA, ACT, GCT, AGA, and GGT. All of these codons contained A3s or T3s. In contrast, pangolin Beta‐CoV, SARS CoV, bat SARS CoV, and MERS CoV showed 8, 8, 9, and 8 over‐biased codons, respectively (Table 3). Therefore, SARS‐CoV‐2 has the largest number of over‐biased codons. The over‐biased codons were similar to that provided for SARS‐CoV‐2 except for CCA and ACT for pangolin Beta‐CoV, CCA, and GGT for SARS CoV, CCA for bat SARS CoV and ATT and CCA for MERS CoV.
In the N gene, the over‐biased codons, SARS‐CoV‐2 showed the highest number of over‐biased codons (six codons), including TTG, CTT, ATT, ACT, GCT, and AGA. In contrast, pangolin Beta‐CoV, SARS CoV, bat SARS CoV, and MERS CoV showed 4, 4, 4, and 5 over‐biased codons, respectively (Table 4). Therefore, SARS‐CoV‐2 has the largest number of over‐biased codons in the N gene. The over‐biased codons were similar to that provided for SARS‐CoV‐2 except TTG for pangolin Beta‐CoV, TTG, and CTT for SARS TTG and CTT for bat SARS CoV and TTG for MERS CoV. For MERS CoV, the over‐biased codons were slightly different and included CTT, ATT, TCT ACT, GCT, TAC, and AGA.
In the M gene, the over‐biased codons were CTT, ATT, GTA, GCT, CCA, GAC, GAA, TGT, CGT, and GGA for SARS‐CoV‐2 (10 codons), CTT, ATT, TCT, TCA, GCT, CCA, and GGA for pangolin Beta‐CoV (seven codons), CTT, ATT, GTA, GCT, CCA, GAC, TGT, and CGT for SARS CoV and bat SARS CoV (eight codons), CTT, ATT, GTA, GCT, CCA, GAC, TGT, GGT, and CGT for MERS CoV (nine codons). GGA and GAA codons were nonbiased codons in all coronaviruses except for the SARS‐CoV‐2 and pangolin Beta‐CoV were over‐biased. In the E gene, the codon usage could be biased by the short length of the E gene that favors excluding its delivered RSCU values.
The frequent negatively biased codons among CoVs include CTG, TCG, AGC, CCG, ACC, ACG, GCG, CGC, and GGG in the S gene, ATA, GTA, TGC, GCG, TGT, AGG and TGC in the N gene and ATA, TGC, and CCC in the M gene.
The number of over‐biased and negatively biased codons were compared in SARS‐CoV‐2, pangolin Beta‐CoV, SARS CoV, Bat CoV, and MERS CoV. SARS‐CoV‐2 almost coding the highest number of over‐biased and negatively biased codons among all of the structural proteins. In the S gene, SARS‐CoV‐2 bears 12 and 19 over‐biased and negatively biased codons, respectively. The SARS‐CoV‐2/SARS over‐biased codons ratio was 1.2, 1.14, and 1.44 for S, N, and M genes, respectively. In addition, The SARS‐CoV‐2/SARS negatively biased codons ratio was 1, 1.2, and 1.44 for S, N, and M genes, respectively. Therefore, the SARS‐CoV‐2 showed the highest number of extreme codon usage patterns of over‐ or under‐biased codons, followed by SARS CoV. The gap between SARS‐CoV‐2 and SARS CoVs is more tighter than the gap of SARS‐CoV‐2 or SARS CoV and the bat SARS CoV, which showed a much lower number of biased codons in comparison with the other viruses. MERS CoV showed a higher number of biased codons only in the N gene, compared with SARS and SARS‐CoV‐2.
The structural genes undertook a homogenous profile of codon usage with little differences among the genes. In contrast, NSP as RdRP and Mpro showed larger variations. RdRP showed three over‐biased codons and eight common under‐biased codons. SARS‐CoV‐2 and pangolin Beta‐CoV showed the highest number of under‐biased codons (12 codons). Compared to 10 codons in SARS and bat SARS and five codons in MERS CoV (Table S1).
In Mpro, the number of over‐ and under‐biased genes were 11, 10, 8, 8, 6 and 15, 15, 14, 11, and 9 for SARS‐CoV‐2, pangolin Beta‐CoV, SARS, bat SARS, and MERS CoVs, respectively. This agrees with the general predicted highest number of biased codons in SARS‐CoV‐2.
3.3. Effective number of codons
ENc implies the effective number of codons and can be used as a measure of codon usage bias. ENc values range from 20 to 61. As the ENc value increases, the codon usage bias is lower. Low ENc value indicates high codon usage bias.
SARS‐CoV‐2 showed the lowest ENc value for all nonstructural and structural genes, compared with pangolins Beta‐CoV, SARS, and bat SARS CoVs (Figure 1). MERS CoV has the lowest ENc value for N and RdRP. The differences between ENc values between SARS‐CoV‐2 and pangolins Beta‐CoV were 0.6, 4.2, 2.9, 0.7, and 0.7 for S, E, M, N, RdRP, and Mpro. These values were the lowest differences compared with the other CoVs.
Figure 1.
