Abstract
The pregnane X receptor (PXR), or nuclear receptor (NR) 1I2, is a ligand-activated NR superfamily member that is enriched in liver and intestine in mammals. Activation of PXR regulates the expression of genes encoding key proteins involved in drug metabolism, drug efflux, and drug transport. Recent mechanistic investigations reveal that post-translational modifications (PTMs), such as phosphorylation, play a critical role in modulating the bimodal function of PXR-mediated transrepression and transactivation of target gene transcription. Upon ligand binding, PXR undergoes a conformational change that promotes dissociation of histone deacetylase–containing multiprotein corepressor protein complexes while simultaneously favoring recruitment histone acetyl transferase–containing complexes. Here we describe a novel adenoviral vector used to deliver and recover recombinant human PXR protein from primary cultures of hepatocytes. Using liquid chromatography and tandem mass spectrometry we report here that PXR is phosphorylated at amino acid residues threonine 135 (T135) and serine 221 (S221). Biochemical analysis reveals that these two residues play an important regulatory role in the cycling of corepressor and coactivator multiprotein complexes. These data further our foundational knowledge regarding the specific role of PTMs, namely phosphorylation, in regulating the biology of PXR. Future efforts are focused on using the novel tools described here to identify additional PTMs and protein partners of PXR in primary cultures of hepatocytes, an important experimental model system.
SIGNIFICANCE STATEMENT
Pregnane X receptor (PXR), or nuclear receptor 1I2, is a key master regulator of drug-inducible CYP gene expression in liver and intestine in mammals. The novel biochemical tools described in this study demonstrate for the first time that in cultures of primary hepatocytes, human PXR is phosphorylated at amino acid residues threonine 135 (T135) and serine 221 (S221). Moreover, phosphorylation of PXR promotes the transrepression of its prototypical target gene CYP3A4 through modulating its interactions with coregulatory proteins.
Introduction
Ligand activation of pregnane X receptor (PXR) facilitates phase I (oxidative metabolism) (Kliewer et al., 1998; Lehmann et al., 1998), phase II (conjugative metabolism) (Sonoda et al., 2002; Chen et al., 2003; Xu et al., 2009; Staudinger et al., 2010), and phase III (transport and efflux) metabolic pathways (Geick et al., 2001; Staudinger et al., 2001; Guo et al., 2002; Kast et al., 2002). Two important PXR target genes whose gene products participate in first-pass metabolism include CYP3A4 (Kliewer et al., 1998; Lehmann et al., 1998) and multidrug resistance 1 (Owen et al., 2004; Burk et al., 2005; Cerveny et al., 2007; Satsu et al., 2008). The CYP3A4 enzyme is responsible for the oxidative metabolism of approximately 50% of all clinically prescribed medications (Guengerich, 1999), whereas P-glycoprotein 1 (P-gp1), the product of the multidrug resistance 1 gene, is an ATP-dependent broadly selective drug efflux pump (Hodges et al., 2011). The promiscuous nature of PXR activation is therefore mirrored in the broad selectivity that characterizes the respective diverse substrate specificities of both CYP3A4 and P-gp1. Through induction of CYP3A4 and P-gp1 activity, particularly in intestine, PXR activation plays a critical role in tumor-acquired drug resistance (Chen et al., 2007) and clinically important drug-drug, herb-drug, and food-drug interactions (Moore et al., 2000; Ding and Staudinger, 2005a; Staudinger et al., 2006).
Similar to other nuclear receptor (NR) superfamily members, PXR activation can serve as a negative regulator of the inflammatory response (Cheng et al., 2010, 2012; Kittayaruksakul et al., 2013; Sun et al., 2015). In fact, patients suffering from irritable bowel syndrome can be treated with a gut-specific PXR agonist, rifaximin, to reduce the inflammatory symptoms associated with this disease (Cheng et al., 2010). PXR activation is important in other human diseases, including inflammatory liver disease (Sun et al., 2015), inflammatory bowel disease (Shah et al., 2007; Cheng et al., 2010, 2012), intrahepatic cholestasis (Xie et al., 2001; Goodwin et al., 2003; Kakizaki et al., 2009, 2011; Geenes et al., 2011), Type 2 diabetes (Hukkanen et al., 2014), and numerous cancers (Kotiya et al., 2016; Wen et al., 2018). Therefore, understanding the precise molecular mechanisms that regulate PXR biology is critical in developing new therapeutic strategies to alter the course of these diseases.
The NR corepressor (NCoR) and the silencing mediator of retinoid and thyroid hormone receptor (SMRT) proteins comprise a family of transcription factor–scaffolding and transcriptional corepressor proteins involved in the formation of multiprotein histone deacetylase–containing protein complexes on the promoters of specific genes encoding inflammatory response proteins and drug-metabolizing enzymes (Perissi et al., 2008; Ghisletti et al., 2009). Several studies indicate a preferential association of PXR with SMRT (Johnson et al., 2006; Konno et al., 2009; Li et al., 2009), whereas other studies indicate that NCoR can associate with PXR in multiprotein corepressor complexes (Ding and Staudinger, 2005c; Lichti-Kaiser et al., 2009a,b). Somewhat paradoxically, the strength of PXR association with SMRT is increased by the presence of ligands (Konno et al., 2009; Li et al., 2009).
The major coactivator proteins implicated in PXR-mediated transactivation include ligand-dependent steroid receptor coactivator (SRC) family members SRC1 and glucocorticoid receptor-interacting protein 1 (GRIP1) as well as peroxisome proliferator-activated receptor-binding protein (PBP). Understanding the precise molecular mechanisms that govern circulation of coregulatory protein exchange in response to xenobiotic stimuli and pathogenic challenge is critical for understanding the biology of PXR-initiated drug-drug interaction, drug toxicity, and tumor-acquired drug resistance.
In the current study, we use a novel adenovirus-mediated recombinant PXR expression, purification, and recovery system used in conjunction with a liquid chromatography–tandem mass spectrometry (LC-MS/MS)-based proteomic strategy (Fig. 1A). Using this experimental approach, we identify specific sites of phosphorylation of recombinant human PXR recovered from primary cultures of hepatocytes. To examine the biologic outcome of the identified site-specific phosphorylation of the human PXR protein, we constructed phosphomimetic mutants T135D and S221D as well as the phosphodeficient mutant T135A. The mutational analysis presented here indicates that modification of PXR at these sites decreases strength of interaction with retinoid X receptor α (RXRα), SRC1, GRIP1, and PBP while simultaneously increasing the strength of PXR interaction with SMRT and NCoR. Taken together, these data indicate that phosphorylation of PXR plays a key role in modulating PXR biology through alterations in coregulatory protein-cofactor cycling.
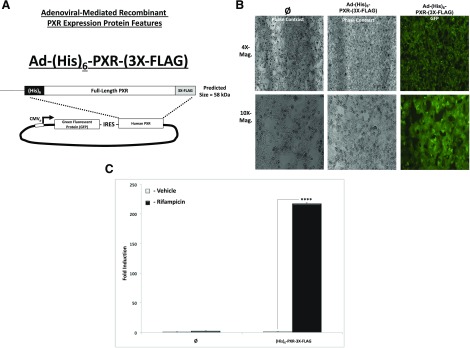
Fig. 1.
Adenoviral vector, cell morphology, GFP protein expression, and Ad-(His)6-PXR-(3X-FLAG) biologic function. Cells were observed and recorded 48 hours post-transduction with the novel adenoviral expression vector (A) using both bright-field and fluorescence microscopy (B) to visualize transduction efficiency and cell viability. (C). Hepa1-6 cells were transduced with blank virus (∅ − GFP) or Ad-(His)6-PXR-(3X-FLAG) for 48 hours (MOI = 10), and cells were treated with either vehicle (0.1% DMSO) or Rif (10 μM) for an additional 24 hours. rt-QPCR was performed, and Cyp3a11 gene expression levels were determined (n = 3). Data are expressed as fold induction ± S.D. compared with 18S ribosomal RNA expression. Asterisk (****) indicates statistically significant induction (P ≤ 0.001). Mag., magnification.
Materials and Methods
Chemicals and Plasmids.
Rifampicin (Rif) was purchased from Sigma-Aldrich, St. Louis, MO. All other reagents, including culture medium for primary hepatocytes and mammalian cell lines, were purchased from Fisher Scientific (St. Louis). Human PXR wild-type and mutant constructs were fused to the VP16 transcriptional activation domain by subcloning into the pVP16 expression vector (Clontech, Mountain View, CA) at EcoRI and BamHI restriction enzyme sites. The GAL4-SRC1, GAL4-GRIP1, GAL4-PBP, GAL4-NCoR, and GAL4-SMRT expression vectors were constructed as previously described (Synold et al., 2001; Ding and Staudinger, 2005a,c; Ding et al., 2006). To generate the GAL4-RXRα ligand-binding–domain (LBD) expression vector, the cDNA-encoding human RXRα was inserted into the GAL4 expression vector using BamHI and EcoRI restriction sites. The pFR-LUC GAL4-dependent luciferase reporter gene, which is responsive to GAL4-fusion proteins, is commercially available (BD Biosciences, San Jose, CA). The CYP3A4 reporter plasmid xenobiotic response element (XREM)-luciferase (Luc) was previously described (Goodwin et al., 1999). The construction of the adenoviral expression construct encoding human PXR [Ad-(His)6-PXR-3X-FLAG] was described previously (Cui et al., 2015). Briefly, this novel adenovirus expresses both GFP and an engineered form of human PXR, with both under the control of an internal ribosomal entry sequence (IRES) (Fig. 1A). This experimental approach allowed us to monitor transduction efficiency, toxicity, and cell morphology. As a negative control group, we used blank unprogrammed adenovirus (∅ = GFP only), whereas the experimental group received virus programmed with GFP plus recombinant human PXR, with both under the control of the IRES.
Site-Directed Mutagenesis.
The identified phosphorylation sites of PXR (T135, S221) were mutated to an aspartic acid (D) as a phosphomimetic mutation or to an alanine (A) as a phosphodeficient mutation. The mutant pCMV-FLAG-hPXR expression vectors were generated by site-directed mutagenesis with the use of the QuikChange Mutagenesis system (Stratagene, La Jolla, CA). Primer sequences used for site-directed mutagenesis are shown in Table 1.
TABLE 1.
