Graphical abstract
Keywords: Epitopes, Binding affinity, Antibody development, Host-pathogen interaction, Electrostatic interactions, Antigenic analysis, Computer simulation, pH effect, Coronavirus, SARS-CoV-2, ACE2, Protein-protein interaction
Highlights
-
•
The RBD of both SARS-CoV-1 and 2 spike proteins do bind to the cell receptor ACE2 with similar affinities although the latter is less specific.
-
•
The complexation of the RBD proteins with ACE2 can happen at both acid and neutral pH regimes.
-
•
The neutralizing monoclonal antibody CR3022 has shown the highest binding affinity under all studied conditions.
-
•
Amino acids substitutions for CR3022 are suggested to improve its binding affinity as a template to design optimized binders.
-
•
The interaction of the RBD proteins with both the studied Abs and ACE2 involves several common titratable amino acids.
Abstract
A new betacoronavirus named SARS-CoV-2 has emerged as a new threat to global health and economy. A promising target for both diagnosis and therapeutics treatments of the new disease named COVID-19 is the coronavirus (CoV) spike (S) glycoprotein. By constant-pH Monte Carlo simulations and the PROCEEDpKa method, we have mapped the electrostatic epitopes for four monoclonal antibodies and the angiotensin-converting enzyme 2 (ACE2) on both SARS-CoV-1 and the new SARS-CoV-2 S receptor binding domain (RBD) proteins. We also calculated free energy of interactions and shown that the S RBD proteins from both SARS viruses binds to ACE2 with similar affinities. However, the affinity between the S RBD protein from the new SARS-CoV-2 and ACE2 is higher than for any studied antibody previously found complexed with SARS-CoV-1. Based on physical chemical analysis and free energies estimates, we can shed some light on the involved molecular recognition processes, their clinical aspects, the implications for drug developments, and suggest structural modifications on the CR3022 antibody that would improve its binding affinities for SARS-CoV-2 and contribute to address the ongoing international health crisis.
1. Introduction
The SARS-CoV-2, virus recently found in Wuhan, Hubei province, China and officially named by the World Health Organization (WHO) (Wu et al., 2020), has already spread through China from all continents (more than 168 countries), with 1,133,758 confirmed cases globally and 62,784 deaths (data as reported by Central European Time 5 April 2020). Due to the pandemic, the disease is not only affecting the health services, but also the economy in a global scale, interfering in the widespread displacement of people, tourism, local and even international markets. Once China’s economy is a worldwide reference, its disruption leads to a global impact in the supply chains and the production itself (Wu et al., 2020; Ahani and Nilashi, 2020; Wu and McGoogan, 2020).
The Coronaviridae family, to which SARS-CoV-2 belongs, includes a large variability of viruses and became recognized in the spring of 2003, when a human coronavirus caused severe acute respiratory syndrome (SARS) (Weiss and Navas-Martin, 2005; Peiris, 2016; Maier et al., 2015; Zhou et al., 2020). Based on phylogenetic analysis, the SARS-CoV-2 is classified as a lineage B betacoronavirus8 and belongs to the same group as SARS-CoV-1 and HKU9-1, the bat coronavirus, demonstrating wide similarity with both genetically (Xu et al., 2020a) (96,2% of sequence identity with HKU9-1) (Ceraolo and Giorgi, 2020). The transmission was confirmed to be human-to-human once several medical care personnel and relatives got infected (Tian et al., 2020; Xu et al., 2020a), but it is believed that it all started with an animal host, may it be a bat or another intermediate host (Zhou et al., 2020; Xu et al., 2020a; Andersen et al., 2020; Shereen et al., 2020). Even though the virus usually does not cause severe damage to the body, as will be explained below, the major concern is its high infectivity and pathogenicity (Tian et al., 2020; Shereen et al., 2020; Li et al., 2020a).
The COVID-19 disease, caused by SARS-CoV-2 virus (Coronaviridae Study Group of the International Committee on Taxonomy of Viruses, 2020), generally causes mild upper respiratory tract infections, resulting in fever and cough, yet it can also affect the lower respiratory tract (Shereen et al., 2020; Huang et al., 2020; Lupia et al., 2020). SARS-CoV-2, on the other hand, usually remains asymptomatic in an early stage and then manifests itself with dyspnea, severe pneumonia and even death (Li et al., 2020b), with fatality rates of about 10 % (Chu et al., 2020). Although many groups of researchers are combining their efforts to solve the mysteries of the new virus, some issues are still uncertain. Examples of these queries are the virus’ incubation period, that may be longer than the 14 days scientists believed it to be previously, and the fatality rates for each age range (Lai et al., 2020).
Despite the fact that the virus’ molecular mechanism is partially unknown, the SARS-CoV-2 has proteins, such as the Spike (S) glycoprotein, that densely decorates the viral external surface and can potentially be a key target for the development of vaccines and therapeutic antibodies (Abs) (Tian et al., 2020; Wrapp et al., 2020; Du et al., 2014; Simmons et al., 2004). Due to the similarity of the receptor binding domain (RBD) in SARS-CoV-2 and SARS-CoV-1, the first strategy that has been used is to search for Abs that succeed interacting with both, once SARS-CoV-1 has been more widely studied. However, preliminary experimental studies have shown that many Abs that successfully interact with SARS-CoV-1 do not bind with SARS-CoV-2 (Tian et al., 2020).
The spike protein, which is responsible for the “corona” (Latin word for crown) appearance in all coronaviruses, is a type I glycoprotein that has an especial role in the interaction between the virus and the host cell. This protein attaches itself to specific cellular receptors and suffers a conformational change that enables the fusion of the virus and the cell (Weiss and Navas-Martin, 2005; Li, 2016; Walls et al., 2020). Studies have shown that the SARS-CoV-2’s S RBD protein interacts strongly with the Angiotensin-converting enzyme 2 (ACE2) (Xu et al., 2020a; Walls et al., 2020; Dimitrov, 2003). Therefore, aiming to develop better diagnosis tools, vaccines and therapeutic Abs, it was measured the competition of mAbs and the ACE2 for the binding to SARS-CoV-2 (named before 2019-nCoV (Tian et al., 2020; Jiang et al., 2020) RBD protein in order to enlighten the binding epitopes of these Abs (Tian et al., 2020; Lai et al., 2020).
The focus of this article is to initially reproduce the observations of previous laboratory experiments by a theoretical approach. Secondly, we aim to contribute with the understanding of the molecular mechanisms involved in the SARS viral infection, and finally to show how to apply this knowledge to design new functional molecules. To achieve these goals, it was tested by constant-pH simulation methods the complexation between the S RBD proteins of SARS-CoV-1 and SARS-CoV-2 with the fragments of the monoclonal Abs (mAbs) 80R, CR3022, m396 and F26G29, measuring their binding affinities and quantifying the titratable amino acids that are involved in these interactions. Thus, using a theoretical method recently proposed to identify “electrostatic epitopes” (Poveda-Cuevas et al., 2020), it is possible to identify the similarities and differences between these molecular complexes, and to map their origin and possible biological implications.
Another aspect discussed in this research is the interaction between the S RBD protein from these viruses and the ACE2 in order to discover if the S RBD protein binds to either of them with higher affinity, because, if so, an antibody (Ab) might have smaller chances of binding. All this information together provided important insights to design more specific and effective neutralizing Abs which is relevant for the future prevention and treatment of this now widespread illness that should be immediately controlled. At the end, a new designed mAb candidate is proposed based on our present in silico findings.
2. Theoretical methods
Computational virology is an emergent research field that takes advantage of the progress from molecular and structural biology, immunology, bioinformatics and related areas to foster the understanding of virus, their evolutionary dynamics in nature, infectivity, pathogenesis, cell/host-tropism, viral assembly and their molecular interactions in general (including how to predict epitopes, how to design specific neutralizing antibodies and basically any drug design & discovery related to viral infections) (Poveda-Cuevas et al., 2020; Ibrahim et al., 2018; Greber, 2019; Sharma et al., 2015; Sato et al., 2013; Backert and Kohlbacher, 2015; Chun et al., 2018; Viso et al., 2018). In particular, structural and interactive aspects can benefit from the solid foundations that computational molecular simulation methods such as Molecular Dynamics (MD) (Frenkel et al., 2001; Rapaport, 2004) and Monte Carlo (MC) (Frenkel et al., 2001; Binder, 1986) have achieved to probe the thermodynamic, dynamics and interactive properties of biomolecules in material science, food and pharma (see Refs. Barroso da Silva et al. (2020); van Gunsteren and Dolenc (2012)) for reviews). Here, we applied a fast constant-pH MC scheme (Teixeira et al., 2010; Barroso da Silva and MacKernan, 2017) for protein-protein studies (Barroso da Silva et al., 2016; Delboni and Barroso da Silva, 2016) to improve our understanding of the molecular interactions involving SARS-CoV-1 and 2 S RBD proteins and to identify key amino acids for the host-pathogen interactions.
