Abstract
Bone loss in response to alcohol intake has previously been hypothesized to be mediated by excessive production of reactive oxygen species via NADPH oxidase (Nox) enzymes. Nox4 is one of several Nox enzymes expressed in bone. We investigated the role of Nox4 in the chondro-osteoblastic lineage of the long bones in mice during normal chow feeding and during chronic ethanol feeding for 90 days. We generated mice with a genotype (PrxCre +/− Nox4 fl/fl) allowing conditional knockout of Nox4 in the limb bud mesenchyme. Adult mice had 95% knockdown of Nox4 expression in the femoral shafts. For mice on regular chow, only whole-body Nox4 knockout mice had clearly increased cortical thickness and bone mineral density in the tibiae. When chronically fed a liquid diet with and without ethanol, conditional Nox4 knockout mice had slightly reduced dimensions of the cortical and trabecular regions of the tibiae (P < 0.1). The ethanol diet caused a significant reduction in cortical bone area and cortical thickness relative to a control diet without ethanol (P < 0.05). The ethanol diet further reduced gene expression of Frizzled related protein (Frzb), myosin heavy chain 3, and several genes encoding collagen and other major structural bone proteins (P < 0.05), whereas the Nox4 genotype had no effects on these genes. In conclusion, Nox4 expression from both mesenchymal and nonmesenchymal cell lineages appears to exert subtle effects on bone. However, chronic ethanol feeding reduces cortical bone mass and cortical gene expression of major structural bone proteins in a Nox4-independent manner.
SIGNIFICANCE STATEMENT
Excessive alcohol intake contributes to osteopenia and osteoporosis, with oxidative stress caused by the activity of NADPH oxidases hypothesized to be a mediator. We tested the role of NADPH oxidase (Nox) 4 in osteoblast precursors in the long bones of mice with a conditional Nox4 knockout model. We found that Nox4 exerted effects independent of alcohol intake, and ethanol effects on bone were Nox4-independent.
Introduction
Chronic alcohol consumption affects bone turnover and predisposes to the development of osteopenia and osteoporosis in humans (Luo et al., 2017; Watt et al., 2019). In rodent models, chronic intake of ethanol also leads to reduction of bone mass (Chen et al., 2011; Mercer et al., 2014; Watt et al., 2018). In osteoblasts, alcohol exposure is associated with increased parameters of oxidative stress [i.e., an excessive, deleterious production of reactive oxygen species (ROS)] (Chen et al., 2008). Dietary antioxidants prevent ethanol-mediated loss of bone mass in vivo (Chen et al., 2011; Alund et al., 2017). We therefore hypothesized that bone loss caused by alcohol intake is mediated by oxidative stress.
NADPH oxidases are enzymes that generate ROS in the form of superoxide and/or hydrogen peroxide. NADPH oxidase (Nox) 2 and Nox4 are both expressed in murine bone. The role of Nox2 and Nox4 in ethanol-mediated bone loss has previously been investigated using whole-body knockout mice. Knockout of neutrophil cytosolic factor 1 (p47phox), which is an essential cofactor for functional Nox2 activity, partly protected against bone loss but only blocked ethanol-induced increases in osteoclastogenesis and bone resorption (Mercer et al., 2014). Whole-body knockout of Nox4 in male mice actually increased trabecular bone loss during ethanol feeding, possibly by impairing normal osteogenic processes, such as mesenchymal stem cell proliferation (Watt et al., 2018). On the other hand, female mice of a different Nox4 knockout genotype maintained on a regular rodent diet have been reported to have increased bone mass and decreased osteoclastogenesis relative to wild-type animals (Goettsch et al., 2013). Whether these apparently disparate effects of knocking out Nox4 are due to different Nox4 knockout models, different dietary regimens, or sex and age differences is currently unknown. To get a better understanding of the role of Nox4 on bone homeostasis, we generated a genetic model (PrxCre +/− Nox4 fl/fl) in which knockdown of Nox4 occurs in cells of the chondroblastic/osteoblastic lineage in the long bones of mice. Both male and female mice were investigated after being maintained on regular mouse chow and after being exposed to 90 days of chronic ethanol feeding using Lieber-DeCarli liquid diets. The tibial bone microstructure and the femoral shaft transcriptome were analyzed.
Materials and Methods
Animals.
PrxCre mice with CRE recombinase under control of a rat Prx1-derived promoter/enhancer [B6.Cg-Tg(Prrx1-cre)1Cjt/J, Cat. 005584] were purchased from The Jackson Laboratory, Bar Harbor, ME. The genotype of hemizygous animals is termed PrxCre +/−. The Nox4 knockout model, in which exons 1 and 2 (relative to NM_0157605) are flanked by LoxP sites (Fig. 1A), was obtained under a Material Transfer Agreement with Drs. Ralf Brandes and Katrin Schröder, University of Frankfurt, Germany and has been previously described (Goettsch et al., 2013). The genotype of homozygous animals is termed Nox4 fl/fl. Experimental conditional knockout and control animals were the progeny of crossing PrxCre +/− Nox4 fl/fl males with Nox4 fl/fl females. Since the Prrx1 gene promoter/enhancer is expressed in the female germline (Logan et al., 2002), whole-body Nox4 knockout alleles, termed Nox4 0, were generated in breedings with PrxCre expressed in the female parent. Experimental whole-body knockout animals were the progeny of crossing Nox4 0/0 males with Nox4 0/0 females. Whole-body Nox4 knockout mice lacking exon 4 of the Nox4 gene and wild-type C57Bl/6J mice of genotypes Nox4 −/− and Nox4 +/+ were purchased from The Jackson Laboratory (Cat. 022996 and 000664, respectively). Genotyping primers used for distinguishing the Nox4 fl/fl allele and the Nox4 + wild-type allele were tttctaagtagccttggctggttgc and tgtcctccaatcatgaaagtg. The Nox4 0 allele was verified with genotyping primers tggtgcacaaccattctagg and tgtcctccaatcatgaaagtg. The presence of the CRE transgene was tested with genotyping primers acctgaagatgttcgcgattatct and accgtcagtacgtgagatatctt. The Institutional Animal Care and Use Committee of Louisiana State University Health Sciences Center approved all animal experiments in accordance with the Guide for the Care and Use of Laboratory Animals (U.S. National Institutes of Health).