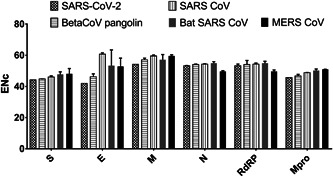
Effective number of codons values for structural (S, E, M, N) and nonstructural genes (RNA‐dependent RNA polymerase and main protease genes) from SARS‐CoV‐2, pangolins Beta‐CoV, SARS CoV, Bat CoV, and MERS CoV. CoV, coronavirus; MERS, Middle East respiratory syndrome; SARS, severe acute respiratory syndrome
4. DISCUSSION
Codon usage bias is used in the analysis of genes composition and conclusion of the forces controlling evolution and functions. 23 , 24 It has been used in the analysis of viral structural 25 , 26 and nonstructural genes. 25 , 27 In this study, the codon usage bias and genomic composition were compared in structural proteins of the three major CoV epidemics—SARS, MERS, and SARS‐CoV‐2.
In correlation with the previous knowledge of CoVs genome composition, AT% was higher than GC% in SARS‐CoV‐2. 28 , 30 In all of the structural genes of SARS‐CoV‐2, either A or T nucleotides were the most predominant nucleotides. In addition, A or T nucleotides were the most predominant nucleotides at the 3rd position of codons. This is in agreement with the previous studies of CoVs. 25 , 31
RNA viruses had evolved high ENc value (>35), implying low codon bias to adapt a wide range of hosts with various codon usage preferences. 29 ENc values above 50, in general, mean low codon usage bias. The codon usage data indicated lower number of ENc values of the SARS‐CoV‐2 compared with SARS, bat SARS, and pangolin CoV. This indicates a higher codon usage bias of SARS‐CoV‐2. Within these CoVs, pangolin CoV had the least ENc differences.
There is a negative correlation between the ENc value and codon usage bias. ENc values indicate higher codon usage bias in SARS‐CoV‐2 compared with SARS and MERS CoVs, due to lower ENc values, which is mostly observed in S, E, and M genes and to a lesser extent in N and RdRP genes. In SARS, bat SARS, and MERS CoVs E gene, ENc was >60, while in SARS‐CoV‐2, the ENc value was decreased by an amount of 18 to be no more than 42. Similarly, the M gene ENc value in SARS‐CoV‐2 was decreased by an amount of 3 to 5. Genes with low expression levels have high ENc values and more rare codons. 32 The expression of highly biased genes is considered as high. 33 The relative expression can be concluded from the ENc value, where small ENc value indicates higher bias and a generally higher level of expression. 34 Thus, the small ENc value is suggesting for higher gene expression. The lower observed ENc, especially for Spike and Envelope genes, values for SARS‐CoV‐2 structural genes are indicative for higher gene expression potency.
Supporting information
Supporting information
Supporting information
Supporting information
Supporting information
Supporting information
Supporting information
Supporting information
ACKNOWLEDGMENT
The authors acknowledge the Deanship of Scientific Research at King Faisal University for the financial support under Research Groups track (Grant No. 1811016).
Kandeel M, Ibrahim A, Fayez M, Al‐Nazawi M. From SARS and MERS CoVs to SARS‐CoV‐2: Moving toward more biased codon usage in viral structural and nonstructural genes. J Med Virol. 2020;92:660–666. 10.1002/jmv.25754
REFERENCES
- 1. Velavan TP, Meyer CG. The COVID‐19 epidemic. Trop Med Int Health. 2020;25:278‐280. 10.1111/tmi.13383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wang Y, Kang H, Liu X, Tong Z. Combination of RT‐qPCR testing and clinical features for diagnosis of COVID‐19 facilitates management of SARS‐CoV‐2 outbreak. J Med Virol. 2020. 10.1002/jmv.25721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Benvenuto D, Giovanetti M, Ciccozzi A, Spoto S, Angeletti S, Ciccozzi M. The 2019‐new coronavirus epidemic: Evidence for virus evolution. J Med Virol. 2020;92:455‐459. 10.1002/jmv.25688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565‐574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Peiris J, Lai S, Poon L, et al. Coronavirus as a possible cause of severe acute respiratory syndrome. The Lancet. 2003;361(9366):1319‐1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Li W, Wong SK, Li F, et al. Animal origins of the severe acute respiratory syndrome coronavirus: insight from ACE2‐S‐protein interactions. J Virol. 2006;80(9):4211‐4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367(19):1814‐1820. [DOI] [PubMed] [Google Scholar]
- 8. Siddell SG, Ziebuhr J, Snijder EJ. Coronaviruses, toroviruses, and arteriviruses. Topley and Wilson's microbiology and microbial infections. New York, NY: John Wiley & Sons; 2005. [Google Scholar]
- 9. Cavanagh D. The coronavirus surface glycoprotein. The coronaviridae. Germany: Springer; 1995:73‐113. [Google Scholar]
