Abstract
Bioactivation of the antidepressant and smoking cessation drug bupropion is catalyzed predominantly by CYP2B6. The metabolite hydroxybupropion derived from t-butylhydroxylation is considered to contribute to the antidepressant and smoking-cessation effects of the parent drug. Bupropion hydroxylation is the canonical in vitro and in vivo probe for CYP2B6 activity. P450 also requires obligate partnership with P450 oxidoreductase (POR). Human CYP2B6 and POR genes are highly polymorphic. Some CYP2B6 variants affect bupropion disposition. This investigation evaluated the influence of several human CYP2B6 and POR genetic variants on stereoselective bupropion metabolism, using an insect cell coexpression system containing CYP2B6, POR, and cytochrome b5. Based on intrinsic clearances (Clints), relative activities for S,S-hydroxybupropion formation were in the order CYP2B6.4 > CYP2B6.1 > CYP2B6.17 > CYP2B6.5 > CYP2B6.6 ≈ CYP2B6.26 ≈ CYP2B6.19 > CYP2B6.7 > CYP2B6.9 > > CYP2B6.16 and CYP2B6.18; relative activities for R,R-hydroxybupropion formation were in the order CYP2B6.17 > CYP2B6.4 > CYP2B6.1 > CYP2B6.5 ≈ CYP2B6.19 ≈ CYP2B6.26 > CYP2B6.6 > CYP2B6.7 ≈ CYP2B6.9 > > CYP2B6.16 and CYP2B6.18. Bupropion hydroxylation was not influenced by POR variants. CYP2B6-catalyzed bupropion hydroxylation is stereoselective. Though Vmax and Km varied widely among CYP2B6 variants, stereoselectivity was preserved, reflected by similar Clint(S,S-hydroxybupropion)/Clint(R,R-hydroxybupropion) ratios (1.8–2.9), except CYP2B6.17, which was less enantioselective. Established concordance between human bupropion hydroxylation in vitro and in vivo, together with these new results, suggests additional CYP2B6 variants may influence human bupropion disposition.
SIGNIFICANCE STATEMENT
Bupropion pharmacokinetics, metabolism, and clinical effects are affected by the CYP2B6*6 polymorphism. Other expressed CYP2B6 polymorphisms had diminished (*5, *6, *7, *9, *19, *26) or defective (*16, *18) in vitro bupropion hydroxylation. P450 oxidoreductase genetic variants had no effect on metabolism, suggesting no clinical consequence of this polymorphism. These CYP2B6 polymorphisms may portend diminished in vivo bupropion hydroxylation and predict additional clinically important variant alleles.
Introduction
Bupropion is approved by the Food and Drug Administration to treat depression (Dhillon et al., 2008) and for smoking cessation therapy (Reid et al., 2016) and is also used for obesity (Wilding, 2017), seasonal affective disorder (Cools et al., 2018), attention deficit hyperactivity disorder (Verbeeck et al., 2017), drug abuse (Lee et al., 2018), and chronic pain (Urits et al., 2019). Bupropion inhibits norepinephrine and dopamine reuptake transporters, increasing extracellular norepinephrine and dopamine concentrations, and is a nicotinic antagonist, blocking nicotinic acetylcholine receptors in the brain.
Bupropion is extensively metabolized by the liver, with less than 1% eliminated unchanged in urine (Welch et al., 1987). The major primary metabolic pathway is t-butyl hydroxylation to hydroxybupropion. Hydroxybupropion plasma exposure exceeds that of bupropion by an order of magnitude. Bupropion is used clinically as a racemate, and metabolism is stereoselective (Xu et al., 2007; Coles and Kharasch, 2008; Kharasch et al., 2008; Masters et al., 2016; Kharasch and Crafford, 2019).
Hydroxybupropion is pharmacologically active (Bondarev et al., 2003; Damaj et al., 2004) and is considered to contribute to the antidepressant and smoking-cessation effects of bupropion (Lee et al., 2007; Zhu et al., 2012; Laib et al., 2014; Malcolm et al., 2015). Hydroxylation of the t-butyl group and subsequent cyclic hemiketal formation creates a second chiral center. Therefore, there is the potential for four diastereomeric hydroxylated metabolites. However, only 2S,3S- and 2R,3R-hydroxybupropion have been found in plasma in humans, presumably due to steric hindrance precluding formation of 2S,3R- and 2R,3S-hydroxybupropion (Suckow et al., 1997). Plasma concentrations of 2R,3R-hydroxybupropion are approximately 20-fold greater than 2S,3S-hydroxybupropion. In contrast, R-bupropion concentrations are only about 2-fold greater than S-bupropion (Kharasch et al., 2008; Masters et al., 2016; Kharasch and Crafford, 2019). 2S,3S-hydroxybupropion was deemed responsible for antidepressant activity and nicotine-cessation activities of bupropion, whereas 2R,3R-hydroxybupropion was inactive toward inhibition of norepinephrine and dopamine reuptake (Lee et al., 2007; Zhu et al., 2012; Laib et al., 2014).
Human bupropion racemate and enantiomers hydroxylation are catalyzed exclusively by liver CYP2B6 (Faucette et al., 2000; Coles and Kharasch, 2008). Bupropion hydroxylation is the standard in vitro and in vivo probe to assess CYP2B6 activity, polymorphisms, and drug interactions (Faucette et al., 2000; Fahmi et al., 2016). The CYP2B6 gene is highly polymorphic; at least 38 allelic variants have been described (https://www.pharmvar.org/gene/CYP2B6), of which 25 are considered important, and eight are common or unique in at least one racial or ethnic population (Zhou et al., 2017). Carriers of certain CYP2B6 allelic variants have altered in vivo hydroxylation of bupropion racemate (Kirchheiner et al., 2003; Chung et al., 2011; Zhu et al., 2012; Benowitz et al., 2013; Høiseth et al., 2015; Gao et al., 2016; Lv et al., 2016; Ma et al., 2018) and enantiomers (Kharasch and Crafford, 2019). CYP2B6*4 and more so CYP2B6*6 are the most studied common variants. CYP2B6*6 homozygotes had decreased bupropion hydroxylation, whereas CYP2B6*4 carriers had somewhat increased hydroxylation. These polymorphisms have been associated with clinical differences in bupropion-mediated smoking cessation rates (Tran et al., 2019).
CYP2B6 metabolizes nearly 8% of marketed drugs, including, for example, methadone, ketamine, efavirenz, cyclophosphamide, and artemisinin. Previous studies showed that the activity of CYP2B6 allelic variants is allele-, substrate-, and expression system–dependent; cannot be extrapolated across substrates or expression systems; and requires substrate-specific evaluation (Coles and Kharasch, 2008; Ariyoshi et al., 2011; Honda et al., 2011; Zhang et al., 2011; Xu et al., 2012; Radloff et al., 2013; Gadel et al., 2015; Wang et al., 2018, 2019). Bupropion hydroxylation in vitro by CYP2B6 variants has been evaluated, including CYP2B6.4, CYP2B6.5, CYP2B6.6, CYP2B6.7, and CYP2B6.9 (Zhang et al., 2011; Xu et al., 2012; Radloff et al., 2013). Nevertheless, expression systems have variably used Escherichia coli, Spodoptera frugiperda (Sf9), and COS-1 cells. Therefore, the first purpose of this investigation was to characterize stereoselective bupropion hydroxylation by a greater number of clinically relevant CYP2B6 variants and by using a standard expression system for all.
P450 catalysis requires an obligate partnership with P450 oxidoreductase (POR), which transfers electrons from NADPH to P450. Cytochrome b5 can also function electively in this capacity. POR plays important roles in metabolism of drugs and steroid hormones. Genetic variation in POR is associated with altered metabolism of endogenous steroids (Burkhard et al., 2017) and xenobiotics (Riddick et al., 2013). Therefore, the second purpose of the present investigation was to determine the influence of POR variants on bupropion hydroxylation by wild-type CYP2B6.