2.1. Molecular systems and their structural modeling
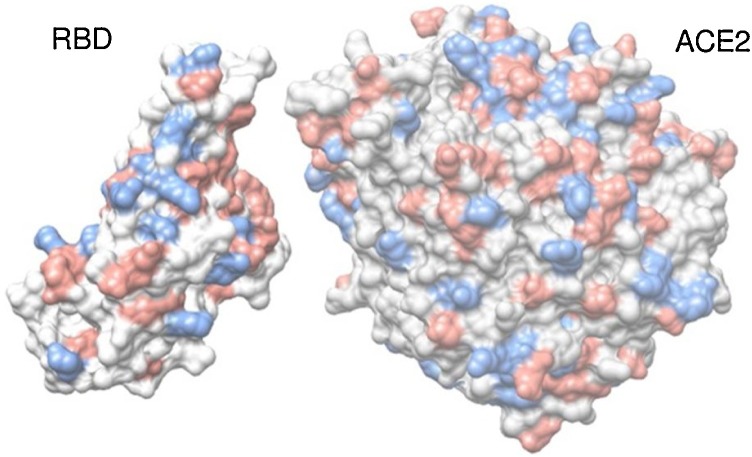
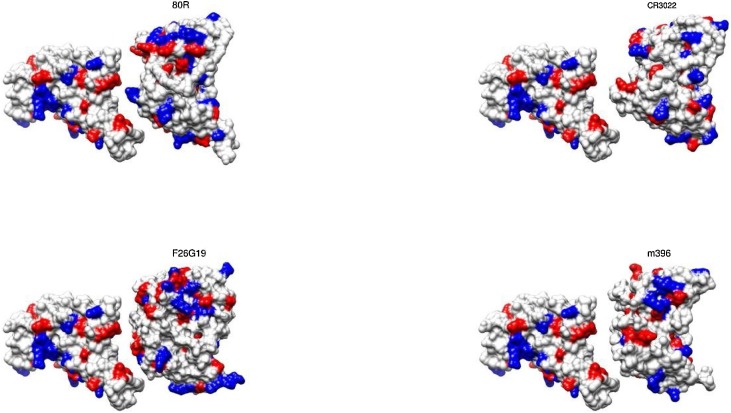
Several molecular systems were investigated in the present study employing the two SARS-CoV-1 and 2 S RBD proteins (see Fig. 1 ) with ACE2 and the fragments of the mAbs 80R, CR3022, m396, and F26G29. Typically, these fragments of mAbs are fusion proteins from variable regions of the heavy and light chains of immunoglobulins connected with a short linker peptide. Additional calculations were carried out for the SARS-CoV-2 S RBD with a new proposed mAb based on CR3022. For most of these macromolecules, three dimensional crystallographic structures are available at the RCSB Protein Data Bank (PDB) (Berman et al., 2000): (a) the SARS-CoV-1 S RBD protein was extracted from the PDB id 2AJF (chain E, resolution 2.9 Å, pH 7.5) where it was found complexed with ACE2 (chain A) − see Fig. 2 ; (b) the fragment of the Ab 80R was taken out from the PDB id 2GHW (chain B, resolution 2.3 Å, pH 4.6); (c) the anti-SARS-CoV-1 m396 Ab was extracted from the PDB id 2G75 (chains A and B, resolution 2.28 Å, pH 8.5) removing part of the chains to keep only the variable regions and the short linker peptide; d) F26G19 Fab was taken out from PDB id 3BGF (chains L and C, resolution 3.0 Å, pH 5.5), following the same procedure used for m396. Missing regions in these proteins were built up using the “UCSF Chimera 1.11.2” interface (Pettersen et al., 2004) of the program “Modeller” with default parameters (Eswar et al., 2006). Fig. 3 shows the molecular structures of the SARS-CoV-2 S RBD complexed with these fragments of the mAbs. The isolated fragments of these mAbs in their final three-dimensional structures as used in this work are given in Figure S2. All PDB files were edited before the calculations. Water molecules and hetero atoms were completely removed from all used files. The “UCSF Chimera 1.11.2” package (Pettersen et al., 2004) was employed for all molecular visualizations and representations too.
Fig. 1.
Crystal structure of the SARS-CoV-1 S RBD (PDB id 2AJF, chain E) and the modeled SARS-CoV-2 S RBD. See text for details regarding the modeling aspects. These macromolecules are shown, respectively, in blue and red in a ribbon representation. The RMSD between these structures is equal to 0.638Å.
Fig. 2.
Crystal structure of SARS-CoV-1 S RBD complexed with ACE2 (PDB id 2AJF). Only standard amino acids of chain A (ACE2) and E (SARS-CoV-1 S RBD) are shown in a molecular representation using spheres for its atoms. Atoms are colored accordingly to their amino acids physical chemical properties: red for acid amino acids, blue for base amino acids and gray for non-titrating amino acids. For a better visualization of the interface, the ACE2 structure was translated ∼12 Å.
Fig. 3.
Molecular structures of the SARS-CoV-2 S RBD complexed with fragments of the investigated monoclonal antibodies. Only standard amino acids are shown in a molecular representation using its surface. Atoms are colored accordingly to their amino acids physical chemical properties: red for acid amino acids, blue for base amino acids and gray for non-titrating amino acids. For a better visualization of the epitope-paratope interface, the structures of the Abs were translated ∼12 Å. The RBD of SARS-CoV-2 S protein is given by the modeled generated at the SWISS-MODEL workspace (YP_009724390.1). The fragments of the mAbs 80R (PDB id 2GHW), F26G19 (PDB id 3BGF) and m396 (PDB id 2G75) are shown using their original PDB structures. CR3022 was modeled as described in the text.
When this study started, no experimental structure was available for the RBD of SARS-CoV-2 S protein. A model was built up at the SWISS-MODEL workspace (YP_009724390.1) based on the NCBI reference sequence NC_045512 (Arnold et al., 2006). The root-mean-square deviation (RMSD) of atomic positions between this modeled structure for the RBD of SARS-CoV-2 S protein and the available one for SARS-CoV-1 (PDB id 2AJF) is 0.638Å. The structural comparison between the RBD proteins of both SARS viruses can be seen in Fig. 1. This field is evolving so fast that new experimental structures are continuously been solved. For example, a cryo-EM structure is now available for the prefusion S glycoprotein (a homotrimer structure) with an incomplete RBD (PDB id 6VSB, resolution 3.46 Å). This study revealed an interesting pivotal movement of the RBD between two transition states (“up” or “down” conformations) with direct implications for the binding of ACE2 and/or Abs (Wrapp et al., 2020). At least two monomeric S units need to be at the “up” conformation and properly rotated to remove the steric hindrance which allows the binding (Wrapp et al., 2020; Yuan et al., 2020). This implies at least a two-step “expose–dock-like” mechanism where the RBD has to be exposed towards the solution in the first step (characterized by important conformational adjustments of the whole homotrimeric structure) followed by the bind or dock phase where with the targeted-RBD can interact with other molecules. Here, the focus is at this second step of this mechanism where we assume that a single targeted-RBD structure is at its final state and prepared for the binding with the ligands (i.e. at least two S monomers were already at the “up” conformation, correctly rotated and there was no clashes between the targeted-RBD with any other parts of the trimeric structure that could prevent the binding) (Wrapp et al., 2020; Yuan et al., 2020). This is the same assumption for the available crystallographic structures (e.g. PDB id 2AJF). The conformational changes between the “up” and “down” conformations (the “expose” phase) are not investigated in this work.
The RMSD between our model and the S RBD (chain A) from this structure is 0.790Å. This number is closer to the RMSD differences between two chains of the same experimental (PDB id 6VSB) trimer structure (e.g. 0.668Å for chain A x chain B, and 0.732Å for chain A x chain C). Even more appropriate for the present discussion was the subsequent contribution by Yuan and co-authors that determined the crystal structure of the complex S RBD-CR3022 (PDB id 6W41, resolution 3.1 Å, pH 4.6) (Yuan et al., 2020). Many other experimental structures for this complex are now available [PDB ids 6YM0 (X-ray, resolution 4.36 Å, pH 8.0), 6YOR (cryo-EM, resolution 3.3 Å), 6YLA (X-ray, resolution 2.42 Å, pH 8.0)]. Such diversity of possible conformations might motivate further studies exploring their effects on the theoretical predictions. Additionally, the existence of such set of experimental structures for the complex RBD-CR3022 without the other parts of the S protein (or even the other chains of the S homotrimer) indicates that the missing structural part is not essential for the complexation at the dock phase of this “expose–dock-like” mechanism. These RMSD values above mentioned for the different chains (∼0.7 Å for PDB id 6VSB) also indicate that the modeled structure for the SARS-CoV-2 virus as used here is reasonable and within the expected conformational fluctuations from any other structure that could have been chosen for this work. Moreover, an intrinsic assumption here is that an experimental structure obtained at a given and specific physical chemical condition (ionic strength, pH, PEG6000 concentration, etc.) is valid in another condition (Poveda-Cuevas et al., 2020).
CR3022 is a particularly successful SARS-CoV-1 neutralizing human mAb first isolated from a convalescent patient by ter Meulen and co-authors (ter Meulen et al., 2006). For the present study, its three dimensional structure (see Fig. 3) was built up at the SWISS-MODEL workspace (Arnold et al., 2006) from the linear sequences of the variable regions of the heavy and light chains that were deposited in GenBank under accession numbers DQ168569 and DQ168570, respectively (ter Meulen et al., 2006). The RMSD between our computed generated model of CR3022 and the crystallographic structure (PDB id 6W41) is 0.537Å. This is again in the range of typical values seen for different structures determined at the same pH condition (e.g. 0.601Å for the comparison between PDB ids 6YM0 and 6YLA).
The linear sequences of the SARS S proteins are available at UniProt with the ids P59594 and P0DTC2 for SARS-CoV-1 and 2, respectively. The S1 subunit that attaches the virion to the cell membrane receptor is the cleaved chain between residues 14 and 667 for SARS-CoV-1 and 13 and 685 for SARS-CoV-2. Fig. 4 shows their pair sequences alignment as obtained by the server EMBOSS Needle (Needleman and Wunsch, 1970) with default settings. The identity and the similarity are, respectively, 64.8 % and 78.6 %. The RBD corresponds to positions 306–527 and 319–541 for SARS-CoV-1 and 2, respectively. Both the identity (I) and the similarity (S) are higher for this specific structural region (I = 73.1 % and S = 82.1 %).
Fig. 4.
Sequences alignment of SARS-CoV-1 and SARS-CoV-2 RBD (S1 subunit) proteins. Symbols between the two pairwise aligned sequences have the usual meaning: a) conservative amino acids where both sequences have the same residues are indicated by “|”; b) Similarities with a high score are marked with “;” and c) the ones with low positive score are indicated by “.”. Gaps are represented by “-”. Numbers are used to guide the identification of the amino acids sequence numbers. See text for more details.
2.2. Molecular simulations
A large diversity of models is available for MD and MC molecular simulations (Barroso da Silva et al., 2020; Kmiecik et al., 2016; Fossepre et al., 2020; Leach, 1996; Barroso da Silva and Dias, 2017). The need to repeat the calculations on several different physical chemical conditions and to obtain free energy of interactions at them drives the options to the so-called cost-effective coarse-grained (CG) models. These CG models offer the possibility to explore the main physical features of a system with a reduced number of parameters and lower computational costs (Poveda-Cuevas et al., 2020; Barroso da Silva et al., 2016; Delboni and Barroso da Silva, 2016). During the last years, a fast constant-pH (CpH) CG model has been devised to successfully study protein-protein interactions of several biological systems (including host-pathogens interactions) (Poveda-Cuevas et al., 2020; Barroso da Silva et al., 2016; Delboni and Barroso da Silva, 2016; Persson et al., 2010; Mendonça et al., 2019). The possibility to fully consider the pH effects makes this modeling approach more appealing and appropriated to address this problem (Poveda-Cuevas et al., 2020; Poveda-Cuevas et al., 2018).