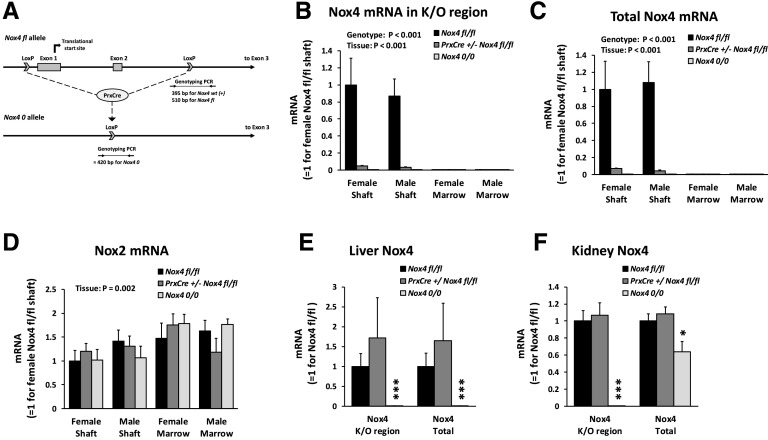
Fig. 1.
Conditional knockdown of Nox4 expression in long bones. (A) Model for PrxCre-mediated excision of exons 1 and 2 of the Nox4 gene. Indicated are genotyping amplicons for distinguishing the various Nox4 alleles. (B–D) The concentrations of Nox4 mRNA from the knockout (K/O) region, total Nox4 mRNA, and Nox2 relative to β-actin mRNA were determined in the femoral shaft and femoral bone marrow from 13-week-old mice by qRT-PCR. There were 5–11 mice per group. (E and F) The concentration of Nox4 mRNA was determined in liver and kidney aliquots from 32-week-old female mice. There were four mice per group. *P < 0.05; ***P < 0.001 vs. the Nox4 fl/fl genotype. PCR, polymerase chain reaction.
Ethanol Feeding.
Ethanol was provided as a variant of the liquid high-fat Lieber-DeCarli diet. The control diet contained 176.2 g/l DYET 710260 Lieber-DeCarli Ethanol Diet and 38.1 g/l DYET 402850 Maltose Dextrin for a protein:carbohydrate:fat caloric ratio of 24:30:47 and a concentration of 1000 kcal/l. The ethanol diets were made by replacing total food calories with ethanol while maintaining the 24:30:47 protein:carbohydrate:fat ratio and 1000 kcal/l. Mice were housed two to a cage. Ethanol diets were provided ad libitum, whereas volumes of the control diet were adjusted weekly to pair-fed mice to ensure the same caloric intake as in the ethanol-fed mice. Feeding was initiated with 13–15-week-old mice. The ethanol concentration was increased from 0% to 25% of dietary calories over 15 days, maintained at 25% for 2 months, and increased to 28% in the last 14 days before euthanasia. The mice were 27–28 weeks old when euthanized. The number of mice fed the ethanol diet were 10 male Nox4 fl/fl, 10 male PrxCre +/− Nox4 fl/fl, nine female Nox4 fl/fl, and nine female PrxCre +/− Nox4 fl/fl. The number of mice fed the control diet were nine male Nox4 fl/fl, nine male PrxCre +/− Nox4 fl/fl, 10 female Nox4 fl/fl, and six female PrxCre +/− Nox4 fl/fl.
Mouse Dissection.
Mice were euthanized by CO2. Body weights and liver weights were recorded. Blood was collected from the right ventricle of the heart. The blood was clotted for 20 minutes at room temperature followed by centrifugation at 2000 rcf for 30 minutes at 4°C. Serum was collected and stored at −80°C. The tibiae were fixed in 10% formalin. For RNA isolation, the femoral shafts and femoral bone marrow were isolated and stored at −80°C for later isolation of high-quality RNA as previously described (Pedersen et al., 2019). Liver aliquots were frozen and stored at −80°C. Additional liver aliquots and a whole kidney were incubated overnight in RNAlater with subsequent storage at −80°C for later isolation of RNA. RNA from the femoral marrow as well as from the kidney and liver was isolated by the TRI Reagent method. RNA from the femoral shaft was isolated by the TRI Reagent–RNeasy hybrid protocol previously described (Pedersen et al., 2019). For later protein isolation, whole femurs were frozen at −80°C. Liver triglyceride content was determined with a colorimetric triglyceride quantification assay kit ab65336 from Abcam (Cambridge, MA).
Serum Markers.
The serum contents of alanine transaminase (ALT), osteocalcin, and carboxy-terminal collagen crosslinks (CTXs) were determined with commercial kits (Cat. 700260; Cayman Chemical, Ann Arbor MI, Cat. 60-1305; Quidel, San Diego, CA, and Cat. LS-F21349; LSBio, Seattle, WA).
Quantitative Reverse Transcription Polymerase Chain Reaction.
Quantitative reverse transcription polymerase chain reaction (qRT-PCR) was conducted as previously described (Pedersen et al., 2019) with RNA samples diluted to a concentration of 5 ng/μl. Primers are listed in Table 1.
TABLE 1.
qRT-PCR primers
| Gene | Gene Symbol | Forward Primer | Reverse Primer |
|---|---|---|---|
| Nox4 knockout region | Nox4 | acc tct gcc tgc tca ttt gg | cct agg ccc aac att tgg tga |
| Total Nox4 | Nox4 | gga tca cag aag gtc cct agc | gtt gag ggc att cac caa gtg |
| Nox2 | Cybb | ggg atg aat ctc agg cca at | gcc gtc cat aca gag tct tc |
| β-Actin | Actb | aga tga ccc aga tca tgt ttg aga | cca gag gca tac agg gac agc |
| GAPDH | Gapdh | cat ctt cca gga gcg aga cc | cct tca agt ggg ccc cg |
| RANKL | Tnfsf11 | cag cat cgc tct gtt cct gta | ctg cgt ttt cat gga gtc tca |
| Cyp2S1 | Cyp2s1 | ttg gca tcc gtt tgc cct at | gag aac atc tcg tag gcc tgg |
| Osteocalcin | Bglap | agc ctt cat gtc caa gca gga g | gac tga ggc tcc aag gta gcg |
| Col2a1 | Col2a1 | ggc cag gat gcc cga aaa tta | cgc acc ctt ttc tcc ctt gt |
| Frzb | Frzb | aca act atg tca tcc ggg ct | tga cgg tgt ccc ttg gaa tg |
Microcomputed Tomography.