Materials and Methods
Materials.
Racemic bupropion hydrochloride was purchased from Sigma Aldrich (St. Louis, MO). The standard rac-hydroxybupropion was purchased from Cerilliant (Round Rock, TX) and rac-hydroxybupropion-d6 from Toronto Research Chemicals (Toronto, ON, Canada). Sf9 cells and Sf-900 III SFM culture media were from ThermoFisher (Waltham, MA). Trichoplusia ni cells and ESF AF culture media were from Expression Systems (Davis, CA). All other reagents were from Sigma Aldrich.
Generation of Baculovirus Constructs and Recombinant Proteins Expression.
Production of recombinant proteins CYP2B6 variants, POR variants, and cytochrome b5 was carried out in insect cells by triple infection as described previously (Gadel et al., 2015; Wang et al., 2018, 2019). All CYP2B6 variants were coexpressed with POR and cytochrome b5. For each variant, protein contents of CYP2B6, POR, and b5 were measured as described previously (Wang et al., 2018, 2019).
Bupropion Metabolism.
All incubations were carried out in triplicate in 96-well polymerase chain reaction plates with raised wells in 100 mM potassium phosphate buffer (pH 7.4), as adapted from published protocols with modifications (Coles and Kharasch, 2008). RS-bupropion substrate (final racemate concentrations of 0, 2, 5, 10, 25, 50, 100, 250, 500, and 1000 µM) was mixed with CYP2B6/POR/b5. Final CYP2B6 concentration was 2.5 pmol/ml, and total reaction volume was 200 µl. After preincubation for 5 minutes at 37°C, the reaction was initiated by adding NADPH regenerating system (final concentrations: 10 mM glucose 6-phosphate, 1 mM β-NADP, 1 U/ml glucose-6-phosphate dehydrogenase, and 5 mM magnesium chloride, preincubated at 37°C for 10 minutes). The reaction was terminated after 10 minutes by adding 40 µl 20% trichloroacetic acid containing 75 ng/ml internal standard rac hydroxybupropion-d6. The plate was centrifuged at 2500 rpm for 5 minutes to remove precipitated proteins, and the supernatant was transferred to a 96-well shallow well plate for liquid chromatography (LC)–tandem mass spectrometry (MS/MS) analysis. Preliminary experiments showed that under the assay conditions bupropion hydroxylation was linear with time and CYP2B6 concentration.
Analysis of Bupropion Hydroxylation by High-Performance Liquid Chromatography–Tandem Mass Spectrometry.
Calibration samples were prepared using standard of rac hydroxybupropion, which is made up of equal amount of the two enantiomers (R,R)- and (S,S)-hydroxybupropion. Aqueous working stock solutions of rac hydroxybupropion were prepared at concentrations of 0.1, 1, and 100 µg/ml by diluting the certified methanolic stock solution (1 mg/ml) in deionized water. The calibration standards were prepared at 2, 5, 10, 50, 100, 500, 1000, and 2000 ng/ml rac hydroxybupropion from the working stock solutions. Two hundred microliters calibration samples were treated with 40 µl 20% trichloroacetic acid containing 75 ng/ml internal standard rac hydroxybupropion-d6 and followed by centrifugation at 2500 rpm for 5 minutes. The supernatant was transferred to a 96-well shallow well plate for LC-MS/MS analysis.
LC-MS/MS analysis was performed on a Shimadzu high-performance liquid chromatography system composed of two LC-20AD XR pumps, DGU20A5R degasser, CBM-20A system controller, CTO-20AC column oven, FCV-11AL solvent selection valve, and a SIL-20AC XR temperature regulated autosampler. The LC system was coupled to an API6500 triple quadrupole tandem mass spectrometer (Applied Biosystems/MDS Sciex, Foster City, CA) operated with Analyst 1.6.2. MultiQuant 3.0.1 (AB Sciex) was used for peak integration, generation of calibration curves, and data analysis.
Bupropion metabolites were analyzed utilizing a Chiralpak α1-acid glycoprotein analytical column (100 × 2.0 mm, 5 µm; Chiral Technologies, Westchester, PA) equipped with a chiral α1-acid glycoprotein guard cartridge (10 × 2.0 mm; Chiral Technologies). A 0.25 µm inline filter was placed prior to the sample entering the column. The column oven was at ambient temperature and the autosampler was at 4°C. The flow rate was 0.22 ml/min with a mobile phase consisting of 20 mM aqueous ammonium formate, pH 5.7 (A), and methanol (B). The sample injection volume was 5 µl, and total run time was 12 minutes. The time program to achieve the separation was as follows: 10% B for 0.5 minutes, linear gradient to 20% B until 1 minute, held at 20% B until 5 minutes, linear gradient to 50% B until 8 minutes, and then re-equilibrated to initial conditions until 12 minutes. Flow was directed into the mass spectrometer at 2.5 minutes and diverted to waste at 11.5 minutes. Under these conditions, the retention time was 6.7 minutes for (R,R)-hydroxybupropion, 4.4 minutes for (S,S)-hydroxybupropion, 6.4 minutes for (R,R)-hydroxybupropion-d6, and 4.4 minutes for (S,S)-hydroxybupropion-d6. The mass spectrometer was operated with a turbo spray ion source in the positive mode (electrospray ionization+) with multiple reaction monitoring. Analytes were detected with the multiple reaction monitoring transition of m/z 256.2 > 238.2 for both (R,R)- and (S,S)-hydroxybupropion and m/z 262.2 > 244.2 for (R,R)- and (S,S)-hydroxybupropion-d6. Operating conditions for analysis on the mass spectrometer were as previously described (Teitelbaum et al., 2016). Global parameters were optimized as follows: curtain gas 20 psig, ion spray voltage 5000 V, source temperature 600°C, Gas 1 40 psig, and Gas 2 40 psig. Dwell times were 500 ms. Interday coefficients of variation for hydroxybupropion were 5% or less at all concentrations.
Data Analysis.
Hydroxybupropion formation by enzyme variants was analyzed by ANOVA with post hoc Dunnet’s test (SigmaPlot 12.5; Systat). The results are means ± 95% confidence intervals. Statistical significance was assigned at P < 0.05. Hydroxybupropion formation versus substrate concentration data were analyzed by nonlinear regression analysis using Michaelis-Menten model. The results are reported as the parameter estimate ± standard error of the estimate. In vitro intrinsic clearance (Clint) was Vmax/Km.