Oligonucleotide primers used in site-directed mutagenesis
| Amino Acid | Oligos for Mutagenesis to A | Oligos for Mutagenesis to D | |
|---|---|---|---|
| T135 | Left primer | 5′ tgaacggacaggggctcagccactggg 3′ | 5′ aagtgaacggacaggggatcagccactgggagtg 3′ |
| Right primer | 5′ cccagtggctgagcccctgtccgttca 3′ | 5′ cactcccagtggctgatcccctgtccgttcactt 3′ | |
| S221 | Left primer | 5′ ttgtagttccagacagcgccatcctccccccg 3′ | 5′ tttgtagttccagacatcgccatcctccccccgc 3′ |
| Right primer | 5′ cggggggaggatggcgctgtctggaactacaa 3′ | 5′ gcggggggaggatggcgatgtctggaactacaaa 3′ | |
Hepa1-6 Cell Culturing and Viral Transduction.
The Hepa1-6 cell line was used because of its proven utility in studies of hepatic gene expression and liver biochemistry (Darlington et al., 1980) and because of the fact that this mouse hepatoma–derived cell line does not express detectable levels of PXR (Cui et al., 2015). Briefly, cells were cultured in Dulbecco’s modified Eagle’s medium, 10% FBS, and penicillin-streptomycin under standard conditions in 3-cM wells in six-well dishes. Cells at 80–90% confluence were transduced (MOI = 10) with either blank virus (∅ = GFP only) or virus that was programmed to express modified PXR. Cells were treated with either vehicle (0.1% DMSO) of Rif (10 μM) for 24 hours. Total RNA was isolated, and real-time quantitative polymerase chain reaction (rt-QPCR) was performed to determine expression levels of Cyp3a11 as described (Staudinger et al., 2003).
Mammalian Two-Hybrid System.
The mammalian two-hybrid system analysis was performed in 96-well plates using CV-1 cells plated at 7000 cells/well. Transfection mixtures contained the GAL4-dependent reporter gene pFR-LUC (20 ng), GAL4-cofactor NR-interaction domain (NRID) (20 ng), VP16-PXR (10 ng), pSV–β-galactosidase (40 ng) (Promega, Madison, Wi), and pBluescript (20 ng) per well. The next day transfected cells were drug-treated for 24 hours. All compounds were added to the culture medium as 1000X stocks dissolved in DMSO. Luciferase and β-galactosidase activities were determined using the Dual-Light Kit per the manufacturer’s instructions (Applied Biosystems, Foster City, CA).
Isolation and Culturing of Primary Hepatocytes.
Primary hepatocytes were isolated from Sprague-Dawley rats at the age of 10–12 weeks with a standard collagenase perfusion procedure as previously described (Staudinger et al., 2003). Potential sex differences were evaluated, and identical results were obtained from both male and female rats. The representative results were acquired from male rats.
Nuclei and Cytosol Fraction Separation.
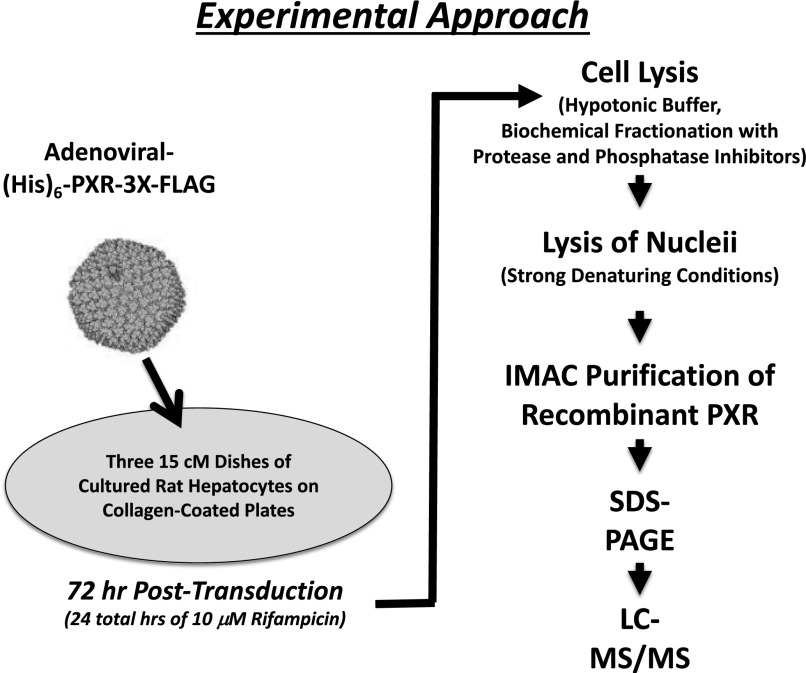
Primary rat hepatocytes were transduced with the Ad-(His)6-PXR-3X-FLAG recombinant virus (MOI = 10) and were cultured for an additional 48 hours post-transduction. Transduced hepatocytes were harvested in 1.5 ml PBS containing a cocktail of protease inhibitors (A32965; Fisher) and centrifuged at 240g for 10 minutes at 4°C. Cell pellets were lysed by adding approximately 5-pellet volumes of hypotonic buffer (10 mM Tris, pH 7.7; 1.5 mM MgCl2; and 1 mM dithiothreitol), and the cell lysates were centrifuged at 106g for 10 minutes at 4°C. The supernatant was saved and was labeled as the cytosol fraction, and the pellet was labeled as the nuclear fraction and was subsequently resuspended under strong denaturing conditions for isolation of human PXR protein for LC-MS/MS analysis using immobilized metal affinity chromatography (IMAC).
Cell-Based Cobalt-Bead Affinity PXR Pull-Down Assay.
Post-translationally modified PXR was purified and enriched from primary cultured rat hepatocytes as previously described (Staudinger et al., 2011). Briefly, 10-cM dishes containing primary cultures of hepatocytes or Hepa1-6 cells were harvested using 1 ml of a strong denaturing lysis buffer, 6 M Guanidine-Cl (pH 8), according to specified experimental treatment. The whole-cell lysates were then applied to 30 μl of TALON Metal Affinity Resin (ClonTech, Palo Alto, CA) cobalt beads and incubated on a rotor at room temperature for 2 hours with shaking. The gathered proteins were collected via centrifugation and washed twice with lysis buffer, three times with 8 M Urea buffer (pH 6.5), and once with 1× PBS. Proteins were removed from the beads using 30 μl of 2× Laemmli buffer and heated at 95°C for 10 minutes for subsequent Western blot or LC-MS/MS analysis.
Fluorescence Microscopy.
Primary cultured hepatocytes were transduced with blank virus (∅ = GFP only) or recombinant PXR that has GFP under the control of an internal ribosomal entry sequence as a marker of transduction efficiency. Twenty-four hours post-transduction, hepatocytes were washed once with 1× PBS. To visualize, hepatocytes were washed three times with 1x PBS and then maintained in Opti-MEM I Reduced Serum Media (Thermo Fisher) during fluorescence protein image analysis. Fluorescent proteins were imaged with a 30× phase-contrast objective, whereas cells were excited at 400 nm to visualize GFP as a qualitative marker of both cell morphology as well as viral transduction efficiency.
Western Blot Analysis.
Western blot analysis was performed as described previously (Xu et al., 2009). Antibodies used include the mouse monoclonal anti-PXR antibody (H-11; Santa Cruz, Dallas, TX), anti-GAL4 (DNA-binding domain), or VP16 monoclonal antibodies (Upstate Biotechnology, Lake Placid, NY) and an anti-actin monoclonal antibody (MP Biomedicals, Irvine, CA).
Liquid Chromatography–Tandem Mass Spectrometry Analysis.
The LC-MS/MS analysis for identification of PXR phosphorylation was performed as described previously (Cui et al., 2015).
Luciferase Reporter Gene Assay.
The XREM-LUC and GAL4-dependent luciferase reporter gene assays were performed as described previously (Ding and Staudinger, 2005c; Cui et al., 2016). Briefly, CV-1 cells were seeded at 1 × 104 cells per well (n = 8) in 96-well plate and transfected using LipofectAMINE 2000. Twenty-four hours post-transfection, cells were treated with either vehicle (0.1% DMSO) 10 μM Rif. The transfection efficiency was quantified according to the activity of β-galactosidase. The relative fold change in reporter gene luciferase units was normalized to β-galactosidase values to reflect transfection efficiency among all experimental groups, respectively.
Statistical Analysis.
Data presented are expressed as the mean ± S.D. Differences between treatment groups were evaluated by one-way ANOVA followed by Tukey’s multiple comparison test or by two-way ANOVA followed by Tukey’s multiple comparison test. Among experimental groups, *P < 0.05, **P < 0.01, and ****P < 0.001 compared with the control cell population (vehicle treatment). Tukey’s or Dunnett’s post hoc comparison test was used to determine statistical differences between groups, and values of P < 0.05 (*), P < 0.01 (**), and P < 0.001 (****) were considered statistically significant.
Results
Recombinant PXR Expression Level, Recovery, and Biologic Function.
To determine the extent to which hepatocyte transduction was robust and cultured cells were morphologically healthy, hepatocytes were visualized using phase-contrast microscopy 24 hours post-transduction. The LC-MS/MS experiments used three 15-cm collagen-coated tissue culture plates of primary rat hepatocytes with approximately 20 million live cells per dish at plating for both the control group (Ø = GFP − blank virus) and the experimental group transduced with Ad-(His)6-hPXR-(3X-FLAG). Figure 1A depicts the relationship between the GFP marker gene, the IRES, and the human PXR transgene delivered using viral transduction. Note that the construct delivers either GFP alone (Ø = GFP − blank virus) or GFP plus the Ad-(His)6-hPXR-(3X-FLAG) transgene. Cell morphology and GFP protein expression were observed and recorded 48 hours post-transduction using both phase-contrast and fluorescence microscopy to broadly visualize transduction efficiency and cell viability. There was no dramatic visual difference in cell viability between the experimental group and control group (Fig. 1B).
We next sought a suitable PXR-deficient model system to examine the transactivation capacity of our recombinant form of human PXR. The expression and activity level of PXR is undetectable in the murine Hepa1-6 hepatoma cell line, and hence the induction of PXR target genes is absent in this model system (unpublished data). The rt-QPCR analysis revealed that Rif-inducible Cyp3a11 gene expression was present only in the PXR viral-transduced experimental group (Fig. 1C). Identical results were obtained utilizing PXR knockout hepatocytes as an experimental model system (unpublished data). Collectively, the data in Figure 1 indicate that the recombinant form of (His)6-hPXR-(3X-FLAG) human PXR protein is biologically functional. Additionally, these data indicate that this novel viral vector can be delivered with high efficiency to both primary hepatocytes as well as to immortalized cell lines without producing marked visual evidence of toxicity in cell-based experimental model systems.