Trabecular and cortical bone morphology of formalin-fixed tibiae were scanned and their morphology assessed using a µCT 40 system from SCANCO Medical AG (Switzerland). All specimens were scanned using a voltage of 55 kVp, a current of 145 µA, an integration time of 200 milliseconds, with a voxel resolution of 6 µm for trabecular bone and 12 µm for cortical bone. Two trabecular regions interior to the cortex and primary spongiosa were analyzed: one segment located 0.4–0.7 mm distal to the proximal growth plate and a second segment located 0.3–1.2 mm distal to the end of the primary spongiosa. A single cortical region was analyzed and was located 4.8–4.2 mm proximal to the tibia-fibula junction. Thresholds to differentiate bone from the surrounding soft tissue were set to 486 and 703 mg hydroxyapatite/ccm for the trabecular and cortical regions, respectively.
RNA Sequencing.
RNA sequencing (RNA-Seq) service was purchased from Genewiz (South Plainfield, NJ). Samples were of male femoral shaft RNA with each sample being a pool of RNA from two mice. The samples were from mice of genotypes Nox4 fl/fl or PrxCre +/− Nox4 fl/fl fed the ethanol or control diets with three samples of each condition. Gene ontology analysis was done using the Gene Ontology Resource with protein annotation through evolutionary relationship overrepresentation test (The Gene Ontology Consortium, 2019).
Western Blotting.
Femur shaft proteins were isolated by a Minute Total Protein Extraction Kit for Bone Tissue (SA-02-BT; Invent Biotechnologies, Inc., Plymouth, MN) using the denaturing buffer from the kit supplemented with protease inhibitors. Protein-bound nitrotyrosine was detected with a rabbit anti-nitrotyrosine antibody (06-284; MilliporeSigma, St. Louis, MO) at a 1:1000 dilution followed by a horseradish peroxidase–conjugated anti-rabbit IgG (NA934VS; GE Healthcare) at a 1:20,000 dilution. Nitrotyrosine bovine serum albumin (89542; Cayman Chemical Company) was a positive control. Blots were developed with SuperSignal West Pico PLUS Chemiluminescent Substrate (34577; Thermo Scientific, Rockford, IL). Proteins on the blotting membrane were visualized by staining with Naphthol Blue Black (N3393; MilliporeSigma).
Statistical Analysis.
Two-way and three-way ANOVAs were carried out. For qRT-PCR data, the ANOVAs were conducted on the cycle threshold values. For RNA-Seq, a two-way ANOVA of log2-transformed normalized counts of each gene was conducted for assessment of main effects of genotype and diet as well as the interaction between genotype and diet. The test probabilities were not adjusted for the multiplicity of genes. Figures illustrate mean and S.E. of the parameters.
Results
The Genetic Model Ensures Conditional Nox4 Knockdown.
Nox4 fl/fl mice have exons 1 and 2 of the Nox4 gene flanked by LoxP sites (Fig. 1A). PrxCre is expressed in limb bud mesenchyme as well as part of the craniofacial mesenchyme (Logan et al., 2002), so PrxCre +/− Nox4 fl/fl mice should have knockdown of Nox4 in these tissues. With a primer set amplifying an amplicon stretching from exon 1 to exon 3 of Nox4 mRNA by qRT-PCR, Nox4 expression from the knockout region in the femoral shaft is indeed reduced by more than 95% and not detected in whole-body knockout mice of genotype Nox4 0/0 (Fig. 1B). There are described human Nox4 mRNA isoforms without exons 1 and 2, such as transcript variant 6 (National Center for Biotechnology Information Reference Sequence NM_001291929.1), in which translation occurs from alternate translation initiation sites. We therefore also designed a qRT-PCR assay for total Nox4 mRNA with an amplicon stretching from exon 3 to exon 5. This assay confirmed that total Nox4 mRNA in the femoral shaft is knocked down by more than 95% in PrxCre +/− Nox4 fl/fl mice (Fig. 1C). The expression of Nox4 in the femoral bone marrow was too low to quantify accurately. Nox2 mRNA expression in bone was not affected by the Nox4 genotype (Fig. 1D). As expected, mice of the PrxCre +/− Nox4 fl/fl genotype do not show reduced Nox4 expression in liver and kidney (Fig. 1, E and F). The kidney is the major organ expressing Nox4, at least in humans (Shiose et al., 2001). Although there is no detection of Nox4 mRNA from the knockout region in the kidney of Nox4 0/0 mice, total kidney Nox4 mRNA was only reduced by 36% (Fig. 1F), suggesting that the majority of kidney Nox4 mRNA lacks exons 1 and 2.
Whole-Body Knockout of Nox4 Increases Thickness and Density of Cortical Bone.
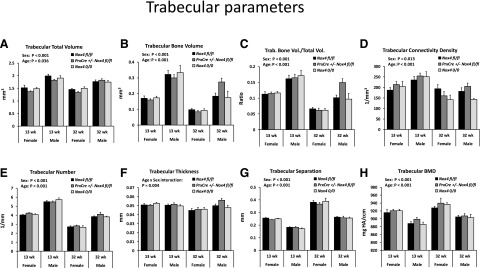
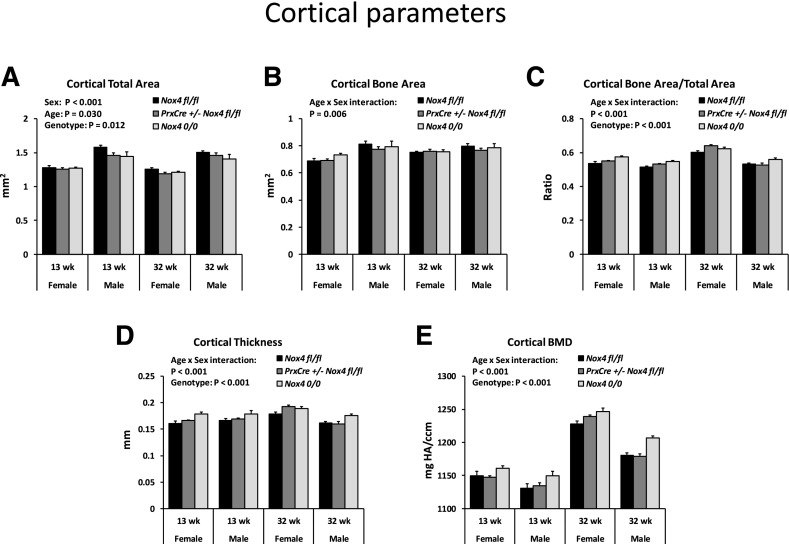
We assessed the effects of conditional and whole-body knockout of Nox4 on the bone microstructure of the tibiae for adult mice fed regular rodent chow (Figs. 2 and 3). We compared mice in early adulthood at 13 weeks of age and in later adulthood at 32 weeks of age. The mice continued gaining weight during this period, from an average of 20.8–27.6 g for females and from 27.6 to 37.5 g for males. At 32 weeks of age, PrxCre +/− Nox4 fl/fl mice had an average lower body weight than Nox4 fl/fl mice in females (26.0 g vs. 30.9 g; P = 0.07) and males (33.8 g vs. 40.0 g; P = 0.05). All trabecular bone parameters were affected by sex (Fig. 2, A–H) with less trabecular bone present in females than males. There was a loss of trabecular bone as the animals aged from 13 to 32 weeks but an increased trabecular bone mineral density during that same period (Fig. 2, A–H). There were no main effects of the Nox4 genotype on the trabecular bone parameters. Cortical bone parameters (Fig. 3) showed significant interactions between age and sex reflecting that females have a more pronounced increase in cortical bone parameters with age than males (Fig. 3, B–E). The Nox4 genotype had small but highly significant (P < 0.001) main effects on cortical bone with Nox4 0/0 mice showing higher levels of cortical bone area/total area, cortical thickness, and cortical bone mineral density than Nox4 fl/fl mice. For 32-week-old female mice, the highest mean levels of cortical bone area/total area and cortical thickness occurred for the PrxCre +/− Nox4 fl/fl genotype, suggesting that local conditional Nox4 knockdown may be enough to affect cortical bone. Otherwise, the Nox 0/0 mice exhibited the highest means, indicating stronger effects for whole-body knockdown than for conditional knockdown of Nox4 expression.
