Abstract
Background
In December, 2019, the newly identified severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged in Wuhan, China, causing COVID-19, a respiratory disease presenting with fever, cough, and often pneumonia. WHO has set the strategic objective to interrupt spread of SARS-CoV-2 worldwide. An outbreak in Bavaria, Germany, starting at the end of January, 2020, provided the opportunity to study transmission events, incubation period, and secondary attack rates.
Methods
A case was defined as a person with SARS-CoV-2 infection confirmed by RT-PCR. Case interviews were done to describe timing of onset and nature of symptoms and to identify and classify contacts as high risk (had cumulative face-to-face contact with a confirmed case for ≥15 min, direct contact with secretions or body fluids of a patient with confirmed COVID-19, or, in the case of health-care workers, had worked within 2 m of a patient with confirmed COVID-19 without personal protective equipment) or low risk (all other contacts). High-risk contacts were ordered to stay at home in quarantine for 14 days and were actively followed up and monitored for symptoms, and low-risk contacts were tested upon self-reporting of symptoms. We defined fever and cough as specific symptoms, and defined a prodromal phase as the presence of non-specific symptoms for at least 1 day before the onset of specific symptoms. Whole genome sequencing was used to confirm epidemiological links and clarify transmission events where contact histories were ambiguous; integration with epidemiological data enabled precise reconstruction of exposure events and incubation periods. Secondary attack rates were calculated as the number of cases divided by the number of contacts, using Fisher's exact test for the 95% CIs.
Findings
Patient 0 was a Chinese resident who visited Germany for professional reasons. 16 subsequent cases, often with mild and non-specific symptoms, emerged in four transmission generations. Signature mutations in the viral genome occurred upon foundation of generation 2, as well as in one case pertaining to generation 4. The median incubation period was 4·0 days (IQR 2·3–4·3) and the median serial interval was 4·0 days (3·0–5·0). Transmission events were likely to have occurred presymptomatically for one case (possibly five more), at the day of symptom onset for four cases (possibly five more), and the remainder after the day of symptom onset or unknown. One or two cases resulted from contact with a case during the prodromal phase. Secondary attack rates were 75·0% (95% CI 19·0–99·0; three of four people) among members of a household cluster in common isolation, 10·0% (1·2–32·0; two of 20) among household contacts only together until isolation of the patient, and 5·1% (2·6–8·9; 11 of 217) among non-household, high-risk contacts.
Interpretation
Although patients in our study presented with predominately mild, non-specific symptoms, infectiousness before or on the day of symptom onset was substantial. Additionally, the incubation period was often very short and false-negative tests occurred. These results suggest that although the outbreak was controlled, successful long-term and global containment of COVID-19 could be difficult to achieve.
Funding
All authors are employed and all expenses covered by governmental, federal state, or other publicly funded institutions.
Introduction
On Dec 31, 2019, Chinese officials reported a cluster of cases of pneumonia in Wuhan, China. The newly discovered severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was identified to be responsible for the ensuing outbreak.1 As of May 13, 2020, more than 4·1 million confirmed cases of COVID-19 have been reported with more than 280 000 attributable deaths worldwide.2
As of March 1, 2020, WHO adhered to the strategic objective to “interrupt human-to-human transmission including reducing secondary infections among close contacts and health-care workers, preventing transmission amplification events, and preventing further international spread”.2 The International Health Regulations (IHR) Emergency Committee stated in its most recent declaration that it “believes that it is still possible to interrupt virus spread, provided that countries put in place strong measures to detect disease early, isolate and treat cases [and] trace contacts”.3 China implemented unprecedented measures to curb the epidemic, including cordoning off entire cities and implementing rigorous contact restrictions.4 In the meantime, countries outside of China attempted to contain the spread of the virus upon detection of travel-associated cases.
Research in context.
Evidence before this study
We searched MEDLINE (via PubMed) for articles published until March 2, 2020, using the key words “novel coronavirus”, “2019-nCoV”, “COVID-19”, “SARS-CoV-2”, “transmission”, “incubation period”, “serial interval”, and “Europe”. Moreover, we screened preprint servers such as medrxiv and SSRN for relevant articles and consulted the webpages of organisations such as WHO, European Centre for Disease Prevention and Control, and the Robert Koch Institute. No reports describing events of human-to-human transmission of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) within Europe other than the Bavarian outbreak were identified.
Added value of this study
Originating from a single travel-associated primary case from China, we describe the first reported cluster of COVID-19 cases with human-to-human transmission of SARS-CoV-2 within Europe and outside of Asia. The outbreak, which resulted in 16 subsequent cases, comprised four generations of virus transmission in only 16 days, corresponding to a median incubation period and serial interval of 4 days. By testing all people who had high-risk contact with confirmed patients with SARS-CoV-2 infection (and people with low-risk contact upon onset of symptoms), we were additionally able to detect and follow up patients with only very mild clinical symptoms of COVID-19 that would have probably remained undetected otherwise. By combining methods of epidemiology and whole genome sequencing, we were able to reconstruct and describe transmission events precisely. Virus transmission before or on the day of symptom onset or during a prodromal phase was substantial in this outbreak. Moreover, we were able to estimate attack rates according to different scenarios of contact intensity. As of May 2, 2020, no further cases associated with the outbreak were detected, suggesting that applied containment measures have worked.
