Abstract
Plants are under relentless challenge by pathogenic bacteria, fungi, and oomycetes, for whom they provide a resource of living space and nutrients. Upon detection of pathogens, plants carry out multiple layers of defense response, orchestrated by a tightly organized network of hormones. In this review, we provide an overview of the phytohormones involved in immunity and the ways pathogens manipulate their biosynthesis and signaling pathways. We highlight recent developments, including the discovery of a defense signaling molecule, new insights into hormone biosynthesis, and the increasing importance of signaling hubs at which hormone pathways intersect.
Introduction
During the course of evolution, pathogenic bacteria, fungi, oomycetes, and their plant hosts have evolved a multi-layered attack and defense relationship, often described as an arms race or zig-zag model. Once microbes pass the outer structural defense barriers of plants, they are likely to be recognized by their microbe-associated molecular patterns (MAMPs), which include bacterial proteins, endotoxins, and the fungal cell wall component chitin. Perception of MAMPs by specific pattern recognition receptors (PRRs) activates a first layer of defense response, called pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI). Several pathogens have developed strategies to suppress PTI using specialized effector proteins that interfere with immune signaling and secure continued virulence. This phenomenon is called effector-triggered susceptibility (ETS). However, many plants have developed strategies to detect and overcome these effectors with so-called R-proteins, which either directly recognize the presence of pathogenic effector proteins or scan whether targets of the effectors have been modified. If successful, this will activate a second layer of response, called effector-triggered immunity (ETI) (Figure 1A) (Chisholm et al., 2006; Jones and Dangl, 2006).
Figure 1. Hormone Pathways Interact and Cause Two Different Kinds of Systemic Immunity.
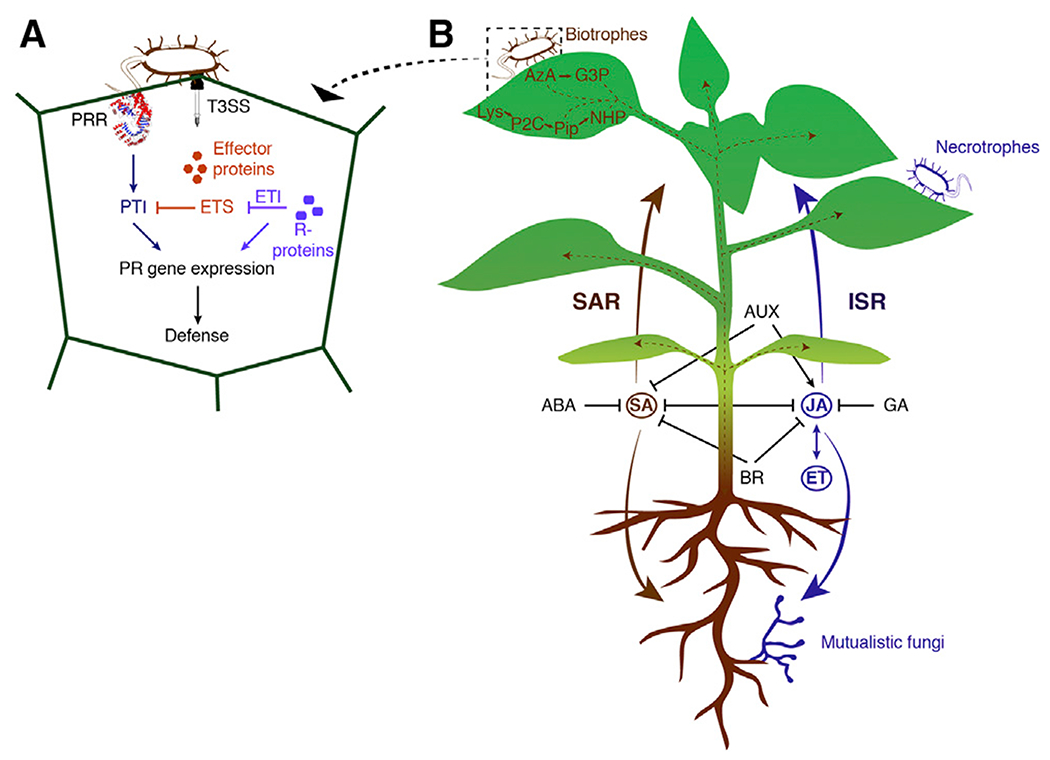
(A) Typical components and steps preceding the establishment of SAR: pattern recognition receptors (PRRs) recognize microbe-associated molecular patterns (MAMPS), leading to PAMP-triggered immunity (PTI). Pathogenic bacteria eject effector proteins into the host using a type III secretion system (T3SS), causing effector-triggered susceptibility (ETS). The plant can counteract effector proteins with R-proteins, leading to effector-triggered immunity (ETI).
(B) Left side, in brown: systemic acquired resistance (SAR) is established against biotrophic pathogens and is controlled by SA. Azelaic acid (AzA), glycerol-3-phosphate (G3P), pipecolic acid (Pip), and N-hydroxypipecolic acid (NHP) are all transported from the infected site to uninfected tissues. Right side, in blue: induced systemic resistance (ISR) requires JA and ET signaling and is found as a response against necrotrophic pathogens but also mutualistic organisms.
Whether ETI is activated depends on the lifestyle of the pathogen. While biotrophic bacteria or fungi feed on living tissue and have developed strategies to exploit resources while keeping the host alive, necrotrophic pathogens usually start by destroying their host with toxins and then consume its contents. Some pathogens, called hemibiotrophs, start with a biotrophic strategy but can live in dead tissue later. ETI is only effective against biotrophs and hemibiotrophs in their early stages and does not work as a strategy against necrotrophs. A common ETI response against biotrophs is the hypersensitive response (HR), in which the plant triggers the death of cells in a localized area surrounding the site of infection, thus containing any spread of the disease. While cell death works as a defense against biotrophs, necrotrophs release specific toxins that induce cell death. After the initial infection, different kinds of systemic immune responses can be activated that protect distant parts in the plant. The so-called systemic acquired resistance (SAR) is typically based on the hormone salicylic acid (SA) and involves activation of PATHOGENESIS-RELATED (PR) genes. Jasmonic acid (JA) causes a different type of systemic resistance, which is called induced systemic resistance (ISR) (Ton et al., 2002). These differences are linked to the nature of the pathogen. Typically, SA will result in the activation of defense genes against biotrophic pathogens, whereas responses against necrotrophs are triggered by JA signaling. Generally, these two hormone pathways are in an antagonistic relationship with one another, a situation that is fine-tuned by other hormones, such as ethylene (ET), abscisic acid (ABA), gibberellins (GAs), and auxin (AUX) (Figure 1B). Thus, manipulation of hormone production, perception, and downstream signaling is an important part of pathogen strategies. We review the current understanding of how pathogens manipulate hormone biosynthesis and signal transduction to remodel plant defense responses. We focus on recent developments in immune signaling research and highlight signaling hubs at which different hormone pathways intersect.