Results
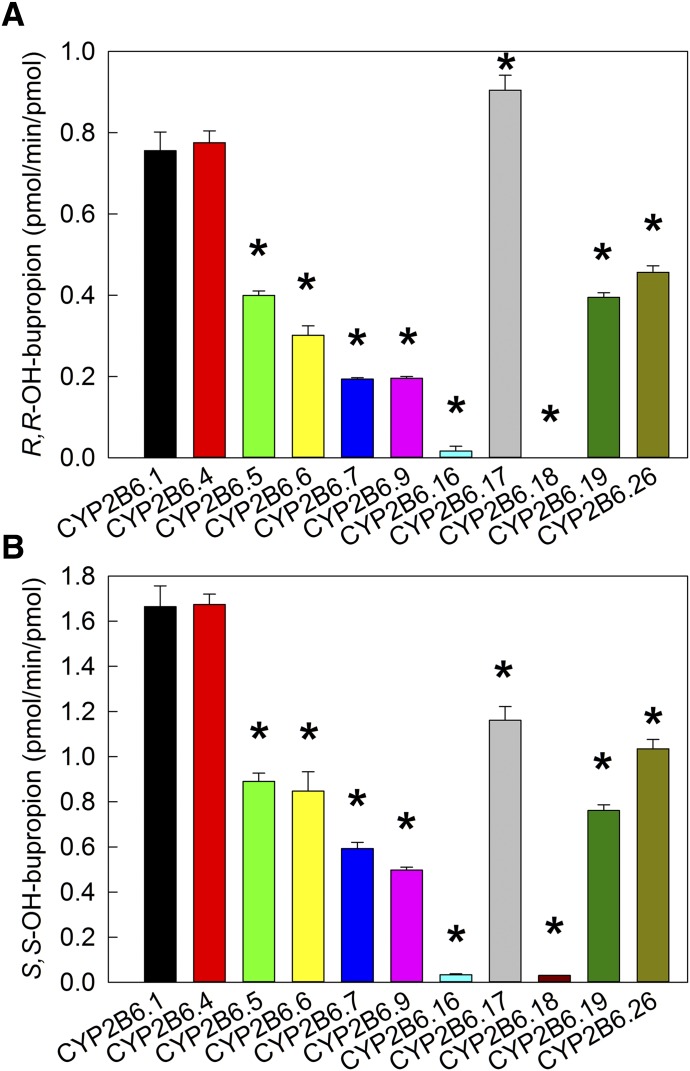
Metabolism of bupropion by CYP2B6 was evaluated first at approximately peak therapeutic concentrations. Based on previous publication (Coles and Kharasch, 2008), 2 µM RS-bupropion (1 µM enantiomers) was considered to be in the range of clinically relevant concentrations. As shown in Fig. 1 for R,R- and S,S-hydroxybupropion formation at 2 µM RS-bupropion substrate, activities of CYP2B6 variants vary over a wide range. For S,S-hydroxybupropion formation, CYP2B6.4 showed similar activity with the wild type, whereas all other variants had diminished activity compared with CYP2B6.1, and CYP2B6.16 and CYP2B6.18 were essentially catalytically inactive. At clinically relevant concentrations, hydroxylation rates were of the order CYP2B6.1 ≈ CYP2B6.4 > CYP2B6.17 ≈ CYP2B6.26 > CYP2B6.5 ≈ CYP2B6.6 ≈ CYP2B6.19 > CYP2B6.7 ≈ CYP2B6.9 > > CYP2B6.16 ≈ CYP2B6.18. For R,R-hydroxybupropion formation, the order is similar except that CYP2B6.17 was more active than CYP2B6.1.
Fig. 1.
Metabolism of RS-bupropion catalyzed by coexpressed recombinant CYP2B6 variants/POR.1/b5 at concentration of 2 µM RS-bupropion (1 µM R- or S-bupropion). Results are the mean ± 95% confidence interval of triplicate determinations. Asterisks denote rates significantly different from the wild-type CYP2B6.1 (P < 0.001). (A) R,R-hydroxybupropion formation from RS-bupropion. (B) S,S-hydroxybupropion formation from RS-bupropion.
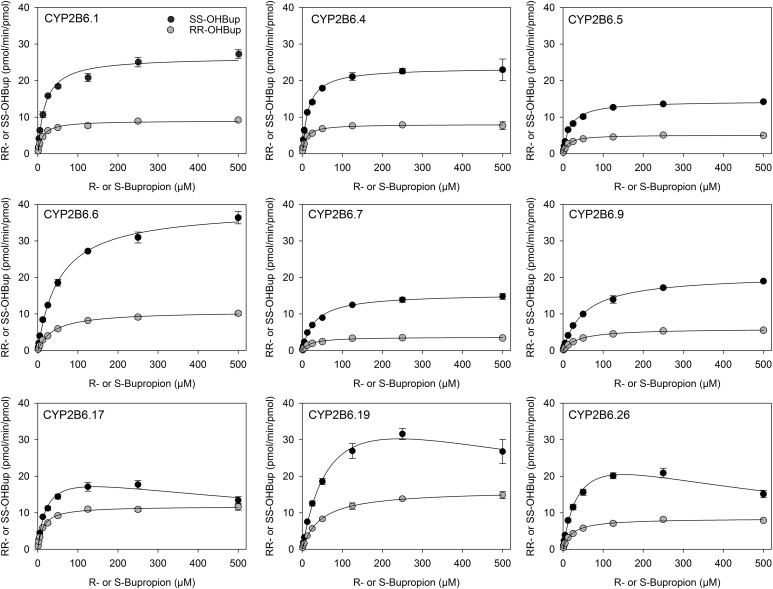
Racemic bupropion hydroxylation catalyzed by wild-type CYP2B6.1 and the catalytically active CYP2B6 variants with coexpressed POR and b5 were determined as a function of bupropion enantiomer concentrations (Fig. 2). For CYP2B6.1 and all CYP2B6 variant proteins, S,S-hydroxyburopion formation exceeded that of R,R-hydroxybupropion. Eadie-Hosfstee plots were generally linear (data not shown). Michaelis-Menten kinetic parameters were determined for all active enzymes (Table 1). There was a 3-fold (S-bupropion) and 4-fold (R-bupropion) range in Vmax and a 5-fold range in Km parameters for both R- and S-bupropion. CYP2B6.19 had the highest Vmax, which was about 2-fold greater than that of CYP2B6.1 (for both S- and R-bupropion), but the Km was also higher (4-fold), so the net effect was a lower in vitro Clint compared with CYP2B6.1. Similarly, CYP2B6.6 Vmax was also greater than that of CYP2B6.1 for S-bupropion, but the Km was also higher, so Clint was lower than that of CYP2B6.1. All variants had Clint values lower than that of CYP2B6.1, except CYP2B6.4 (R- and S-bupropion) and CYP2B6.17 (R-bupropion). Based on intrinsic clearances, relative activities for S,S-hydroxybupropion formation were in the order CYP2B6.4 ≥ CYP2B6.1 > CYP2B6.17 > CYP2B6.5 > CYP2B6.6 ≈ CYP2B6.26 ≈ CYP2B6.19 > CYP2B6.7 > CYP2B6.9 > > CYP2B6.16 ≈ CYP2B6.18, and relative activities for R,R-hydroxybupropion formation were in the order CYP2B6.17 ≈ CYP2B6.4 ≈ CYP2B6.1 > CYP2B6.5 ≈ CYP2B6.19 ≈ CYP2B6.26 > CYP2B6.6 > CYP2B6.7 ≈ CYP2B6.9 > > CYP2B6.16 ≈ CYP2B6.18.
Fig. 2.
Hydroxybupropion formation from RS-bupropion catalyzed by co-expressed CYP2B6 variants, wild-type P450 oxidoreductase and cytochrome b5. (●) S,S-hydroxybupropion from RS-bupropion, (○) R,R-hydroxybupropion from RS-bupropion. Results are the mean ± 95% confidence interval of triplicate determinations. Lines are predicted concentrations based on kinetic parameters obtained by nonlinear regression analysis of measured concentrations.
TABLE 1.