A sketch of the simulations model is given in Fig. 5 . The S RBD proteins and the fragments of the mAbs were modeled as rigid bodies (i.e. bond lengths, angles, and dihedral angles are kept fixed) formed by a set of amino acids placed at positions given by their three-dimensional structures as described above. This additional approximation is justified by the prohibitive computational costs of constant-pH methods with pH-dependent conformational changes (Barroso da Silva and Dias, 2017; Barroso da Silva et al., 2019; Chen and Roux, 2015). Moreover, it is known that SARS-CoV-1 S protein does not exhibit large conformational changes upon the binding to ACE2 at least (Kirchdoerfer et al., 2018).
Fig. 5.
A sketch of the simulation model system for the constant-pH Monte Carlo simulations. A SARS-CoV-2 S RBD and the fragment of the mAb 80R (as given by the PDB id 2GHW) represented by a collection of charged Lennard-Jones spheres of radii Ri and valences zi mimicking amino acids are surrounded by counter ions and added salt, implicitly described by the inverse Debye length κ. The solvent is represented by its static dielectric constant ε. Positive and negatively charged protein amino acids are represented in blue and red, respectively. The macromolecules's centers of mass are separated by a distance r. The cylindrical simulation box is defined by the length lcyl and radius rcyl. Translation (back and forward) and rotation (in all directions) possible movements are illustrated by the gray arrows while the protonation/deprotonation processes are indicated by the dashed arrows labeled with H+.
Each group of atoms that define an amino acid is converted in a single charged Lennard-Jones (LJ) sphere of radius (Ri) and valence zi. This CG process turns a protein atomistic structure as a collection of charged LJ particles representing their amino acids. The centers-of-masses of the beads (mimicking amino acids) are used to place them accordingly to the coordinates given by the three-dimensional structures. The values of Ri for each type of amino acids were taken from Ref. Persson et al. (2010). The valences of all ionizable residues are a function of the solution pH. The fast proton titration scheme (FPTS) (Teixeira et al., 2010; Barroso da Silva and MacKernan, 2017; Barroso da Silva and Dias, 2017) was employed both to initially assign these valences zi’s for the amino acids and to let them vary during the simulation sampling at a given pH. This method has proved to predict pK a’s with a very good accuracy at low computational costs (Barroso da Silva and MacKernan, 2017). The fundamental physical chemical basis of this titration scheme, its numerical implementation, benchmarks, discussions related to its approximations, pros and cons can be found in previous publications (Teixeira et al., 2010; Barroso da Silva and MacKernan, 2017; Barroso da Silva and Dias, 2017; Barroso da Silva et al., 2017).
As illustrated in Fig. 5, two proteins are placed in an electroneutral open cylindrical simulation box, and free to translate back and forward along the axis in which their centers are laying, rotate in any direction and titrate. In this example, these two proteins are the modeled three dimensional structure of the SARS-CoV-2 S RBD and the crystallographic structure of the fragment of the mAb 80R. Unless otherwise specified, simulation runs were carried out with a cell of radius (rcyl) and height (lcyl) equals to 150 and 200 Å, respectively. The static dielectric constant was set to 78.7 (assuming a temperature of 298 K). Counter-ions and added salt particles were represented implicitly using a screening term, i.e., for two ionizable amino acids i and j, the screening is given by [exp(-κrij)] where κ is the modified inverse Debye length, and rij is the interparticle separation distance (Poveda-Cuevas et al., 2020; Barroso da Silva et al., 2016; Delboni and Barroso da Silva, 2016; Barroso da Silva et al., 2018). Additionally, a simplified simulation box with only one protein present was used to characterize the titration properties of a single macromolecule.
The electrostatic interactions between any two ionizable amino acids of valences and are given by:
| (1) |
where is the dielectric constant of the vacuum ( C2/Nm2), is the dielectric constant of the medium (we used 78.7 to mimic an aqueous solution) and C is the elementary charge. See Refs. Poveda-Cuevas et al. (2020); Barroso da Silva et al. (2016); Delboni and Barroso da Silva (2016); Barroso da Silva et al. (2018) for more details. Ionizable amino acids have their charged defined by the FPTS (Teixeira et al., 2010; Barroso da Silva and MacKernan, 2017). All the others are fixed neutral.
Protein-protein interactions are also controlled by other physical contributions (van der Waals interactions, hydrophobic effect, and excluded volume repulsion). (Poveda-Cuevas et al., 2020; Barroso da Silva et al., 2016; Delboni and Barroso da Silva, 2016). A simple and effective way to include their effects is by means of a LJ term [u vdw(rij)] between the beads (amino acids) (Poveda-Cuevas et al., 2020). Mathematically, for any two beads (charged or neutral ones) i and j, u vdw(rij) is given by
| (2) |
where σij (= Ri + Rj) is the separation distance of two amino acids i and j at contact. For instance, σij for the pair VAL-GLU is 7.2 Å (= RVAL + RGLU, where RVAL =3.4 Å and RGLU =3.8 Å – see Ref. Persson et al. (2010). The possibility to use different sizes for these beads allows the incorporation of non-specific contributions from the hydrophobic effect in the model (Delboni and Barroso da Silva, 2016). This should preserve the macromolecular hydrophobic moments (Eisemberg et al., 1982) and contributes to guide a correct docking orientation at short separation distances (Poveda-Cuevas et al., 2020).
The term εLJ regulates the strength of the attractive forces in the system (Poveda-Cuevas et al., 2020; Barroso da Silva et al., 2016; Delboni and Barroso da Silva, 2016). Typically, εLJ is assumed to be universal for any biomolecular system and equals to 0.124 kJ/mol (Barroso da Silva et al., 2016; Delboni and Barroso da Silva, 2016; Persson et al., 2010; Hyltegren et al., 2020). This should correspond to a Hamaker constant of ca. 9kBT (kB = 1.380 × 10−23 m2 kgs-2 K-1 is the Boltzmann constant, and T is the temperature in Kelvin) for amino acid pairs (Delboni and Barroso da Silva, 2016; Persson et al., 2010; Kurut et al., 2012). However, this value might result in both an over or an underestimation of the attraction depending on the biomolecular system (Barroso da Silva et al., 2016; Delboni and Barroso da Silva, 2016; Hyltegren et al., 2020). For instance, εLJ equals to 1.7kBT (a value 34 times greater than 0.124 kJ/mol) was necessary to reproduce experimental data for the histatin-5 adsorption to a hydrophilic silica surface (Hyltegren et al., 2020). Conversely, the? ?-lactoglobulin–lactoferrin complexation seems to be overestimated by the usual value of εLJ (Delboni and Barroso da Silva, 2016). Consequently, our research strategy has been to adopt the consensus value of 0.05kBT (=0.124 kJ/mol) for εLJ. This also implies that the outcomes should be interpreted with relative caution bearing in mind all the intrinsic approaches assumed in the modeling. The direct impact is seen in the free energy derivatives as discussed later at the results section.
Combining Eqs. (1) and (2), the total system's interaction energy for a given configuration can be written as:
| (3) |
where are amino acid positions and is their total number. This includes both charged and neutral beads.
This model was solved by Metropolis MC simulations that were performed at physiological ionic strength (150 mM) and different pH conditions. The choice to simulate at pH 4.6 and 7.0 was motivated by the needs to understand the low and neutral pH conditions (e.g. low pH of endosomes). Furthermore, it seems controversial in the experimental works if the acidification is essential or not for uptake of cell-free SARS virus (Simmons et al., 2004; Li, 2016; Dimitrov, 2003). The exact value of the acid pH condition is unknown. We made a choice to use the pH value of the crystallographic environment more acid among the studied structures (pH 4.6 for PDB ids 2GHW and 6W41). This also made possible to easily investigate the behavior of the systems at intermediate conditions by interpolation from the present outcomes.
After the proper equilibration of the simulated molecular systems, long production runs were carried out. Simulations whose focus was on titration properties [Z(pH) and pK as] required 108 MC steps. Conversely, runs to measure the free energy of interactions [Δw(r)] were calculated from radial distribution functions [Δw(r)=-ln g(r), where? ? = 1/kBT] that demanded even longer runs with at least 3.0 109 MC steps. These are massive simulations and very costly in terms of cpu time even at the CG representation. Four main factors contribute with this high cpu costs: a) the free energy barriers of the systems; b) the electrostatic coupling between a large number of titratable groups; c) the need to populate all the histogram bins used for the g(r) during the sampling; d) the reduction of the statistical noises in the calculated? ?w(r). (Poveda-Cuevas et al., 2020; Barroso da Silva et al., 2016; Delboni and Barroso da Silva, 2016). Standard deviations were controlled by means of the use of 5 replicates per simulated system as done before for the study of flaviviruses (Poveda-Cuevas et al., 2020).
2.3. Electrostatic epitopes determined by the PROCEEEDpKa method
Antibody-antigen recognition is a challenger and intensive research field. (Poveda-Cuevas et al., 2020; Regenmortel, 2014; Ramaraj et al., 2012; O’Kennedy et al., 2017). It is a molecular process that involves different physical intermolecular interactions. Electrostatic interactions deserve a special attention in this process for several reasons [e.g. is long range nature, the fact that the interface antibody-antigen has a peculiar electrostatic pattern (richer in titratable groups) that is different than other general protein-protein interfaces (Ramaraj et al., 2012), etc.] (Poveda-Cuevas et al., 2020; Poveda-Cuevas et al., 2018). Such facts contribute to shift the canonical view of the “lock and key” (with a clear focus on the protein surface) to a broader definition that led to the “electrostatic epitopes” (EE) concept (Poveda-Cuevas et al., 2020). This means that inner titratable groups (not only the ones at the epitope-paratope interface) can also participate in the interplay of interactions with Abs.