Fig. 2.
Sex and age affect trabecular bone of mice maintained on standard chow. Males and females of genotypes Nox4 fl/fl, PrxCre Nox4 fl/fl, and Nox4 0/0 at 13 and 32 weeks of age were analyzed. There were 5–12 mice per group. Three-way ANOVAs were conducted with sex, age, and genotype as the three factors. The panels indicate significant main effects and important interaction effects. (A–H) BMD, bone mineral density; Trab., trabecular; Vol., volume.
Fig. 3.
Age, sex, and the Nox4 genotype affect cortical bone of mice maintained on standard chow. Males and females of genotypes Nox4 fl/fl, PrxCre Nox4 fl/fl, and Nox4 0/0 at 13 and 32 weeks of age were analyzed. There were 5–12 mice per group. Three-way ANOVAs were conducted with sex, age, and genotype as the three factors. The panels indicate significant main effects and important interaction effects. (A–E) BMD, bone mineral density;
Chronic Alcohol Consumption Affects Body Weight, Liver Weight, and Bone Turnover.
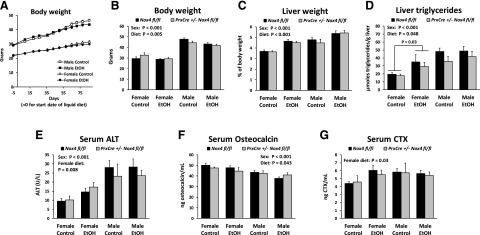
We wished to determine effects of conditional Nox4 knockdown on bone characteristics after chronic alcohol consumption for 3 months. In the past, our laboratory has typically provided alcohol to mice as liquid Lieber-DeCarli–based diets in which carbohydrates are exchanged for alcohol (Mercer et al., 2014; Watt et al., 2018). Unfortunately, the high content of fat and simple carbohydrates in control diets without alcohol tend to cause as much hepatic damage as the alcohol-containing diets, and it becomes difficult to separate alcohol from carbohydrate effects [discussed previously (Shearn et al., 2018)]. For the current study, we therefore decided to replace a fraction of the whole control diet with alcohol and thereby maintain the same protein:carbohydrate:fat ratio in alcohol and control diets. We achieved nearly identical average caloric intakes in alcohol-fed and control-fed animals (14.2 kcal/mouse per day in both alcohol-fed and control-fed males, 12.6 kcal/mouse per day for ethanol-fed females and 12.7 kcal/mouse per day for control-fed females).
Animals gained weight throughout the feeding period (Fig. 4A). The average weights of ethanol- and control-fed animals started to diverge after 2 months, resulting in significantly lower body weight for the ethanol diet when the mice were euthanized (Fig. 4B). The ethanol diet also led to a highly significant elevation of the liver weight relative to body weight (Fig. 4C). The content of hepatic triglycerides and serum ALT, which is a marker of hepatic injury, was increased in ethanol-fed females but not males (Fig. 4, D and E).
Fig. 4.
Chronic ethanol feeding for 3 months affects body composition and bone turnover. (A) The diagram illustrates the time course of the body weights for males and females fed either the ethanol diet or control diet for 3 months. (B–D) Body weight, relative liver weight, and the concentration of liver triglycerides were determined at the end of the chronic ethanol feeding experiment. (E–G) Serum concentrations of ALT, Osteocalcin and CTX were determined. Three-way ANOVAs were conducted with sex, diet, and genotype as the three factors. The panels indicate significant main effects. For ALT and CTX, a separate two-way ANOVA for just the data for females was calculated. EtOH, ethanol.
The concentration of serum osteocalcin, a marker of bone formation, was slightly lower in alcohol-fed animals (Fig. 4F). The bone resorption marker CTX was elevated in alcohol-fed females but not males (Fig. 4G). The effects observed on these parameters are all changes typically observed with chronic alcohol consumption. Although the sex also had clear effect on the parameters, the genotype did not affect any of them significantly.
Chronic Alcohol Consumption and Nox4 Depletion Have Independent Effects on Tibial Bone Structure.