Kinetic parameters for (S,S)- and (R,R)-hydroxybupropion formation from RS-bupropion by CYP2B6 variants
| CYP2B6 variant | (S,S)-hydroxybupropion formation from RS-bupropion | (R,R)-hydroxybupropion formation from RS-bupropion | ||||
|---|---|---|---|---|---|---|
| Vmax | Km | Clinta | Vmax | Km | Clinta | |
| pmol/min per picomole | µM | ml/min per nanomole | pmol/min per picomole | µM | ml/min per nanomole | |
| CYP2B6.1 | 26.4 ± 0.5 | 18.2 ± 1.5 | 1.45 (100) | 9.0 ± 0.1 | 11.4 ± 0.8 | 0.79 (100) |
| CYP2B6.4 | 23.5 ± 0.4 | 14.5 ± 1.0 | 1.62 (112) | 8.0 ± 0.1 | 9.3 ± 1.6 | 0.86 (109) |
| CYP2B6.5 | 14.4 ± 0.2 | 17.3 ± 0.8 | 0.83 (57) | 5.1 ± 0.1 | 12.2 ± 0.7 | 0.42 (53) |
| CYP2B6.6 | 39.0 ± 0.6 | 53.0 ± 2.7 | 0.74 (51) | 10.8 ± 0.2 | 39.3 ± 2.0 | 0.27 (34) |
| CYP2B6.7 | 15.6 ± 0.2 | 31.1 ± 1.5 | 0.50 (34) | 3.7 ± 0.1 | 21.4 ± 1.5 | 0.17 (22) |
| CYP2B6.9 | 20.7 ± 0.2 | 53.3 ± 2.1 | 0.39 (27) | 6.0 ± 0.6 | 37.7 ± 1.3 | 0.16 (20) |
| CYP2B6.16b | 0.74 ± 0.03 | 0.72 ± 0.02 | ||||
| CYP2B6.17 | 22.3 ± 1.1 | 21.3 ± 2.5 | 1.05 (72) | 11.8 ± 0.2 | 13.1 ± 0.8 | 0.90 (114) |
| CYP2B6.18b | 0.91 ± 0.03 | 0.78 ± 0.01 | ||||
| CYP2B6.19 | 49.2 ± 4.1 | 75.6 ± 11.0 | 0.65 (45) | 16.2 ± 0.2 | 45.4 ± 2.1 | 0.36 (46) |
| CYP2B6.26 | 31.7 ± 2.1 | 41.5 ± 5.3 | 0.76 (52) | 8.5 ± 0.1 | 22.0 ± 1.4 | 0.39 (49) |
Values in brackets represent percentages of CYP2B6.1 Clint.
For CYP2B6.16 and CYP2B6.18, rates were measured at a fixed substrate concentration of 1000 µM RS-bupropion.
Bupropion hydroxylation by CYP2B6.1 is stereoselective, with S,S-hydroxybupropion formation greater than that of R,R-hydroxybupropion. The ratios of Vmax(S,S-hydroxybupropion)/Vmax(R,R-hydroxybupropion) and Clint(S,S-hydroxybupropion)/Clint(R,R-hydroxybupropion) for CYP2B6.1 were 2.9 and 1.8, respectively, consistent with a previous report (Coles and Kharasch, 2008). All CYP2B6 variants showed similar stereoselectivity, with the Vmax(S,S-hydroxybupropion)/Vmax(R,R-hydroxybupropion) ratio 1.9–4.2. The Clint(S,S-hydroxybupropion)/Clint(R,R-hydroxybupropion) ratio for most 2B6 variants was also similar to the wild type, 1.8–2.9, except that CYP2B6.17 (1.2) was lower.
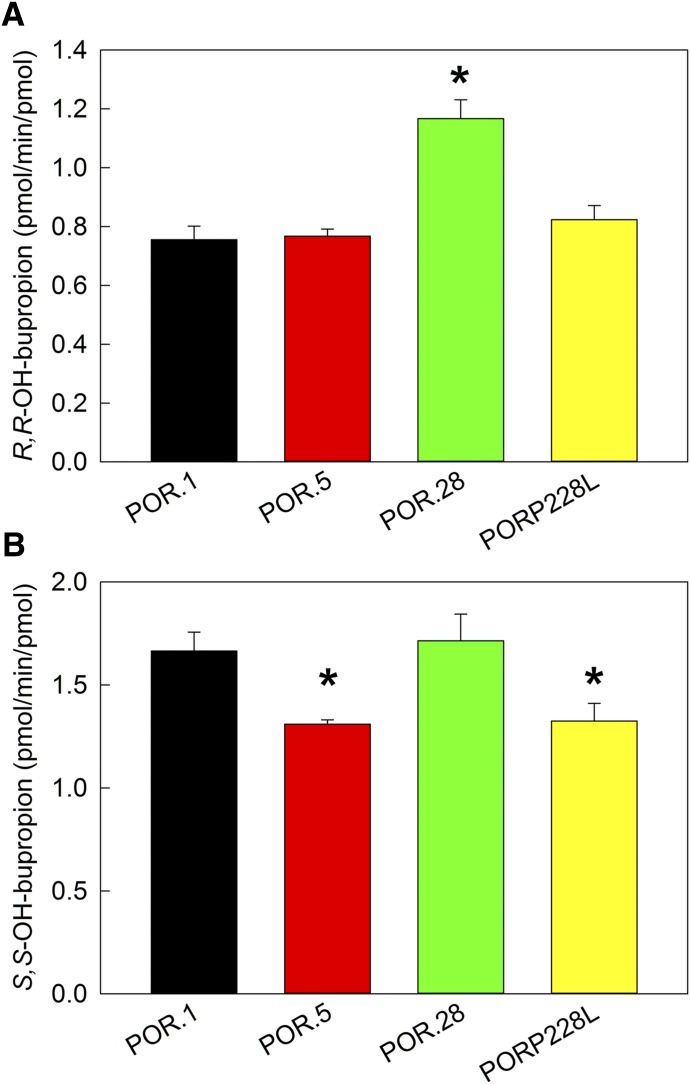
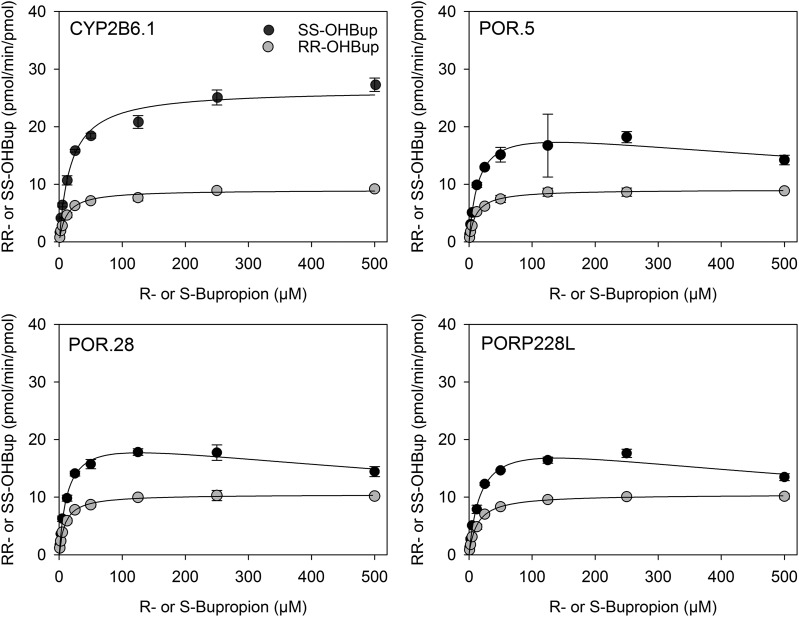
The activity of POR variants was determined with coexpressed wild-type CYP2B6 and cytochrome b5. In contrast to P450, POR variants caused minor, albeit statistically significant changes to bupropion (2 µM) hydroxylation compared with wild type POR (Fig. 3). Bupropion hydroxylation by POR variants over the full substrate concentration range is shown in Fig. 4 and Table 2. The kinetic parameters for the three POR variants POR.5, POR.28, and PORP228L are similar to the wild type.
Fig. 3.
Metabolism of RS-bupropion catalyzed by coexpressed recombinant CYP2B6.1/POR variants/b5 at concentration of 2 µM RS-bupropion (1 µM R- or S-bupropion). Results are the mean ± 95% confidence interval of triplicate determinations. Asterisks denote rates significantly different from the wild-type POR (P < 0.001 for R-bupropion and P = 0.002 for S-bupropion). (A) R,R-hydroxybupropion formation from RS-bupropion. (B) S,S-hydroxybupropion formation from RS-bupropion.