The EEs are the core idea of the PROCEEDpKa method (Poveda-Cuevas et al., 2020) where pKa shifts are used to identify the key amino acids responsible for a host-pathogen association. It applies the fact that the location of these shifts is a practical mean to probe molecular interactions as before demonstrated (Srivastava et al., 2017). Moreover, this can be easily measured during computer simulations of a protein-protein complexation. The predictive properties of this powerful tool have been previously i) statistically analyzed for flaviviruses, ii) compared to other bioinformatic tools (that often ignore that pH and ionic strength can drastically affect the complexation process) and iii) discussed in details in a preceding work (Poveda-Cuevas et al., 2020). The capacity of this method to test EE for specific mAbs makes it even more appealing for the present study where four known mAbs should be investigated. For the sake of convenience, predicted EE for the studied systems were graphically compared at the sequence level. The pairwise sequence alignments were generated by the server EMBOSS Needle (Needleman and Wunsch, 1970) with default settings.
3. Results and discussion
3.1. Free energy of interactions of SARS spike RBD proteins
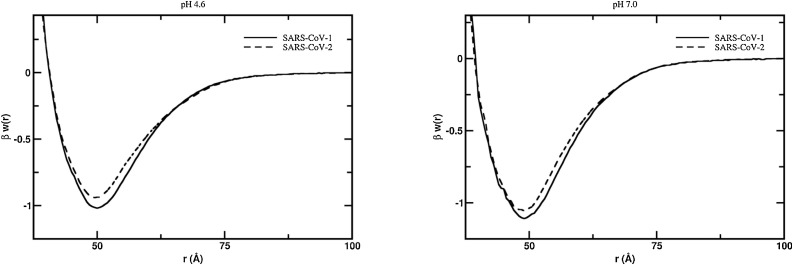
One of the central questions in the understanding of the COVID-19, its pathology including the high transmissibility, and the possible therapeutic interventions to control the epidemics spreading is to decipher and prevent the molecular interactions between the S protein and ACE2 (Dimitrov, 2003; Zhu et al., 2013; Wevers and van der Hoek, 2010; Böttcher-Friebertshäuser et al., 2018; Heald-Sargent and Gallagher, 2012; Li et al., 2005). This SARS S protein-ACE2 complexation is the first step toward infecting the cell by the virus. Several studies have shown that both SARS-CoV-1 and SARS-CoV-2 viruses share the function interaction with this cell receptor (i.e. ACE2) (Zhou et al., 2020; Dimitrov, 2003; Li et al., 2005; Hoffmann et al., 2020). We investigated the association pathway for the binding of the S RBD proteins to ACE2 for both SARS-CoV-1 and SARS-CoV-2 viruses by means of constant-pH MC simulations at two different solution pH values (4.6 and 7.0). The calculated free energy of interactions as given by the potentials of mean force [βw(r)] for these studied pH conditions at physiological ionic strength are given at Fig. 6 . Despite similar binding affinities observed in the present theoretical calculations (as seen in Fig. 6) and in the laboratory experiments (Tian et al., 2020), the SARS-CoV-1 S RBD protein has a small tendency to bind to the ACE2 at both pH regimes. This agrees quite well with the experimental results measured by the biolayer interferometry binding (BLI) assay as reported by Walls and co-authors using the functional subunit of the S protein responsible for binding to the host cell receptor (Walls et al., 2020). The measured binding affinity (KD) was 5.0 ± 0.1 nM for the system SARS-CoV-1 S RDB (also referred to as the domain B (Kirchdoerfer et al., 2018)–ACE2 and 1.2 ± 0.1 nM for the SARS-CoV-2 S RBD–ACE2. Yet, other experimental measurements using the S1 domain (this is the subunit that contains both the RBD and the N-terminal domain (Kirchdoerfer et al., 2018) might suggest an inverted behavior where SARS-CoV-2 S1 domain would have a tendency for a stronger bind to ACE2 (KD = 15.0 ± 0.1 nM) (Walls et al., 2019) in comparison to SARS-CoV-1 S (KD = 15.2 nM) (Tian et al., 2020). This small experimental difference of 0.2 nM could be due to several reasons including the experimental uncertainties that were not reported in Tian’s work (Tian et al., 2020). In contrast to these results, another recent study (Wrapp et al., 2020) advocated that SARS-CoV-2 has greater binding affinity for ACE2 than SARS-CoV-1. Even so, both the present theoretical and previously reported experimental data do agree that SARS-CoV-1 S RBD and SARS-CoV-2 S RBD have similar attraction to the ACE2. This high binding affinity implies that all human organs rich on ACE2 (oral and nasal mucosa, lung, stomach, small intestine, colon, skin, lymph nodes, thymus, bone marrow, spleen, liver, kidney, and brain) (Hamming et al., 2004; Xu et al., 2020b) can be easily infected. A clear opportunity for the virus is the lung alveolar epithelial cells and enterocytes of the small intestine, where ACE2 is abundant (Hamming et al., 2004).
Fig. 6.
Free energy profiles for the interaction of RBD proteins with ACE2. The simulated free energy of interactions [Δw(r)] between the centers of mass of the RBD proteins from both SARS-CoV-1 and SARS-CoV-2 and the ACE2 at different solution pH conditions. Salt concentration was fixed at 150 mM. The structures of these macromolecules were extracted from the PDB id 2AJF for SARS-CoV-1 S RBD and ACE. SARS-CoV-2 S RBD was built-up by modeling as described in the text. Simulations started with the two molecules placed at random orientation and separation distance. Results for the systems SARS-CoV-1 and SARS-CoV-2 are show as continuous and dashed lines, respectively.
Note that the simulations were performed with a single RBD protein in the absence of the full structure of the S protein and the others two chains of the homotrimeric S glycoprotein. This brings with it the evidence that the other structural parts of the S1 subunit, the S2 subunit and the two other chains are not essential for the individual pair of RBD–ACE2 complexation (assuming that the targeted-RBD has no steric hindrance as discussed above). Also, this observation supports the argument that the dissociation of the S1 subunit complexed with ACE2 can happen without the interruption of the infection. This also allows the S2 subunit to transit from a metastable prefusion to its post-fusion state as a second step in the viral infection (Kirchdoerfer et al., 2018; Song et al., 2018).
At pH 4.6 which is closer to the low pH that occur outside the cell (Li, 2016; Millet and Whittaker, 2015), the association between the SARS-CoV-1 S RBD protein and ACE2 showed a free energy depth [βwmin] of −1.02 at the separation distance of 50.0 Å (see Fig. 6a). Conversely, for SARS-CoV-2 S RBD, βwmin is −0.95 at the separation distance of 49.5 Å. The estimated standard deviations are 0.01kBT for all studied cases. Such computed measurements of βw(r) obtained by CG models that smooth the free-energy landscape must be interpreted with care as we already have pointed out above. By one side, it can be used a simple thermodynamic criterion that a negative free energy value (βwmin<0) would result in a molecular complex. Conversely, any free energy value smaller than the thermal energy (1 kBT) would indicate an unstable association. The observed difference (βwmin) of 0.07 between the two RDB proteins is greater than the estimated statistical errors. However, we prefer to interpret such small differences as tendencies of the system. Based on previous studies (Poveda-Cuevas et al., 2020; Barroso da Silva et al., 2016; Delboni and Barroso da Silva, 2016; Mendonça et al., 2019; Barroso da Silva et al., 2018) where we have observed either that the computed complexation was weaker than the experimental measurements or too much stronger, we feel safer to use this data (and the others discussed below) in relative terms (i.e. comparing different situations). This allows us to successfully predict experimental observations respecting the limits of such CG models (Poveda-Cuevas et al., 2020; Barroso da Silva et al., 2016; Delboni and Barroso da Silva, 2016; Mendonça et al., 2019). The similarities between their free energy minima are relatively amplified at pH 4.6 (Δβwmin = 0.07 as seen above) in comparison with pH 7.0 where βwmin is −1.11 and −1.06 (Δβwmin = 0.05). Both viral S RBD proteins have their affinities to ACE2 slightly raised when pH is increased from the acid to the neutral regime. This somewhat higher binding affinity at neutral pH suggests that the role of pH for RBD proteins constantly under debate at the literature (Simmons et al., 2004; Dimitrov, 2003; Li et al., 2005; Kielian and Helenius, 1985) might not be so critical for the infection. It also reinforces the possibility that the viral cell invasion is not a pH-dependent process. Indeed, it seems that pH is more relevant for the next steps to continue the viral infection and not at the first entry level. This possible non pH-depend process might increase the opportunities for the SARS viruses to easily infect cells and therefore to contribute to its high infectivity. This might limit the use of chloroquine as an efficient drug against COVID-19 since its first action is to increase the cell pH. At least a neutral pH solution will not prevent the binding of the RBD to ACE2. On the opposite as it can favor this affinity.