Identification of PXR Phosphorylation Sites Using LC-MS/MS in Primary Hepatocytes.
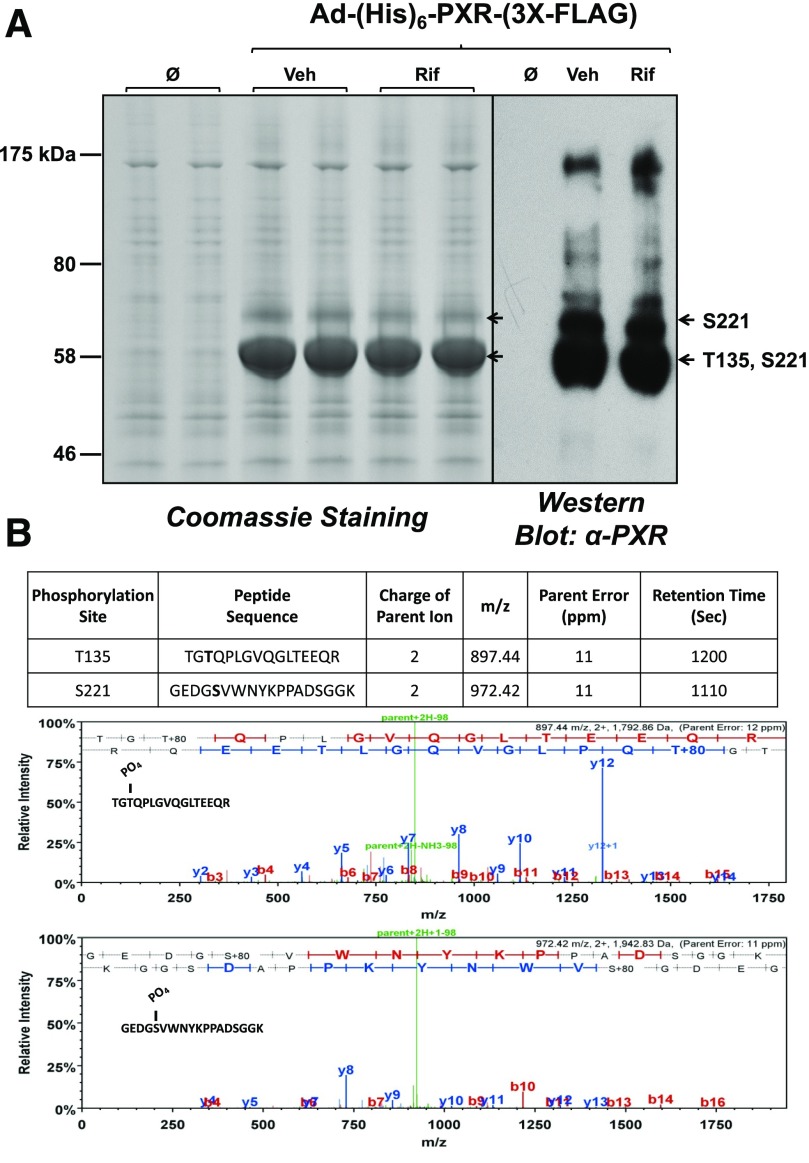
There are several critical limitations of phosphopeptide-mapping experiments using the LC-MS/MS approach. One limitation is the relatively low endogenous expression and low stoichiometry of the phosphomodified protein of interest. Moreover, the inherent LC-MS/MS machine-specific detection limits can also produce difficulties in detection of phosphomodified peptides. To overcome these obstacles, we used an adenoviral delivery method to achieve high-level expression and recovery of recombinant human 6X-histidine–tagged PXR [(His)6-PXR-(3X-FLAG)]. Because we were primarily interested in the PTMs of nuclear PXR protein, we isolated intact nuclei using a hypotonic buffer in the presence of high concentrations of both protease and phosphatase inhibitors. Our overall experimental approach is outlined in Figure 2. After biochemical fractionation of cytosolic and nuclear fractions, the recombinant nuclear PXR protein was isolated using IMAC under strong denaturing buffer conditions. The resulting protein isolates were resolved using 4%–15% gradient SDS-PAGE and subjected to Coomassie staining (Fig. 3A, left panel). This experimental approach achieved high-level PXR enrichment and a relatively low level of background or nonspecific protein binding. Western blot analysis using a monoclonal antibody against human PXR further confirmed the expression and efficient recovery of the recombinant and likely modified PXR protein (Fig. 3A, right panel). The total coverage achieved in our LC-MS/MS analysis of IMAC-enriched tryptic peptides derived from recombinant (His)6-PXR-(3X-FLAG) was approximately 60% (unpublished data). Using this approach, we identified two novel phosphorylation sites in the PXR protein to include threonine 135 (T135) and serine 221 (S221). The mass spectrometric parameters for the identification of PXR phosphorylation sites are listed in Figure 3B. It is worth noting here that we always detected T135 together with S221 phosphorylation in the primary faster-migrating PXR-immunoreactive band, whereas the slightly slower migrating PXR-immunoreactive band was reliable and repeatedly contained the S221 phosphorylation event alone (Fig. 3A).
Fig. 2.
Experimental strategy for adenoviral expression and recovery of human PXR protein from primary cultures of hepatocytes for LC-MS/MS analysis. Three 15-cm dishes of primary cultured rat hepatocytes were transduced with Ad-(His)6-PXR-3X-FLAG (MOI = 10) for 48 hours. Cells were treated with either vehicle (0.1% DMSO) or Rif (10 μM) for an additional 24 hours. Cells were lysed in a hypotonic buffer, and the nuclear fraction was isolated. Isolated nuclei were lysed under strong denaturing conditions, and nuclear PXR was isolated using IMAC as described in Materials and Methods.
Fig. 3.
SDS-PAGE analysis of isolated nuclear protein and LC-MS/MS analysis of the recombinant human PXR protein. (A) Isolated nuclei were lysed under strong denaturing conditions, and following IMAC protein isolates were resolved using 4%–20% SDS-PAGE. Coomassie staining was performed (left panel) and Western blot analysis was performed (right panel) using an anti-PXR monoclonal antibody (H-11; Sant Cruz). (B) LC-MS/MS analysis was performed on immunoreactive PXR protein as indicated (arrows). Two novel PXR phosphorylated peptides were detected as indicated in the figure (bottom panel). Threonine 135 (T135) phosphorylation on human PXR was identified according to the assignment of multiple product ions (b and y ions) in the MS2 scan of the precursor ion at m/z 897.44 to the PXR tryptic peptide sequence, with a mass addition of 80 at the threonine residue (phosphate group). Serine 221 (S221) phosphorylation on human PXR was identified according to the assignment of multiple product ions (b and y ions) in the MS2 scan of the precursor ion at m/z 972.42 to the PXR tryptic peptide sequence, with a mass addition of 80 at the serine residue. Veh, vehicle.
Constitutive Phosphorylation Inhibits the Transactivation Capacity of PXR.
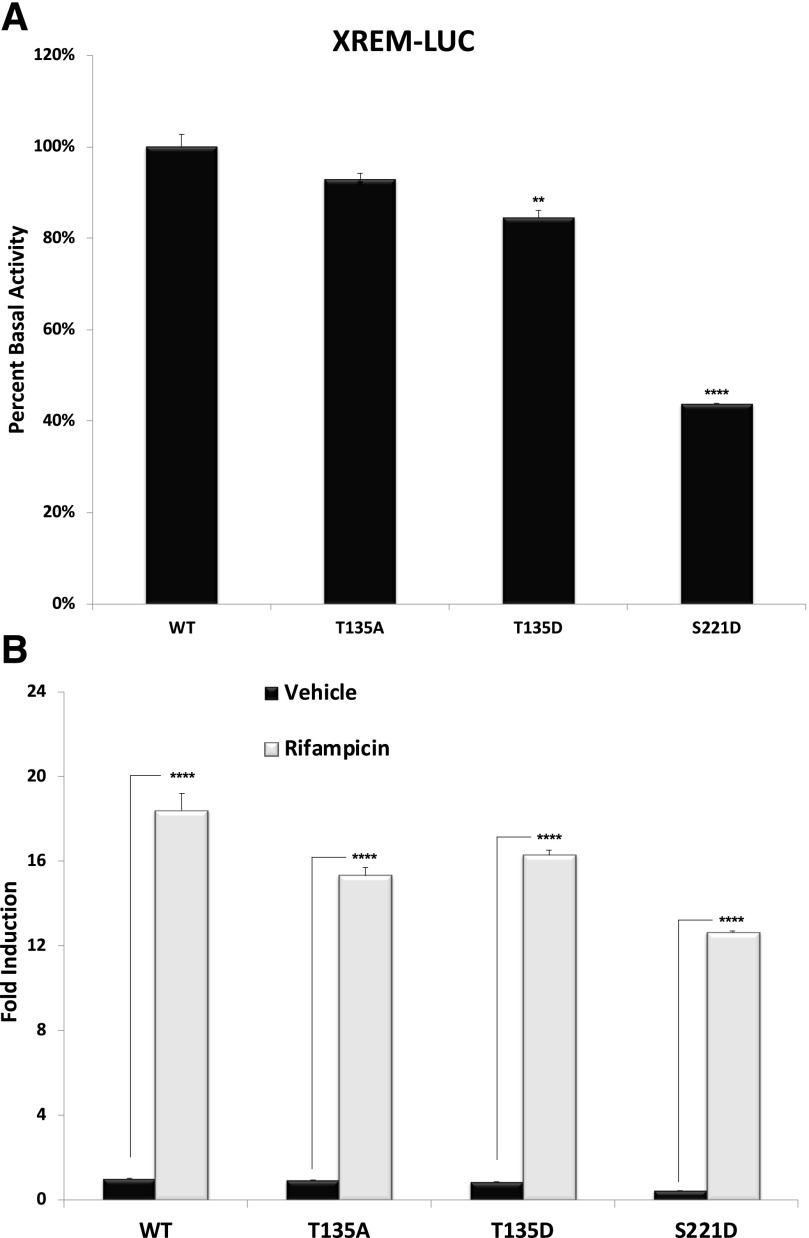
To examine the biologic significance of the two novel identified sites of PXR phosphorylation, we constructed two phosphomimetic mutant PXR proteins: T135D and S221D. We also constructed a T135 phosphodeficient mutant (T135A), and all three mutant proteins were inserted into a mammalian expression vector that uses the cytomegalovirus promoter to drive expression in transfected cell lines. Unfortunately, we were unable to generate the phosphodeficient mutation at S221 or the double phosphodeficient mutant because of a technical difficulty in modifying a guanine-enriched region on template DNA. We thus sought to determine the extent to which these three mutant PXR proteins are altered in their ability to support either basal or ligand-inducible transactivation capacity using the CYP3A4-derived XREM-LUC reporter gene assays in CV-1 cells.