Fig. 4.
Hydroxybupropion formation from RS-bupropion catalyzed by co-expressed CYP2B6.1, P450 oxidoreductase variants and cytochrome b5. (●) S,S-hydroxybupropion from RS-bupropion, (○) R,R-hydroxybupropion from R-bupropion. Results are the mean ± 95% confidence interval of triplicate determinations. Lines are predicted concentrations based on kinetic parameters obtained by nonlinear regression analysis of measured concentrations.
TABLE 2.
Kinetic parameters for (S,S)- and (R,R)-hydroxybupropion formation from RS-bupropion by POR variants
| POR variant | (S,S)-hydroxybupropion formation from RS-bupropion | (R,R)-hydroxybupropion formation from RS-bupropion | ||||
|---|---|---|---|---|---|---|
| Vmax | Km | Clinta | Vmax | Km | Clint a | |
| pmol/min per picomole | µM | ml/min per nanomole | pmol/min per picomole | µM | ml/min per nanomole | |
| POR.1 | 26.4 ± 0.5 | 18.2 ± 1.5 | 1.45 (100) | 9.0 ± 0.1 | 11.4 ± 0.8 | 0.79 (100) |
| POR.5 | 21.0 ± 1.6 | 15.5 ± 3.1 | 1.35 (93) | 9.1 ± 0.1 | 10.5 ± 0.7 | 0.87 (110) |
| POR.28 | 21.4 ± 0.7 | 13.3 ± 1.1 | 1.61 (111) | 10.5 ± 0.1 | 8.9 ± 0.5 | 1.18 (149) |
| POR P228L | 21.3 ± 0.8 | 18.8 ± 1.8 | 1.13 (78) | 10.5 ± 0.1 | 13.0 ± 0.5 | 0.81 (103) |
Values in brackets represent percentages of CYP2B6.1 Clint.
Discussion
The first major observation was that CYP2B6 allelic variants have altered and diverse activities toward racemic bupropion hydroxylation compared with wild type CYP2B6, for both R- and S-bupropion. CYP2B6 variants selection was based on allele frequency and consequence. The 516G>T (Q172H) and 785G>T (K262R) polymorphisms are common, and variants with these mutations (2B6*4, *6, *7, *9, *16, *19, *26) were studied, even though some have minor population frequency. The other three variants (*5, *17, *18) have relatively high allele frequency. At clinically relevant concentrations, CYP2B6.5, CYP2B6.6, CYP2B6.7, CYP2B6.9, CYP2B6.19, and CYP2B6.26 were less active than wild type CYP2B1.1. CYP2B6.4 had activity similar to CYP2B1.1, whereas Clint was slightly higher than wild type for both enantiomers. CYP2B6.7 and CYP2B6.9 had the lowest activities of the enzymatically active variants, with 20%–35% of wild type Clint. CYP2B6.16 and CYP2B6.18 were essentially inactive.
Bupropion hydroxylation by CYP2B6 is stereoselective. Although Vmax and Km vary over a wide range among the variants, stereoselectivity was retained in most variants, reflected by similar ratios of Vmax(S,S-hydroxybupropion)/Vmax(R,R-hydroxybupropion) and Clint(S,S-hydroxybupropion)/Clint(R,R-hydroxybupropion). CYP2B6.17 was the only variant with a moderate difference in enantioselectivity compared with CYP2B6.1.
Only limited kinetic data are available on bupropion hydroxylation by CYP2B6 variants, and no data report on enantiomer metabolism. Table 3 summarizes the previously reported activity of expressed CYP2B6 variants toward RS-bupropion hydroxylation (Zhang et al., 2011; Xu et al., 2012; Radloff et al., 2013) and the present results. For CYP2B6.6, our results are comparable with previous studies, and lower Clint for CYP2B6.6 is consistently reported. CYP2B6.6 had both increased Vmax (1.5-fold for S,S-hydroxybupropion and 1.2-fold for R,R- hydroxybupropion) and increased Km (2.9-fold for S,S-hydroxybupropion and 3.4-fold for R,R-hydroxybupropion) compared with CYP2B6.1. Zhang et al. reported that E. coli–expressed CYP2B6.6 had 1.8-fold greater kcat versus wild type and 4-fold greater Km, although the protein expression system was different than used herein (Zhang et al., 2011). In contrast, the one previous investigation of CYP2B6.4 reported substantially lower Clint, whereas we found normal to slightly greater activity (Zhang et al., 2011).
TABLE 3.
Summary of reported bupropion hydroxylation
Relative activities are shown as percentage of CYP2B6 variants Clint compared with the wild type (100 × Clint(variant)/Clint(wild type)).
| Reference | Zhang et al. (2011) | Xu et al. (2012) | Radloff et al. (2013) | This study | |
|---|---|---|---|---|---|
| Expression system | E. coli | Sf9 | COS-1 | T. ni | |
| RS | RS | RS | S,S | R,R | |
| CYP2B6.1 | 100 | 100 | 100 | 100 | 100 |
| CYP2B6.4 | 35 | 112 | 109 | ||
| CYP2B6.5 | 47 | 44 | 57 | 53 | |
| CYP2B6.6 | 43 | 65 | 81 | 51 | 34 |
| CYP2B6.7 | 79 | 34 | 22 | ||
| CYP2B6.9 | 38 | 27 | 20 | ||
| CYP2B6.17 | 72 | 114 | |||
| CYP2B6.19 | 45 | 46 | |||
| CYP2B6.26 | 52 | 49 | |||
CYP2B6 is an unusual P450 isoform, in that activity of variants is clearly substrate-dependent, and generalizations about catalytic consequence or clinical implications of CYP2B6 polymorphisms are not possible. CYP2B6.4 (785G>T, K262R) had greater activity in metabolism of methadone (Gadel et al., 2015), efavirenz (Wang et al., 2019), artemether (Honda et al., 2011), and selegiline (Watanabe et al., 2010), and decreased activity with cyclophosphamide (Ariyoshi et al., 2011; Raccor et al., 2012), ifosfamide (Zanger and Klein, 2013; Calinski et al., 2015), ketamine (Wang et al., 2018), and nicotine (Bloom et al., 2019). Bupropion metabolism in the present study, using the CYP2B6.4 expression system used to study many of these other substrates, was slightly (10%) but not substantially greater than CYP2B6.1. CYP2B6.5 (1459C>T, R487C) was less active than CYP2B6.1 in metabolizing bupropion, as well as methadone (Gadel et al., 2015), ketamine (Wang et al., 2018), cyclophosphamide (Raccor et al., 2012), artmether (Honda et al., 2011) and nicotine (Bloom et al., 2019). It had moderately decreased activity in S-efavirenz hydroxylation and slightly increased activity in R-efavirenz hydroxylation (Wang et al., 2019). CYP2B6.9 (516G>T, Q172H) was substantially less active than CYP2B6.1 in bupropion metabolism, as well as methadone, ketamine, efavarenz, and nicotine, and moderately less (30%) with artemether, but more active than CYP2B6.1 in ifosfamide bioactivation (Honda et al., 2011; Calinski et al., 2015; Gadel et al., 2015; Wang et al., 2018, 2019; Bloom et al., 2019). CYP2B6.6 (516G>T-785A>G, Q172H-K262R) has been studied extensively in vitro and in vivo due to its high frequency of occurrence and therapeutic significance. CYP2B6.6 was less active than wild type in metabolism of bupropion, as well as methadone, ketamine, efavirenz, bupropion, ifosfamide, and nicotine, but more active in metabolizing cyclophosphamide and artemether (Ariyoshi et al., 2011; Honda et al., 2011; Calinski et al., 2015; Gadel et al., 2015; Wang et al., 2018, 2019; Bloom et al., 2019). CYP2B6.16 and CYP2B6.18, which were essentially inactive toward buproiopion, are also inactive with methadone, ketamine, and efavirenz (Gadel et al., 2015; Wang et al., 2018, 2019). These two variants are expressed as apoproteins only, without detectable cofactor heme bound. The mutation shared in the two variants, 983T>C (I328T), is located in the J-helix, and the structural change caused by this mutation is relayed to C- and I-helices, which are directly involved in ligand/heme recognition (Kobayashi et al., 2014). As a result, heme binding is disrupted, leading to abolition of catalytic activity. The common polymorphisms of CYP2B6 include 516G>T, 785G>T, 983T>C, and 1459C>T. Based on in vitro studies with substrates evaluated so far, 516G>T, 983T>C, and 1459C>T can generally be considered loss of function variants.