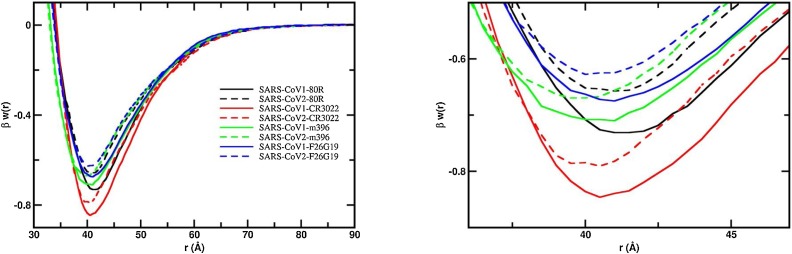
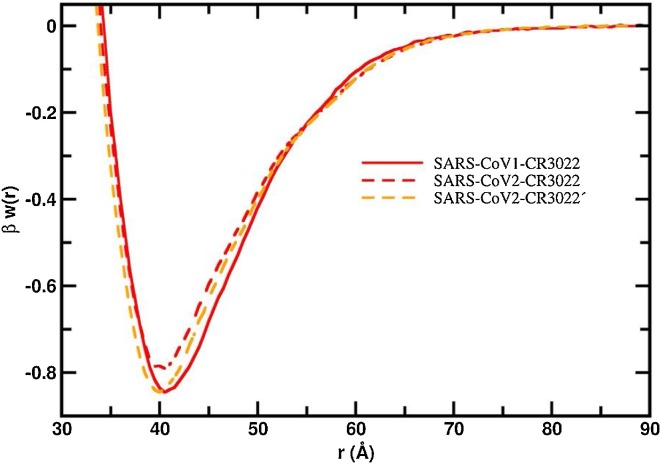
Another important aspect is the evaluation of putative mAbs that could bind to the RBD of the new SARS-CoV-2. Following the work of Tian and collaborators (Tian et al., 2020), we investigated the interactions between the two SARS S RBD proteins with some of the most potent SARS-CoV-1 S RBD specific neutralizing antibodies (80R, F26G19, m396, CR3022). Fig. 7 shows the free energy profiles at the acid regime and physiological salt conditions. For all studied fragments of Abs, a relatively stronger attraction is always observed for the S RBD protein from the SARS-CoV-1 interacting with any of these mAbs. This can be better seen in Fig. 7b where the region around the well depth is highlighted. The lowest observed binding affinities are observed for the system SARS-CoV-2-F26G19 (blue dashed line, βwmin = −0.63) followed by SARS-CoV-2-80R (black dashed line, βwmin = −0.66) and SARS-CoV-2-m396 (green dashed line, βwmin = −0.67). The difference between SARS-CoV-1-80R and SARS-CoV-2-m396 (Δβwmin = 0.01) is within the estimated statistical errors. The most promising complexation was found for the SARS-CoV-2 RBD-CR3022 (red dashed line, βwmin = −0.79) which is in good agreement with the experimental measurements (Tian et al., 2020). In fact, this was the only mAb that could bind potently with SARS-CoV-2 RBD (KD of 6.3 nM determined by BLI assay) in the experiments performed by Tian and co-authors using the Ab isolated from the blood of a convalescent SARS patient (Tian et al., 2020). This cross-reactivity binding between SARS-CoV-1 and 2 of CR3022 has been confirmed by other recent experimental studies (Yuan et al., 2020). Nevertheless, CR3022 could not neutralize SARS-CoV-2 in the experimental in vitro study (which does not exclude the possibility for in vivo neutralization) (Yuan et al., 2020). The Ab m396 only showed an insignificant binding at the highest measured concentration of 2 μM in the experimental studies (Tian et al., 2020).
Fig. 7.
Free energy profiles for the interaction of RBD proteins with monoclonal antibodies. The simulated free energy of interactions [Δw(r)] between the centers of mass of the RBD proteins from both SARS-CoV-1 and SARS-CoV-2 and the monoclonal antibodies at pH 4.6. Salt concentration was fixed at 150 mM. See text for details about the structures of these macromolecules. Simulations started with the two molecules placed at random orientation and separation distance. Results for SARS-CoV-1 and SARS-CoV-2 are show as continuous and dashed lines, respectively. Different line colors are used for each fragment of the Abs: 80R (black), CR3022 (red), m396 (green) and F29G19 (blue). (a) Left panel: Full plot. (b) Right panel: The well depth region of the βw(r) for each studied complex.
Using the free energy minima values observed in the simulations, we can order the binding affinities for the RBD proteins from the lower to the higher as SARS-CoV-2-F26G19 (βwmin = −0.63) < SARS-CoV-2-80R (βwmin = −0.66) < SARS-CoV-2-m396 (βwmin = −0.67) = SARS-CoV-1-F26G19 (βwmin = −0.67) < SARS-CoV-1-m396 (βwmin = −0.71) < SARS-CoV-1-80R (βwmin = −0.73) < SARS-CoV-2-CR3022 (βwmin = −0.79) < SARS-CoV-1-80R (βwmin = −0.85). As mentioned above, the values of βwmin should be used in relative terms. Moreover, the work of Tian and co-authors suggested that only for CR3022 it was experimentally measured a reasonable binding (Tian et al., 2020). The combination of these information could indicate that a threshold of −0.67KBT for βwmin can be used to better refine the theoretical binding predictions of these macromolecular complexations (i.e. all viral protein-protein systems with a value of βwmin smaller than −0.67KBT are expected to experience binding in vivo at least). Table S1 summarizes the values of βwmin given between parenthesis.
It should be noted that the attraction between the S RBD proteins and ACE2 (βwmin equals to −1.02 and −0.95 for SARS-CoV-1 and SARS-CoV-2, respectively) is always stronger than what was calculated to any studied mAb including to the CR3022 (βwmin equals to −0.85 and −0.79 for SARS-CoV-1 and SARS-CoV-2, respectively) for both SARS viruses (see Table S1). The same tendency was experimentally verified (Tian et al., 2020). It was measured by BLI assay a KD of 6.3 nM for the binding of CR3022 to SARS-CoV-2 S RBD which corresponds to a fraction of 0.41 of the KD measured for the binding of ACE2 to the same RBD (KD equals to 15.2 nM) (Tian et al., 2020).
3.2. Physical chemistry properties
Next, we explored basic physical chemical aspects that could offer a simple and quick reasoning to understand the above free energy results and eventually be used as descriptors to scan databases of mAbs to filter promising ideal candidates. Although different driven forces can result in protein-protein complexation (Barroso da Silva et al., 2016; Delboni and Barroso da Silva, 2016), pH and charge-charge interactions seems especially important for viral proteins (Poveda-Cuevas et al., 2020; Poveda-Cuevas et al., 2018; Yoshida et al., 2019; Yang et al., 2004; Tan et al., 2005; Lamarre and Talbot, 1989; Jaume et al., 2011). Indeed, the protein net charges numbers (Z) obtained as function of the solution pH show that the SARS-CoV-2 S RDB protein is always slightly more positively charged than SARS-CoV-1 S RDB protein at the same physical chemical environment (Z equals to 5.2 and 5.5, respectively, for them at pH 4.6) − see Table 1 . Since all studied fragments of Abs are also positively charged at pH 4.6 (Z equals to 9.1, 4.2, 2.7, 5.8 for 80R, CR3022, m396 and F26G19, respectively), it can be easily seen that the order observed for the binding affinities above in the free energy analyses do follow a simple charge-charge rule for the mAbs with similar surface area (A∼10,000 Å2). For the SARS-CoV-1 S RDB protein, from the weaker to the stronger repulsive cases in terms of the Coulomb contributions (Zi*Zj assuming the same Bjerrum length, salt screening and separation distances (Barroso da Silva et al., 2016; Delboni and Barroso da Silva, 2016; Jönsson et al., 2007), the predicted order for the binding affinity is 80R (5.2*9.1 = 47.3) < F26G19 (5.2*5.8 = 30.2) < CR3022 (5.2*4.2 = 21.8). This agrees with the previous free energy analyses (see above). As large is A, larger is the attractive van der Waals interactions that can overcome the charge-charge repulsion. This can also explain why m396 (that is smaller and has roughly half of A) is less attracted to the RBD proteins even being slightly less positively charged (Z equals to 2.7 at pH 4.6) than the others (Z∼5−6). Similarly, this is the physical reason to understand the stronger binding affinity that ACE2 (A = 25,290Å2) has to the S RBD proteins. Although ACE2 (Z = 5.4) and F26G19 (Z = 5.8) have similar Zs, the molecular surface of ACE2 is ca. 2.5 times larger than F26G29 (A = 10,120 Å2). We tested the van der Waals (vdw) contribution comparing? ?w(r) for CR3022 and m396 in a model where all electrostatic interactions where completely switched off and only vdw interactions are considered. This test-case system is shown in Figure S2. It can be seen that m396 in this hypothetical test does have a weaker binding affinity to SARS-CoV-2 S RBD protein in comparison to CR3022 confirming the arguments above presented.
Table 1.
Main physical chemistry properties of the studied proteins. Protein net charge numbers (Z) for the investigated proteins at physiological ionic strength (150 mM) and two different pH solution conditions (4.6 and 7.0). The macromolecular area (A) is given in Å2 as calculated by “UCSF Chimera” package. (Pettersen et al., 2004).
| pH | RBD SARS-CoV-1 | RBD SARS-CoV-2 | 80R | CR3022 | m396 | F26G19 | ACE2 |
|---|---|---|---|---|---|---|---|
| 4.6 | 5.2 | 5.5 | 9.1 | 4.2 | 2.7 | 5.8 | 5.4 |
| 7.0 | 2.0 | 2.2 | 5.1 | 1.0 | −2.6 | 0.1 | −23.0 |
| A | 8889 | 9079 | 9767 | 9831 | 5362 | 10,120 | 25,290 |
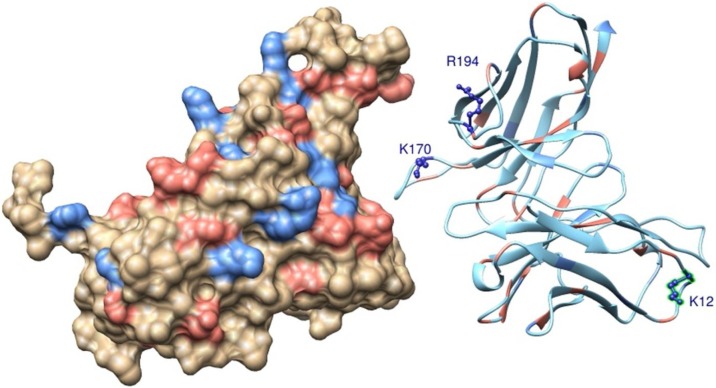
3.3. Insights to design a more efficient monoclonal antibody
Combining the findings above reported with a theoretical alanine scanning scheme employed to determine the contribution of specific titratable group to the complexation process, we identified three possible mutations that can improve the binding affinity of CR3022 to SARS-CoV-2 S RBD. The suggested mutations are K12E, K170A and R194A. These amino acids (K12, K170 and R194) can be seen in Fig. 8 at the wild type structure of CR3022. The main physical chemical reasoning to design this new functional molecule was to reduce the net charge of CR3022 in general together with a decrease of the repulsion for groups that are closely located at the host-pathogen interface. Two amino acids substitutions (K170A and R194A) are suggested at this biological interface while the other one (K12E) is more peripheral (see Fig. 8). Doing such mutations, the Z of the new molecule (labeled CR3022′) drops down from +4.2 to +1.2 at pH 4.6 and from +1.0 to −3.0 at pH 7.0.
Fig. 8.
Molecular structures of a possible SARS-CoV-2 S RBD complexed with CR3022. Standard amino acids of SARS-CoV-2 S RBD (molecule at left) and CR3022 (molecule at right) are shown in a molecular representation using spheres and ribbons, respectively. Atoms are colored accordingly to their amino acids physical chemical properties: red for acid amino acids, blue for base amino acids and wheat/green for non-titrating amino acids. For a better visualization of the interface, the two macromolecules were placed ∼12 Å apart from each other. Suggested residues to be mutated to improve the functional properties of CR3022 are indicated by the labeled amino acids (K12, K170, R194) are represented using the ball-and-stick model.