Mammalian expression vectors encoding wild-type or the indicated mutant PXR proteins were cotransfected together with the XREM-LUC reporter gene into cultured CV-1 cells. Twenty-four hours post-transfection, cells were treated with either vehicle (0.1% DMSO) or Rif (10 μM) for an additional 24 hours. The relative basal expression and fold induction of reporter gene levels were determined. With respect to T135, both the phosphomimetic (T135D) and the phosphodeficient (T135A) mutations somewhat reduced basal expression of PXR-dependent reporter gene activity. In contrast, the S221D phosphomimetic mutation produced a very highly significant reduction in basal reporter gene activity (Fig. 4A), indicating a strong repression of basal PXR transactivation capacity in this cell line. The S221D phosphomimetic mutation also produced the most significant repression of Rif-inducible reporter gene activity (Fig. 4B). The results obtained here are in line with previous findings that PXR phosphorylation leads to a suppressive effect of its transactivation capacity (Ding and Staudinger, 2005a,c; Lichti-Kaiser et al., 2009a,b; Pondugula et al., 2009a,b).
Fig. 4.
Phospho-deficient and phosphomimetic mutations of human PXR in the XREM-LUC reporter gene assay. Cultures of CV-1 cells were transfected with plasmids encoding wild-type and mutant PXR proteins as indicated. (A) Twenty-four hours post-transfection, the basal levels of XREM-LUC reporter gene activity were measured. (B) After an additional 24 hours of treatment with 10 μM Rif, the inducible levels of XREM-LUC reporter gene activity were measured. All data (n = 4) are normalized to the vehicle-treated wild-type PXR group (n = 4) and, further, relative to β-galactosidase activity. Data are reported as the mean ± S.D., in which P < 0.05, P < 0.01 (**), and P < 0.001 (****). WT, wild type.
Validation of the Mammalian Two-Hybrid System Protein-Protein Interaction Panel.
PXR-mediated transcriptional events require the recruitment of a variety of positive and negative regulatory proteins in a highly coordinated and orchestrated manner. The precise molecular mechanisms that control the promoter-dependent circulation of coregulatory multiprotein complexes likely involve numerous finely tuned PTMs interacting with each other in a yet-to-be discovered manner. The extent to which any specific phosphorylation event contributes to the coregulatory protein exchange with respect to PXR is currently unknown or is not well-characterized.
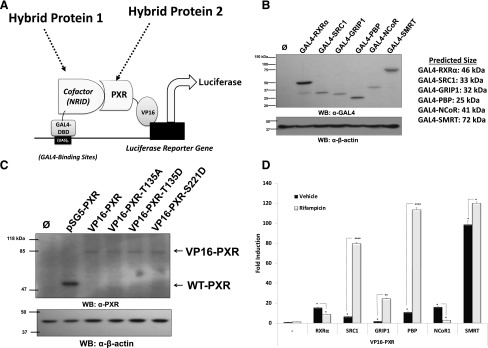
We therefore sought to construct a mammalian two-hybrid system to assess the effect of these specific phosphomimetic and phosphodeficient mutations in PXR to potentially examine any alterations in their coregulatory protein-protein interaction profile. To accomplish this, we fused the GAL4 DNA-binding domain (DBD) to the NRID, respectively, from cDNA sequences derived from RXRα, SRC1, GRIP1, PBP, NCoR, and SMRT proteins. Using this experimental approach, we have created a custom panel of known PXR-associated coregulatory proteins for experimental examination. We next fused cDNAs encoding wild-type and mutant full-length PXR proteins in-frame to the strong ligand-independent transcriptional activation domain derived from the herpes viral protein, VP16. All constructs described here were subsequently verified by DNA sequence analysis on both strands to ensure the integrity of their respective open-reading frames.
This two-hybrid strategy renders activation of the GAL4-dependent pFR-LUC reporter gene reliant upon protein-protein interaction between these two hybrid fusion proteins. In other words, the strength of protein-protein interaction is described by the amount of reporter gene activity in this model system. In addition to examining basal strength of protein-protein interaction, this experimental approach also allows for the direct examination of the effects of ligand on the strength of interaction between PXR and our custom panel of well-known coactivator and corepressor proteins.
Western blot analysis using a monoclonal anti-GAL4 antibody against cell extracts from CV-1 cells transfected indicated that all GAL4-cofactor NRID fusion proteins were expressed at their predicted mass, respectively (Fig. 5B). Western blot analysis with a monoclonal antibody directed against PXR indicates that all of the PXR proteins were expressed at equivalent levels at the appropriate size when expressed in CV-1 cells by transient transfection (Fig. 5C).
Fig. 5.
Validation of a panel of PXR-associated coregulatory proteins in the mammalian two-hybrid system. (A) The NRID derived from known PXR-associated coregulatory proteins, including its obligate dimeric partner RXRα; coactivator proteins SRC1, GRIP1, and PBP; and corepressor proteins SMRT and NCoR, were fused to the GAL4 DBD. (B) Expression levels of the GAL4-cofactor NRID were confirmed using anti-GAL4 antibody in Western blot analysis. (C) Expression levels of the wild-type and mutant PXR VP16-fusion proteins were confirmed using anti-PXR antibodies in Western blot analysis. All constructs described here were analyzed for their open-reading frame integrity using DNA sequence analysis, respectively. (D) The interaction between wild-type VP16-PXR and GAL4 cofactors was analyzed. Forty-eight hours post-transfection, reporter gene activity was determined in the presence and absence (0.1% DMSO) of 10 μM Rif for an additional 24 hours. The blank (−) represents CV-1 cells transfected with luciferase and β-gal reporter genes only treated with vehicle or Rif as indicated. WB- Western Blot, UAS)5- Upstream Activating Sequence. Data are normalized to β-galactosidase and are reported as the avg. ± S.D. (n = 4), in which P < 0.05 (*), P < 0.01 (**), and P < 0.001 (****).
Our initial efforts focused on an examination of the strength of interaction between GAL4-RXRα, GAL4-SRC1, GAL4-GRIP1, GAL4-PBP, GAL4-NCoR, and GAL4-SMRT and wild-type VP16-PXR. Twenty-four hours post-transfection, CV-1 cells were treated either with 0.1% DMSO (vehicle) or Rif (10 μM) for an additional 24 hours. The reporter gene activity was measured in cell extracts using standard methods. The wild-type VP16-PXR fusion protein interacted strongly with all tested GAL4-NRID coregulatory proteins in the absence of ligand (Fig. 5D). As expected, treatment with Rif increased the strength of interaction between PXR and coactivator proteins, including SRC1, GRIP1, and PBP. Also as expected, the addition of Rif resulted in decrease in the strength of PXR interaction and both RXRα and NCoR. In contrast, the strength of interaction between VP16-PXR and GAL4-SMRT (NRID) was increased by ligand activation. Although this is a somewhat counterintuitive observation, since SMRT is a corepressor protein, this result is consistent with other studies of PXR-SMRT protein-protein interaction (Johnson et al., 2006; Konno et al., 2009).
PXR-RXRα Heterodimerization.
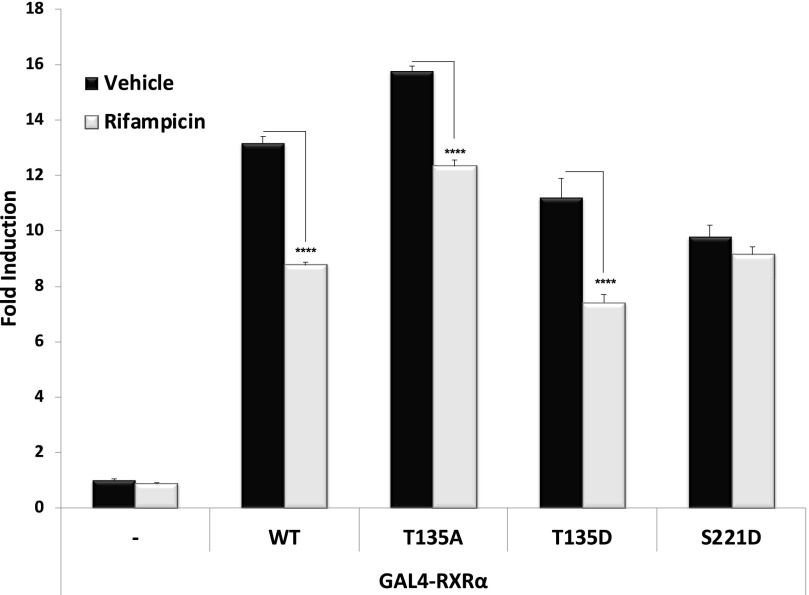
The heterodimerization of PXR and RXRα is the first wave of action for ligand-mediated transactivation of PXR target genes. A previous study in our laboratory demonstrated that phosphorylation at certain sites on PXR impairs the heterodimerization of PXR-RXRα (Lichti-Kaiser et al., 2009a). We therefore next sought to determine the extent to which the phosphomutant PXR proteins were able to undergo heterodimerization with RXRα. CV-1 cells were cotransfected with the expression vector encoding the GAL4-RXRα-LBD fusion protein together with the wild-type and mutant forms of VP16-PXR. Twenty-four hours post-transfection, cells were treated with 0.1% DMSO (Vehicle) or Rif (10 μM) for an additional 24 hours. The luciferase activity of the GAL4-dependent reporter gene was measured to evaluate the relative strength interaction between PXR and RXRα. Both T135D and S221D phosphomimetic mutations reduce the ability of PXR to dimerize with RXRα, whereas the T135A phosphodeficient mutation increased the strength of interaction between these two proteins (Fig. 6).