Implications of all the available evidence
In the beginning of an epidemic caused by a newly discovered virus, the assessment of key epidemiological parameters such as attack rates, incubation period, and serial interval is important, as it provides essential information for estimating the potential size of the epidemic and developing containment measures. Detailed reports on transmission events of SARS-CoV-2, especially in a well described setting, could improve the understanding of transmissibility and further spread of the virus.
On Jan 27, 2020, the Bavarian Health and Food Safety Authority, Germany, was informed of the first human case of infection with SARS-CoV-2 in a German national working for a company in the greater Munich area. The primary case in this satellite outbreak of COVID-19 is a person from Shanghai, China, who had been in contact with their parents from Wuhan before visiting Germany for a business meeting in the aforementioned company. Between Jan 27 and Feb 11, 16 cases of COVID-19 were identified in this cluster.
Management and investigation of the outbreak was immediately initiated on Jan 27 to identify further cases and contacts and to understand transmission events and parameters that are relevant for successful containment, such as incubation period and secondary attack rates.
Methods
Study design and participants
This outbreak investigation was done in Bavaria, Germany, as a collaboration of state (Bavarian Health and Food Safety Autority) and national level (Robert Koch Institute) public health authorities and four public health laboratories. It included all people with confirmed SARS-CoV-2 infection and contacts (ie, people who had contact with a person with confirmed SARS-CoV-2 infection) linked to patient 0 of the Bavarian cluster.
The outbreak investigation was conducted as part of the authoritative, official tasks of the county health departments as well as the state health department of the Bavarian Health and Food Safety Authority, supported by the Robert Koch Institute. As conducted in response to a public health emergency, this study was exempt from institutional review board approval.
Contact classification and management
On Jan 27, 2020, all employees of the affected company were informed about the potential risk of COVID-19 infection. International public health authorities were informed via the Early Warning and Response System or IHR National Focal points. Employees were actively queried for any contact with the first two known cases. New cases were asked about professional and private contacts. Contacts were classified as high risk if they had cumulative face-to-face contact with a patient with laboratory-confirmed SARS-CoV-2 infection for at least 15 min, had direct contact with secretions or body fluids of a patient with confirmed COVID-19, or, in the case of health-care workers, had worked within 2 m of a patient with confirmed COVID-19 without personal protective equipment. All other contacts were classified as low-risk contacts.
High-risk contacts were ordered to stay in home quarantine for 14 days after the last known contact event with a confirmed case. Their health status was monitored daily, usually by self-report. Laboratory testing for SARS-CoV-2 was done at the beginning and end of home quarantine periods, irrespective of the presence of symptoms. If a contact tested positive for SARS-CoV-2, they were immediately hospitalised and isolated. Low-risk contacts were asked to self-monitor their health status and report any symptoms. In both high-risk and low-risk contacts, further laboratory testing was triggered upon onset of any symptoms.
Case interviews
Case interviews were done in a two-stage procedure. In the first stage, patients with confirmed SARS-CoV-2 infection or their household members were interviewed to determine date of symptom onset, links between cases, contact events during the incubation period, and contact classification. In a second stage, in-depth interviews with ten patients with COVID-19 were done by teams of two professionals (one physician and one epidemiologist; SB, KP, NM, NZ, and TSB) using a semi-structured questionnaire. Additional topics included characteristics of symptoms and details on type, setting, and environment of contact with other patients and with high-risk contacts. The in-depth interview with the primary case from Shanghai, China, was done twice (Jan 30 and 31, 2020) and supported by a Chinese native speaker (CW). We defined fever or cough as specific symptoms; a prodromal phase (with non-specific symptoms) was defined as presence of symptoms other than fever and cough for at least 1 day before the onset of specific symptoms. The day of symptom onset was defined as the day when any symptom (specific or non-specific) occurred, and thus could coincide with the start of the prodromal phase. The potential infectious period was defined as the period 2 days before onset of any kind of symptoms (specific or non-specific) until 14 days later.
Laboratory testing
Laboratory testing involved two swabs (nasopharyngeal and oropharyngeal, pooled) that were stored in viral transport medium and cooled. RNA was extracted using the QiAamp Bio Robot Kit (Qiagen; Hilden, Germany) on a Hamilton Microlab Star as recommended by the manufacturer. Real-time RT-PCR was done with the QuantiTect Virus +Rox Vial Kit (Qiagen, Hilden, Germany) on the Bio-Rad CFX96 Touch Real-Time PCR Detection System. Primer and probes were used as described by Corman and colleagues5 and provided by Tib-Molbiol (Berlin, Germany). The reference laboratory worked exactly as described in Corman and colleagues.5 Whole genome sequencing involved Roche KAPA HyperPlus library preparation and sequencing on Illumina NextSeq and MiSeq instruments as well as RT-PCR product sequencing on Oxford Nanopore MinION using the primers described in Corman and colleagues.6 Patient 1 was sequenced on all three platforms; patients 2–7 were sequenced on Illumina NextSeq, both with and without RT-PCR product sequencing with primers as in Corman and colleagues;6 and patients 8–11, 14, and 16 were sequenced on Oxford Nanopore MinION. Sequencing of patient 15 was not successful. Sequence gaps were filled by Sanger sequencing.
Secondary attack rate among contacts
We calculated secondary attack rates among contacts and case–contact pairs. A case–contact pair is the connection between a given contact and a potentially infectious case with contact time of at least 15 min; the number of case–contact pairs is higher than the number of contacts because a single contact person could have had contact with more than one case. To calculate secondary attack rates, we considered four distinct groups: (1) a household cluster where one household member had COVID-19 and all household members were quarantined together in one hospital room; (2) any other household contacts (ie, anyone sharing living space with a patient with COVID-19); (3) non-household high-risk contacts; (4) known low-risk contacts. Secondary attack rates were calculated as number of cases divided by number of contacts or case–contact pairs, using Fisher's exact test for the 95% CIs.
Role of the funding source
The sponsors of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report. The corresponding author had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Results
Details on SARS-CoV-2 importation to Germany have been previously described by Rothe and colleagues.7 In brief, the primary case (patient 0) stated that her parents, who normally live in Wuhan, had arrived for a visit in Shanghai on Jan 16. Both parents recalled cold-like symptoms the week before, and one parent showed fatigue and loss of appetite while visiting. Patient 0, who was an employee of the Chinese branch of a German company based in greater Munich, travelled from Shanghai to Munich by aeroplane on Jan 19, 2020, to facilitate workshops and attend meetings in the company building. The day after arrival (Jan 20, 2020), patient 0 felt chest and back aches—which she reported to be unusual—and took a single dose of medicine containing paracetamol. The patient reported fatigue during her whole stay in Germany and attributed the symptom to jetlag. After an overnight flight back to Shanghai on Jan 22, the patient felt feverish. With a self-measured temperature of 38·6°C and cough on Jan 24, the patient visited a physician's office on Jan 25. The patient tested positive for SARS-CoV-2 on Jan 26 and was hospitalised the next day. The clinical situation in both parents also deteriorated during the primary case's stay in Germany and both were laboratory-confirmed with COVID-19 later.7 The German company was informed of the primary case's infection in the morning of Jan 27, 2020, and immediately informed its employees as well as the local health authority.
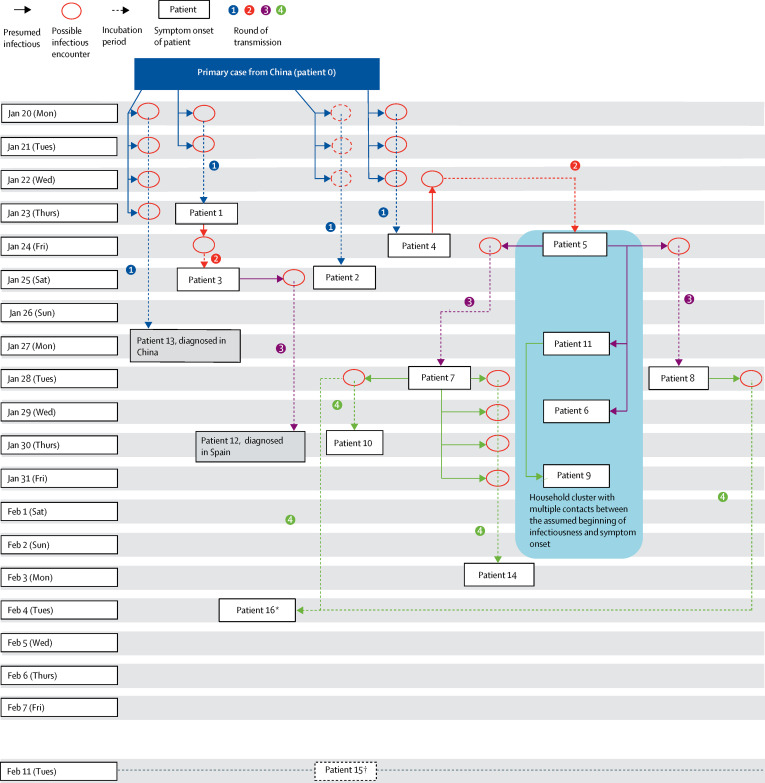
The initial testing by RT-PCR of high-risk contacts between Jan 27–29 identified patients 1–4 as first-generation cases (figure ; table 1 ).7 All four patients were immediately hospitalised and isolated. Their immediate contacts, including people they had contact with 2 days before symptom onset, were traced. A 14-day home quarantine was ordered for all newly identified high-risk contacts, starting at the day of the last known contact event with a case during the potential infectious period. Contacts were actively followed up on a daily basis. All high-risk contacts were instructed to minimise contact with other people, including household members in home quarantine. The affected company site was closed on the company's own initiative until Feb 11, 2020, and on-site disinfection measures were applied.
Figure.
Transmission chain of COVID-19 satellite outbreak in Bavaria, Germany, in January–February, 2020.
Boxes denote the day of symptom onset of cases, transmission rounds (arrows) are numbered and displayed in different colours. Red circles indicate the encounter when transmission is likely to have occurred; transmission from patient 0 to patient 2 is confirmed by whole genome sequencing, but no specific encounter could be identified. Potential presymptomatic infectious encounters are only included if no other encounter could be identified. Dotted arrows indicate the incubation period (transmission event until presentation of first symptoms), solid arrows lead from source cases to likely infectious encounters with recipient cases. For cases, the infectious period was assumed to start 2 days before symptom onset. SARS-CoV-2=severe acute respiratory syndrome coronavirus 2. *Also met patient 8 on Jan 28, but transmission was more likely through patient 7. †Asymptomatic household contact of case 2, with contact assumed Jan 25–28. Tested positive for SARS-CoV-2 on Feb 11.
Table 1.
Characteristics of laboratory-confirmed cases in the Bavarian COVID-19 outbreak in January–February, 2020
| Date of symptom onset | Most likely predecessor (primary case or other patient) | Most likely dates of infection (other possible dates) | Incubation period, days |
Transmission forwards to successor case* |
Self-reported symptoms† | ||||
|---|---|---|---|---|---|---|---|---|---|
| Asymptomatic | Presymptomatic | On date of symptom onset | In the prodromal phase | ||||||
| Patient 1 | Jan 23 | Primary case | Jan 20–21 | 2–3 (assumed 2·5) | No | No | No | No | On day of symptom onset: sore throat Further symptoms: cold-like symptoms, fatigue, chills, fever, cough, headache, joint pain, muscle pain, shortness of breath, and diarrhoea. |
| Patient 2 | Jan 25 | Primary case | Unknown (Jan 20–22) | 3–5 (assumed 4) | Unknown | Unknown | Unknown | Unknown | On day of symptom onset: cold-like symptoms and mild headacheFurther symptoms: mild earache, chills, fatigue, mild sore throat, blocked nose, loose stool, and shortness of breath. |
| Patient 3 | Jan 25 | Patient 1 | Jan 24 | 1 | No | No | Patient 12 | Patient 12 | On day of symptom onset: fatigue, blocked nose, sinus congestion, headache and swollen lymph nodesFurther symptoms: chest pain, cough, and loose stool |
| Patient 4 | Jan 24 | Primary case | Jan 20 (Jan 21–22) | 2–4 (assumed 4) | No | Patient 5 | No | No | On day of symptom onset: chillsFurther symptoms: fatigue, blocked nose, and sinus congestion |
| Patient 5 | Jan 24 | Patient 4 | Jan 22 | 2 | No | Patient 8 (possible) | Patient 6 (possible); patient 7; patient 8 (possible); patient 11 (possible) | No | On day of symptom onset: fever, limb pain, nausea, vomiting, cough, and feverFurther symptoms: fatigue, loss of appetite, and chest pain |
| Patient 6 | Jan 29 | Patient 5 | Unknown | Unknown | NA | NA | NA | NA | Fever, vomiting, and nausea |
| Patient 7 | Jan 28 | Patient 5 | Jan 24 | 4 | No | Patient 14 (possible) | Patient 10; patient 14 (possible); patient 16 (option 1) | No | On day of symptom onset: cough and blocked noseFurther symptoms: fatigue, headache, fever, nosebleed, and pneumonia |
| Patient 8 | Jan 28 | Patient 5 | Jan 24 (Jan 22–23) | 4 | No | No | Patient 16 (option 2) | Patient 16 (option 2) | On day of symptom onset: neck painFurther symptoms: headache and fatigue |
| Patient 9 | Jan 31 | Patient 11 | Unknown | Unknown | NA | NA | NA | NA | Fever, cough, vomiting, and diarrhoea |
| Patient 10 | Jan 30 | Patient 7 | Jan 28 | 2 | NA | NA | NA | NA | On day of symptom onset: shortness of breathFurther symptoms: cold-like-symptoms, night sweat, cough, and pneumonia |
| Patient 11 | Jan 27 | Patient 5 | Unknown | Unknown | No | Patient 9 (possible) | Patient 9 (possible) | No | Fever, limb pain, nausea, vomiting, back pain, and fatigue |
| Patient 12 (diagnosed in Spain) | Jan 30 | Patient 3 | Jan 25 | 5 | NA | NA | NA | NA | Blocked nose |
| Patient 13 (diagnosed in China) | Jan 27 | Primary case | Jan 20–23 | 4–7 (assumed 5·5) | NA | NA | NA | NA | Cough, general symptoms |
| Patient 14 | Feb 3 | Patient 7 | Jan 28–31 | 3–6 (assumed 4·5) | NA | NA | NA | NA | On day of symptom onset: feverFurther symptoms: mild cough, fatigue, mild headache, and loose stool |
| Patient 15 | NA | Patient 2 | Jan 23–28 | Unknown | NA | NA | NA | NA | Asymptomatic |
| Patient 16 | Feb 4 | Patient 7 or Patient 8 | Jan 28 | 7 | NA | NA | NA | NA | Blocked nose |
NA=not applicable.
Asymptomatic indicates transmission through a patient who never developed any symptoms during infection; pre-symptomatic indicates transmission through a patient who developed symptoms only after the transmission to another person; at day of symptom onset indicates transmission through a patient on the date of symptom onset, including both specific (fever and cough) and non-specific symptoms; in the prodromal phase indicates transmission through a patient during the phase where only non-specific symptoms (ie, other than fever or cough) were present.
Symptoms are divided into those on the day of symptom onset and further symptoms that developed after the day of symptom onset. Where no division is specified, all symptoms were present from day of symptom onset.
By Feb 19, 2020, 16 subsequent cases had been identified: four female and 12 male. All patients had been registered as high-risk contacts of the primary case or subsequent cases before being identified. The median age of the 16 patients was 35 years (IQR 27–42; range 2–58). Ten patients (patients 1–5, 7, 8, 10, 13, and 16) as well as the primary case (patient 0) are employees of the company. A Chinese colleague (patient 13) of patient 0 accompanied them in multiple activities while in Germany. Patient 13 travelled back to China with patient 0, developed symptoms on Jan 27, and tested SARS-CoV-2 positive a few days later.
Patient 1 was an employee who attended a 1-h business meeting with patient 0 and two other colleagues on Jan 20, 2020. The meeting took place in a small room (approximately 12 m2); patient 1 sat next to patient 0, and the two other colleagues sat at the opposite side of the table. The two other colleagues did not test positive during follow-up. Patient 1 had another brief contact event with patient 0 on Jan 21, and developed a sore throat on Jan 23 (figure). Over the following weekend (Jan 25–26), they developed cold-like symptoms with self-measured fever up to 39°C and mild productive cough. On Jan 27, patient 1 felt well enough to go to work. There, patient 1 learned about patient 0's infection and was tested positive on the same day.
Patient 2 was not aware of any direct person-to-person contact with patient 0; however, virus sequence analysis supports the assumption that patient 0 transmitted the virus to patient 2. Patient 3 also did not have direct contact with patient 0; however, patient 3 had contact with patient 1 on Jan 24 (figure), when both worked simultaneously on the same computer for a short period of time. Thus, transmission from patient 1 to patient 3 most likely took place the day after symptom onset (Jan 23). On Jan 25, the day of symptom onset of patient 3, patient 3 had a private meeting with patient 12, sitting next to him for approximately 90 min. Afterwards, they spent the rest of the evening together at patient 3's home, joined by patient 3's partner, who did not test positive during follow-up. Patient 12 departed for vacation to Spain 3 days later (Jan 28). After Spanish authorities were informed, patient 12 was isolated in hospital on Jan 30 and diagnosed with COVID-19.
Patient 4 had contact with patient 0 on Jan 20, 21, and 22, and reported chills on Jan 24. They had subsequent mild symptoms with slight malaise and slight nose and sinus congestion and were isolated on Jan 28. Patient 5 did not meet patient 0 but did meet patient 4 on Jan 22. Their only encounter was a canteen visit, sitting back to back, when patient 5 turned to patient 4 to borrow the salt shaker from their table. The encounter was 2 days before symptom onset in patient 4. Presymptomatic transmission from patient 4 to patient 5 is strongly supported by virus sequence analysis (table 2 ): a nonsynonymous nucleotide polymorphism (a G6446A substitution) was found in the virus from patients 4 and 5 onwards but not in any cases detected before this point (patients 1–3). Later cases with available specimens, all containing this same substitution, were all traced back to patient 5. The possibility that patient 4 could have been infected by patient 5 was excluded by detailed sequence analysis:8 patient 4 had the novel G6446A virus detected in a throat swab and the original 6446G virus detected in her sputum, whereas patient 5 had a homogeneous virus population containing the novel G6446A substitution in the throat swab.
Table 2.
Genome single nucleotide polymorphisms compared with reference sequence
|
Gene and position* |
GenBank accession number |
||||||
|---|---|---|---|---|---|---|---|
| Non-coding gene, position 241 |
ORF1ab |
S |
|||||
| Position 3037 | Position 6446 | Position 9891 | Position 22323 | Position 23403 | |||
| Nucleotide in reference | C | C | G | C | C | A | .. |
| Patient 1 | T | T | G | C | C | G (Asp>Gly) | MT270101 |
| Patient 2 | T | T | G | C | C | G (Asp>Gly) | MT270112 |
| Patient 3 | T | T | G | C | C | G (Asp>Gly) | MT270103 |
| Patient 4 | T | T | A (Val>Ile) or G | Y (for T, Ala>Val) | C | G (Asp>Gly) | MT270102 |
| Patient 5 | T | T | A (Val>Ile) | C | C | G (Asp>Gly) | MT270105 |
| Patient 6 | T | T | A (Val>Ile) | C | C | G (Asp>Gly) | MT270104 |
| Patient 7 | T | T | A (Val>Ile) | C | C | G (Asp>Gly) | MT270106 |
| Patient 8 | T | T | A (Val>Ile) | C | C | G (Asp>Gly) | MT270107 |
| Patient 9 | T | T | A (Val>Ile) | C | T (Ser>Phe) | G (Asp>Gly) | MT270108 |
| Patient 10 | T | T | A (Val>Ile) | C | C | G (Asp>Gly) | MT270110 |
| Patient 11 | T | T | A (Val>Ile) | C | C | G (Asp>Gly) | MT270109 |
| Patient 14 | T | T | A (Val>Ile) | C | C | G (Asp>Gly) | MT270111 |
| Patient 16 | T | T | A (Val>Ile) | C | C | G (Asp>Gly) | MT270113 |
No specimens were available for sequencing for patients 12, 13, and 15. A=adenine. G=guanine, T=thymine. C=cytosine.
Positions relative to EPI_ISL_402125 (the genome with closest similarity presently available on GISAID). Information in parentheses are amino acid exchanges in case of non-silent mutations.
The household of patient 5 consisted of five members, who were all hospitalised and kept together in one room in the hospital after patient 5 was confirmed positive. Three members—patients, 11, 6, and 9—developed symptoms and tested SARS-CoV-2 positive, whereas one member had no symptoms and did not test positive based on RT-PCR during follow-up. Patient 11 became symptomatic first; however, her first two tests were negative. Patient 9 became symptomatic last, with the virus showing an additional C22323T substitution. This same mutation was found as a minority virus population of approximately 4% reads also in the sputum of patient 11, but in none of the other household members. Because patient 11 had become symptomatic 4 days before patient 9, patient 11 infected patient 9.
Patient 7 met patient 5 for a 1·5-h meeting with a distance of approximately 1·5 m on Jan 24, the day of symptom onset in patient 5. Symptoms in patient 7 started 4 days later, on Jan 28, when patient 7 had a 1-h meeting with patient 10, in which patient 16 participated. Patients 8 and 5 had regular daily meetings at work, including on Jan 22, 23, and 24. Because the onset of symptoms for patient 5 was on Jan 24, transmission from patient 5 to patient 8 could therefore be presymptomatic or on the day of symptom onset. Patients 8 and 10 were identified as cases when a large number of employees, including both high-risk and low-risk contacts, were invited to be sampled at the company during the 3 days following the initial discovery of the cluster (Jan 29–31). Retrospectively, during detailed interviews, both patients recalled mild non-specific symptoms before testing, but no symptoms on the day of testing.
Patient 14 was a household member of patient 7 and both spent multiple days together after patient 7 was sent to home quarantine. Patient 15 is a household member of patient 2 and was asymptomatic. Patients 15 and 16 initially tested negative between Jan 29 and 31 at the company and tested positive when the test was repeated at the end of the follow-up period, thus 12–14 days after the last exposure to a case. Retrospectively, patient 16 had experienced a short period of very mild rhinorrhoea from Feb 4 and had not perceived this as a relevant symptom. Two possible transmission options could have occurred for patient 16, through patients 7 and 8, both on Jan 28. Patient 16 participated in the same meeting in which patient 10 was probably infected by patient 7. Patient 16 had also met patient 8 on the same day in a meeting, at a distance greater than 1 m; notably, Jan 28 was also the day of patient 8's symptom onset. Transmission history cannot be resolved because viral sequences in patients 7, 8, 10, and 16 are identical (table 2).
Notification via IHR focal points resulted in identification of patient 12 by Spanish authorities. Extensive contact tracing involved the international flights from Munich to Shanghai (patient 0 on Jan 22, 2020) and from Munich to Tenerife (patient 12 on Jan 28, 2020). As of May 2, no further cases have been identified among flight passengers or other (personal) contacts.
All cases except for patient 15 were symptomatic, even if the symptoms were mostly mild (table 1). However, no transmission was documented from asymptomatic patient 15 to the patient's contacts until the end of the observation period (beginning of March, 2020). Presymptomatic transmission was the only possible explanation for the transmission from patient 4 to patient 5. It was also a possibility in the transmission from patient 5 to patient 8 and could not be ruled out for the household clusters (ie, transmission from patient 5 to patients 11 and 6 and transmission from patient 11 to patient 9) or transmission from patient 7 to patient 14. Patients 4 and 5 had symptom onset on the same day, which is only plausible if patient 4 shed virus 2 days before symptom onset and patient 5 had a 2-day incubation period. Four transmission events occurred certainly on the day of symptom onset (from patient 3 to patient 12, from patient 5 to patient 7, from patient 7 to patient 10, and from patient 7 or 8 to patient 16) and for one transmission event (patient 5 to patient 8), the only other possibility is presymptomatic transmission (table 1; figure). Five patients were possibly infected when they had contact with their source case at the day of the source case's symptom onset (patients 5 to 8, 5 to 11, 5 to 6, 11 to 9, and 7 to 14; table). Transmission during the prodromal phase of the illness occurred from patient 0 to patients 1, 2, and 4, as well as from patient 3 to patient 12. The incubation period ranged from 1 to 7 days. When using the most likely duration of the incubation periods or, in case of two equally likely durations, the mean duration, the median incubation period was 4·0 days (IQR 2·3–4·3). The median serial interval was also 4·0 days (IQR 3·0–5·0).
After the infection of patient 0 was confirmed, and after identification of every new case, high-risk contacts were identified and put under home quarantine. By Feb 19, 2020, 241 high-risk contacts had been identified: four household contacts of patient 5 who were isolated together with the patient in a single hospital room, 20 household contacts of other cases, and 217 close, non-household contacts (table 3 ). Of the family isolated in one room, three (75%) members subsequently became cases, resulting in a secondary attack rate of 75·0% (95% CI 19·0–99·0; table 3). Among 20 household contacts of other patients who lived with the patients until they were isolated in hospital, two further cases were identified, resulting in a secondary attack rate of 10·0% (1·2–32·0). Because these 20 household contacts had 80 contact days (mean 4 days), the secondary attack rate can also be expressed as one infection per 40 household contact days. Among 217 further high-risk contacts, 11 transmissions occurred, leading to a secondary attack rate of 5·1% (2·6–8·9). No cases occurred among the 108 identified low-risk contacts (table 3).
Table 3.
Secondary attack rates among high-risk and low-risk contacts in the Bavarian COVID-19 outbreak in January–February, 2020
| Number of contacts | Number of cases originating from these contacts | Secondary attack rate (95% CI) | ||
|---|---|---|---|---|
| High risk | ||||
| Household contacts | ||||
| Shared isolation in a hospital room | 4 | 3 | 75·0% (19·0–99·0) | |
| Together until isolation of case | 20 | 2 | 10·0% (1·2–32·0) | |
| Other close unprotected contact | 217 | 11 | 5·1% (2·6–8·9) | |
| Case–contact pairs* | 249 | 11 | 4·4% (2·2–7·8) | |
| Low risk | ||||
| Distant unprotected contact | 108 | 0 | 0·0% (0·0–3·4) | |
A case–contact pair is the connection between a given contact person and a potentially infectious case with contact time of at least 15 min. The number of case–contact pairs is higher than the number of contacts because a single contact person could have had contact with more than one case.
Over the four generations of transmission in this outbreak, the virus acquired two mutations, both of them non-silent (table 2). The G6446A exchange leads to a valin-to-isoleucine change in the betacoronavirus-specific marker domain in non-structural protein 3. No structure and function is known for this domain in SARS coronavirus.9 The C22323T exchange causes a serine-to-phenylalanine change in the spike protein S1 domain, which is outside of the receptor binding domain.10 The directly observed substitution rate was two substitutions per 29 903 nucleotides per 11 days, equalling 2·2 × 10−3 substitutions per site, per year.
Discussion
This case series of the Bavarian cluster constitutes the first documented chain of multiple human-to-human transmissions of SARS-CoV-2 outside of Asia. Due to the particular setting centred around a business company with encounters tractable through electronic calendars, the timing and setting of most contact events were well defined. The sensitising and close monitoring of people involved might have triggered sensitive reporting of prodromal symptoms. This might explain a rather short median incubation period of 4·0 days in our study, which is substantially shorter than the 5·2 days calculated from data of the first 425 confirmed cases in China11 but similar to the findings of Guan and colleagues.12 For comparison, the incubation period for SARS was 6·4 days, with a range of 1–14 days.13
The short incubation period combined with a median serial interval of the same length correspond to transmission early in the course of disease, or even before disease onset, which is corroborated by studies of viral shedding in the same patients.8 High-level replication with frequent virus isolation from the pharynx stands by contrast with the SARS coronavirus—genetically similar to SARS-CoV-2—that was shed at lower concentrations from the upper respiratory tract and was effectively transmitted roughly a week after symptom onset.11 This delayed shedding in SARS caused substantial problems with the sensitivity of RT-PCR based on upper respiratory tract swabs. In our study, two tests from patient 11 came back negative although the patient had already developed symptoms, suggesting that these results were false negative. Nevertheless, it is evident from our study that RT-PCR on throat swabs can discover asymptomatic or oligosymptomatic people who shed the virus.
The present investigation enables us to determine secondary attack rates based on closely monitored high-risk contacts. The secondary attack rate thereby decreases with the intensity of contact: among members of the cohorted household, the secondary attack rate was 75%, but decreased to 10% among household contacts that were only together until isolation of the case. Although the experience with the cohorted family is based on a single observation, it shows the high risk of close household contacts—ie, sharing a room—of a case in home isolation and calls for strong measures within households to prevent transmission, particularly if susceptible people are present. Among 217 non-household high-risk contacts that were cumulatively followed up because of direct contact with a confirmed case, 11 were infected. The resulting secondary attack rate of 5% seems low, indicating little spread in this cluster. However, more effective spread of the virus might have been prevented by the proactive quarantine of high-risk contacts later identified as cases while still being presymptomatic or mildly symptomatic, as well as the proactive closure of the affected company. Transmission originating from people with more distinct respiratory symptoms might have resulted in a higher number of secondary cases.
Although two patients developed signs of pneumonia later in the course of disease, all others had a light course of disease and all patients recovered fully.8 However, it should be taken into account that the outbreak occurred in a population of working-age, generally healthy individuals. The overall clinical picture might have been different in a population including older individuals, or those with underlying chronic diseases. Another limitation of our study is that, naturally, not all infectious encounters could be reconstructed. Because the company held larger business and social events during the exposure period, it is possible that an infectious case might have met a successor case so briefly that neither of the two remembered the encounter. Some transmission events were nevertheless confirmed by viral sequencing and population analysis. The contact investigation and interviews followed a general hypothesis of direct human-to-human transmission, and we believe that the concurring epidemiological and genetic results in reconstructing the transmission network support this assumption.
In conclusion, while COVID-19 cases presented partially with mild, non-specific symptoms in the Bavarian outbreak studied here, we found that infectiousness before symptom onset, on the day of symptom onset, and during mild prodromal symptoms was substantial and poses a huge challenge on the implementation of public health measures. Additionally, the incubation period was often very short and false negative tests can occur. Thus, although the outbreak was controlled and therefore might have granted valuable time before more intense transmission occurred in Germany, successful long-term and global containment of COVID-19 could be difficult to achieve.
Acknowledgments
Acknowledgments
We thank all participants for sharing information necessary for the outbreak investigation and management. Moreover, we thank the company affected by the outbreak for a very good collaboration and great support with implementing the public health measures. We thank all county health authorities involved in the outbreak management as well as the Spanish health authorities for managing patient 12 and their contacts. Thanks also to Marie Reupke, Jörg Lekschas, and Joachim-Martin-Mehlitz (Robert Koch Institute) for their assistance in giving legal advice as well as Juliana Breitenberger, Linda Ploß, Christine Hartberger, Sabine Lohrer, Jasmin Fräßdorf, and Evelyn Bauermeister (Bavarian Health and Food Safety Authority) for expert technical assistance. Furthermore, we acknowledge the Cambridge High Performance Computing Service. Contributions by CD and VMC were funded by the German Ministry of Health (Konsiliarlabor für Coronaviren) and the German Center for Infection Research. SB, KP, and NM are fellows of the Postgraduate Training for Applied Epidemiology, supported financially by Robert Koch Institute. TW and TSB are fellows of the European Centre for Disease Prevention and Control (ECDC) Fellowship Programme, supported financially by the ECDC. The views and opinions expressed herein do not state or reflect those of ECDC. ECDC is not responsible for the data and information collation and analysis and cannot be held liable for conclusions or opinions drawn.
Contributors
MMB and UB contributed to the study design and literature search. WH, AS, CD, and AZ provided supervision. MMB, UB, VMC, MH, KK, DVM, SB, NA, RK, UE, BT, AD, KB, VF, AB, SH, SI, BW, AG, KP, NM, NZ, TSB, WC, AR, MadH, UR, OH, JS, TV, BM, RW, MA, MW, UP, AS, and CD contributed to data collection. MMB, UB, VMC, KK, DVM, SB, TW, KP, NM, JS, TV, BM, AS, and CD contributed to data analysis. MMB, UB, VMC, KK, DVM, SB, KP, NM, JS, TV, BM, WH, AS, and CD contributed to data interpretation. MMB, UB, VMC, MH, RK, and CD wrote the manuscript. MMB, UB, and MadH, produced the figures. MH, KK, DVM, SB, TW, NA, RK, UE, BT, AD, KB, VF, AB, SH, SI, BW, AG, KP, NM, NZ, TSB, WC, AR, MadH, UR, OH, JS, TV, BM, RW, MA, MW, UP, BL, WH, AS, and AZ reviewed the manuscript.
Declaration of interests
We declare no competing interests.
References
- 1.Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO Coronavirus disease 2019 (COVID-19) situation report—114. May 13, 2020. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports
- 3.WHO Statement on the second meeting of the International Health Regulations (2005) Emergency Committee regarding the outbreak of novel coronavirus (2019-nCoV) Jan 30, 2020. https://www.who.int/news-room/detail/30-01-2020-statement-on-the-second-meeting-of-the-international-health-regulations-(2005)-emergency-committee-regarding-the-outbreak-of-novel-coronavirus-(2019-ncov)
- 4.Novel Coronavirus Pneumonia Emergency Response Epidemiology Team The epidemiological characteristics of an outbreak of 2019 novel coronavirus diseases (COVID-19) in China. Zhonghua Liu Xing Bing Xue Za Zhi. 2020;41:145–151. (in Chinese). [Google Scholar]
- 5.Corman VM, Landt O, Kaiser M, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Quick J. nCoV-2019 sequencing protocol. Jan 22, 2020. https://www.protocols.io/view/ncov-2019-sequencing-protocol-bbmuik6w
- 7.Rothe C, Schunk M, Sothmann P, et al. Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. N Engl J Med. 2020;382:970–971. doi: 10.1056/NEJMc2001468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wolfel R, Corman VM, Guggemos W, et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020 doi: 10.1038/s41586-020-2196-x. published online April 1. [DOI] [PubMed] [Google Scholar]
- 9.Lei J, Kusov Y, Hilgenfeld R. Nsp3 of coronaviruses: structures and functions of a large multi-domain protein. Antiviral Res. 2018;149:58–74. doi: 10.1016/j.antiviral.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wan Y, Shang J, Graham R, Baric RS, Li F. Receptor recognition by novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS. J Virol. 2020;94:e00127–e00130. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Q, Guan X, Wu P, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med. 2020;94:e00127–e00220. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guan WJ, Ni ZY, Hu Y, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020 doi: 10.1056/NEJMoa2002032. published online Feb 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.WHO . World Health Organization; Geneva: 2003. Consensus document on the epidemiology of severe acute respiratory syndrome (SARS) [Google Scholar]