The second major observation in this investigation was that the three POR polymorphisms evaluated had very minor influence on bupropion metabolism. POR transfers electrons from NADPH to the P450 heme, an essential step in P450-catalyzed metabolism. Influence of POR polymorphism on drug metabolism is substrate- and P450 isoform-dependent. POR.5 (859G>C) had minimal or no effect on bupropion metabolism and 25% less activity in a previous bupropion study (Chen et al., 2012). In contrast, POR.5 had substantially lower activity with CYP3A4 and testosterone and with CYP17A1 and androgen biosynthesis (Chen et al., 2012; Pandey and Sproll, 2014). POR.28 had a small (R-bupropion) or no (S-bupropion) influence on CYP2B6.1-catalyzed bupropion metabolism.
There is high concordance between POR and CYP2B6 genetic variant effects on bupropion metabolism in vitro and in vivo. POR*28 had little effect on bupropion hydroxylation in vitro, or in vivo, based on plasma hydroxybupropion/bupropion area under the curve (AUC) ratios and urine hydroxybupropion formation clearance (Gao et al., 2016; Lv et al., 2016; Kharasch and Crafford, 2019). Accordingly, the present results would also suggest little influence of POR*5 and P228L polymorphisms on bupropion hydroxylation in vivo. The influence of CYP2B6*6, which consistently results in diminished bupropion hydroxylation in vitro (Table 3), is highly concordant in vivo. Plasma racemic hydroxybupropion/bupropion AUC ratios after single-dose bupropion were lower in CYP2B6*6 carriers than noncarriers (Chung et al., 2011) and lower in CYP2B6*6/*6 and CYP2B6*1/*6 versus CYP2B6*1/*1 genotypes (Gao et al., 2016; Lv et al., 2016), and hydroxybupropion concentrations were lower in CYP2B6*6 heterozygotes than wild types (Ma et al., 2018). Assessed stereoselectively, plasma AUC ratios and urine hydroxybupropion formation clearances for both bupropion enantiomers, after a single bupropion dose, were lower in CYP2B6*6/*6 but not CYP2B6*1/*6 compared with CYP2B6*1/*1 genotypes (Kharasch and Crafford, 2019). At steady state, plasma racemic hydroxybupropion/bupropion AUC ratios in CYP2B6 slow (*6/*6, *18/*18 genotypes) and intermediate (*6 and *18 heterozygotes) metabolizers were 34% and 54%, respectively, of those in normal metabolizers (*1/*1, *1/*4, *1/*5, *1/*22, *22/*22), and hydroxybupropion plasma concentrations were 40% and 20% lower in slow and intermediate than normal metabolizers (Zhu et al., 2012). Steady-state ratios were also lower in CYP2B6*6 carriers than wild types (Benowitz et al., 2013). Steady-state ratios were also lower than wild types in CYP2B6*6/*6 and CYP2B6*1/*6 genotypes, and plasma hydroxybupropion concentrations were less in CYP2B6*6/*6 but not CYP2B6*1/*6 genotypes (Høiseth et al., 2015). The influence of CYP2B6 polymorphisms on steady-state hydroxylation of bupropion enantiomers has not been reported. CYP2B6*18 coded for markedly deficient activity in vitro, and at steady state, hydroxybupropion/bupropion AUC ratios were lower in CYP2B6*18 carriers than wild types and even less than in CYP2B6*6 carriers (Benowitz et al., 2013). In vitro–in vivo concordance is less consistent for CYP2B6*4 than for decreased metabolism polymorphisms, which may be influenced by the rarity of CYP2B6*4. CYP2B6.4 had minimally increased bupropion hydroxylation in vitro and numerically but not significantly higher plasma hydroxybupropion/bupropion AUC ratios in CYP2B6*4 hererozygotes for the racemate (Kirchheiner et al., 2003; Zhu et al., 2012; Ma et al., 2018; Kharasch and Crafford, 2019) and enantiomers (Kharasch and Crafford, 2019). Based on these patterns and the present in vitro results, it could be expected that other CYP2B6 polymorphisms coding for diminished (*5, *6, *7, *9, *19, *26) or defective (*16) in vitro bupropion hydroxylation might result in diminished in vivo bupropion hydroxylation.
There are acknowledged potential limitations to this investigation. First, the purpose was only to test singular P450 and POR variants, not combinations; hence P450 and POR variants coexpressed in combination was not evaluated. Second, racemic bupropion was used as the substrate, rather than single enantiomers, because bupropion is used clinically as a racemate and results were intended to more accurately reflect and inform on clinical metabolism. Third, for the P450 variants, the P450:POR ratio varied, ranging from 1:1.3 to 1:4.5. P450 and POR form a 1:1 complex, and if POR content equals or exceeds P450 content all constructs should be fully catalytically competent and extra POR beyond 1:1 should not further increase activity. For the POR variants, the P450:POR ratio exceeded 1, except for POR*5 (1:0.6). Although it is possible that POR.5 expression affected apparent activity, it was not different than wild type for S-bupropion and only marginally less with R-bupropion. In addition, there are several types of P450 expression systems, including yeast, bacteria, mammalian, and insect cells, using transient or stable transfection, with or without coexpressed POR and b5, and expression systems can influence apparent catalytic activity (Wang et al., 2018). Yeast and bacteria are straightforward but require exogenous POR and b5, and bacterial expression usually requires sequence modification of mammalian P450s. Mammalian systems (e.g., monkey kidney COS cells and human embryonic kidney HEK cells) contain POR and b5 and allow easy P450 expression; protein expression and integrity can vary and thus influence apparent catalytic activity. Baculovirus-mediated expression in insect cells is a mature technology and allows simultaneous and flexible expression of individual genetic variants, using separate or combination multiprotein virus cassettes.
The newly identified CYP2B6 genetic polymorphisms in bupropion hydroxylation may also portend clinical consequence because there are important relationships between CYP2B6 genetics and clinical outcomes of bupropion pharmacotherapy. Bupropion hydroxylation is a bioactivation pathway, and hydroxybupropion, specifically S,S-hydroxybupropion, is considered to contribute to bupropion antismoking (Bondarev et al., 2003; Damaj et al., 2004; Lee et al., 2007; Zhu et al., 2012; Carroll et al., 2014; Malcolm et al., 2015) and antidepressant effects (Carroll et al., 2014; Laib et al., 2014). Bupropion-treated smokers who were CYP2B6 1459C>T carriers (*5, *7) reported greater cravings for cigarettes and had higher relapse rates compared with noncarriers (Lerman et al., 2002). Lower plasma racemic hydroxybupropion concentrations, related to CYP2B6 slow and intermediate metabolizer status, were associated with a lesser incidence of smoking abstinence (Zhu et al., 2012), although not all studies agree (Lee et al., 2007). Among depressed patients, lower plasma racemic hydroxybupropion concentrations were associated with lesser improvement in depression (Laib et al., 2014). The incidence of other CYP2B6 polymorphisms coding for deficient (*5, *6, *7, *9, *19, *26) or defective (*16) in vitro bupropion hydroxylation on clinical bupropion effectiveness is unknown.