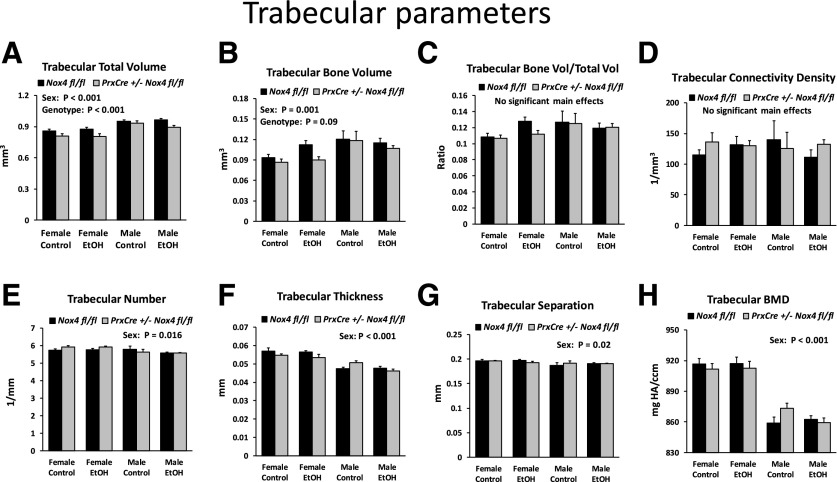
The bone microstructure of tibiae from the mice in the ethanol feeding experiment was analyzed by µCT. Trabecular microstructure below the primary spongiosa showed the same significant sex differences for trabecular total volume, trabecular bone volume, bone volume/total volume, trabecular volume, and trabecular separation as observed in mice maintained on the regular chow diet. However, the diet or genotype had no statistically significant main effects (unpublished data). Since the density of trabecular bone turned out to be relatively low in this region, we also analyzed a region closer to the proximal growth plate (Fig. 5, A–H). The ethanol diet still showed no significant effect. However, trabecular total volume was slightly but significantly (P < 0.001) reduced in the pair-fed PrxCre +/− Nox4 fl/fl mice on the control diet. There was also a trend (P = 0.09) for reduced trabecular bone volume in the pair-fed PrxCre +/− Nox4 fl/fl mice. Finally, there was a significant interaction between genotype and sex (P = 0.027) for trabecular thickness, reflecting a slight reduction in thickness in pair-fed PrxCre +/− Nox4 fl/fl females but not males.
Fig. 5.
Trabecular data for mice from the chronic ethanol feeding experiment. (A–H) Trabecular bone parameters for a region of the tibiae 0.4–0.7 mm distal to the proximal growth plate. Three-way ANOVAs were conducted with sex, diet, and genotype as the three factors. The panels indicate significant main effects. BMD, bone mineral density; EtOH, ethanol;
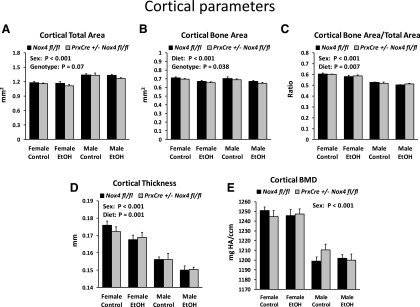
Cortical bone of the tibiae showed the same sex differences as those observed in 32-week-old mice on the regular chow diet (i.e., a smaller total cortical area but thicker and denser cortex in females) (Fig. 6, A, D, and E). The ethanol diet significantly reduced bone area, bone area/total area, and cortical thickness (Fig. 6, B–D). The PrxCre +/− Nox4 fl/fl genotype resulted in slightly but significantly reduced cortical bone area (Fig. 6B). There was also a trend (P = 0.07) for reduced total cortical area (Fig. 6A).
Fig. 6.
Cortical data for mice from the chronic ethanol feeding experiment. (A–E) Cortical bone parameters for the tibiae. Three-way ANOVAs were conducted with sex, diet, and genotype as the three factors. The panels indicate significant main effects. BMD, bone mineral density; EtOH, ethanol.
We conclude that the chronic ethanol consumption decreased cortical bone mass, whereas the conditional knockdown of Nox4 tended to decrease the dimensions of cortical bone and the trabecular region close to the growth plate.
Chronic Alcohol Consumption Reduces Gene Expression of Major Structural Bone Proteins.
We assessed the effects of chronic alcohol consumption and the conditional Nox4 knockdown on the femoral shaft transcriptome by RNA-Seq. Because of cost considerations, we conducted RNA-Seq for only mice of one sex and chose males since they gave the higher RNA yield. The samples for RNA-Seq were of good quality with an average RNA integrity number from 7.9 to 8.8. The expression of 18,044 genes was analyzed for animals of the genotypes PrxCre +/− Nox4 fl/fl and Nox4 fl/fl fed either the ethanol diet or the control diet (Supplemental File 1). The two most highly expressed genes in Nox4 fl/fl males fed the control diet were the major bone collagen genes Col1a1 and Col1a2. Surprisingly, the fourth most highly expressed gene was Hbb-bs encoding the s form of the beta polypeptide of adult hemoglobin. This expression must come from bone marrow erythrocyte precursors. Thus, although the RNA isolated from the femoral shafts is highly enriched in bone RNA, there is also a non-negligible contribution of RNA from bone marrow cells adhering to the femur shaft.
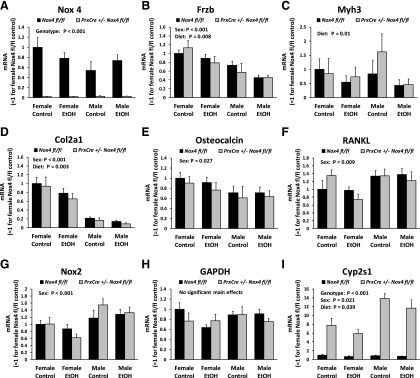
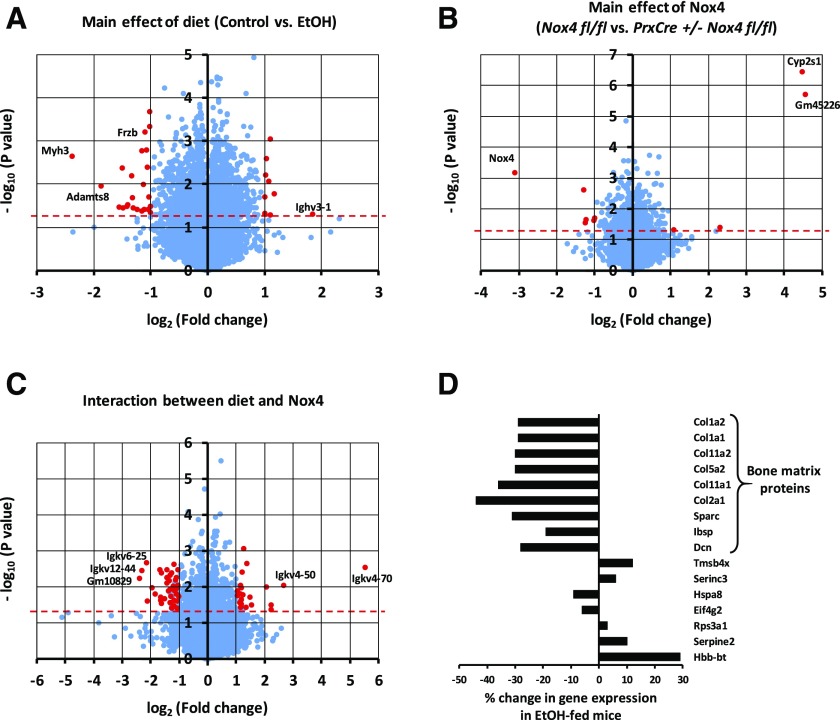
From two-way ANOVAs conducted for each gene, 2117 and 635 genes showed significance (P < 0.05) for the main effects of the diet and the genotype, respectively, whereas 945 genes had a significant interaction between diet and genotype. Expression of the housekeeping gene Gapdh was not significantly affected by the treatments as measured by RNA-Seq or qRT-PCR (Fig. 8H). Since 902 genes can be expected to show significance at P < 0.05 by chance when 18,044 genes are analyzed, the ethanol diet affects gene expression of more genes than could be expected by chance. For most of these genes, ethanol exerts a relatively modest, less than 2-fold, change in gene expression. Only 35 genes are regulated significantly (P < 0.05) and by at least 2-fold (Fig. 7A). Six of these are immunoglobulin genes, suggesting ethanol effects on bone marrow cells. The strongest downregulation by ethanol (5.2-fold, P = 0.002) was for the gene for myosin heavy chain 3 (Myh3), of which certain mutations can cause skeletal abnormalities (Carapito et al., 2016). Another gene known to have direct bone effects is Frzb-encoding Frizzled-related protein (Enomoto-Iwamoto et al., 2002). It was downregulated 2.1-fold (P = 0.0006) by the chronic ethanol feeding. Ethanol regulation of Myh3 and Frzb was confirmed by qRT-PCR (Figs. 8, B and C).