The binding affinity of this computer-designed molecule was tested. The calculated? ?w(r) for this new fragment of mAb is given in Fig. 9 . As it can be seen, CR3022′ is now able to bind with an equivalent binding affinity to what was observed for the SARS-CoV-1S RBD-CR3022 system. βwmin decreased from −0.79 to −0.85 recovering the value found for the SARS-CoV-1 S RBD-CR3022 case − see Fig. 7 and Table S1. Therefore, this is a promising designed mAb candidate to be carefully and systematically examined in further experimental assays. It can also be used as a template to design functional peptides and/or other inhibitors for preventing the interactions between the virus and ACE2.
Fig. 9.
Free energy profile for the interaction of SARS-CoV-2 S RBD proteins with a new monoclonal antibody. The simulated free energy of interactions [Δw(r)] between the centers of mass of the SARS-CoV S RBD protein and CR3022’ at pH 4.6 (dashed line in orange). Salt concentration was fixed at 150 mM. See text for details about the structures of these macromolecules. Simulations started with the two molecules placed at random orientation and separation distance. The results for SARS-CoV-1-CR3022 and SARS-CoV-2-CR3022 (continuum and dashed lines in red) are also shown for comparison. This data was extracted from Fig. 6.
3.4. Estimates of the antigenic regions by pKa shifts – the PROCEEDpKa method
To refine this analysis at the sequence level, the PROCEEDpKa method (Poveda-Cuevas et al., 2020) was employed to determine the EE of the RBD proteins for the most relevant studied complexes. Three questions should be addressed here: 1) if SARS-CoV-1 and 2 S RBD proteins share a common binding region when they bind to ACE2; 2) if these viral RBD proteins interact with the mAbs using a similar epitope-paratope interface; 3) if the interaction with CR3022 and CR3022’ involves the same EE. The data to answer such questions is given in Fig. 10, Fig. 11 .
Fig. 10.
Electrostatic epitopes. Primary sequences of the SARS-CoV-1 S RBD and the SARS-CoV-2 S RBD with the interface with ACE2 and the estimated antigenic regions (shown in blue) for 80R and CR3022 by the electrostatic method. Data obtained using the threshold |ΔpKa|>0.01. Symbols between the two pairwise aligned sequences have the usual meaning: a) conservative amino acids where both sequences have the same residues are indicated by “|”; b) Similarities with a high score are marked with “;” and c) the ones with low positive score are indicated by “.”. Gaps are represented by “-”. Numbers are used to guide the identification of the amino acids sequence numbers.
Fig. 11.
Electrostatic epitopes. Primary sequence of the SARS-CoV-2 S RBD with the estimated antigenic regions (shown in blue) for CR3022 and CR3022’ by the electrostatic method. Data for CR3022 is the same shown in Fig. 10. All other details are also as in Fig. 10.
In Fig. 10, the primary sequences of SARS-CoV-1 and 2 S RBD proteins are plotted together with the estimated ionizable amino acids of the interface with ACE2 and the antigenic regions as defined by the PROCEEDpKa method. By the different distribution of amino acids identified as EE (shown in blue), it can be seen that the electrostatic method is sensitive to the structures and their titratable groups that can produce electrical perturbations on their partners when they are interacting as demonstrated before (Poveda-Cuevas et al., 2020). The patterns observed for both viral SARS proteins are similar (i.e. they share a common region when they bind to ACE2 as observed in some experimental structures (Yuan et al., 2020) although some interesting observations can be made. Comparing the number of ionizable residues involved in the interactions for the RBD proteins of SARS-CoV-1 and ACE2 with the pair SARS-CoV-2 S RBD-ACE2, we can see an increase from 30 to 40 with a high number of common cases (22 amino acids) where the same amino acid interacts with ACE2 for both viral proteins. Most of the differences are observed for neighbor groups (e.g. “AWERKKISN” for SARS-CoV-1 and “AWNRKRISN” for SARS-CoV-2) indicating that the same biological interface was explored by the two viral RBD proteins in spite of their structural differences. The RBD protein responsible for COVID-19 clearly has more titratable residues interacting with ACE2 than its precursor. This observation suggests that its binding to ACE2 might be less specific than what happens for SARS-CoV-1. As such, the presence of an Ab may not completely block the SARS-CoV-2 S RBD-ACE2 interaction. In general, as seen in this Figure, most of the titratable groups from the viral RBD proteins involved in the binding to ACE2 are also the antigenic regions of the studied fragments of mAbs.
Virtually the same number of ionizable groups are seen at the antigenic regions for RBD proteins from SARS-CoV-1 (25 aa) and SARS-CoV-2 (24 aa) when interacting with 80R. The number of common cases is 12 while some regions are more affected by their structural differences (e.g. “DYSVLYNSTFFSTFKCYG” for SARS-CoV-1 and “DYSVLYNSASFSTFKCYG” for SARS-CoV-2). A replacement of an amino acid from the same physical chemical group (e.g. D by E) can be enough to result in different interactions (e.g. “KGDDVRQIA” for SARS-CoV-1 and “RGDEVRQIA” for SARS-CoV-2). CR3022 perturbed more titratable groups: 27 for SARS-CoV-1 and 33 for SARS-CoV-2 (22 % more). Using a different criterion to determine the epitopes, Yuan and co-authors also observed an increase of 15 % in the number of titratable residues at the epitope for SARS-CoV-2 (Yuan et al., 2020). Taking into account what was hypothesized for ACE2 above, this might be an addition contribution to improve the capability of this mAb to interact and inhibit the RBD proteins. In fact, the experimental work of Tian et al. (2020) do show that the CR3022 binding to SARS-CoV-2 S RBD is not affected by ACE2. This might be the molecular basis for this behavior. We are careful with the use of stronger statements here due to the limitation of the theoretical approach. Several additional issues remain to be further investigated.
Finally, we compared the EE predictions for CR3022’ (34 aa) with CR3022 (33 aa) interacting with SARS-CoV-2 S RBD protein − see Fig. 11. The predicted EEs for the interaction with CR3022’ are essentially the same ones observed for CR3022 (27 common aa). This implies that the suggested mutations here do not affected the antigenic regions. Another particularly interesting feature of this computer-designed molecule is that the number of EEs shared with ACE2 has increased from 18 (for CR3022) to 27 (for CR3022’). This might amplify the potential of this mAb candidate to better block the virus-host cell interaction.
4. Conclusions
Free energies of interactions were calculated for several molecular complexes involving the RBD of SARS-CoV-1 and 2 spike proteins. The present theoretical results confirmed that both RBD proteins have similar binding affinity to the ACE2 as previously reported in experimental studies. This is observed at both acid and neutral pH regimes which probably indicates that the medium pH is not so relevant for the beginning of the viral cell invasion. pH seems to be more important for the next steps of the viral infection and not at the first entry level. This has a direct implication for the drug development since the proposal of some like chloroquine is to raise cell pH.
Analyzing the interactions between these RBD proteins and the SARS-CoV-1 S RBD specific neutralizing mAbs (80R, F26G19, m396, CR3022) allowed us to reproduce the experimental results. The only mAb with measured affinities for the SARS-CoV-2 S RBD protein by BLI assay was CR3020 (Tian et al., 2020) which was also the one with higher affinity quantified in the present theoretical study. Moreover, we could map their electrostatic epitopes and identify that all mAbs tend to share the same titratable residues, and they are like the residues involved in the interaction with ACE2. However, the RBD protein responsible for COVID-19 clearly has more titratable residues interacting with ACE2 than its precursor suggesting that its binding to ACE2 might be less specific. This can explain the general difficulty that mAbs can experience to completely block the SARS-CoV-2 S RBD-ACE2 interaction.
Charge-charge interactions were found to be a good simple descriptor for a fast screening to the designing of improved mAb for diagnostics, therapeutics and vaccines. Our theoretical approach, while still being further developed, has identified three amino acids substitution that can increase the binding affinity of CR3022 to the RBD protein responsible for the present pandemic. These results can contribute to guide the design of new functional and high specific mAbs providing a cost-and-time-effective computational framework towards the development of better diagnostic strategies and an effective treatment and/or vaccine for COVID-19.
CRediT authorship contribution statement
Carolina Corrêa Giron: Visualization, Investigation, Writing - original draft. Aatto Laaksonen: Data curation, Methodology, Writing - review & editing, Supervision. Fernando L. Barroso da Silva: Conceptualization, Investigation, Methodology, Software, Data curation, Writing - review & editing, Supervision.
Acknowledgments
This work has been supported in part by the “Fundação de Amparo à Pesquisa do Estado de São Paulo” [Fapesp 2015/16116-3 (F.L.B.d.S.)] and the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) [(F.L.B.d.S.)]. F.L.B.d.S. is also deeply thankful to resources provided by the Swedish National Infrastructure for Computing (SNIC) at NSC. It is also a pleasure to acknowledge initial discussions with Zhenlin Yang (Zhongshan Hospital, Fudan University, China) that also provided us some preliminary modeled structures used for our first tests. A. Laaksonen acknowledges Swedish Science Council for financial support, and partial support from a grant from Ministry of Research and Innovation of Romania (CNCS - UEFISCDI, project number PN-III-P4-ID-PCCF-2016-0050, within PNCDI III).