Abbreviations
- AUC
area under the curve
- CLint
intrinsic clearance
- LC
liquid chromatography
- MS/MS
tandem mass spectrometry
- P450
cytochrome P450
- POR
P450 oxidoreductase
- Sf9
Spodoptera frugiperda
Authorship Contributions
Participated in research design: Wang, Kharasch.
Conducted experiments: Wang, Neiner.
Contributed new reagents or analytic tools: none.
Performed data analysis: Wang, Neiner, Kharasch.
Wrote or contributed to the writing of the manuscript: Wang, Kharasch.
Footnotes
This work was supported by the National Institutes of Health [Grants R01-DA14211 and R01-DA042985] and by the Washington University in St. Louis Department of Anesthesiology Russell B. and Mary D. Shelden fund.
References
- Ariyoshi N, Ohara M, Kaneko M, Afuso S, Kumamoto T, Nakamura H, Ishii I, Ishikawa T, Kitada M. (2011) Q172H replacement overcomes effects on the metabolism of cyclophosphamide and efavirenz caused by CYP2B6 variant with Arg262. Drug Metab Dispos 39:2045–2048. [DOI] [PubMed] [Google Scholar]
- Benowitz NL, Zhu AZ, Tyndale RF, Dempsey D, Jacob P., III (2013) Influence of CYP2B6 genetic variants on plasma and urine concentrations of bupropion and metabolites at steady state. Pharmacogenet Genomics 23:135–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom AJ, Wang PF, Kharasch ED. (2019) Nicotine oxidation by genetic variants of CYP2B6 and in human brain microsomes. Pharmacol Res Perspect 7:e00468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondarev ML, Bondareva TS, Young R, Glennon RA. (2003) Behavioral and biochemical investigations of bupropion metabolites. Eur J Pharmacol 474:85–93. [DOI] [PubMed] [Google Scholar]
- Burkhard FZ, Parween S, Udhane SS, Flück CE, Pandey AV. (2017) P450 oxidoreductase deficiency: analysis of mutations and polymorphisms. J Steroid Biochem Mol Biol 165:38–50. [DOI] [PubMed] [Google Scholar]
- Calinski DM, Zhang H, Ludeman S, Dolan ME, Hollenberg PF. (2015) Hydroxylation and N-dechloroethylation of Ifosfamide and deuterated Ifosfamide by the human cytochrome p450s and their commonly occurring polymorphisms. Drug Metab Dispos 43:1084–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll FI, Blough BE, Mascarella SW, Navarro HA, Lukas RJ, Damaj MI. (2014) Bupropion and bupropion analogs as treatments for CNS disorders. Adv Pharmacol 69:177–216. [DOI] [PubMed] [Google Scholar]
- Chen X, Pan LQ, Naranmandura H, Zeng S, Chen SQ. (2012) Influence of various polymorphic variants of cytochrome P450 oxidoreductase (POR) on drug metabolic activity of CYP3A4 and CYP2B6. PLoS One 7:e38495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung JY, Cho JY, Lim HS, Kim JR, Yu KS, Lim KS, Shin SG, Jang IJ. (2011) Effects of pregnane X receptor (NR1I2) and CYP2B6 genetic polymorphisms on the induction of bupropion hydroxylation by rifampin. Drug Metab Dispos 39:92–97. [DOI] [PubMed] [Google Scholar]
- Coles R, Kharasch ED. (2008) Stereoselective metabolism of bupropion by cytochrome P4502B6 (CYP2B6) and human liver microsomes. Pharm Res 25:1405–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cools O, Hebbrecht K, Coppens V, Roosens L, De Witte A, Morrens M, Neels H, Sabbe B. (2018) Pharmacotherapy and nutritional supplements for seasonal affective disorders: a systematic review. Expert Opin Pharmacother 19:1221–1233. [DOI] [PubMed] [Google Scholar]
- Damaj MI, Carroll FI, Eaton JB, Navarro HA, Blough BE, Mirza S, Lukas RJ, Martin BR. (2004) Enantioselective effects of hydroxy metabolites of bupropion on behavior and on function of monoamine transporters and nicotinic receptors. Mol Pharmacol 66:675–682. [DOI] [PubMed] [Google Scholar]
- Dhillon S, Yang LP, Curran MP. (2008) Bupropion: a review of its use in the management of major depressive disorder. Drugs 68:653–689. [DOI] [PubMed] [Google Scholar]
- Fahmi OA, Shebley M, Palamanda J, Sinz MW, Ramsden D, Einolf HJ, Chen L, Wang H. (2016) Evaluation of CYP2B6 induction and prediction of clinical drug-drug interactions: considerations from the IQ consortium induction working group-an industry perspective. Drug Metab Dispos 44:1720–1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faucette SR, Hawke RL, Lecluyse EL, Shord SS, Yan B, Laethem RM, Lindley CM. (2000) Validation of bupropion hydroxylation as a selective marker of human cytochrome P450 2B6 catalytic activity. Drug Metab Dispos 28:1222–1230. [PubMed] [Google Scholar]
- Gadel S, Friedel C, Kharasch ED. (2015) Differences in methadone metabolism by CYP2B6 variants. Drug Metab Dispos 43:994–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao LC, Liu FQ, Yang L, Cheng L, Dai HY, Tao R, Cao SP, Wang D, Tang J. (2016) The P450 oxidoreductase (POR) rs2868177 and cytochrome P450 (CYP) 2B6*6 polymorphisms contribute to the interindividual variability in human CYP2B6 activity. Eur J Clin Pharmacol 72:1205–1213. [DOI] [PubMed] [Google Scholar]
- Høiseth G, Haslemo T, Uthus LH, Molden E. (2015) Effect of CYP2B6*6 on steady-state serum concentrations of bupropion and hydroxybupropion in psychiatric patients: a study based on therapeutic drug monitoring data. Ther Drug Monit 37:589–593. [DOI] [PubMed] [Google Scholar]
- Honda M, Muroi Y, Tamaki Y, Saigusa D, Suzuki N, Tomioka Y, Matsubara Y, Oda A, Hirasawa N, Hiratsuka M. (2011) Functional characterization of CYP2B6 allelic variants in demethylation of antimalarial artemether. Drug Metab Dispos 39:1860–1865. [DOI] [PubMed] [Google Scholar]
- Kharasch ED, Crafford A. (2019) Common polymorphisms of CYP2B6 influence stereoselective bupropion disposition. Clin Pharmacol Ther 105:142–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharasch ED, Mitchell D, Coles R. (2008) Stereoselective bupropion hydroxylation as an in vivo phenotypic probe for cytochrome P4502B6 (CYP2B6) activity. J Clin Pharmacol 48:464–474. [DOI] [PubMed] [Google Scholar]
- Kirchheiner J, Klein C, Meineke I, Sasse J, Zanger UM, Mürdter TE, Roots I, Brockmöller J. (2003) Bupropion and 4-OH-bupropion pharmacokinetics in relation to genetic polymorphisms in CYP2B6. Pharmacogenetics 13:619–626. [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Takahashi O, Hiratsuka M, Yamaotsu N, Hirono S, Watanabe Y, Oda A. (2014) Evaluation of influence of single nucleotide polymorphisms in cytochrome P450 2B6 on substrate recognition using computational docking and molecular dynamics simulation. PLoS One 9:e96789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laib AK, Brünen S, Pfeifer P, Vincent P, Hiemke C. (2014) Serum concentrations of hydroxybupropion for dose optimization of depressed patients treated with bupropion. Ther Drug Monit 36:473–479. [DOI] [PubMed] [Google Scholar]