Fig. 8.
qRT-PCR gene expression analysis replicates findings from RNA-Seq. Gene expression was determined for RNA isolated from the femur shafts of mice from the chronic ethanol feeding experiments by qRT-PCR. The expression levels were normalized to the concentration of total RNA. The expression levels were determined for total Nox4 mRNA (A), Frzb (B), Myh3 (C), Col2a1 (D), osteocalcin (E), RANKL (F), Nox2 (G), GAPDH (H), and Cyp2s1 (I). 3-way ANOVAs were conducted with sex, diet, and genotype as the three factors. The panels indicate significant main effects. EtOH, ethanol.
Fig. 7.
RNA-Seq analysis of femoral shafts of mice from the chronic ethanol feeding experiment. RNA-Seq was conducted on femoral shaft RNA isolated from male mice. A two-way ANOVA was conducted for each gene with diet and genotype as the two factors. The test probabilities were not adjusted for the multiplicity of genes. Volcano plots depicting the test probability for each gene against the fold change in gene expression are shown for the main effect of diet (A), the main effect of genotype (B), and the interaction effect between diet and genotype (C). Points above the red horizontal dashed line represent genes with test probabilities P < 0.05. Red points indicate genes with at least a 2-fold change in gene expression and a test probability P < 0.05. (D) Among the 100 genes with the highest expression levels in Nox4 fl/fl mice on the control diet, the change in gene expression caused by ethanol is depicted for the 16 genes that show a significant (P < 0.05) main diet effect. EtOH, ethanol.
Nox4 itself is the most downregulated gene for the PrxCre +/− Nox4 fl/fl genotype (Fig. 7B). Two genes, Cyp2s1 and Gm45226, show strong upregulation for the PrxCre +/− Nox4 fl/fl genotype. The regulation of Nox4 and Cyp2s1 was confirmed by qRT-PCR (Fig. 8, A and I). The Cyp2s1 and Gm45336 genes are located on chromosome 7, as is Nox4. We speculated that the upregulation could be due to excision of cis-regulatory inhibitors for Cyp2s1 and Gm45336 expression located between the LoxP sites of the Nox4 gene rather than a decrease in Nox4 activity. This was tested by comparing Cyp2s1 induction in Nox4 0/0 mice to induction in the Nox4 knockout (Nox4 −/−) animals previously used (Watt et al., 2018), in which exon 4 required for enzymatic activities was deleted rather than exons 1 and 2. However, none of the whole-body knockout models showed Cyp2s1 induction (Supplemental Fig. 1A). We next speculated that the PrxCre +/− genotype itself could lead to Cyps1 induction. We determined Cyp2s1 expression in the femur shaft in a subset of 13-week-old male mice kept on normal chow. There was only Cyp2s1 induction in the conditional knockout genotype PrxCre +/− Nox4 fl/fl but not in the whole-body knockout Nox4 0/0 genotype (Supplemental Fig. 1B). This demonstrates that it is the presence of the PrxCre transgene construct rather than decreased Nox4 activity that increases Cyp2s1 expression. Apart from these genes mentioned above, the Nox4 genotype had little overall effect on gene expression. There were 84 genes showing significant (P < 0.05) interaction between genotype and diet with at least a 2-fold difference in the diet-mediated change for the Nox4 fl/fl and the PrxCre +/− Nox4 fl/fl genotype. No gene ontology classes were found to be significantly overrepresented. Presumably from bone marrow cells adhering to the shaft, there were five Igkv immunoglobulin genes with different variable regions appeared to have a statistically significant interaction effect (Fig. 7C). However, this number is not substantially different from the 3.8 (= 76 * 0.05) genes of the 76 Igkv genes quantified in the RNA-Seq data set that by chance can be expected to show statistical significance at P < 0.05.
Previous research from our group has indicated that expression of osteocalcin is downregulated, whereas expression of Nox2 and RANKL is upregulated by ethanol feeding in growing female rodents fed liquid diets from ages 6 to 14 weeks (Chen et al., 2011; Mercer et al., 2014; Watt et al., 2018). The RNA-Seq data set as well as qRT-PCR showed no significant main diet effect for these three genes in the current experimental design in male mice when ethanol exposure was begun during adulthood at age 13 weeks. However, sex did have a significant effect on these genes (Fig. 8, E–G).
Because most significant effects mediated by the ethanol diet are less than a 2-fold change, we speculated that such modest changes in highly expressed genes could be biologically relevant. Looking at the 100 most highly expressed genes for the Nox4 fl/f mice fed the control diet, there were 16 genes with a significant main diet effect (Fig. 7D) as compared with five genes expected by chance. Compared with the mouse reference genome, the list shows a 26× and >100× overrepresentation of genes involved in ossification and collagen fibril organization, respectively. It included all six collagen genes among the top 100 genes as well as three other genes encoding structural bone matrix proteins [osteonectin (Sparc), integrin binding sialoprotein (Ibsp), and decorin (Dcn))\]. All these bone matrix genes are downregulated 19%–44% by ethanol. We have previously observed downregulation of Col1a1 expression by ethanol in wild-type mice, and in this study, we further confirmed the ethanol-mediated downregulation of Col2a1 expression by qRT-PCR (Fig. 8D). We conclude that ethanol downregulates the expression of Myh3, Frzb, and several genes encoding major structural bone proteins in the femoral shaft.