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.virusres.2020.198021.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- Ahani A., Nilashi M. Coronavirus outbreak and its impacts on global economy: the role of social network sites. J. Soft Comput. Decis. Support Syst. 2020;7(2):19–22. [Google Scholar]
- Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020 doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold K., Bordoli L., Kopp J. Schwede, T. The SWISS-MODEL Workspace: A Web-Based Environment for Protein Structure Homology Modelling. Bioinformatics. 2006;22(2):195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- Backert L., Kohlbacher O. Immunoinformatics and Epitope Prediction in the Age of Genomic Medicine. Genome Med. 2015;7(1):119. doi: 10.1186/s13073-015-0245-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barroso da Silva F.L., Dias L.G. Development of Constant-PH simulation methods in implicit solvent and applications in biomolecular systems. Biophys. Rev. 2017;9(5):699–728. doi: 10.1007/s12551-017-0311-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barroso da Silva F.L., Pasquali S., Derreumaux P., Dias L.G. Electrostatics Analysis of the Mutational and PH Effects of the N-Terminal Domain Self-Association of the Major Ampullate Spidroin. Soft Matter. 2016;12:5600–5612. doi: 10.1039/c6sm00860g. [DOI] [PubMed] [Google Scholar]
- Barroso da Silva F.L., Derreumaux P., Pasquali S. Fast coarse-grained model for RNA titration. J. Chem. Phys. 2017;146(3):035101+. doi: 10.1063/1.4972986. [DOI] [PubMed] [Google Scholar]
- Barroso da Silva F.L., Derreumaux P., Pasquali S. Protein-RNA complexation driven by the charge regulation mechanism. Biochem. Biophys. Res. Commun. 2018;298(2):264–273. doi: 10.1016/j.bbrc.2017.07.027. [DOI] [PubMed] [Google Scholar]
- Barroso da Silva F.L., Sterpone F., Derreumaux P. OPEP6: a new Constant-PH molecular dynamics simulation scheme with OPEP coarse-grained force field. J. Chem. Theory Comput. 2019 doi: 10.1021/acs.jctc.9b00202. [DOI] [PubMed] [Google Scholar]
- Barroso da Silva F.L., Carloni P., Cheung D., Cottone G., Donnini S., Foegeding E.A., Gulzar M., Jacquier J.C., Lobaskin V., MacKernan D., Mohammad Hosseini Naveh Z., Radhakrishnan R., Santiso E.E. Understanding and controlling food protein structure and function in foods: perspectives from experiments and computer simulations. Annu. Rev. Food Sci. Technol. 2020;11(1):365–387. doi: 10.1146/annurev-food-032519-051640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder K., editor. Monte Carlo Methods in. Statistical Physics. Springer-Verlag; 1986. [Google Scholar]
- Böttcher-Friebertshäuser E., Garten W., Klenk H.D., editors. Activation of Viruses by Host Proteases. Springer International Publishing; Cham: 2018. [DOI] [Google Scholar]
- Ceraolo C., Giorgi F.M. Genomic variance of the 2019‐nCoV coronavirus. J. Med. Virol. 2020;92(5):522–528. doi: 10.1002/jmv.25700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Roux B. Constant-PH Hybrid Nonequilibrium Molecular Dynamics−Monte Carlo Simulation Method. J. Chem. Theory Comput. 2015;11:3919–3931. doi: 10.1021/acs.jctc.5b00261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu D.K.W., Pan Y., Cheng S.M.S., Hui K.P.Y., Krishnan P., Liu Y., Ng D.Y.M., Wan C.K.C., Yang P., Wang Q., Peiris M., Poon L.L.M. Molecular diagnosis of a novel coronavirus (2019-NCoV) causing an outbreak of pneumonia. Clin. Chem. 2020;66(4):549–555. doi: 10.1093/clinchem/hvaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun S., Muthu M., Gopal J., Paul D., Kim D.H., Gansukh E., Anthonydhason V. The Unequivocal Preponderance of Biocomputation in Clinical Virology. RSC Adv. 2018;8(31):17334–17345. doi: 10.1039/C8RA00888D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronaviridae Study Group of the International Committee on Taxonomy of Viruses The species severe acute respiratory syndrome-related coronavirus: classifying 2019-NCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5(4):536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delboni L., Barroso da Silva F.L. On the complexation of whey proteins. Food Hydrocoll. 2016;55:89–99. [Google Scholar]
- Dimitrov D.S. The secret life of ACE2 as a receptor for the SARS virus. Cell. 2003;115(6):652–653. doi: 10.1016/S0092-8674(03)00976-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., Zhao G., Yang Y., Qiu H., Wang L., Kou Z., Tao X., Yu H., Sun S., Tseng C.-T.K., Jiang S., Li F., Zhou Y.A. Conformation-Dependent Neutralizing Monoclonal Antibody Specifically Targeting Receptor-Binding Domain in Middle East Respiratory Syndrome Coronavirus Spike Protein. J. Virol. 2014;88(12):7045–7053. doi: 10.1128/JVI.00433-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisemberg D., Weiss R.M., Terwilliger T.C., Wilcox W. Hydrophobic Moments and Protein Structure. Faraday Symp Chem Soc. 1982;17:109–120. [Google Scholar]
- Eswar N., Webb B., Marti-Renom M.A., Madhusudhan M.S., Eramian D., Shen M., Pieper U., Sali A. Comparative Protein Structure Modeling Using Modeller. Curr. Protoc. Bioinforma. 2006;15(1) doi: 10.1002/0471250953.bi0506s15. 5.6.1–5.6.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fossepre, M., Laaksonen, A., Lyubartsev, A., Mocci, F., Naômé, A., Vercauteren, D. Biomolecular Modeling across Spatial & Temporal Scales. 2.
- Frenkel D., Frenkel D., Smit B. edição: 2. Academic Press; San Diego: 2001. Understanding Molecular Simulation: From Algorithms to Applications. [Google Scholar]
- Greber U.F., editor. Vol. 1215. Springer International Publishing; Cham: 2019. Physical virology: virus structure and mechanics. (Advances in Experimental Medicine and Biology). [DOI] [Google Scholar]
- Hamming I., Timens W., Bulthuis M., Lely A., Navis G., van Goor H. Tissue Distribution of ACE2 Protein, the Functional Receptor for SARS Coronavirus. A First Step in Understanding SARS Pathogenesis. J. Pathol. 2004;203(2):631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heald-Sargent T., Gallagher T. Ready, Set, Fuse! The Coronavirus Spike Protein and Acquisition of Fusion Competence. Viruses. 2012;4(4):557–580. doi: 10.3390/v4040557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020 doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyltegren K., Polimeni M., Skepö M., Lund M. Integrating all-atom and coarse-grained simulations—toward understanding of IDPs at surfaces. J. Chem. Theory Comput. 2020;16(3):1843–1853. doi: 10.1021/acs.jctc.9b01041. [DOI] [PubMed] [Google Scholar]
- Ibrahim B., McMahon D.P., Hufsky F., Beer M., Deng L., Mercier P.L., Palmarini M., Thiel V., Marz M. A new era of virus bioinformatics. Virus Res. 2018;251:86–90. doi: 10.1016/j.virusres.2018.05.009. [DOI] [PubMed] [Google Scholar]
- Jaume M., Yip M.S., Cheung C.Y., Leung H.L., Li P.H., Kien F., Dutry I., Callendret B., Escriou N., Altmeyer R., Nal B., Daeron M., Bruzzone R., Peiris J.S.M. Anti-severe acute respiratory syndrome coronavirus spike antibodies trigger infection of human immune cells via a PH- and cysteine protease-independent fc r pathway. J. Virol. 2011;85(20):10582–10597. doi: 10.1128/JVI.00671-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S., Shi Z., Shu Y., Song J., Gao G.F., Tan W., Guo D. A distinct name is needed for the new coronavirus. Lancet. 2020;395(10228):949. doi: 10.1016/S0140-6736(20)30419-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jönsson B., Lund M., Barroso da Silva F.L. Food Colloids. 2007. Electrostatics in macromolecular solutions; pp. 129–154. Chapter 9. [DOI] [Google Scholar]
- Kielian M., Helenius A. PH-Induced Alterations in the Fusogenic Spike Protein of Semliki Forest Virus. J. Cell Biol. 1985;101(6):2284–2291. doi: 10.1083/jcb.101.6.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchdoerfer R.N., Wang N., Pallesen J., Wrapp D., Turner H.L., Cottrell C.A., Corbett K.S., Graham B.S., McLellan J.S., Ward A.B. Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis. Sci. Rep. 2018;8(1):15701. doi: 10.1038/s41598-018-34171-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kmiecik S., Gront D., Kolinski M., Wieteska L., Dawid A.E., Kolinski A. Coarse-grained protein models and their applications. Chem. Rev. 2016;116(14):7898–7936. doi: 10.1021/acs.chemrev.6b00163. [DOI] [PubMed] [Google Scholar]
- Kurut A., Persson B.A., AAkesson T., Forsman J., Lund M. Anisotropic interactions in protein mixtures: self assembly and phase behavior in aqueous solution. J. Phys. Chem. Lett. 2012;3(6):731–734. doi: 10.1021/jz201680m. [DOI] [PubMed] [Google Scholar]
- Lai C.-C., Shih T.-P., Ko W.-C., Tang H.-J., Hsueh P.-R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus Disease-2019 (COVID-19): the epidemic and the challenges. Int. J. Antimicrob. Agents. 2020;55(3) doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamarre A., Talbot P.J. Effect of PH and temperature on the infectivity of human coronavirus 229E. Can. J. Microbiol. 1989;35(10):972–974. doi: 10.1139/m89-160. [DOI] [PubMed] [Google Scholar]
- Leach A.R. 1st ed. Longman; Singapore: 1996. Molecular Modelling – Principles and Applications. [Google Scholar]
- Li F. Structure, Function, and Evolution of Coronavirus Spike Proteins. Annu. Rev. Virol. 2016;3(1):237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Zhang C., Sui J., Kuhn J.H., Moore M.J., Luo S., Wong S.-K., Huang I.-C., Xu K., Vasilieva N., Murakami A., He Y., Marasco W.A., Guan Y., Choe H., Farzan M. Receptor and viral determinants of SARS-Coronavirus adaptation to human ACE2. EMBO J. 2005;24(8):1634–1643. doi: 10.1038/sj.emboj.7600640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Geng M., Peng Y., Meng L., Lu S. Molecular immune pathogenesis and diagnosis of COVID-19. J. Pharm. Anal. 2020 doi: 10.1016/j.jpha.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Fan Y., Lai Y., Han T., Li Z., Zhou P., Pan P., Wang W., Hu D., Liu X., Zhang Q., Wu J. Coronavirus infections and immune responses. J. Med. Virol. 2020;92(4):424–432. doi: 10.1002/jmv.25685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupia T., Scabini S., Mornese Pinna S., Di Perri G., De Rosa F.G., Corcione S. 2019 novel coronavirus (2019-NCoV) outbreak: a new challenge. J. Glob. Antimicrob. Resist. 2020;21:22–27. doi: 10.1016/j.jgar.2020.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier H.J., Bickerton E., Britton P., editors. Vol. 1282. Springer New York; New York, NY: 2015. Coronaviruses: methods and protocols. (Methods in Molecular Biology). [DOI] [Google Scholar]