- 10. Kandeel M, Al‐Taher A, Li H, Schwingenschlogl U, Al‐Nazawi M. Molecular dynamics of Middle East Respiratory Syndrome Coronavirus (MERS CoV) fusion heptad repeat trimers. Comput Biol Chem. 2018;75:205‐212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Risco C, Antón IM, Enjuanes L, Carrascosa JL. The transmissible gastroenteritis coronavirus contains a spherical core shell consisting of M and N proteins. J Virol. 1996;70(7):4773‐4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ruch T, Machamer C. The coronavirus E protein: assembly and beyond. Viruses. 2012;4:363‐382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Neuman BW, Kiss G, Kunding AH, et al. A structural analysis of M protein in coronavirus assembly and morphology. J Struct Biol. 2011;174(1):11‐22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Al Hajjar S, Memish ZA, McIntoshc K. Middle East respiratory syndrome coronavirus (MERS‐CoV): a perpetual challenge. Ann Saudi Med. 2013;33(5):427‐436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bahir I, Fromer M, Prat Y, Linial M. Viral adaptation to host: a proteome‐based analysis of codon usage and amino acid preferences. Mol Syst Biol. 2009;5(1):311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. van Hemert F, van der Kuyl AC, Berkhout B. Impact of the biased nucleotide composition of viral RNA genomes on RNA structure and codon usage. J Gen Virol. 2016;97(10):2608‐2619. [DOI] [PubMed] [Google Scholar]
- 17. Nakamura Y, Gojobori T, Ikemura T. Codon usage tabulated from international DNA sequence databases: status for the year 2000. Nucleic Acids Res. 2000;28(1):292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chaney JL, Clark PL. Roles for synonymous codon usage in protein biogenesis. Annu Rev Biophys. 2015;44:143‐166. [DOI] [PubMed] [Google Scholar]
- 19. Supek F. The code of silence: Widespread associations between synonymous codon biases and gene function. J Mol Evol. 2016;82(1):65‐73. [DOI] [PubMed] [Google Scholar]
- 20. CLC Genomics Workbench 12.0 (QIAGEN, Aarhus, Denmark). www.qiagenbioinformatics.com, 2018.
- 21. Peden JF. Analysis of codon usage (Doctoral dissertation). University of Nottingham; 2000.
- 22. Behura SK, Severson DW. Codon usage bias: causative factors, quantification methods and genome‐wide patterns: with emphasis on insect genomes. Biol Rev. 2013;88(1):49‐61. [DOI] [PubMed] [Google Scholar]
- 23. Kandeel M, Elshazly K, El‐Deeb W, Fayez M, Ghonim I. Species specificity and host affinity rather than tissue tropism controls codon usage pattern in respiratory mycoplasmosis. J Camel Pract Res. 2019;26(1):29‐40. [Google Scholar]
- 24. Gumpper RH, Li W, Luo M. Constraints of viral RNA synthesis on codon usage of negative‐strand RNA virus. J Virol. 2019;93(5):e01775‐01718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sheikh A, Al‐Taher A, Al‐Nazawi M, Al‐Mubarak AI, Kandeel M. Analysis of preferred codon usage in the coronavirus N genes and their implications for genome evolution and vaccine design. J Virol Methods. 2020;277:113806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Makhija A, Kumar S. Analysis of synonymous codon usage in spike protein gene of infectious bronchitis virus. Can J Microbiol. 2015;61(12):983‐989. [DOI] [PubMed] [Google Scholar]
- 27. Alnazawi M, Altaher A, Kandeel M. Comparative genomic analysis MERS CoV isolated from humans and camels with special reference to virus encoded helicase. Biol Pharm Bull. 2017;40(8):1289‐1298. [DOI] [PubMed] [Google Scholar]
- 28. Gu W, Zhou T, Ma J, Sun X, Lu Z. Analysis of synonymous codon usage in SARS Coronavirus and other viruses in the Nidovirales. Virus Res. 2004;101(2):155‐161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jenkins GM, Holmes EC. The extent of codon usage bias in human RNA viruses and its evolutionary origin. Virus Res. 2003;92(1):1‐7. [DOI] [PubMed] [Google Scholar]
- 30. Zhou T, Gu W, Ma J, Sun X, Lu Z. Analysis of synonymous codon usage in H5N1 virus and other influenza A viruses. Biosystems. 2005;81(1):77‐86. [DOI] [PubMed] [Google Scholar]
- 31. Kandeel M, Altaher A. Synonymous and biased codon usage by MERS CoV papain‐like and 3CL‐proteases. Biol Pharm Bull. 2017;40(7):1086‐1091. [DOI] [PubMed] [Google Scholar]
- 32. Wang L, Xing H, Yuan Y, et al. Genome‐wide analysis of codon usage bias in four sequenced cotton species. PLoS One. 2018;13(3):e0194372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nair RR, Raveendran NT, Dirisala VR, et al. Mutational pressure drives evolution of synonymous codon usage in genetically distinct oenothera plastomes. Iran J Biotechnol. 2014;12(4):58‐72. [Google Scholar]
- 34. Zhang R, Zhang L, Wang W, et al. Differences in codon usage bias between photosynthesis‐related genes and genetic system‐related genes of chloroplast genomes in cultivated and wild solanum species. Int J Mol Sci. 2018;19(10):3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information
Supporting information
Supporting information
Supporting information
Supporting information
Supporting information
Supporting information