Discussion
The role of Nox4 in bone morphology and bone turnover has previously been addressed by using whole-body Nox4 knockout mice. Female Nox4 knockout mice with the same genotype as the Nox4 0/0 animals showed increased trabecular width, trabecular thickness, and trabecular density in the distal femur (Goettsch et al., 2013). In the context of ethanol feeding, our laboratory found decreased bone mass in both trabecular and cortical bone of the tibiae in males of a different Nox4 knockout strain (Watt et al., 2018). Our µCT data of the tibiae in the current study suggest that these seemingly contrasting effects of Nox4 deletion on bone morphology may actually both occur. Thus, Nox4 0/0 mice kept on regular chow had increased cortical thickness and bone mineral density compared with Nox4 fl/fl mice. On the other hand, PrxCre +/− Nox4 fl/fl mice fed the liquid Lieber-DeCarli diets tended to have a reduced bone mass for both the trabecular area near the growth plate and for the cortical region as compared with those of the Nox4 fl/fl mice. The latter may represent a liquid diet/genotype interaction. For mice on the chow diet, the Nox4 0/0 seemed to have larger effects on cortical bone than the PrxCre +/− Nox4 fl/fl genotype (Fig. 2). This suggests that these morphologic changes are primarily due to knockout of Nox4 in cells that are not of the mesenchymal lineage, such as osteoclasts, as previously suggested (Goettsch et al., 2013). Females in general had smaller dimensions of tibial bone but higher bone mineral density. Aging from 13 to 32 weeks of age led to a decrease in trabecular bone and an increase in bone mineral density (Fig. 2).
We used an ethanol feeding model in which the ratios between protein, fat, and carbohydrate were the same in ethanol and control-fed mice. The ethanol intake had clear effects on body weight and relative liver weight (Fig. 3). There were also small but highly significant effects on cortical bone (Fig. 5). Surprisingly, there were none of the ethanol effects on the trabecular bone morphology that we found in our previous studies (Mercer et al., 2014; Watt et al., 2018). This discrepancy may be due to a higher ethanol concentration in the previous studies (30% vs. 25%–28%) or due to the exchange of only carbohydrate calories with ethanol in previous research. As in our previous study with whole-body Nox4 knockout animals (Watt et al., 2018), both ethanol intake and Nox4 depletion led to reduced cortical bone mass. Females responded more to the ethanol diet in terms of liver triglycerides, serum ALT, and serum CTX. Yet, for bone morphology, there were no pronounced sex differences in the response to ethanol.
Interpretation of gene expression data from the femoral shafts needs to take into account the part of the RNA that originates from bone marrow cells. Only if a gene shows clearly higher expression in RNA isolated from the femoral shaft than in RNA isolated from the marrow can the gene unequivocally be concluded to be expressed in the shaft. We have observed approximately 50-fold higher Frzb expression in the femoral shaft than in the marrow and essentially no Myh3 expression in the marrow (unpublished data). Thus, the regulation of Frzb and Myh3 depicted in Fig. 7B is a shaft phenomenon.
Apart from downregulation of Nox4 expression and the strong upregulation of Cyp2s1 and Gm45336 in PrxCre +/− Nox4 fl/fl animals that seems regulated by the PrxCre transgene construct itself, there were hardly any effects of the conditional Nox4 knockdown on gene expression in the femoral shaft. However, the RNA-Seq data set also revealed that the most highly expressed NADPH oxidase gene was Nox2 (Cybb), with approximately 650 times higher expression than Nox4. Nox4 deletion may thus only lead to a marginal decrease in NADPH oxidation. The lack of strong effects of Nox4 depletion on the shaft gene expression suggests that the observed bone morphology effects in the conditional Nox4 knockout mice are mediated by mesenchyme-derived cells acting earlier in development and/or residing outside of the bone matrix itself.
In contrast to previous studies, we did not observe ethanol-mediated effects on Nox2 or RANKL mRNA expression in the femoral shaft. RANKL expression in the femoral bone marrow was likewise unaffected by ethanol (unpublished data). A recently implemented technical improvement in our laboratory was the ability to consistently isolate RNA of high quality from mouse bones (Pedersen et al., 2019), thus reducing the risk of finding spurious changes due to variations in the RNA integrity. In combination with the higher number of animals in the ethanol feeding experiment compared with previous studies, the lack of induction of these two genes by the current ethanol diet is a robust finding. It is possible that RANKL, an inducer of bone resorption, is upregulated in other bones leading to the increased serum CTX observed in females. Lack of ethanol regulation of Nox2 and RANKL in the femur shaft may also reflect a different diet composition with a lower blood ethanol concentration resulting from a final dietary dose of 28% of calories compared with 30% of calories in other studies. In a 1-week ethanol binge-drinking model of mice, we recently did observe a robust induction of both femoral shaft and femoral bone marrow RANKL mRNA (unpublished data).
Since Nox4 had surprisingly little effect on gene expression in the femoral shaft, we also tested whether Nox4 and ethanol feeding affected ROS in the shaft. As an indicator of ROS, nitrotyrosine determined by immunohistochemistry was previously observed to become increased with chronic alcohol feeding of female mice, particularly in the bone marrow under the tibial growth plate (Alund et al., 2017). We quantified nitrotyrosine in femoral shaft proteins from male mice from the chronic ethanol feeding study using highly sensitive Western blots allowing detection of <2 ng of nitrotyrosine bovine serum albumin. The levels of nitrotyrosine were very low and not clearly affected by either ethanol feeding or conditional knockdown of Nox4 (Supplemental Fig. 2). It is possible that the lower concentration of dietary alcohol in the current study compared with previous studies results in lower blood ethanol concentrations and thus less generation of ROS. On the other hand, the data suggest that the observed ethanol effects on bone structure and femoral gene expression are independent of local ROS in the bone matrix, at least ROS leading to protein nitrotyrosinylation.