Fig. 6.
Strength of interaction between PXR and its obligate dimeric partner RXRα. Wild-type and indicated mutant PXR expression vectors were transfected along with GAL4-RXRα LBD and the XREM-LUC reporter gene. Twenty-four hours post-transfection, cells were treated with Rif (10 μM) or vehicle (0.1% DMSO) for an additional 24 hours. Forty-eight hours post-transfection, reporter gene activity was determined. Data are normalized to β-galactosidase and are reported as the avg. ± S.D. (n = 4), in which P < 0.001 (****). WT, wild type.
Strength of Association with Coactivator NRIDs.
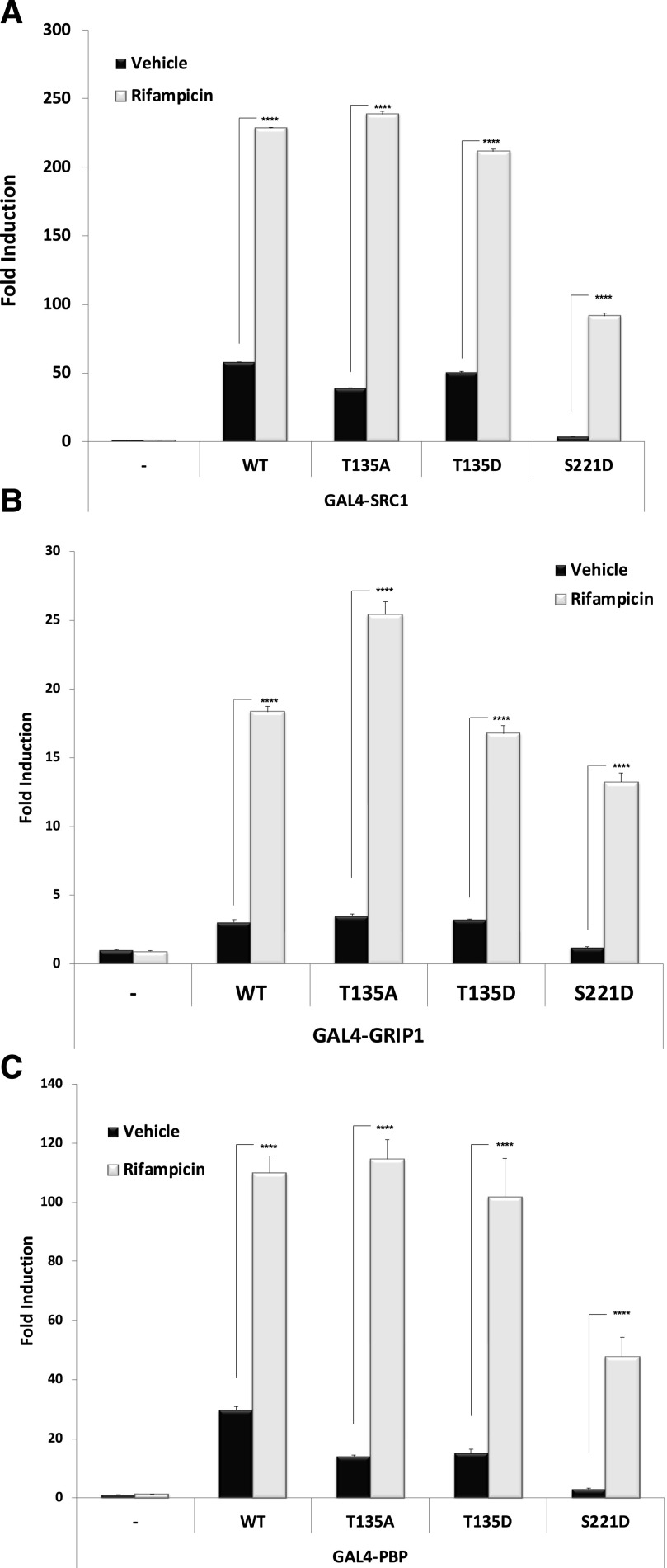
We next sought to determine the extent to which the wild-type and mutant PXR proteins are able to physically associate with NRIDs derived from well-known canonical PXR-associated coactivator proteins SRC1, GRIP1, and PBP. Both the phosphomimetic and phosphodeficient mutations at position T135 slightly but significantly reduced the strength of ligand-independent interaction with SRC1, whereas the ligand-dependent increase in interaction was largely preserved. In contrast, the S221D phosphomimetic mutant strongly inhibited the strength of both ligand-independent and ligand-dependent interaction with coactivator protein SRC1 (Fig. 7A). Similar to SRC1, the S221D strength of interaction with coactivator NRIDs derived from GRIP1 (Fig. 7B) and PBP was most significantly diminished (Fig. 7C). It is notable that the T135D phosphomimetic mutation did not have very significant impact on the strength of ligand-dependent association with SRC1-, GRIP1-, or PBP-NRIDs.
Fig. 7.
Strength of interaction between PXR and coactivator proteins. Wild-type and indicated mutant PXR expression vectors were transfected along with (A) GAL4-SRC1, (B) GAL4-GRIP1, and (C) GAL4-PBP together with the XREM-LUC reporter gene. Twenty-four hours post-transfection, cells were treated with Rif (10 μM) or vehicle (0.1% DMSO) for an additional 24 hours. Forty-eight hours post-transfection, reporter gene activity was determined. Data are normalized to β-galactosidase activity and are reported as the avg. ± S.D. (n = 4), in which P < 0.001 (****). WT, wild type.
Strength of Association with SMRT- and NCoR-NRIDs.
Together, the SMRT and NCoR proteins collectively form a small family of transcriptional corepressor proteins. Current dogma suggests that SMRT is the preferred partner for PXR (Johnson et al., 2006; Konno et al., 2009), whereas in some instances PXR interaction with NCoR is implicated in PXR-mediated transrepression phenomena (Ding and Staudinger, 2005a,b,c). In an effort to determine the extent to which the two identified phosphorylation events alter PXR association with the NRIDs derived from either SMRT or NCoR, respectively, we next examined their interaction using our mammalian two-hybrid analysis.
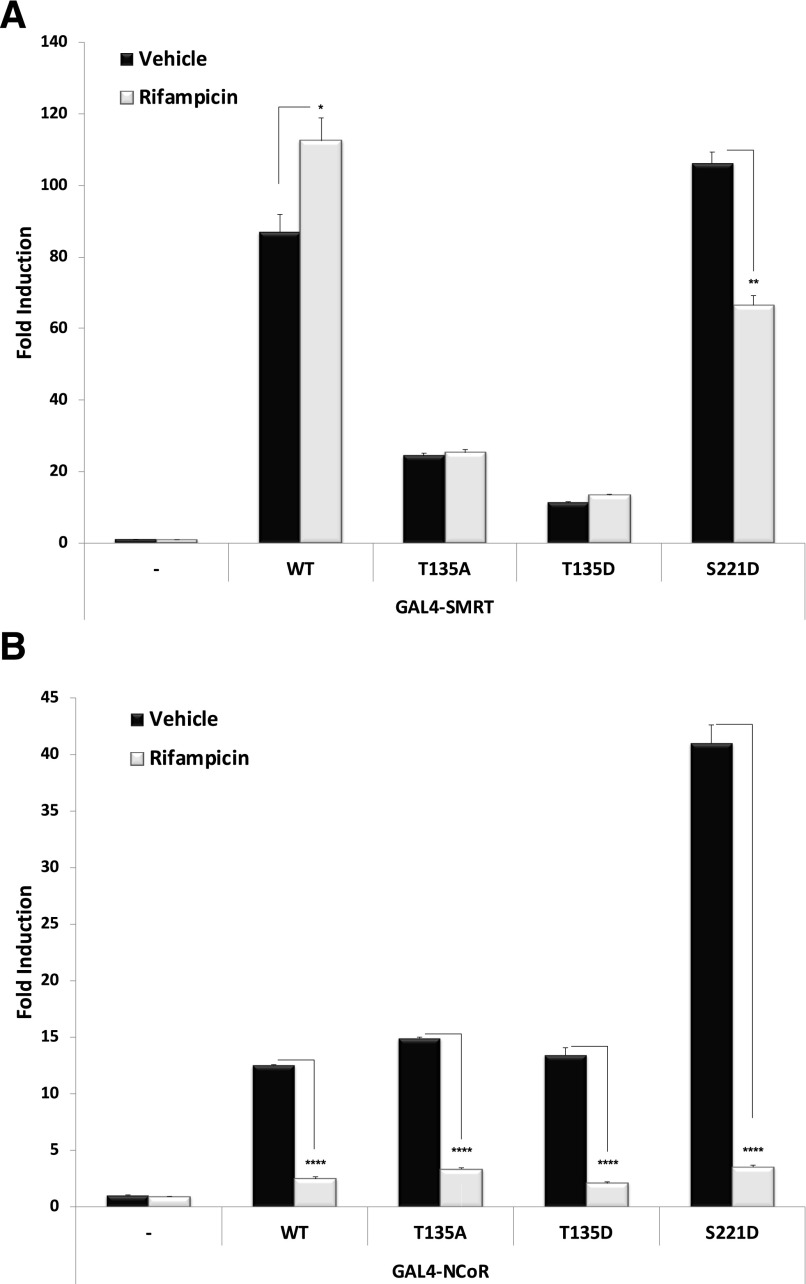
Both the phosphodeficient (T135A) and phosphomimetic (T135D) mutations produced highly significant reductions in the strength of interaction between PXR and the SMRT-NRID (Fig. 8A). Moreover, the addition of Rif to the system had no significant effect on the strength of association between either of the two T135 mutant proteins (T135A and T135D) and the SMRT-NRID. In contrast, the S221D phosphomimetic mutation modestly but significantly strengthened basal PXR-SMRT-NRID protein-protein interaction in the absence of ligand. Of note, the strength of association between S221D and SMRT was strongly reduced by the addition of ligand, an effect opposite to that of the ligand-dependent increase in the strength of interaction between the SMRT-NRID and wild-type PXR.
Fig. 8.