- Mendonça D.C., Macedo J.N., Guimarães S.L., Barroso da Silva F.L., Cassago A., Garratt R.C., Portugal R.V., Araujo A.P.U. A Revised Order of Subunits in Mammalian Septin Complexes. Cytoskeleton. 2019;76(9–10):457–466. doi: 10.1002/cm.21569. [DOI] [PubMed] [Google Scholar]
- Millet J.K., Whittaker G.R. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015;202:120–134. doi: 10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needleman S.B., Wunsch C.D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 1970;48(3):443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- O’Kennedy R., Fitzgerald S., Murphy C. Don’t Blame It All on Antibodies – The Need for Exhaustive Characterisation, Appropriate Handling, and Addressing the Issues That Affect Specificity. TrAC Trends Anal. Chem. 2017;89:53–59. doi: 10.1016/j.trac.2017.01.009. [DOI] [Google Scholar]
- Peiris J.S.M. Clinical Virology. ASM Press; Washington, DC, USA: 2016. Coronaviruses; pp. 1243–1265. [DOI] [Google Scholar]
- Persson B., Lund M., Forsman J., Chatterton D.E.W., Torbjörn\AAkesson Molecular Evidence of Stereo-Specific Lactoferrin Dimers in Solution. Biophys. Chem. 2010;3(3):187–189. doi: 10.1016/j.bpc.2010.06.005. [DOI] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Greenblatt G.S., Meng E.C., Ferrin T.E. UCSF Chimera: A Visualization System for Exploratory Research and Analysis. J Comp Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Poveda-Cuevas S.A., Etchebest C., Barroso da Silva F.L. Insights into the ZIKV NS1 virology from different strains through a fine analysis of physicochemical properties. ACS Omega. 2018;3(11):16212–16229. doi: 10.1021/acsomega.8b02081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poveda-Cuevas S.A., Etchebest C., Barroso da Silva F.L. Identification of Electrostatic Epitopes in Flavivirus by Computer Simulations: The PROCEEDpKa Method. J. Chem. Inf. Model. 2020;60(2):944–963. doi: 10.1021/acs.jcim.9b00895. [DOI] [PubMed] [Google Scholar]
- Ramaraj T., Angel T., Dratz E.A., Jesaitis A.J., Mumey B. Antigen–antibody interface properties: composition, residue interactions, and features of 53 non-redundant structures. Biochim. Biophys. Acta. 2012;1824(3):520–532. doi: 10.1016/j.bbapap.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapaport D.C. 2nd ed. Cambridge University Press; 2004. The Art of Molecular Dynamics Simulation. [DOI] [Google Scholar]
- Regenmortel M.H.V.V. Specificity, Polyspecificity, and Heterospecificity of Antibody-Antigen Recognition. J. Mol. Recognit. 2014;27(11):627–639. doi: 10.1002/jmr.2394. [DOI] [PubMed] [Google Scholar]
- Sato H., Yokoyama M., Toh H. Genomics and computational science for virus research. Front. Microbiol. 2013;4 doi: 10.3389/fmicb.2013.00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma D., Priyadarshini P., Vrati S. Unraveling the Web of Viroinformatics: Computational Tools and Databases in Virus Research. J. Virol. 2015;89(3):1489–1501. doi: 10.1128/JVI.02027-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shereen M.A., Khan S., Kazmi A., Bashir N., Siddique R. COVID-19 Infection: Origin, Transmission, and Characteristics of Human Coronaviruses. J. Adv. Res. 2020;24:91–98. doi: 10.1016/j.jare.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barroso da Silva F.L., MacKernan D. Benchmarking a fast proton titration scheme in implicit solvent for biomolecular simulations. J. Chem. Theory Comput. 2017;13(6):2915–2929. doi: 10.1021/acs.jctc.6b01114. [DOI] [PubMed] [Google Scholar]
- Simmons G., Reeves J.D., Rennekamp A.J., Amberg S.M., Piefer A.J., Bates P. Characterization of Severe Acute Respiratory Syndrome-Associated Coronavirus (SARS-CoV) Spike Glycoprotein-Mediated Viral Entry. Proc. Natl. Acad. Sci. 2004;101(12):4240–4245. doi: 10.1073/pnas.0306446101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W., Gui M., Wang X., Xiang Y. Cryo-EM Structure of the SARS Coronavirus Spike Glycoprotein in Complex with Its Host Cell Receptor ACE2. PLoS Pathog. 2018;14(8):e1007236. doi: 10.1371/journal.ppat.1007236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava D., Santiso E., Gubbins K., Barroso da Silva F.L. Computationally Mapping PKa Shifts Due to the Presence of a Polyelectrolyte Chain around Whey Proteins. Langmuir. 2017;33(42):11417–11428. doi: 10.1021/acs.langmuir.7b02271. [DOI] [PubMed] [Google Scholar]
- Tan J., Verschueren K.H.G., Anand K., Shen J., Yang M., Xu Y., Rao Z., Bigalke J., Heisen B., Mesters J.R., Chen K., Shen X., Jiang H., Hilgenfeld R. PH-Dependent Conformational Flexibility of the SARS-CoV Main Proteinase (Mpro) Dimer: Molecular Dynamics Simulations and Multiple X-Ray Structure Analyses. J. Mol. Biol. 2005;354(1):25–40. doi: 10.1016/j.jmb.2005.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira A.A., Lund M., Barroso da Silva F.L. Fast proton titration scheme for multiscale modeling of protein solutions. J. Chem. Theory Comput. 2010;6(10):3259–3266. doi: 10.1021/ct1003093. [DOI] [PubMed] [Google Scholar]
- ter Meulen J., van den Brink E.N., Poon L.L.M., Marissen W.E., Leung C.S.W., Cox F., Cheung C.Y., Bakker A.Q., Bogaards J.A., van Deventer E., Preiser W., Doerr H.W., Chow V., T.; de Kruif J., Peiris J.S.M., Goudsmit J. Human monoclonal antibody combination against SARS coronavirus: synergy and coverage of escape mutants. PLoS Med. 2006;3(7):e237. doi: 10.1371/journal.pmed.0030237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X., Li C., Huang A., Xia S., Lu S., Shi Z., Lu L., Jiang S., Yang Z., Wu Y., Ying T. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg. Microbes Infect. 2020;9(1):382–385. doi: 10.1080/22221751.2020.1729069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gunsteren W.F., Dolenc J. Thirty-five years of biomolecular simulation: development of methodology, force fields and software. Mol. Simul. 2012;38(14–15):1271–1281. doi: 10.1080/08927022.2012.701744. [DOI] [Google Scholar]
- Viso J.F., Belelli P., Machado M., González H., Pantano S., Amundarain M.J., Zamarreño F., Branda M.M., Guérin D.M.A., Costabel M.D. Multiscale Modelization in a Small Virus: Mechanism of Proton Channeling and Its Role in Triggering Capsid Disassembly. PLoS Comput. Biol. 2018;14(4) doi: 10.1371/journal.pcbi.1006082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Xiong X., Park Y.-J., Tortorici M.A., Snijder J., Quispe J., Cameroni E., Gopal R., Dai M., Lanzavecchia A., Zambon M., Rey F.A., Corti D., Veesler D. Unexpected receptor functional mimicry elucidates activation of coronavirus fusion. Cell. 2019;176(5) doi: 10.1016/j.cell.2018.12.028. 1026–1039.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020 doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss S.R., Navas-Martin S. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol. Mol. Biol. Rev. 2005;69(4):635–664. doi: 10.1128/MMBR.69.4.635-664.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wevers B.A., van der Hoek L. Renin–Angiotensin system in human coronavirus pathogenesis. Future Virol. 2010;5(2):145–161. doi: 10.2217/fvl.10.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM Structure of the 2019-NCoV Spike in the Prefusion Conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., McGoogan J.M. Characteristics of and Important Lessons From the Coronavirus Disease 2019 (COVID-19) Outbreak in China: Summary of a Report of 72 314 Cases From the Chinese Center for Disease Control and Prevention. JAMA. 2020 doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- Wu Y., Ho W., Huang Y., Jin D.-Y., Li S., Liu S.-L., Liu X., Qiu J., Sang Y., Wang Q., Yuen K.-Y., Zheng Z.-M. SARS-CoV-2 is an appropriate name for the new coronavirus. Lancet. 2020;395(10228):949–950. doi: 10.1016/S0140-6736(20)30557-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Chen P., Wang J., Feng J., Zhou H., Li X., Zhong W., Hao P. Evolution of the novel coronavirus from the ongoing wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci. China Life Sci. 2020;63(3):457–460. doi: 10.1007/s11427-020-1637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Zhong L., Deng J., Peng J., Dan H., Zeng X., Li T., Chen Q. High expression of ACE2 receptor of 2019-NCoV on the epithelial cells of oral mucosa. Int. J. Oral Sci. 2020;12(1):8. doi: 10.1038/s41368-020-0074-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z.-Y., Huang Y., Ganesh L., Leung K., Kong W.-P., Schwartz O., Subbarao K., Nabel G.J. PH-dependent entry of severe acute respiratory syndrome coronavirus is mediated by the spike glycoprotein and enhanced by dendritic cell transfer through DC-SIGN. J. Virol. 2004;78(11):5642–5650. doi: 10.1128/JVI.78.11.5642-5650.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K., Kuroda D., Kiyoshi M., Nakakido M., Nagatoishi S., Soga S., Shirai H., Tsumoto K. Exploring Designability of Electrostatic Complementarity at an Antigen-Antibody Interface Directed by Mutagenesis, Biophysical Analysis, and Molecular Dynamics Simulations. Sci. Rep. 2019;9(1) doi: 10.1038/s41598-019-40461-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Wu N.C., Zhu X., Lee C.-C.D., So R.T.Y., Lv H., Mok C.K.P., Wilson I.A. A Highly Conserved Cryptic Epitope in the Receptor-Binding Domains of SARS-CoV-2 and SARS-CoV. Science. 2020 doi: 10.1126/science.abb7269. eabb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L.A. Pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Liu Q., Du L., Lu L., Jiang S. Receptor-binding domain as a target for developing SARS vaccines. J. Thorac. Dis. 2013;5(2) doi: 10.3978/j.issn.2072-1439.2013.06.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.