In the current study, we did observe the previously noticed downregulation of Col1a1 as well as of five other highly expressed collagen genes. Downregulation of collagen genes and other genes for structural bone proteins might be one mechanism whereby ethanol decreases bone quality. Whether the downregulation of Frzb and Myh3 expression has consequences for the bone morphology will have to be determined in future research. The Nox4 genotype had no significant effect on any of these genes.
We did not address the influence of sex on global gene expression in the femoral shaft. However, there was a significant main effect of sex for six of the eight genes we analyzed by qRT-PCR, suggesting that sex has a strong influence on the femoral gene expression.
In summary, we conclude that chronic ethanol feeding in mice decreases cortical bone mass and reduces expression of Frzb, Myh3, and genes for major structural bone proteins independent of Nox4 gene expression within the osteoblast lineage.
Abbreviations
- ALT
alanine transaminase
- Col
collagen
- CTX
carboxy-terminal collagen crosslink
- Myh3
myosin heavy chain 3
- µCT
microcomputed tomography
- Nox
NADPH oxidase
- qRT-PCR
quantitative reverse transcription polymerase chain reaction
- RANKL
receptor activator of nuclear factor κ-Β ligand
- RNA-Seq
RNA sequencing
- ROS
reactive oxygen species
Authorship Contributions
Participated in research design: Pedersen, Watt, Ronis.
Conducted experiments: Pedersen, Osborn, Robertson, Williams, Watt, Denys.
Contributed new reagents or analytic tools: Schröder.
Performed data analysis: Pedersen, Robertson, Ronis.
Wrote or contributed to the writing of the manuscript: All authors.
Footnotes
This work was supported by National Institutes of Health grants National Institute on Alcohol Abuse and Alcoholism R37 [AA018282] (to M.J.R.), National Institute on Alcohol Abuse and Alcoholism F32 [AA026480] (to J.W.), National Institute on Alcohol Abuse and Alcoholism [5T35AA021097 07] (to A.C.R.), and Postbaccalaureate Research Education Program [R25GM12189] (A.W.).
The authors declare that they have no conflicts of interest.
 This article has supplemental material available at jpet.aspetjournals.org.
This article has supplemental material available at jpet.aspetjournals.org.
References
- Alund AW, Mercer KE, Pulliam CF, Suva LJ, Chen J-R, Badger TM, Ronis MJJ. (2017) Partial protection by dietary antioxidants against ethanol-induced osteopenia and changes in bone morphology in female mice. Alcohol Clin Exp Res 41:46–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carapito R, Goldenberg A, Paul N, Pichot A, David A, Hamel A, Dumant-Forest C, Leroux J, Ory B, Isidor B, et al. (2016) Protein-altering MYH3 variants are associated with a spectrum of phenotypes extending to spondylocarpotarsal synostosis syndrome. Eur J Hum Genet 24:1746–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J-R, Lazarenko OP, Shankar K, Blackburn ML, Lumpkin CK, Badger TM, Ronis MJJ. (2011) Inhibition of NADPH oxidases prevents chronic ethanol-induced bone loss in female rats. J Pharmacol Exp Ther 336:734–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J-R, Shankar K, Nagarajan S, Badger TM, Ronis MJJ. (2008) Protective effects of estradiol on ethanol-induced bone loss involve inhibition of reactive oxygen species generation in osteoblasts and downstream activation of the extracellular signal-regulated kinase/signal transducer and activator of transcription 3/receptor activator of nuclear factor-kappaB ligand signaling cascade. J Pharmacol Exp Ther 324:50–59. [DOI] [PubMed] [Google Scholar]
- Enomoto-Iwamoto M, Kitagaki J, Koyama E, Tamamura Y, Wu C, Kanatani N, Koike T, Okada H, Komori T, Yoneda T, et al. (2002) The Wnt antagonist Frzb-1 regulates chondrocyte maturation and long bone development during limb skeletogenesis. Dev Biol 251:142–156. [DOI] [PubMed] [Google Scholar]
- Goettsch C, Babelova A, Trummer O, Erben RG, Rauner M, Rammelt S, Weissmann N, Weinberger V, Benkhoff S, Kampschulte M, et al. (2013) NADPH oxidase 4 limits bone mass by promoting osteoclastogenesis. J Clin Invest 123:4731–4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan M, Martin JF, Nagy A, Lobe C, Olson EN, Tabin CJ. (2002) Expression of Cre Recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis 33:77–80. [DOI] [PubMed] [Google Scholar]
- Luo Z, Liu Y, Liu Y, Chen H, Shi S, Liu Y. (2017) Cellular and molecular mechanisms of alcohol-induced osteopenia. Cell Mol Life Sci 74:4443–4453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer KE, Sims CR, Yang CS, Wynne RA, Moutos C, Hogue WR, Lumpkin CK, Suva LJ, Chen J-R, Badger TM, et al. (2014) Loss of functional NADPH oxidase 2 protects against alcohol-induced bone resorption in female p47phox-/- mice. Alcohol Clin Exp Res 38:672–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen KB, Williams A, Watt J, Ronis MJ. (2019) Improved method for isolating high-quality RNA from mouse bone with RNAlater at room temperature. Bone Rep 11:100211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearn CT, Pulliam CF, Pedersen K, Meredith K, Mercer KE, Saba LM, Orlicky DJ, Ronis MJ, Petersen DR. (2018) Knockout of the Gsta4 gene in male mice leads to an altered pattern of hepatic protein carbonylation and enhanced inflammation following chronic consumption of an ethanol diet. Alcohol Clin Exp Res 42:1192–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiose A, Kuroda J, Tsuruya K, Hirai M, Hirakata H, Naito S, Hattori M, Sakaki Y, Sumimoto H. (2001) A novel superoxide-producing NAD(P)H oxidase in kidney. J Biol Chem 276:1417–1423. [DOI] [PubMed] [Google Scholar]
- The Gene Ontology Consortium (2019) The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res 47 (D1):D330–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt J, Alund AW, Pulliam CF, Mercer KE, Suva LJ, Chen J-R, Ronis MJJ. (2018) NOX4 deletion in male mice exacerbates the effect of ethanol on trabecular bone and osteoblastogenesis. J Pharmacol Exp Ther 366:46–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt J, Schuon J, Davis J, Ferguson TF, Welsh DA, Molina PE, Ronis MJJ. (2019) Reduced serum osteocalcin in high-risk alcohol using people living with HIV does not correlate with systemic oxidative stress or inflammation: data from the New Orleans Alcohol Use in HIV Study. Alcohol Clin Exp Res 43:2374–2383. [DOI] [PMC free article] [PubMed] [Google Scholar]