- Lee AM, Jepson C, Hoffmann E, Epstein L, Hawk LW, Lerman C, Tyndale RF. (2007) CYP2B6 genotype alters abstinence rates in a bupropion smoking cessation trial. Biol Psychiatry 62:635–641. [DOI] [PubMed] [Google Scholar]
- Lee NK, Jenner L, Harney A, Cameron J. (2018) Pharmacotherapy for amphetamine dependence: a systematic review. Drug Alcohol Depend 191:309–337. [DOI] [PubMed] [Google Scholar]
- Lerman C, Shields PG, Wileyto EP, Audrain J, Pinto A, Hawk L, Krishnan S, Niaura R, Epstein L. (2002) Pharmacogenetic investigation of smoking cessation treatment. Pharmacogenetics 12:627–634. [DOI] [PubMed] [Google Scholar]
- Lv J, Hu L, Zhuo W, Zhang C, Zhou H, Fan L. (2016) Effects of the selected cytochrome P450 oxidoreductase genetic polymorphisms on cytochrome P450 2B6 activity as measured by bupropion hydroxylation. Pharmacogenet Genomics 26:80–87. [DOI] [PubMed] [Google Scholar]
- Ma H, Zhang W, Yang X, Zhang Y, Wei S, Zhang H, Ma Y, Dang H. (2018) Effects of genetic polymorphisms of CYP2B6 on the pharmacokinetics of bupropion and hydroxybupropion in healthy Chinese subjects. Med Sci Monit 24:2158–2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malcolm E, Carroll FI, Blough B, Damaj MI, Shoaib M. (2015) Examination of the metabolite hydroxybupropion in the reinforcing and aversive stimulus effects of nicotine in rats. Psychopharmacology (Berl) 232:2763–2771. [DOI] [PubMed] [Google Scholar]
- Masters AR, Gufford BT, Lu JB, Metzger IF, Jones DR, Desta Z. (2016) Chiral plasma pharmacokinetics and urinary excretion of bupropion and metabolites in healthy volunteers. J Pharmacol Exp Ther 358:230–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey AV, Sproll P. (2014) Pharmacogenomics of human P450 oxidoreductase. Front Pharmacol 5:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raccor BS, Claessens AJ, Dinh JC, Park JR, Hawkins DS, Thomas SS, Makar KW, McCune JS, Totah RA. (2012) Potential contribution of cytochrome P450 2B6 to hepatic 4-hydroxycyclophosphamide formation in vitro and in vivo. Drug Metab Dispos 40:54–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radloff R, Gras A, Zanger UM, Masquelier C, Arumugam K, Karasi JC, Arendt V, Seguin-Devaux C, Klein K. (2013) Novel CYP2B6 enzyme variants in a Rwandese population: functional characterization and assessment of in silico prediction tools. Hum Mutat 34:725–734. [DOI] [PubMed] [Google Scholar]
- Reid RD, Pritchard G, Walker K, Aitken D, Mullen KA, Pipe AL. (2016) Managing smoking cessation. CMAJ 188:E484–E492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddick DS, Ding X, Wolf CR, Porter TD, Pandey AV, Zhang QY, Gu J, Finn RD, Ronseaux S, McLaughlin LA, et al. (2013) NADPH-cytochrome P450 oxidoreductase: roles in physiology, pharmacology, and toxicology. Drug Metab Dispos 41:12–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suckow RF, Zhang MF, Cooper TB. (1997) Enantiomeric determination of the phenylmorpholinol metabolite of bupropion in human plasma using coupled achiral-chiral liquid chromatography. Biomed Chromatogr 11:174–179. [DOI] [PubMed] [Google Scholar]
- Teitelbaum AM, Flaker AM, Kharasch ED. (2016) Development and validation of a high-throughput stereoselective LC-MS/MS assay for bupropion, hydroxybupropion, erythrohydrobupropion, and threohydrobupropion in human plasma. J Chromatogr B 1017–1018:101–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran AX, Ho TT, Varghese Gupta S. (2019) Role of CYP2B6 pharmacogenomics in bupropion-mediated smoking cessation. J Clin Pharm Ther 44:174–179. [DOI] [PubMed] [Google Scholar]
- Urits I, Peck J, Orhurhu MS, Wolf J, Patel R, Orhurhu V, Kaye AD, Viswanath O. (2019) Off-label antidepressant use for treatment and management of chronic pain: evolving understanding and comprehensive review. Curr Pain Headache Rep 23:66. [DOI] [PubMed] [Google Scholar]
- Verbeeck W, Bekkering GE, Van den Noortgate W, Kramers C. (2017) Bupropion for attention deficit hyperactivity disorder (ADHD) in adults. Cochrane Database Syst Rev 10:CD009504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang PF, Neiner A, Kharasch ED. (2018) Stereoselective ketamine metabolism by genetic variants of cytochrome P450 CYP2B6 and cytochrome P450 oxidoreductase. Anesthesiology 129:756–768. [DOI] [PubMed] [Google Scholar]
- Wang PF, Neiner A, Kharasch ED. (2019) Efavirenz metabolism: influence of polymorphic CYP2B6 variants and stereochemistry. Drug Metab Dispos 47:1195–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Sakuyama K, Sasaki T, Ishii Y, Ishikawa M, Hirasawa N, Hiratsuka M. (2010) Functional characterization of 26 CYP2B6 allelic variants (CYP2B6.2-CYP2B6.28, except CYP2B6.22). Pharmacogenet Genomics 20:459–462. [DOI] [PubMed] [Google Scholar]
- Welch RM, Lai AA, Schroeder DH. (1987) Pharmacological significance of the species differences in bupropion metabolism. Xenobiotica 17:287–298. [DOI] [PubMed] [Google Scholar]
- Wilding JP. (2017) Combination therapy for obesity. J Psychopharmacol 31:1503–1508. [DOI] [PubMed] [Google Scholar]
- Xu C, Ogburn ET, Guo Y, Desta Z. (2012) Effects of the CYP2B6*6 allele on catalytic properties and inhibition of CYP2B6 in vitro: implication for the mechanism of reduced efavirenz metabolism and other CYP2B6 substrates in vivo. Drug Metab Dispos 40:717–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Loboz KK, Gross AS, McLachlan AJ. (2007) Stereoselective analysis of hydroxybupropion and application to drug interaction studies. Chirality 19:163–170. [DOI] [PubMed] [Google Scholar]
- Zanger UM, Klein K. (2013) Pharmacogenetics of cytochrome P450 2B6 (CYP2B6): advances on polymorphisms, mechanisms, and clinical relevance. Front Genet 4:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Sridar C, Kenaan C, Amunugama H, Ballou DP, Hollenberg PF. (2011) Polymorphic variants of cytochrome P450 2B6 (CYP2B6.4-CYP2B6.9) exhibit altered rates of metabolism for bupropion and efavirenz: a charge-reversal mutation in the K139E variant (CYP2B6.8) impairs formation of a functional cytochrome p450-reductase complex. J Pharmacol Exp Ther 338:803–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Ingelman-Sundberg M, Lauschke VM. (2017) Worldwide distribution of cytochrome P450 alleles: a meta-analysis of population-scale sequencing projects. Clin Pharmacol Ther 102:688–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu AZ, Cox LS, Nollen N, Faseru B, Okuyemi KS, Ahluwalia JS, Benowitz NL, Tyndale RF. (2012) CYP2B6 and bupropion’s smoking-cessation pharmacology: the role of hydroxybupropion. Clin Pharmacol Ther 92:771–777. [DOI] [PMC free article] [PubMed] [Google Scholar]