SA: Defense against Biotrophs
SA is a key hormone in plant immunity, mediating the so-called SAR, and has been established as a major hormone triggering responses against pathogens with a biotrophic lifestyle. SA is derived from the metabolite chorismate. During defense in Arabidopsis, SA is preferably produced via the isochorismate pathway (Wildermuth et al., 2001). Chorismate is first converted to isochorismate by the enzyme ISOCHORISMATE SYNTHASE (ICS), and isochorismate is then converted into SA. The nature of this final reaction has been a longstanding mystery. In certain bacteria, this step is carried out by the enzyme ISOCHORISMATE PYRUVATE LYASE (IPL). Plants, however, do not have the IPL enzyme. Two recent reports have resolved most of this question. The enzyme AVRPPHB SUSCEPTIBLE 3 (PBS3) conjugates isochorismate and glutamate to isochorismoyl-glutamate (Rekhter et al., 2019; Torrens-Spence et al., 2019). The two reports slightly differ on the precise fate of this reaction product. While both groups agree that isochorismoyl-glutamate is unstable and spontaneously decomposes into enolpyruvyl-N-glutamate and SA, the latter report states that this step can be catalyzed by the enzyme ENHANCED PSEUDOMONAS SUSCEPTIBILTY 1 (EPS1) at a rate four orders of magnitude higher than the spontaneous formation. EPS1 appears to be a protein with an altered BAHD acyltransferase fold but can only be found in Brassicaceae. It is possible that Brassicaceae have evolved an enzyme that accelerates the spontaneous, last step in SA biosynthesis.
During defense, SA accumulates at the site of infection and, although it is not a mobile signal, contributes to amplification of defense responses in systemic tissue (Métraux et al., 1990). In addition, the signal can move to neighboring plants when SA is converted to the biologically inactive, volatile ester methyl salicylate (MeSA) (Park et al., 2007). The very first step of SA biosynthesis is a target for the biotrophic fungus Ustilago maydis. Its effector Cmu1 converts chorismate into prephenate, depleting the pool of chorismate available for isochorismate production (Djamei et al., 2011). Other pathogens, such as the oomycete Phytophthora sojae and fungus Verticillium dahliae encode their own ICS enzymes to stimulate SA production in order to antagonize anti-necrotrophic JA signaling (Liu et al., 2014) (Figure 2A). As a signaling molecule, SA is perceived by several members of the NON-EXPRESSOR OF PATHOGENESIS-RELATED GENES (NPR) family, which are in direct control of gene expression. NPR1 is considered the master regulator of SA signaling. In 2012, the NPR1 homologs NPR3 and NPR4 and, in the same year, NPR1 were discovered to be the receptors for SA (Fu et al., 2012; Wu et al., 2012). More recent research has indicated that NPR3 and NPR4 act as transcriptional co-repressors and function independently from the co-activator NPR1 (Ding et al., 2018). NPR1 is regulated by the opposing activities of S-nitrosoglutathione (GSNO) and thioredoxins (Tada et al., 2008). S-nitrosylation by GSNO oligomerizes NPR1 and retains it in the cytoplasm in an inactive state. SA induces the thioredoxin-catalyzed monomerization of NPR1 and its subsequent relocation into the nucleus. Nuclear NPR1 can, for example, together with the TGA transcription factors, activate defense genes, such as PATHOGENESIS-RELATED 1 (PR-1), a molecular marker for the plant immune response. Given its importance, NPR1 activity is a target for several pathogenic effectors. The necrotrophic fungus Cochliobolus victoriae effector victorin binds to the active site of THIOREDOXIN-h5 and therefore impedes monomerization and activation of NPR1 (Lorang et al., 2012). XopJ, an effector from the bacterial pathogen Xanthomonas euvesitorica targets the protease RPT6 that is involved in NPR1 turnover (Üstün et al., 2013). In addition, proper turnover of phosphorylated NPR1 is required for complete establishment of defense response, and syringolin A produced by Pseudomonas syringae pv. syringae acts as a proteasome inhibitor (Schellenberg et al., 2010). However, not until 2017 was a bacterial effector shown to directly bind NPR1. The P. syringae type III effector AvrPtoB directly targets NPR1, an interaction that is stabilized by SA. AvrPtoB has E3 ligase activity and promotes NPR1’s degradation by the proteasome (Chen et al., 2017) (Figure 3A).
Figure 2. Pathogens Manipulate Hormone Biosynthesis Pathways at Multiple Points.
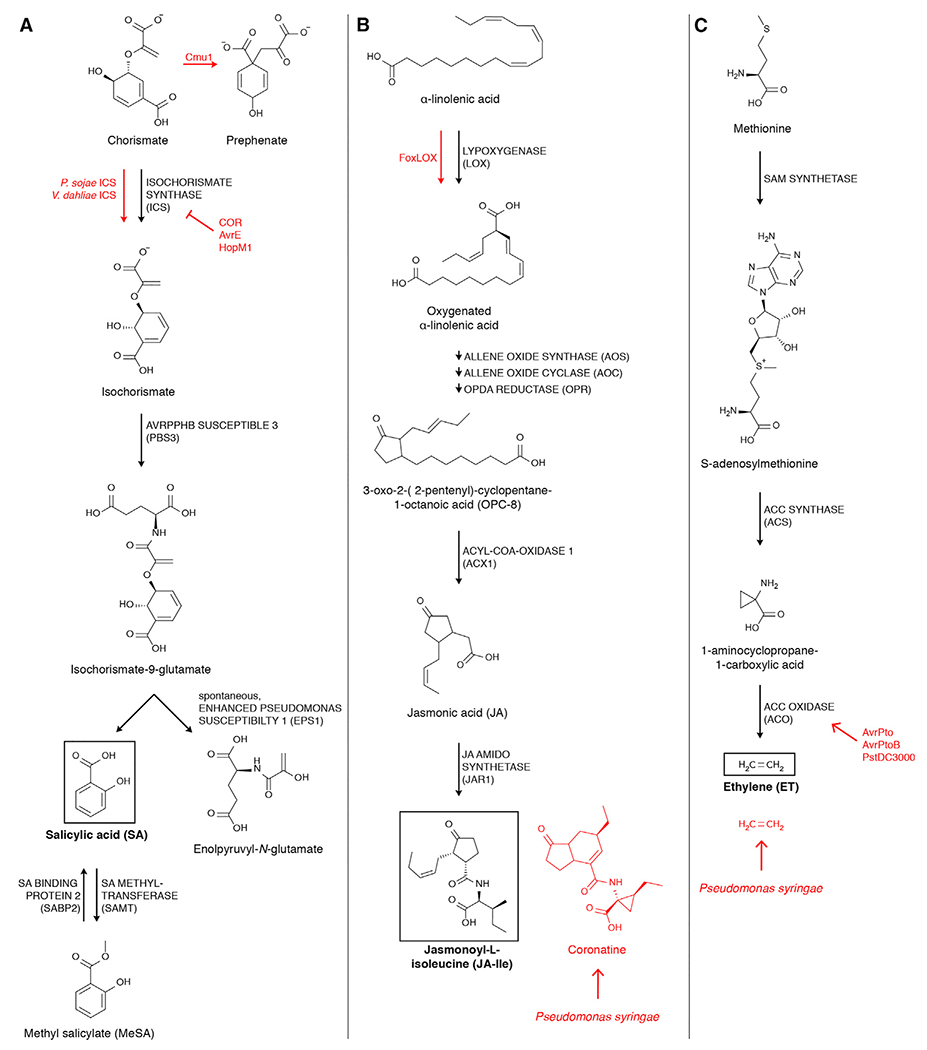
(A) Salicylic acid is abbreviated as SA. Cmu1 depletes the SA precursor chorismate by turning it into prephenate. COR, AvrE, and HopM1 interrupt SA production by blocking the ICS enzyme. P. sojae and V. dahliae have their own ICS enzymes to stimulate SA production.
(B) Jasmonic acid is abbreviated as JA. The pathogenic FoxLOX enzyme and the JA-Ile mimic coronatine both stimulate JA signaling.
(C) Ethyleneis abbreviated as ET. AvrPto, AvrPtoB, and Pseudomonas syringae pv. tomato DC3000 (Pst DC3000) stimulate ET production through upregulation of ACC OXIDASE. Some Pseudomonas syringae strains produce ET.
Figure 3. Pathogens Interfere with Signaling Pathways to Remodel Hormone Responses.
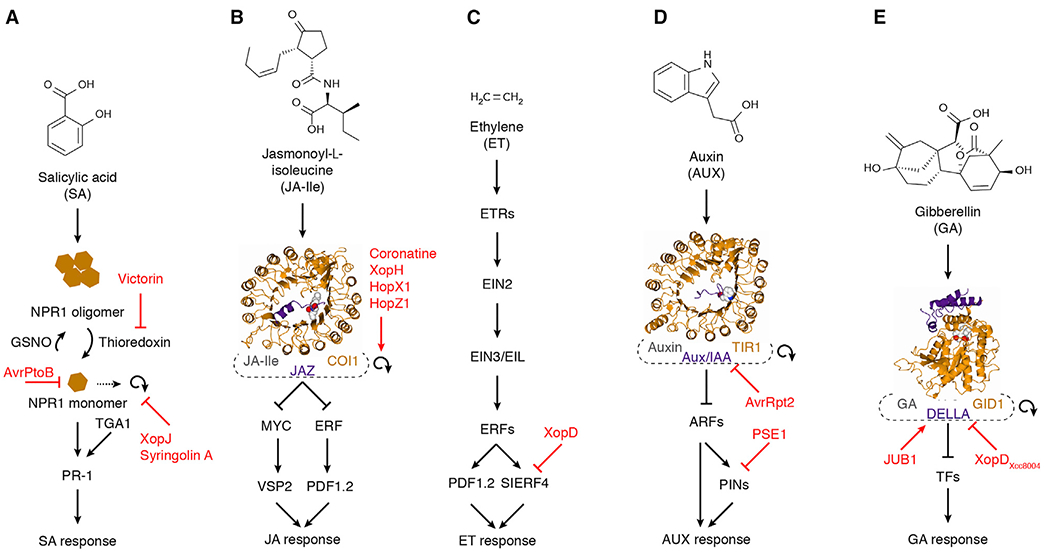
(A) Salicylic acid (SA). Victorin blocks NPR1 activation by inhibiting thioredoxin. XopJ and syringolin block necessary turnover of phosphorylated NPR1. AvrPtoB directly targets NPR1 for degradation.
(B) Jasmonic acid (JA). The JA-Ile mimic coronatine as well as the effector proteins XopH, HopX1, and HopZ1 induce JA signaling by causing turnover of the COI1-JAZ receptor complex.
(C) Ethylene (ET). XopD blocks ET signaling by inhibiting SIERF4.
(D) Auxin (AUX). AvrRpt2 initiates AUX signaling through degradation of Aux/IAA proteins. PSE1 changes AUX levels through altered distribution of PIN proteins.
(E) Gibberellin (GA). XopDXcc8004 blocks GA signaling by interfering with DELLA degradation, and JUB1 accumulates DELLAs through suppression of GA biosynthesis.
N-Hydroxypipecolic Acid: Priming and SAR
SAR signaling also works through nitric oxide (NO), the downstream molecules azelaic acid (AzA) and glycerol-3-phosphate (G3P), and several reactive oxygen species (ROS). Upon contact with pathogens, NO and ROS are activated in a synergistic relationship at the site of infection, in which NO mitigates the damaging roles of ROS via peroxynitrite (ONOO−1). ONOO−1 is the product of NO reacting with superoxide and is, unlike in animals, not a cell death mediator in plants. ONOO−1 keeps superoxide from being converted into hydrogen peroxide by the enzyme superoxide dismutase (Delledonne et al., 2001). ROS oxidize unsaturated fatty acids derived from membrane lipids, creating AzA. AzA upregulates genes for G3P-producing enzymes. G3P then induces SAR. The signal requires another component based on lysine-derived molecules. Upon pathogen infection, lysine is converted into Δ1-piperideine-2 carboxylic acid (P2C) by the enzyme AGD2-LIKE DEFENSE RESPONSE PROTEIN 1 (ALD1) and further processed to pipecolic acid (Pip) by SAR-DEFICIENT 4 (SARD4) and probably a yet undiscovered enzyme (Ding et al., 2016; Hartmann et al., 2017; Návarová et al., 2012). Návarová et al. (2012) showed that ALD1 is required for Pip accumulation and establishment of SAR. It was later found that Pip influences SA-dependent and SA-independent priming of defense responses dependent on the enzyme FLAVIN-DEPENDENT-MONOOXYGENASE1 (FMO1) (Bernsdorff et al., 2016). sard4 knockout plants show reduced accumulation of Pip in leaves distant to the site of infection but still retained SAR, suggesting that locally accumulated Pip triggers a systemic defense response (Ding et al., 2016; Hartmann et al., 2017). It was recently discovered by two different groups that Pip is converted into N-hydroxypipecolic acid (NHP) by FMO1 and that Pip fails to induce SAR in fmo1 mutant plants (Chen et al., 2018; Hartmann et al., 2018). NHP, and not Pip, is therefore now viewed as the SAR triggering molecule. NHP directly triggers activation of genes acting at the onset of SAR, likely to set defense preparedness in a systemic manner. AzA, G3P, Pip, and NHP are all transported from the infected to uninfected tissues. This onset of SAR constitutes a possible interaction point with SA signaling where systemic SA activity relies on amplification through a pre-established NHP signal. The interaction between SA signaling and the Pip pathway has been demonstrated on the transcriptional level (Bernsdorff et al., 2016), and npr1 mutant plants fail to establish resistance normally transduced through Pip (Návarová et al., 2012). Therefore, NPR1 might also be a merging point for integration of NHP-based defense priming. The origin of ROS that is needed to oxidize the fatty acids to generate AzA is an unclear element in this signaling environment, although the enzyme FMO1 was proposed as a potential source (Kachroo and Kachroo, 2018). The catalytic cycle of FMO1 includes the conversion of FAD-OH to FAD, which produces a water molecule but is prone to leakage of hydrogen peroxide or a superoxide radical (Ziegler, 2002).
JA: Defense against Necrotrophs and Herbivores
JA is usually produced during a defense response against necrotrophic pathogens and herbivores and is an antagonist of the SA response. JA is derived from the fatty acid α-linolenic acid, which is first released from phospholipids by the chloroplastic phospholipase A1 DEFECTIVE IN ANTHER DEHISCIENCE 1 (DAD1) and then oxygenated by the enzyme LYPOXYGENASE (LOX). The oxygenated form, OPDA, is then converted into JA. The endogenous bioactive form of jasmonate is not JA itself but (+)-7-iso-Jasmonoyl-L-isoleucine (JAIle), which is produced from JA by the enzyme JA AMIDO SYNTHETASE (JAR1) (Fonseca et al., 2009). The biosynthesis pathway is especially exploited by biotrophic pathogens that stimulate JA signaling to downregulate the SA response. For example, the hemibiotrophic fungus Fusarium oxysporum expresses its own LOX enzyme to upregulate JA signaling (Brodhun et al., 2013). Some P. syringae pathovars produce coronatine, a molecular mimic of JA-Ile (Mitchell, 1982) (Figure 2B). This triggers JA signaling and suppresses the SA response (Zheng et al., 2012) and in particular leads to the reopening of closed stomata to allow bacterial entry. The receptor for JA-Ile is a complex of the F-box protein CORONATINE INSENSITIVE 1 (COI1) and JASMONATE ZIM DOMAIN (JAZ). JAZ proteins repress the transcription factor MYC2, an activator of jasmonate response genes (Fernández-Calvo et al., 2011). The presence of JA-Ile induces recruitment of JAZ proteins into a complex with COI1 and its subsequent degradation (Thines et al., 2007). JA-Ile and the bacterial mimic coronatine bind at the same position in the receptor, but coronatine is more tightly accommodated in the binding pocket, leading to a 10-fold higher affinity than JAIle. The sugar inositol pentakisphosphate (InsP5) is an essential component of the receptor complex (Sheard et al., 2010). In a newly discovered way to remodel hormone balance, one bacterial effector directly manipulates InsP homeostasis. The Xanthomonas protein XopH converts inositol hexakisphosphate (InsP6) into InsP5, which leads to an upregulation of JA signaling, ultimately antagonizing SA responses (Blüher et al., 2017). Interestingly, COI1 responds to different ligands in different species, and a recent report has demonstrated that the bioactive signaling molecule in the bryophyte Marchantia polymorpha is not JA-Ile but in fact the precursor OPDA (Monte et al., 2018). In vascular plants, protection against herbivores or necrotrophs and COI1-dependent gene expression can be mediated by OPDA when JA-Ile is unavailable, whereas JA-Ile is indispensable for certain developmental processes such as pollen maturation (Stintzi et al., 2001). The antagonistic nature of the JA-SA relationship is exploited at the gene regulatory level as well. The P. syringae effector HopZ1a directly targets the receptor complex and acetylates the ZIM domain of JAZ proteins, thereby causing their proteasomal degradation through an unknown mechanism (Jiang et al., 2013). Shortly thereafter, the P. syringae effector protein HopX1 was reported to act as a protease that degrades JAZ proteins (Gimenez-Ibanez et al., 2014). Another branch of JA signaling works through the ETHYLENE RESPONSE FACTOR (ERF) transcription factor family and PLANT DEFENSIN 1.2 (PDF1.2). JAZ proteins also repress ERF transcription factors, a regulator that is also part of the ET signaling pathway. Therefore, two branches in the JA signaling pathway exist, antagonistically regulated by MYC2 and ERF1 (Figure 3B). Thus, activation of JA can either get synergistically transduced with the ET response or act independently of ET signaling through the MYC2 part of the JA pathway.
ET Acts Together with JA
ET is a hydrocarbon gas that is involved in plant immunity. ET is derived from the amino acid methionine. The enzyme S-ADENOSYLMETHIONINE (SAM) SYNTHASE converts methionine into S-adenosylmethionine, which is then converted into 1-aminocyclopropane-1-carboxylic acid (ACC) by ACC SYNTHASE. The final step is the conversion of 1-aminocyclopropane-1-carboxylic acid into ET, which is carried out by the enzyme ACC OXIDASE (Figure 2C). ET itself is perceived by several receptor proteins, ETHYLENE RESPONSE 1 and 2 (ETR1 and ETR2), ETHYLENE RESPONSE SENSOR 1 and 2 (ERS1 and ERS2), and ETHYLENE INSENSITIVE 4 (EIN4). These proteins are histidine kinases, the activation of which inhibits the downstream serine/threonine kinase CONSTITUTIVE TRIPLE RESPONSE 1 (CTR1). In the absence of ET, CTR1 phosphorylates the C-terminal domain of ETHYLENE INSENSITIVE 2 (EIN2). Subsequently, dephosphorylated EIN2 will translocate into the nucleus and stabilize ETHYLENE INSENSITIVE 3 (EIN3) and EIN3-LIKE (EIL) transcription factors that activate ET-controlled genes. EIN3 binds a primary ET response element present in the ERF promoters, activating the transcription of ERF genes. This in turn will activate both transcription factors PDF1.2 and SOLANUM LYCOPERSICUM ETHYLENE RESPONSE FACTOR 4 (SIERF4) (Merchante et al., 2013) (Figure 3C). Pathogens exploit the fact that ET and JA have a synergistic role, which ultimately leads to an antagonistic role between ET and SA. Therefore, ET signaling is a target of many pathogens. As a matter of fact, many pathovars of P. syringae produce ET themselves and others have found ways of forcing the host to increase ET production. The Pseudomonas effectors AvrPto and AvrPtoB trigger ET production in tomato (Cohn and Martin, 2005), an effect that might work indirectly through the previously discussed downregulation of SA signaling by AvrPtoB that was found in tobacco (Chen et al., 2017). ET has been shown to be critical for Xanthomonas euvesitorica-elicited symptom development but not for pathogen inhibition. X. euvesitorica causes bacterial leaf spot and deploys the effector protein XopD, which has a dual function. Like many hemibiotrophic effector proteins, XopD represses SA-dependent gene expression and SA production (Kim et al., 2008). To further delay chlorosis and necrosis in the host, however, XopD also suppresses ET signaling by directly targeting and desumoylating the transcription factor SIERF4 (Kim et al., 2013).
AUX
AUX plays a rather peripheral role in plant defense signaling, as it is usually involved in growth processes. There are five different pathways of AUX production in plants and four of those have in common that they derive AUX from the amino acid tryptophan. After production, AUX can be converted into several conjugates, which are likely not biologically active themselves but serve as storage molecules. In the cell, expression of AUX response genes is generally repressed by the Aux/IAA family of transcription regulators. AUX binds to the F-box protein TRANSPORT INHIBITOR RESPONSE 1 (TIR1) and stabilizes a complex with the Aux/IAA repressor and a ubiquitin ligase (Dharmasiri et al., 2005; Kepinski and Leyser, 2005). Consequently, the entire complex is degraded, killing off the repressor and subsequently activating AUX response genes (Figure 3D). The sugar InsP6 is a cofactor of the TIR receptor complex (Tan et al., 2007), similar to the previously mentioned InsP5 molecule that serves as cofactor in the JA receptor.
The presence of AUX renders plants more susceptible to biotrophic pathogens, and exogenous AUX application leads to increased virulence and disease (Mutka et al., 2013; Navarro et al., 2006; Wang et al., 2007). AUX signaling often gets down-regulated upon infection with biotrophic pathogens, and the signaling repressor Aux/IAA is stabilized by SA. This is likely due to the synergistic role that AUX has to JA signaling, and therefore, together with JA, AUX promotes resistance against necrotrophic pathogens. A few pathogens have been found to exploit this relationship. The P. syringae effector protein AvrRpt2 promotes degradation of Aux/IAA proteins, and therefore initiates AUX signaling (Cui et al., 2013), and the Phytophthora parasitica effector PSE1 acts by local modulation of AUX levels through altered distribution of PIN-FORMED (PIN) proteins, a family of AUX efflux transporters (Evangelisti et al., 2013). In addition to these strategies, several pathogens can produce AUX themselves (Kunkel and Harper, 2018).
GAs
GAs are a class of hormones that are typically involved in various developmental processes such as stem elongation, fruit senescence, and breaking seed dormancy. GAs are produced from the precursor geranylgeranyl diphosphate in four steps, and so far, more than 130 different GAs have been discovered, though only a few have known biological activity (Yamaguchi, 2008). GA perception in plants is carried out through the receptor protein GIBBERELLIN INSENSITIVE DWARF 1 (GID1) (Ueguchi-Tanaka et al., 2005). GA responses are repressed by DELLA proteins. The presence of GA will lead to a complex of GA, GID1, and DELLA, and the entire complex is subject to proteasomal degradation (Figure 3E) (Sasaki et al., 2003). Based on the findings that SA responses against biotrophic pathogens are increased in della mutants, it is thought that pathogens target GA signaling because of its interactions with the more important defense pathways (Navarro et al., 2008). In addition, the competition of DELLA and MYC2 proteins for binding to JAZ proteins is likely an important intersection of the GA and JA pathways. Activation of GA signaling and subsequent degradation of DELLA will allow MYC2 to interact with JAZ, consequently blocking JA activation (Hou et al., 2010). It is telling that GAs were initially named after the necrotrophic fungus Gibberella fujikuroi, now reclassified as Fusarium fujikuroi, which was associated with unusual elongation of rice (Yabuta and Sumiki, 1938). The fungus represses JA activation by secretion of a GA mimic. On the other side, XopDXcc8004, an effector from Xanthomonas campestris, blocks GA-mediated DELLA degradation, ultimately limiting the SA response (Tan et al., 2014). However, this is counteracted by the plant protein JUNGBRUNNEN 1 (JUB1), which accumulates DELLA proteins through suppression of GA biosynthesis and by transcriptional activation of DELLA genes (Shahnejat-Bushehri et al., 2016).
ABA
The hormone ABA is traditionally associated with abiotic stress response. Based on the observation that plants treated with ABA have increased susceptibility to biotrophic fungi, for example Phytophthora infestans (Henfling et al., 1980) and Peronospora tabacina (Salt et al., 1986), it is assumed that ABA has an antagonistic role to SA. In turn, ABA signaling or biosynthesis mutants are more susceptible to the necrotrophic fungi Alternaria brassicicola and Pythium irregulare (Adie et al., 2007). In addition, it has been demonstrated that an ABA biosynthesis mutant is less susceptible to the biotroph P. syringae, which can be reverted by exogenous application of ABA (Fan et al., 2009). In particular, the bacterial effector AvrPtoB reportedly modifies ABA signaling (de Torres-Zabala et al., 2007), which might be an indirect observation originating from AvrPtoB’s direct targeting of NPR1, the master regulator of SA signaling (Chen et al., 2017).
Cytokinins
Cytokinins (CKs) are mainly known for their role in cell division, but their control over senescence has suggested possible exploitation by plant pathogens. A well-studied case of bacterial CK production is the bacterial phytopathogen Rhodococcus fascians, which is able to produce three different CKs in order to force continued tissue proliferation in infected areas (Pertry et al., 2009). CK production has also been found in the biotrophic fungi Pyrenopeziza brassicae, Cladosporium fulvum, and Blumeria graminis as well as in the hemibiotrophs Phyllosticta brassicae and Venturia inaequalis (Murphy et al., 1997). Senescence is delayed at the infection site, leading to so-called green island formation. In addition to these effects, the P. syringae effector protein HopQ1 works through activation of the CK pathway in order to suppress immunity mediated by FLAGELLIN-SENSITIVE 2 (FLS2), a PRR that recognizes bacterial flagellin (flg22) (Hann et al., 2014).
BRs: Balancing Growth and Defense
BRs are a class of growth hormones. The most potent BR, brassinolide, is derived from the sterol campesterol. Brassinolide binds to a 70-amino acid island domain of the dual-specificity receptor kinase BRASSINOSTEROID INSENSITIVE 1 (BRI1), which contains a large extracellular domain composed of 25 leucine-rich repeats (Hothorn et al., 2011). BR binding activates the kinase function of the receptor, and through a phosphorylation cascade, the GLYCOGEN SYNTHASE KINASE 3 (GSK3)-like kinase BRASSINOSTEROID INSENSITIVE 2 (BIN2) is inactivated. BIN2 usually phosphorylates and inhibits the BR transcription factors BRASSINAZOLE-RESISTANT 1 (BZR1) and BRI1-EMS-SUPPRESSOR 1 (BES1). The presence of BR, therefore, keeps BZR1 and BES1 dephosphorylated, allowing them to activate BR-specific genes (He et al., 2002). The fact that BRI1 interacts with BRI1-ASSOCIATED RECEPTOR KINASE 1 (BAK1), which in turn interacts with the flg22 receptor FLS2, has led to early speculation that BAK1 might manage the trade-off between growth and defense as flg22 responses can be inhibited through BR. In 2012, two different groups published reports about the interaction. While one side concluded that the BR-triggered inhibition of flg22 is independent of BAK1 (Albrecht et al., 2012), the other group reported concentration-dependent effects because BRI1 is able to recruit BAK1 away from MAMP defense receptors. In their model, the amount of available BAK1 protein pool would ultimately be the deciding factor over the growth-defense trade-off (Belkhadir et al., 2012). Given this prominent role, it is not surprising that BAK1 itself is the target of effector proteins, and P. syringae AvrPto and AvrPtoB bind to BAK1 to suppress immune signaling (Cheng et al., 2011; Shan et al., 2008).
Studies in rice have revealed that BR treatment makes the host more susceptible to the root pathogens Pythium graminicola and Meloidogyne graminicola. P. graminicola hijacks the rice BR machinery and initiates negative cross talk with the SA and GA pathways (De Vleesschauwer et al., 2012), and M. graminicola engages in downregulation of the JA pathway after BR treatment (Nahar et al., 2013). BR signaling also intersects with the GA pathway through interaction between the BR transcription factor BZR1 and DELLA proteins (Gallego-Bartolomé et al., 2012), which are negative regulators of GA signaling. In addition, BZR1 induces expression of several WRKY transcription factors, which downregulate early immune response, and in that regard, BZR1 directly interacts with WRKY40 (Lozano-Durán et al., 2013). Recently, a new concept has been pushed forward, demonstrating that the BR transcription factors are directly regulated by ROS. Hydrogen peroxide causes oxidative modification on the transcription factor BZR1, which increases its interaction with AUX and light pathway regulators such as ARF6 and PIF4, respectively (Song et al., 2019). Another recent study reported that the interaction between BIN2 and the transcription factor BES1 is ROS regulated and that BIN2 activity is oxygen dependent (Tian et al., 2018).
Signaling Hubs
Exposure to biotrophic and necrotrophic bacteria at the same time requires fine tuning of defense strategies, and the activity of hemibiotrophs that switch their lifestyle, requires a correspondingly quick response by the plant. These defense strategies need to be merged with other environmental challenges, such as temperature, light, and drought, all of which affect growth. Several regulatory hubs have emerged that allow the initiation or interaction of several different hormone pathways. Among the most investigated of these are certainly the DELLA proteins, which interact with three different hormone pathways, downregulating BR and GA signaling, while stimulating JA response. Given the importance of DELLAs, it is somehow surprising that so far only the bacterial effector XopDXcc8004 and counteracting plant protein JUB1 have been found to manipulate the concentration of DELLA in the cell (Figure 4A).
Figure 4. Hormone Pathways Merge at Several Signaling Hubs.
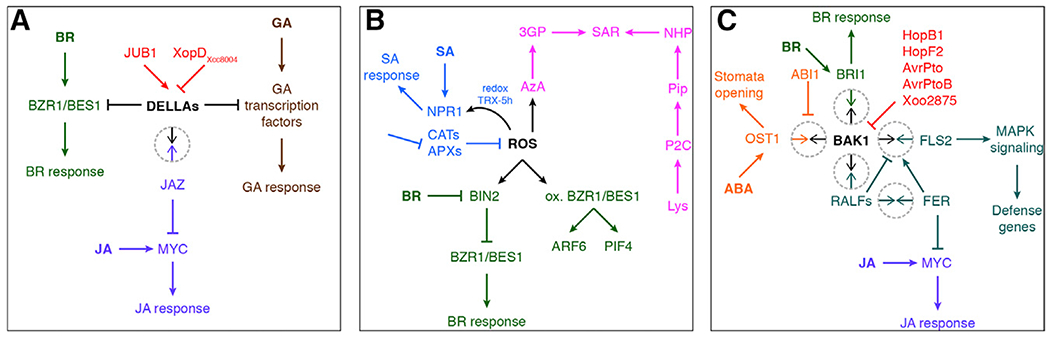
(A) DELLA proteins integrate signals from the JA, GA, and BR pathways.
(B) ROS trigger SAR priming and SA response and have both inhibitory and activating effects on the BR pathway.
(C) BAK1 is a co-receptor of many receptors and thus integrates signals from the BR, JA, and ABA pathways as well as immune signaling through peptides. Protein-protein interactions in (A) and (C) are shown as encircled arrows.
During the establishment of SAR, the SA transcription factor NPR1 not only functions as a likely crossing point of SA signaling and Pip activity but is also impacted by ROS, which directly influences NPR because its protein activity requires redox-triggered monomerization. In turn, SA activity inhibits ROS-degrading enzymes. Direct interaction of ROS with components of the BR signaling pathway is a novel concept that appears in part contradictory. On one hand, BIN2 activity seems to be stimulated by oxygen, which would deactivate BR signaling by inhibition of the transcription factors BZR1 and BES1. On the other hand, hydrogen peroxide directly stimulates the interaction of BZR1 to the light pathway regulators ARF6 and PIF4 (Figure 4B). In that regard, ROS might narrow down BR signaling to a limited number of targets using a limited pool of BZR1 and BES1.
Another prominent interaction point is BAK1, a co-receptor of many other receptors, including multiple PRRs (Chinchilla et al., 2009). As a co-receptor for both the flg22 receptor FLS2 and the BR receptor BRI1, BAK1’s role in a trade-off between defense and growth appears obvious. However, BAK1 is also embedded in the interaction between the peptide RAPID ALKALINIZATION FACTOR (RALF) and its receptor FERONIA (FER). FER suppresses JA signaling and is controlled by RALF peptides. RALFs are small peptide hormones that inhibit root growth by negatively regulating cell expansion and were found to induce mitogen-activated protein (MAP) kinase activation (Bedinger et al., 2010; Bergonci et al., 2014). RALF can bind directly to BAK1 (Dressano et al., 2017) but also inhibits BAK1’s binding to FLS2, an interaction that is conversely stabilized by FER. Signaling of several other immunity involved peptides have been found to be dependent on BAK1, such as Pep1 (Schulze et al., 2010), PIP1 (Hou et al., 2014), and SCOOP12 (Gully et al., 2019). In addition to those interactions, BAK1 also regulates the ABA-induced stomatal closure in guard cells via its interaction with OPEN STOMATA 1 (OST1), and bak1 mutant plants display ABA insensitivity in stomatal closure. Given BAK1’s importance, it is not surprising that to date 5 effector proteins have been identified that target its activity (Figure 4C).
Conclusion and Future Directions
Several exciting discoveries in the recent years have both refined and repositioned our understanding about how plants integrate different defense signals. An outstanding finding in 2018 has been the identification of NHP as the molecule that induces the onset of SAR. It has been known before that Pip is required for initiation of SAR, and it has now become clear that Pip is the precursor of the actual SAR inducing molecule NHP. In addition to NHP, one of these studies identified an NHP glucoside conjugate (N-OGlc-Pip). It is not entirely clear whether this conjugate is a storage form of NHP or if it also acts as a signaling molecule. It has been suggested that the NHP-producing enzyme FMO1 is a potential feeding source of the ROS-NO cycle driving AzA/3GP. Since there is no direct proof for H2O2 production by FMO1, and NHP rescues the SAR-deficient fmo1 mutant, this idea has to be considered speculation at this point. Considering the recent publications about the direct influence of ROS in the BR signaling pathway, it will be exciting to see if future publications will expand the already general role of ROS in signaling. Another interesting question is whether there is functional overlap between FMO1 and other FMO proteins. The Arabidopsis genome encodes for 29 FMO genes, and there are 3 different clades of FMO enzymes, one of which comprises the YUCCA proteins that are involved in AUX biosynthesis (Schlaich, 2007). Although FMO1 is located in a separate clade, its sequence identity to YUC6, which synthesizes AUX from indole-3-pyruvic acid (IPA), is as high as 71%.
An intriguing and novel concept is the alteration of receptor sugar cofactors to manipulate hormone signaling. The phytase activity of Xanthomonas effector XopH produces the cofactor InsP5, which is required for the activity of the JA receptor. However, InsP6, XopH’s substrate, is not only the cofactor of the AUX receptor but is also required for activity of the YopJ family of bacterial effector proteins. This raises the question whether XopH’s activity is only designed to alter hormone homeostasis or if it is embedded in a competition with other pathogens that use InsP6 as well. One might even speculate that a class of effector proteins exists where InsP5 acts as a cofactor, and XopH is a tool to alter sugar pools to their favor.
Lastly, the nature of signal integration at the BAK1 receptor is still elusive. Do BAK1 ligands and co-receptors have different affinities to BAK1 and are these biologically important? And to what degree does the amount of available BAK1 protein at different developmental stages and environmental situations contribute?
It has become increasingly clear that integration of different hormone signaling pathways occurs at more levels than previously thought. On one hand, this allows plants to exert more complex hormone communication and, on the other hand, puts selection pressure on pathogens to manipulate every one of these interaction nodes. While production of either the hormone itself or a molecular mimic seems to be rare, many strategies either induce or block turnover of central regulators or receptor proteins. It should be pointed out that the relationships between signaling pathways are not always as static and straightforward as described here, but they highly depend on the environmental context of the plant. It is also noteworthy that many attack and defense strategies might have escaped our attention because they simply do not produce a disease phenotype. Thus, protein mass spectroscopy and metabolomics will hopefully aid to uncover more host targets and counterstrategies in the future.
ACKNOWLEDGMENTS
Our studies of plant hormones have been supported by NIH grants R01 GM52413 and R01 GM094428. We are currently supported by NIH grant R35 GM122604. Apologies are offered to those colleagues whose work was not cited due to space constraints.
REFERENCES
- Adie BAT, Pérez-Pérez J, Pérez-Pérez MM, Godoy M, Sánchez-Serrano JJ, Schmelz EA, and Solano R (2007). ABA is an essential signal for plant resistance to pathogens affecting ja biosynthesis and the activation of defenses in Arabidopsis. Plant Cell 19, 1665–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht C, Boutrot F, Segonzac C, Schwessinger B, Gimenez-Ibanez S, Chinchilla D, Rathjen JP, deVries SC, and Zipfel C (2012). Brassinosteroids inhibit pathogen-associated molecular pattern-triggered immune signaling independent of the receptor kinase BAK1. Proc. Natl. Acad. Sci. USA 109, 303–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedinger PA, Pearce G, and Covey PA (2010). RALFs: peptide regulators of plant growth. Plant Signal. Behav. 5, 1342–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkhadir Y, Jaillais Y, Epple P, Balsemão-Pires E, Dangl JL, and Chory J (2012). Brassinosteroids modulate the efficiency of plant immune responses to microbe-associated molecular patterns. Proc. Natl. Acad. Sci. USA 109, 297–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergonci T, Ribeiro B, Ceciliato PH, Guerrero-Abad JC, Silva-Filho MC, and Moura DS (2014).Arabidopsis thaliana RALF1 opposes brassinosteroid effects on root cell elongation and lateral root formation. J. Exp. Bot. 65, 2219–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernsdorff F, Döring AC, Gruner K, Schuck S, Bräutigam A, and Zeier J (2016). Pipecolic acid orchestrates plant systemic acquired resistance and defense priming via salicylic acid-dependent and -independent pathways. Plant Cell 28, 102–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blüher D, Laha D, Thieme S, Hofer A, Eschen-Lippold L, Masch A, Balcke G, Pavlovic I, Nagel O, Schonsky A, et al. (2017). A 1-phytase type III effector interferes with plant hormone signaling. Nat. Commun. 8,2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodhun F, Cristobal-Sarramian A, Zabel S, Newie J, Hamberg M, and Feussner I (2013). An iron 13S-lipoxygenase with an a-linolenic acid specific hydroperoxidase activity from Fusarium oxysporum. PLoS One 8, e64919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Chen J, Li M, Chang M, Xu K, Shang Z, Zhao Y, Palmer I, Zhang Y, McGill J, et al. (2017).Abacterialtype III effector targets the master regulator of salicylic acid signaling, NPR1, to subvert plant immunity. Cell Host Microbe 22, 777–788. e7. [DOI] [PubMed] [Google Scholar]
- Chen YC, Holmes EC, Rajniak J, Kim JG, Tang S, Fischer CR, Mudgett MB, and Sattely ES (2018). N-hydroxy-pipecolic acid is a mobile metabolite that induces systemic disease resistance in Arabidopsis. Proc. Natl. Acad. Sci. USA 115, E4920–E4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng W, Munkvold KR, Gao H, Mathieu J, Schwizer S, Wang S, Yan YB, Wang J, Martin GB, and Chai J (2011). Structural analysis of Pseudomonas syringae AvrPtoB bound to host BAK1 reveals two similar kinase-interacting domains in a type III effector. Cell Host Microbe 10, 616–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D, Shan L, He P, deVries S, and Kemmerling B (2009). One for all: the receptor-associated kinase BAK1. Trends Plant Sci. 14, 535–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm ST, Coaker G, Day B, and Staskawicz BJ (2006). Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124, 803–814. [DOI] [PubMed] [Google Scholar]
- Cohn JR, and Martin GB (2005). Pseudomonas syringae pv. tomato type III effectors AvrPto and AvrPtoB promote ethylene-dependent cell death in tomato. Plant J. 44, 139–154. [DOI] [PubMed] [Google Scholar]
- Cui F, Wu S, Sun W, Coaker G, Kunkel B, He P, and Shan L (2013). The Pseudomonas syringae type III effector AvrRpt2 promotes pathogen virulence via stimulating Arabidopsis auxin/indole acetic acid protein turnover. Plant Physiol. 162, 1018–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Torres-Zabala M, Truman W, Bennett MH, Lafforgue G, Mansfield JW, Rodriguez Egea P, Bogre L, and Grant M (2007). Pseudomonas syringae pv. tomato hijacks the Arabidopsis abscisic acid signalling pathway to cause disease. eMbO J. 26, 1434–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vleesschauwer D, Van Buyten E, Satoh K, Balidion J, Mauleon R, Choi IR, Vera-Cruz C, Kikuchi S, and Höfte M (2012). Brassinosteroids antagonize gibberellin- and salicylate-mediated root immunity in rice. Plant Physiol. 158, 1833–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delledonne M, Zeier J, Marocco A, and Lamb C (2001). Signal interactions between nitric oxide and reactive oxygen intermediates in the plant hypersensitive disease resistance response. Proc. Natl. Acad. Sci. USA 98, 13454–13459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, and Estelle M (2005). The F-boxprotein TIR1 is an auxin receptor. Nature 435, 441–445. [DOI] [PubMed] [Google Scholar]
- Ding P, Rekhter D, Ding Y, Feussner K, Busta L, Haroth S, Xu S, Li X, Jetter R, Feussner I, et al. (2016). Characterization of a pipecolic acid biosynthesis pathway required for systemic acquired resistance. Plant Cell 28, 2603–2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Sun T, Ao K, Peng Y, Zhang Y, Li X, and Zhang Y (2018). Opposite roles of salicylic acid receptors NPR1 and NPR3/NPR4 in transcriptional regulation of plant immunity. Cell 173, 1454–1467. e15. [DOI] [PubMed] [Google Scholar]
- Djamei A, Schipper K, Rabe F, Ghosh A, Vincon V, Kahnt J, Osorio S, Tohge T, Fernie AR, Feussner I, et al. (2011). Metabolic priming by a secreted fungal effector. Nature 478, 395–398. [DOI] [PubMed] [Google Scholar]
- Dressano K, Ceciliato PHO, Silva AL, Guerrero-Abad JC, Bergonci T, Ortiz-Morea FA, Bürger M, Silva-Filho MC, and Moura DS (2017). BAK1 is involved in AtRALF1-induced inhibition of root cell expansion. PLoS Genet. 13, e1007053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelisti E, Govetto B, Minet-Kebdani N, Kuhn ML, Attard A, Ponchet M, Panabières F, and Gourgues M (2013). The Phytophthora parasitica RXLR effector penetration-specific effector 1 favours Arabidopsis thaliana infection by interfering with auxin physiology. New Phytol. 199, 476–489. [DOI] [PubMed] [Google Scholar]
- Fan J, Hill L, Crooks C, Doerner P, and Lamb C (2009). Abscisic acid has a key role in modulating diverse plant-pathogen interactions. Plant Physiol. 150, 1750–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Calvo P, Chini A, Fernández-Barbero G, Chico JM, Gimenez-Ibanez S, Geerinck J, Eeckhout D, Schweizer F, Godoy M, Franco-Zorrilla JM, et al. (2011). The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23, 701–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca S, Chini A, Hamberg M, Adie B, Porzel A, Kramell R, Miersch O, Wasternack C, and Solano R (2009). (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat. Chem. Biol. 5, 344–350. [DOI] [PubMed] [Google Scholar]
- Fu ZQ, Yan S, Saleh A, Wang W, Ruble J, Oka N, Mohan R, Spoel SH, Tada Y, Zheng N, et al. (2012). NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 486, 228–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Bartolomé J, Minguet EG, Grau-Enguix F, Abbas M, Locascio A, Thomas SG, Alabadi D, and Blázquez MA (2012). Molecular mechanism for the interaction between gibberellin and brassinosteroid signaling pathways in Arabidopsis. Proc. Natl. Acad. Sci. USA 109, 13446–13451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimenez-Ibanez S, Boter M, Fernández-Barbero G, Chini A, Rathjen JP, and Solano R (2014). The bacterial effector HopX1 targets JAZ transcriptional repressors to activate jasmonate signaling and promote infection in Arabidopsis. PLoS Biol. 12, e1001792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gully K, Pelletier S, Guillou MC, Ferrand M, Aligon S, Pokotylo I, Perrin A, Vergne E, Fagard M, Ruelland E, et al. (2019). The SCOOP12 peptide regulates defense response and root elongation in Arabidopsis thaliana. J. Exp. Bot. 70, 1349–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hann DR, Domínguez-Ferreras A, Motyka V, Dobrev PI, Schornack S, Jehle A, Felix G, Chinchilla D, Rathjen JP, and Boller T (2014). The Pseudomonas type III effector HopQ1 activates cytokinin signaling and interferes with plant innate immunity. New Phytol. 201, 585–598. [DOI] [PubMed] [Google Scholar]
- Hartmann M, Kim D, Bernsdorff F, Ajami-Rashidi Z, Scholten N, Schreiber S, Zeier T, Schuck S, Reichel-Deland V, and Zeier J (2017). Biochemical principles and functional aspects of pipecolic acid biosynthesis in plant immunity. Plant Physiol. 174, 124–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann M, Zeier T, Bernsdorff F, Reichel-Deland V, Kim D, Hohmann M, Scholten N, Schuck S, Bräutigam A, Hölzel T, et al. (2018). Flavin monooxygenase-generated N-hydroxypipecolic acid is a critical element of plant systemic immunity. Cell 173, 456–469. e16. [DOI] [PubMed] [Google Scholar]
- He JX, Gendron JM, Yang Y, Li J, and Wang ZY (2002). The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 99, 10185–10190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henfling J, Bostock R, and Kuc J (1980). Effect of abscisic acid on rishitin and lubimin accumulation and resistance to Phytophthora infestans and Cladosporium cucumerinum in potato tuber tissue slices. Phytopathology 70, 1074–1078. [Google Scholar]
- Hothorn M, Belkhadir Y, Dreux M, Dabi T, Noel JP, Wilson IA, and Chory J (2011). Structural basis of steroid hormone perception by the receptor kinase BRI1. Nature 474, 467–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou S, Wang X, Chen D, Yang X, Wang M, Turrà D, Di Pietro A, and Zhang W (2014). The secreted peptide PIP1 amplifies immunity through receptor-like kinase 7. PLoS Pathog. 10, e1004331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X, Lee LYC, Xia K, Yan Y, and Yu H (2010). DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev. Cell 19, 884–894. [DOI] [PubMed] [Google Scholar]
- Jiang S, Yao J, Ma KW, Zhou H, Song J, He SY, and Ma W (2013). Bacterial effector activates jasmonate signaling by directly targeting JAZ transcriptional repressors. PLoS Pathog. 9, e1003715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JDG, and Dangl JL (2006). The plant immune system. Nature 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Kachroo P, and Kachroo A (2018). Plants pack a quiver full of arrows. Cell Host Microbe 23, 573–575. [DOI] [PubMed] [Google Scholar]
- Kepinski S, and Leyser O (2005). The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435, 446–451. [DOI] [PubMed] [Google Scholar]
- Kim JG, Stork W, and Mudgett MB (2013). Xanthomonas type III effector XopD desumoylates tomato transcription factor SlERF4 to suppress ethylene responses and promote pathogen growth. Cell Host Microbe 13, 143–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JG, Taylor KW, Hotson A, Keegan M, Schmelz EA, and Mudgett MB (2008). XopD SUMO protease affects host transcription, promotes pathogen growth, and delays symptom development in Xanthomonas-infected tomato leaves. