Dear Editor:
Recent correspondence in this Journal has highlighted the current threat posed by recently-emerging corona virus disease 2019 (COVID-19) in the world.1 The COVID-19 is infection caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and is characterized by fever, dry cough, weak, and so on.2 , 3 SARS-CoV-2 has already caused a global pandemic. By 26 Apr, 2020, the spread of SARS-CoV-2 has led to more than 3.0 million infections and above 200,000 deaths;4 thus, its outbreak has become a global public health problem. Recently, COVID-19 epidemic in China has been well controlled, whereas the risk of imported COVID-19 cases has increased dramatically.5 As of April 26, 2020, a total of 1,636 abroad imported patients were reported in China.6 However, limited studies on full-length genome characterization of SARS-CoV-2 from COVID-19 cases imported from abroad. Here, we characterized the genotype and mutation characteristics of SARS-CoV-2 isolated from eight imported cases from abroad in Yunnan, China.
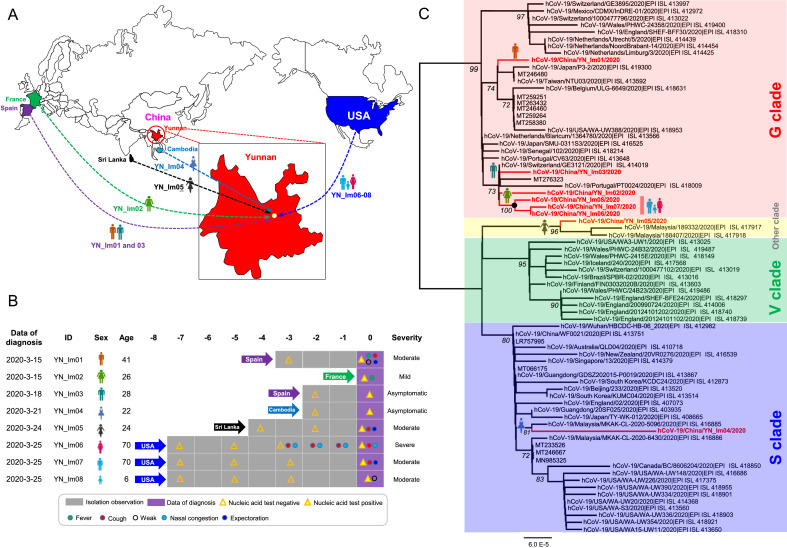
Eight COVID-19 patients imported from overseas were admitted to Yunnan Provincial Infectious Disease Hospital from March 15, 2020 to March 26, 2020. The epidemiological history and respiratory symptoms of the eight patients were summarized in Fig. 1 A and 1B. The 8 patients include 4 males and 4 females, with ages ranging from 6 years to 70 years old. No patient has ever been to Wuhan city in China. Two cases YN_Im01 and YN_Im03 were from Spain to Yunnan, YN_Im02 from France, YN_Im04 from Cambodia, YN_Im05 from Sri Lanka, and YN_Im06-08, a family cluster of COVID-19 patients from the United States (Fig. 1A). Six cases showed different degrees of respiratory symptoms before hospitalization. YN_Im06 was severe, YN_Im01, YN_Im05, YN_Im07, and YN_Im08 were moderate, YN_Im02 was mild, and YN_Im03 and YN_Im04 were asymptomatic according to the latest COVID-19 diagnostic criteria (5th edition) published by the National Health Commission of China (Fig. 1B).
Fig. 2.
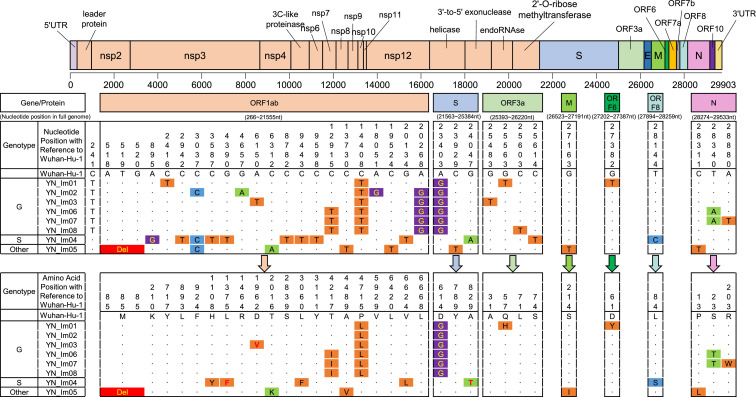
Nucleotide and amino acid substitution differences across the whole genome among clades G, V, S and other strains. Nucleotide and amino acid positions are numbered with a reference to SARS-CoV-2 strain Wuhan-Hu-1 identified earliest in Wuhan seafood market, in Hubei, China.
Fig. 1.
Maps of the study region, epidemiological characteristics, phylogenetic analysis based on the complete genome sequences of the SARS-CoV-2 from eight COVID-19 cases imported from abroad in Yunnan, China. (A) Eight cases of imported SARS-CoV-2 infection in travelers returning to Yunnan from overseas in 2020. (B) Demographic characteristics and respiratory symptoms. (C) Maximum-likelihood tree based on the complete SARS-CoV-2 genomic sequence obtained from positive serum samples was constructed using IQ-tree software. The genotypes of SARS-CoV-2 were divided into G, S, V and other. Virus clades are shown at right.
So far, three main clades involving G, V, and S have been identified based on marker mutations in the complete SARS-CoV-2 genome according to the latest genotyping rules recommended by the GISAID databas.7 G clade containing D614G variant in S protein is predominant in Europe, V clade possessing G251V mutation in ORF3 is more common in Asia and Europe, and S clade having L84S substitution in ORF8 is move prevalent in North America.8 In this study, eight complete genome sequences of SARS-CoV-2 isolated from sputum samples were successfully amplified and sequenced with 38 overlapping fragments. Dataset comprise SARS-CoV-2 full-length sequences of representative clade G, V, and S as previously reported, and reference sequences with the highest similarity (12 sequences) based on BLAST in Genbank using the eight sequences obtained in this study as the query set. Further, phylogenetic trees for SARS-CoV-2 full-length nucleotide sequences were constructed based on the obtained datasets with the maximum-likelihood method using IQ-tree. Phylogenetic analyses revealed that the six isolates, including one from France (YN_Im02), two from Spain (YN_Im01 and YN_Im03), and three from the United States (YN_Im06-08) were clustered as G clade with a high bootstrap value of 99%, one strain from Cambodia (YN_Im04) was grouped into S clade with a bootstrap value of 80%, and the remaining one from Sri Lanka was classified within other clade, a large unclassified sequences because lack the signature variants (Fig. 1C). Of note, the three sequences YN_Im06-08 isolated from a family cluster of SARS-CoV-2 infection formed a close monophyletic subclade supported by a bootstrap value of 100% and had 99.99% nucleotide identity, indicating the three sequences originate from the same strain.
To further characterize the characteristics of virus variation, the sequence analyses based on SARS-CoV-2 full-length nucleotide and amino acid sequences was performed using the strain Wuhan-Hu-1 (Genbank no. NC_045512) identified earliest in Wuhan seafood market, in Hubei, China as the reference strain for nucleotide and amino acid positions.9 The results revealed that 15, 12 and 10 nucleotide mutations to clades G, S, and other, respectively, were mapped across the SARS-CoV-2 full-length genome. Corresponding to these nucleotide substitutions, 8, 6, and 5 non-synonymous amino acid substitutions were detected in clades G, S, and other, respectively. Of note, all clade G strains possessed another P4715L marker substitution in nsp12 besides the signature mutation D614G variant in S protein. YN_Im05 strain belonging to other clade have a unique 3-nucleotide deletion between 518 and 520 nt that was not found in G and S clades, leading to a methionine deletion at position 58 in leader protein. Moreover, three novel mutations, including D1962V in nsp3 from the strain YN_Im03, L1375F in nsp3 and A829T in S protein from the isolate YN_Im04 were first identified in this study according to the comparison with 11,231 genomic sequences available at GISAID on 4/26/2020.10 Interestingly, S23T and R203W mutations located in N protein were identified in the three isolates from a family cluster of SARS-CoV-2 infection. Given that the three COVID-19 patients were diagnosed on the same day and the viruses originated from the same strain, indicated that the strain is replicating and mutating rapidly in different individuals.
In summary, we characterized the full-length genomes of SARS-CoV-2 strains from eight COVID-19 cases imported from abroad in Yunnan, China. Our results showed that the predominant SARS-CoV-2 clade was G (6 cases), followed by S clade (one case) and unclassified clade (one case). Further, comparative genomic analyses revealed that a novel signature amino acid substitution P4715L in nsp12 was found in the G clade strains. Moreover, three novel mutations, including D1962V in nsp3, L1375F in nsp3, and A829T in S protein were first identified in this study. The present study highlights the urgent need for continuous molecular screening and epidemic surveillance for SARS-CoV-2 among COVID-19 individuals imported from abroad to prevent future outbreaks of SARS-CoV-2 infection in China.
Declaration of Competing Interest
The authors declare no competing financial interests.
Acknowledgments
Financial support
This work was supported by the Yunnan Provincial major science and technology special project “New coronavirus infection emergency prevention and control plan project” (202003AC100007), Kunming Science and technology plan project (2020-1-N-023), National Natural Science Foundation of China (NSFC) (82041005), National Science and technology basic resources investigation project (2019FY101400), the Reserve Talents Project for Young and Middle-Aged Academic and Technical Leaders of Yunnan Province (2019HB012), and Youth Talent Program of Yunnan “Ten-thousand Talents Program” (YNWR-QNBJ-2018-054).
Acknowledgments
We thank the members of the Yunnan Infectious Disease Hospital for the data and sample collection.
References
- 1.Lv B., Li Z., Chen Y., Long C., Fu X. Global COVID-19 fatality analysis reveals Hubei-like countries potentially with severe outbreaks. J Infect. 2020 doi: 10.1016/j.jinf.2020.03.029. pii: S0163-4453(20)30157-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang C., Horby P.W., Hayden F.G., Gao G.F. A novel coronavirus outbreak of global health concern. Lancet. 2020;395(10223):470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.https://virusncov.com/
- 5.Chen L., Cai J., Lin Q., Xiang B., Ren T. Imported COVID-19 cases pose new challenges for China. J Infect. 2020 doi: 10.1016/j.jinf.2020.03.048. pii: S0163-4453(20)30179-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.http://www.nhc.gov.cn/xcs/yqtb/202004/ff7a70c762824574a928fa58e91d8744.shtml.
- 7.https://sars-cov-2.netlify.app/images/GISAID_hCoV-19_Analysis_Update_2020-04-04_1500UTC.pdf.
- 8.Francisco Díez-Fuertes, María Iglesias-Caballero, Sara Monzón, Pilar Jiménez, Sarai Varona, Isabel Cuesta, et al. Phylodynamics of SARS-CoV-2 transmission in Spain. bioRxiv2020; bioRxiv preprint doi: https://doi.org/10.1101/2020.04.20.050039. [DOI] [PMC free article] [PubMed]
- 9.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]