Strength of interaction between PXR and corepressor proteins. Wild-type and indicated mutant PXR expression vectors were transfected along with (A) GAL4-SMRT or (B) GAL4-NCoR together with the XREM-LUC reporter gene. Twenty-four hours post-transfection cells were treated with Rif (10 μM) or vehicle (0.1% DMSO) for an additional 24 hours. Forty-eight hours post-transfection, reporter gene activity was determined. Data are normalized to β-galactosidase activity and are reported as the avg. ± S.D. (n = 4), in which P < 0.05 (*), P < 0.01 (**), and P < 0.001 (****). WT, wild type.
Neither phosphodeficient (T135A) nor phosphomimetic (T135D) mutant PXR proteins differed significantly from ligand-independent and ligand-dependent interaction with interaction with the NCoR-NRID when compared with wild-type PXR (Fig. 8B). Remarkably, the phosphomimetic S221D mutation produced a very strong and highly significant increase in the strength of interaction with NCoR-NRID.
Discussion
NR family members are evolutionarily conserved transcription factors that share a similar modular structural arrangement comprised of an N-terminal DBD, a C-terminal LBD, and a flexible hinge region located in between the DBD and LBD regions of the protein. Gene repression by nuclear receptors is a spatial and temporal event that is precisely carried out by the circulation of specific coregulatory proteins to include NcoR- and SMRT-containing multiprotein complexes. Ligand-mediated gene activation is accompanied by dissociation of the corepressor multiprotein complex followed by the recruitment of coactivator multiprotein protein complexes, whereas termination of gene expression commands the proteasomal degradation of the NR coactivator machinery. This last event is accompanied by the physical interaction of the promoter with a newly synthesized NR-corepressor multiprotein complex. This process is therefore cyclic in nature and is thought to be required for maximal gene expression by NR superfamily members in general (Perissi and Rosenfeld, 2005). Post-translational modifications often regulate and fine tune the biology of NR proteins in a collaborative fashion.
PXR expression is highly enriched in liver and intestine in mammals (Kliewer et al., 1998; Lehmann et al., 1998). In comparison with many other NR superfamily members, the PXR LBD has a relatively large volume and an extremely flexible ligand-binding pocket (Watkins et al., 2003; Watkins et al., 2001). This large flexible LBD enables the accommodation of a very broad structural spectrum of compounds to trigger PXR transactivation capacity. Hence, PXR is activated by a myriad of steroid hormones, bile acids, environmental toxins, numerous prescription drugs, key constituents of herbal remedies, several pesticides, and additional xenobiotic chemicals (Staudinger et al., 2001, 2006; Moore et al., 2002, 2003; Lichti-Kaiser and Staudinger, 2008).
After ligand binding, the PXR protein undergoes a conformational change that favors dissociation of the corepressor complex followed closely by recruitment of histone acetyl transferase–containing coactivator multiprotein complexes to initiate full transcriptional activation of its target genes (Lichti-Kaiser and Staudinger, 2008; Lichti-Kaiser et al., 2009a,b). Multiple individual studies observe crosstalk between PTMs at the level of PXR, which regulates many aspects of its biology. Previous studies from our laboratory demonstrate that SUMOylation of PXR can suppress inflammatory gene activation and promote the recognition of ubiquitinated PXR destined for proteasomal degradation (Cui et al., 2015). PXR-mediated transcription activity involves a series of protein-protein interaction events to include the heterodimerization of PXR-RXRα and the circulation of coregulatory proteins. PXR heterodimerization with RXRα is considered the first step in response to ligand binding and is essential for subsequent gene activation. It is well-established that site-specific phosphorylation of PXR contributes to the modulation of its transactivation capacity through modulating a broad range of its biologic activities to include heterodimerization with RXRα, DNA-binding to its response element, and modulation of coregulatory protein-protein interactions (Ding and Staudinger, 2005a,b,c; Lichti-Kaiser et al., 2009a,b; Pondugula et al., 2009a,b; Doricakova et al., 2013). The ligand-dependent dissociation of PXR from RXRα deserves further scrutiny in light of the increasingly complex nature of the release of NR-associated corepressor protein complexes. We are mindful of the fact that most of the specific phosphorylation events targeting PXR examined to date have been determined to function as negative effectors of PXR-transactivation capacity.
Several groups have used in vitro analysis to detect putative sites of PXR phosphorylation that were subsequently analyzed in immortalized cell lines. A myriad of kinases have been reported to drive phosphorylation of PXR to include 70-kDa ribosomal S6 kinase (p70 S6K) (Lichti-Kaiser et al., 2009a; Pondugula et al., 2009a,b), cyclic AMP-dependent protein kinase A (Ding and Staudinger, 2005c; Lichti-Kaiser et al., 2009a,b), protein kinase C (Ding and Staudinger, 2005c; Lichti-Kaiser et al., 2009b), cyclin-dependent kinase 2 (Lin et al., 2008; Sugatani et al., 2010, 2012; Elias et al., 2014), and cyclin-dependent kinase 5 (Dong et al., 2010). All of these previous studies relied primarily upon systematic in vitro phosphorylation analysis followed by experiments in immortalized cell lines. It was our experimental goal in this study to try to directly examine rather than infer PTMs of recombinant PXR protein that was recovered from primary hepatocytes rather than using in vitro or immortalized cell-line model systems.
Utilizing LC-MS/MS–based proteomic approach is a trending and attractive experimental strategy to monitor PTMs. However, because of the low stoichiometry of phosphorylation in vivo and also because of the inherently low limit of detection inherent in the LC-MS/MS approach, we have developed an innovative adenoviral expression construct to achieve high expression and subsequent recovery of PXR protein from cultures of primary hepatocytes. This approach will allow us to continue to examine the myriad of PTMs on PXR moving forward in both cell-based and live animal models. The likely involvement of numerous PTMs in regulating the PXR transactivation and transrepression capacity has been highlighted in numerous recent studies (Lichti-Kaiser et al., 2009a,b; Hu et al., 2010; Biswas et al., 2011; Elias et al., 2014; Cui et al., 2015, 2016; Pasquel et al., 2016; Sugatani et al., 2016). The key PTMs described to date that regulate PXR biology include phosphorylation, acetylation, ubiquitination, and SUMOylation. Moreover, it is clear that the interactions between these PTMs can further modulate receptor biology, such as with phosphorylation-dependent SUMOylation and described interactions between acetylation and SUMOylation (Cui et al., 2015, 2016). Previous studies from our laboratory have demonstrated that increased cAMP-dependent protein kinase A and protein kinase C signaling inhibits PXR transactivation capacity through modulating PXR heterodimerization with RXRα and with coregulatory proteins (Ding and Staudinger, 2005c; Lichti-Kaiser et al., 2009a,b).
In the current study, we have identified two novel phosphorylation sites of PXR at T135 and S221. It is noteworthy that the T135 amino acid residue is located in the hinge region that connects DBD and LBD of PXR. The hinge region controls the particular orientation of the DBD and LBD upon ligand binding, which eventually leads to nuclear localization and activation of PXR (McKenna et al., 1999; Nagy and Schwabe, 2004). Several lines of evidence suggested that phosphorylation at the hinge region of many nuclear receptors contributes to their heterodimerization and transactivation capacity (Delmotte et al., 1999; Blanquart et al., 2004). Importantly, the S221 amino acid residue is located within the LBD of PXR. The LBD of NRs is considered to be a molecular switch that, when liganded, produces a conformational change that transforms PXR from a repressive protein into a transcriptional activator by recruiting different coregulatory protein complexes (Pawlak et al., 2012). However, unlike the LBD of a canonical NR superfamily member, which is composed of 10 α helices and three β strands and forms a three-layer sandwich; the LBD of PXR has two additional β1 and β1′ strands. These additional strands are thought to adapt to and therefore bind a broad spectrum of PXR ligands (Watkins et al., 2001; Xiao et al., 2002). This phenomenon has been referred to as directed promiscuity (Chrencik et al., 2005).
Our current investigation employing the phosphomimetic and phosphodeficient mutant PXR proteins indicates that PXR phosphorylation significantly impairs its transactivation capacity, particularly phosphorylation at S221. In our view, it is significant that the location of S221 is in the additional β1′ strands of PXR LBD. Therefore, phosphorylation at this particular serine may be involved in the feedback response to terminate the ligand-activated PXR transactivation capacity to subsequently promote PXR-mediated transrepression. In that respect, it is extremely interesting that the S221D mutation strongly enhanced the association of PXR with the NCoR corepressor protein, a well-characterized transrepressor of key inflammatory response genes. Therefore, PXR phosphorylation at S221 may be a key step in clinically observed drug-induced inhibition of the inflammatory response in liver and intestine (Shah et al., 2007; Cui et al., 2015; Sun et al., 2015; Staudinger, 2019). In contrast, the mutations at T135 significantly interrupt the PXR-SMRT protein-protein interaction while showing comparatively low or no significant effect upon PXR association with NCoR or coactivator proteins RXRα, SRC1, GRIP1, and PBP. Collectively, these findings suggest that the phosphorylation of PXR observed at position T135 likely involves alternative mechanisms or different biologic outcomes not involving coregulatory protein circulation. It is now well-accepted that phosphorylation suppresses PXR activity through interrupting its heterodimerization and interactions with coregulatory proteins. In response to specific cellular signals, PXR phosphorylation inhibits its transcription activity through the action of reducing the capability to form a heterodimer with RXRα, disrupting the interaction between PXR and coactivator proteins and likely through affecting the translocation of PXR into the nucleus.
Acknowledgments
The authors would like to thank Amanda Bearden for help in the preparation of this manuscript and Nadezhda Galeva and Todd D. Williams from the Mass Spectrometry Laboratory at the University of Kansas.
Abbreviations
- DBD
DNA-binding domain
- GRIP1
glucocorticoid receptor-interacting protein 1
- hPXR
human PXR
- IMAC
immobilized metal affinity chromatography
- IRES
internal ribosomal entry sequence
- LBD
ligand-binding domain
- LC-MS/MS
liquid chromatography–tandem mass spectrometry
- Luc
luciferase
- MOI
multiplicity of infection
- NCoR
NR corepressor 1
- NR
nuclear receptor
- NRID
NR-interaction domain
- PBP
peroxisome proliferator-activated receptor-binding protein
- P-gp1
P-glycoprotein 1
- PTM
post-translational modification
- PXR
pregnane X receptor
- SMRT
silencing mediator of retinoid and thyroid hormone receptor
- SRC
steroid receptor coactivator
- Rif
rifampicin
- rt-QPCR
real-time quantitative polymerase chain reaction
- RXRα
retinoid X receptor α
- XREM
xenobiotic response element
Authorship Contributions
Participated in research design: Cui, Agbas, Staudinger.
Conducted experiments: Cui, Shen, Agbas, Staudinger.
Performed data analysis: Cui, Tompkins. Cameron-Carter, Staudinger.
Wrote or contributed to the writing of the manuscript: Cui, Tompkins, Cameron-Carter, Staudinger.
Footnotes
This work was supported by the National Institutes of Health (NIH) National Institute of Digestive, Diabetic, and Kidney Diseases (NIDDK) [Grant R01DK090558] to J.S., the University of Kansas Strategic Initiative Grant [INS0073115] to J.S., and an award from the Centers of Biomedical Research Excellence (COBRE) and the NIH National Institute of General Medical Sciences [Grant P20 GM103549] to J.S. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
References
- Biswas A, Pasquel D, Tyagi RK, Mani S. (2011) Acetylation of pregnane X receptor protein determines selective function independent of ligand activation. Biochem Biophys Res Commun 406:371–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanquart C, Mansouri R, Paumelle R, Fruchart JC, Staels B, Glineur C. (2004) The protein kinase C signaling pathway regulates a molecular switch between transactivation and transrepression activity of the peroxisome proliferator-activated receptor alpha. Mol Endocrinol 18:1906–1918. [DOI] [PubMed] [Google Scholar]
- Burk O, Arnold KA, Nussler AK, Schaeffeler E, Efimova E, Avery BA, Avery MA, Fromm MF, Eichelbaum M. (2005) Antimalarial artemisinin drugs induce cytochrome P450 and MDR1 expression by activation of xenosensors pregnane X receptor and constitutive androstane receptor. Mol Pharmacol 67:1954–1965. [DOI] [PubMed] [Google Scholar]
- Cerveny L, Svecova L, Anzenbacherova E, Vrzal R, Staud F, Dvorak Z, Ulrichova J, Anzenbacher P, Pavek P. (2007) Valproic acid induces CYP3A4 and MDR1 gene expression by activation of constitutive androstane receptor and pregnane X receptor pathways. Drug Metab Dispos 35:1032–1041. [DOI] [PubMed] [Google Scholar]
- Chen C, Staudinger JL, Klaassen CD. (2003) Nuclear receptor, pregname X receptor, is required for induction of UDP-glucuronosyltranferases in mouse liver by pregnenolone-16 alpha-carbonitrile. Drug Metab Dispos 31:908–915. [DOI] [PubMed] [Google Scholar]
- Chen Y, Tang Y, Wang MT, Zeng S, Nie D. (2007) Human pregnane X receptor and resistance to chemotherapy in prostate cancer. Cancer Res 67:10361–10367. [DOI] [PubMed] [Google Scholar]
- Cheng J, Shah YM, Gonzalez FJ. (2012) Pregnane X receptor as a target for treatment of inflammatory bowel disorders. Trends Pharmacol Sci 33:323–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Shah YM, Ma X, Pang X, Tanaka T, Kodama T, Krausz KW, Gonzalez FJ. (2010) Therapeutic role of rifaximin in inflammatory bowel disease: clinical implication of human pregnane X receptor activation. J Pharmacol Exp Ther 335:32–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrencik JE, Orans J, Moore LB, Xue Y, Peng L, Collins JL, Wisely GB, Lambert MH, Kliewer SA, Redinbo MR. (2005) Structural disorder in the complex of human pregnane X receptor and the macrolide antibiotic rifampicin. Mol Endocrinol 19:1125–1134. [DOI] [PubMed] [Google Scholar]
- Cui W, Sun M, Galeva N, Williams TD, Azuma Y, Staudinger JL. (2015) SUMOylation and ubiquitylation circuitry controls pregnane X receptor biology in hepatocytes. Drug Metab Dispos 43:1316–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui W, Sun M, Zhang S, Shen X, Galeva N, Williams TD, Staudinger JL. (2016) A SUMO-acetyl switch in PXR biology. Biochim Biophys Acta 1859:1170–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darlington GJ, Bernhard HP, Miller RA, Ruddle FH. (1980) Expression of liver phenotypes in cultured mouse hepatoma cells. J Natl Cancer Inst 64:809–819. [PubMed] [Google Scholar]
- Delmotte MH, Tahayato A, Formstecher P, Lefebvre P. (1999) Serine 157, a retinoic acid receptor alpha residue phosphorylated by protein kinase C in vitro, is involved in RXR.RARalpha heterodimerization and transcriptional activity. J Biol Chem 274:38225–38231. [DOI] [PubMed] [Google Scholar]
- Ding X, Lichti K, Staudinger JL. (2006) The mycoestrogen zearalenone induces CYP3A through activation of the pregnane X receptor. Toxicol Sci 91:448–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X, Staudinger JL. (2005a) Induction of drug metabolism by forskolin: the role of the pregnane X receptor and the protein kinase a signal transduction pathway. J Pharmacol Exp Ther 312:849–856. [DOI] [PubMed] [Google Scholar]
- Ding X, Staudinger JL. (2005b) The ratio of constitutive androstane receptor to pregnane X receptor determines the activity of guggulsterone against the Cyp2b10 promoter. J Pharmacol Exp Ther 314:120–127. [DOI] [PubMed] [Google Scholar]
- Ding X, Staudinger JL. (2005c) Repression of PXR-mediated induction of hepatic CYP3A gene expression by protein kinase C. Biochem Pharmacol 69:867–873. [DOI] [PubMed] [Google Scholar]
- Dong H, Lin W, Wu J, Chen T. (2010) Flavonoids activate pregnane x receptor-mediated CYP3A4 gene expression by inhibiting cyclin-dependent kinases in HepG2 liver carcinoma cells. BMC Biochem 11:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doricakova A, Novotna A, Vrzal R, Pavek P, Dvorak Z. (2013) The role of residues T248, Y249 and T422 in the function of human pregnane X receptor. Arch Toxicol 87:291–301. [DOI] [PubMed] [Google Scholar]
- Elias A, High AA, Mishra A, Ong SS, Wu J, Peng J, Chen T. (2014) Identification and characterization of phosphorylation sites within the pregnane X receptor protein. Biochem Pharmacol 87:360–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geenes VL, Dixon PH, Chambers J, Raguz S, Marin JJ, Bhakoo KK, Williamson C. (2011) Characterisation of the nuclear receptors FXR, PXR and CAR in normal and cholestatic placenta. Placenta 32:535–537. [DOI] [PubMed] [Google Scholar]
- Geick A, Eichelbaum M, Burk O. (2001) Nuclear receptor response elements mediate induction of intestinal MDR1 by rifampin. J Biol Chem 276:14581–14587. [DOI] [PubMed] [Google Scholar]
- Ghisletti S, Huang W, Jepsen K, Benner C, Hardiman G, Rosenfeld MG, Glass CK. (2009) Cooperative NCoR/SMRT interactions establish a corepressor-based strategy for integration of inflammatory and anti-inflammatory signaling pathways. Genes Dev 23:681–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin B, Gauthier KC, Umetani M, Watson MA, Lochansky MI, Collins JL, Leitersdorf E, Mangelsdorf DJ, Kliewer SA, Repa JJ. (2003) Identification of bile acid precursors as endogenous ligands for the nuclear xenobiotic pregnane X receptor. Proc Natl Acad Sci USA 100:223–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin B, Hodgson E, Liddle C. (1999) The orphan human pregnane X receptor mediates the transcriptional activation of CYP3A4 by rifampicin through a distal enhancer module. Mol Pharmacol 56:1329–1339. [DOI] [PubMed] [Google Scholar]
- Guengerich FP. (1999) Cytochrome P-450 3A4: regulation and role in drug metabolism. Annu Rev Pharmacol Toxicol 39:1–17. [DOI] [PubMed] [Google Scholar]
- Guo GL, Staudinger J, Ogura K, Klaassen CD. (2002) Induction of rat organic anion transporting polypeptide 2 by pregnenolone-16alpha-carbonitrile is via interaction with pregnane X receptor. Mol Pharmacol 61:832–839. [DOI] [PubMed] [Google Scholar]
- Hodges LM, Markova SM, Chinn LW, Gow JM, Kroetz DL, Klein TE, Altman RB. (2011) Very important pharmacogene summary: ABCB1 (MDR1, P-glycoprotein). Pharmacogenet Genomics 21:152–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Xu C, Staudinger JL. (2010) Pregnane X receptor is SUMOylated to repress the inflammatory response. J Pharmacol Exp Ther 335:342–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hukkanen J, Hakkola J, Rysä J. (2014) Pregnane X receptor (PXR)--a contributor to the diabetes epidemic? Drug Metabol Drug Interact 29:3–15. [DOI] [PubMed] [Google Scholar]
- Johnson DR, Li CW, Chen LY, Ghosh JC, Chen JD. (2006) Regulation and binding of pregnane X receptor by nuclear receptor corepressor silencing mediator of retinoid and thyroid hormone receptors (SMRT). Mol Pharmacol 69:99–108. [DOI] [PubMed] [Google Scholar]
- Kakizaki S, Takizawa D, Tojima H, Horiguchi N, Yamazaki Y, Mori M. (2011) Nuclear receptors CAR and PXR; therapeutic targets for cholestatic liver disease. Front Biosci 16:2988–3005. [DOI] [PubMed] [Google Scholar]
- Kakizaki S, Takizawa D, Tojima H, Yamazaki Y, Mori M. (2009) Xenobiotic-sensing nuclear receptors CAR and PXR as drug targets in cholestatic liver disease. Curr Drug Targets 10:1156–1163. [DOI] [PubMed] [Google Scholar]
- Kast HR, Goodwin B, Tarr PT, Jones SA, Anisfeld AM, Stoltz CM, Tontonoz P, Kliewer S, Willson TM, Edwards PA. (2002) Regulation of multidrug resistance-associated protein 2 (ABCC2) by the nuclear receptors pregnane X receptor, farnesoid X-activated receptor, and constitutive androstane receptor. J Biol Chem 277:2908–2915. [DOI] [PubMed] [Google Scholar]
- Kittayaruksakul S, Zhao W, Xu M, Ren S, Lu J, Wang J, Downes M, Evans RM, Venkataramanan R, Chatsudthipong V, et al. (2013) Identification of three novel natural product compounds that activate PXR and CAR and inhibit inflammation. Pharm Res 30:2199–2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliewer SA, Moore JT, Wade L, Staudinger JL, Watson MA, Jones SA, McKee DD, Oliver BB, Willson TM, Zetterström RH, et al. (1998) An orphan nuclear receptor activated by pregnanes defines a novel steroid signaling pathway. Cell 92:73–82. [DOI] [PubMed] [Google Scholar]
- Konno Y, Kodama S, Moore R, Kamiya N, Negishi M. (2009) Nuclear xenobiotic receptor pregnane X receptor locks corepressor silencing mediator for retinoid and thyroid hormone receptors (SMRT) onto the CYP24A1 promoter to attenuate vitamin D3 activation. Mol Pharmacol 75:265–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotiya D, Jaiswal B, Ghose S, Kaul R, Datta K, Tyagi RK. (2016) Role of PXR in hepatic cancer: its influences on liver detoxification capacity and cancer progression. PLoS One 11:e0164087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann JM, McKee DD, Watson MA, Willson TM, Moore JT, Kliewer SA. (1998) The human orphan nuclear receptor PXR is activated by compounds that regulate CYP3A4 gene expression and cause drug interactions. J Clin Invest 102:1016–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CW, Dinh GK, Chen JD. (2009) Preferential physical and functional interaction of pregnane X receptor with the SMRTalpha isoform. Mol Pharmacol 75:363–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichti-Kaiser K, Brobst D, Xu C, Staudinger JL. (2009a) A systematic analysis of predicted phosphorylation sites within the human pregnane X receptor protein. J Pharmacol Exp Ther 331:65–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichti-Kaiser K, Staudinger JL. (2008) The traditional Chinese herbal remedy tian xian activates pregnane X receptor and induces CYP3A gene expression in hepatocytes. Drug Metab Dispos 36:1538–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichti-Kaiser K, Xu C, Staudinger JL. (2009b) Cyclic AMP-dependent protein kinase signaling modulates pregnane x receptor activity in a species-specific manner. J Biol Chem 284:6639–6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Wu J, Dong H, Bouck D, Zeng FY, Chen T. (2008) Cyclin-dependent kinase 2 negatively regulates human pregnane X receptor-mediated CYP3A4 gene expression in HepG2 liver carcinoma cells. J Biol Chem 283:30650–30657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna NJ, Lanz RB, O’Malley BW. (1999) Nuclear receptor coregulators: cellular and molecular biology. Endocr Rev 20:321–344. [DOI] [PubMed] [Google Scholar]
- Moore JT, Moore LB, Maglich JM, Kliewer SA. (2003) Functional and structural comparison of PXR and CAR. Biochim Biophys Acta 1619:235–238. [DOI] [PubMed] [Google Scholar]
- Moore LB, Goodwin B, Jones SA, Wisely GB, Serabjit-Singh CJ, Willson TM, Collins JL, Kliewer SA. (2000) St. John’s wort induces hepatic drug metabolism through activation of the pregnane X receptor. Proc Natl Acad Sci USA 97:7500–7502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore LB, Maglich JM, McKee DD, Wisely B, Willson TM, Kliewer SA, Lambert MH, Moore JT. (2002) Pregnane X receptor (PXR), constitutive androstane receptor (CAR), and benzoate X receptor (BXR) define three pharmacologically distinct classes of nuclear receptors. Mol Endocrinol 16:977–986. [DOI] [PubMed] [Google Scholar]
- Nagy L, Schwabe JW. (2004) Mechanism of the nuclear receptor molecular switch. Trends Biochem Sci 29:317–324. [DOI] [PubMed] [Google Scholar]
- Owen A, Chandler B, Back DJ, Khoo SH. (2004) Expression of pregnane-X-receptor transcript in peripheral blood mononuclear cells and correlation with MDR1 mRNA. Antivir Ther 9:819–821. [PubMed] [Google Scholar]
- Pasquel D, Doricakova A, Li H, Kortagere S, Krasowski MD, Biswas A, Walton WG, Redinbo MR, Dvorak Z, Mani S. (2016) Acetylation of lysine 109 modulates pregnane X receptor DNA binding and transcriptional activity. Biochim Biophys Acta 1859:1155–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlak M, Lefebvre P, Staels B. (2012) General molecular biology and architecture of nuclear receptors. Curr Top Med Chem 12:486–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perissi V, Rosenfeld MG. (2005) Controlling nuclear receptors: the circular logic of cofactor cycles. Nat Rev Mol Cell Biol 6:542–554. [DOI] [PubMed] [Google Scholar]
- Perissi V, Scafoglio C, Zhang J, Ohgi KA, Rose DW, Glass CK, Rosenfeld MG. (2008) TBL1 and TBLR1 phosphorylation on regulated gene promoters overcomes dual CtBP and NCoR/SMRT transcriptional repression checkpoints. Mol Cell 29:755–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pondugula SR, Brimer-Cline C, Wu J, Schuetz EG, Tyagi RK, Chen T. (2009a) A phosphomimetic mutation at threonine-57 abolishes transactivation activity and alters nuclear localization pattern of human pregnane x receptor. Drug Metab Dispos 37:719–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pondugula SR, Dong H, Chen T. (2009b) Phosphorylation and protein-protein interactions in PXR-mediated CYP3A repression. Expert Opin Drug Metab Toxicol 5:861–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satsu H, Hiura Y, Mochizuki K, Hamada M, Shimizu M. (2008) Activation of pregnane X receptor and induction of MDR1 by dietary phytochemicals. J Agric Food Chem 56:5366–5373. [DOI] [PubMed] [Google Scholar]
- Shah YM, Ma X, Morimura K, Kim I, Gonzalez FJ. (2007) Pregnane X receptor activation ameliorates DSS-induced inflammatory bowel disease via inhibition of NF-kappaB target gene expression. Am J Physiol Gastrointest Liver Physiol 292:G1114–G1122. [DOI] [PubMed] [Google Scholar]
- Sonoda J, Xie W, Rosenfeld JM, Barwick JL, Guzelian PS, Evans RM. (2002) Regulation of a xenobiotic sulfonation cascade by nuclear pregnane X receptor (PXR). Proc Natl Acad Sci USA 99:13801–13806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudinger JL. (2019) Clinical applications of small molecule inhibitors of Pregnane X receptor. Mol Cell Endocrinol 485:61–71. [DOI] [PubMed] [Google Scholar]
- Staudinger JL, Ding X, Lichti K. (2006) Pregnane X receptor and natural products: beyond drug-drug interactions. Expert Opin Drug Metab Toxicol 2:847–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudinger JL, Goodwin B, Jones SA, Hawkins-Brown D, MacKenzie KI, LaTour A, Liu Y, Klaassen CD, Brown KK, Reinhard J, et al. (2001) The nuclear receptor PXR is a lithocholic acid sensor that protects against liver toxicity. Proc Natl Acad Sci USA 98:3369–3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudinger JL, Madan A, Carol KM, Parkinson A. (2003) Regulation of drug transporter gene expression by nuclear receptors. Drug Metab Dispos 31:523–527. [DOI] [PubMed] [Google Scholar]
- Staudinger JL, Xu C, Biswas A, Mani S. (2011) Post-translational modification of pregnane x receptor. Pharmacol Res 64:4–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudinger JL, Xu C, Cui YJ, Klaassen CD. (2010) Nuclear receptor-mediated regulation of carboxylesterase expression and activity. Expert Opin Drug Metab Toxicol 6:261–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugatani J, Noguchi Y, Hattori Y, Yamaguchi M, Yamazaki Y, Ikari A. (2016) Threonine-408 regulates the stability of human pregnane X receptor through its phosphorylation and the CHIP/chaperone-autophagy pathway. Drug Metab Dispos 44:137–150. [DOI] [PubMed] [Google Scholar]
- Sugatani J, Osabe M, Kurosawa M, Kitamura N, Ikari A, Miwa M. (2010) Induction of UGT1A1 and CYP2B6 by an antimitogenic factor in HepG2 cells is mediated through suppression of cyclin-dependent kinase 2 activity: cell cycle-dependent expression. Drug Metab Dispos 38:177–186. [DOI] [PubMed] [Google Scholar]
- Sugatani J, Uchida T, Kurosawa M, Yamaguchi M, Yamazaki Y, Ikari A, Miwa M. (2012) Regulation of pregnane X receptor (PXR) function and UGT1A1 gene expression by posttranslational modification of PXR protein. Drug Metab Dispos 40:2031–2040. [DOI] [PubMed] [Google Scholar]
- Sun M, Cui W, Woody SK, Staudinger JL. (2015) Pregnane X receptor modulates the inflammatory response in primary cultures of hepatocytes. Drug Metab Dispos 43:335–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Synold TW, Dussault I, Forman BM. (2001) The orphan nuclear receptor SXR coordinately regulates drug metabolism and efflux. Nat Med 7:584–590. [DOI] [PubMed] [Google Scholar]
- Watkins RE, Maglich JM, Moore LB, Wisely GB, Noble SM, Davis-Searles PR, Lambert MH, Kliewer SA, Redinbo MR. (2003) 2.1 A crystal structure of human PXR in complex with the St. John’s wort compound hyperforin. Biochemistry 42:1430–1438. [DOI] [PubMed] [Google Scholar]
- Watkins RE, Wisely GB, Moore LB, Collins JL, Lambert MH, Williams SP, Willson TM, Kliewer SA, Redinbo MR. (2001) The human nuclear xenobiotic receptor PXR: structural determinants of directed promiscuity. Science 292:2329–2333. [DOI] [PubMed] [Google Scholar]
- Wen J, Lv Z, Ding H, Fang X, Sun M. (2018) Association between PXR polymorphisms and cancer risk: a systematic review and meta-analysis. Biosci Rep 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L, Cui X, Madison V, White RE, Cheng KC. (2002) Insights from a three-dimensional model into ligand binding to constitutive active receptor. Drug Metab Dispos 30:951–956. [DOI] [PubMed] [Google Scholar]
- Xie W, Radominska-Pandya A, Shi Y, Simon CM, Nelson MC, Ong ES, Waxman DJ, Evans RM. (2001) An essential role for nuclear receptors SXR/PXR in detoxification of cholestatic bile acids. Proc Natl Acad Sci USA 98:3375–3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Wang X, Staudinger JL. (2009) Regulation of tissue-specific carboxylesterase expression by pregnane x receptor and constitutive androstane receptor. Drug Metab Dispos 37:1539–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]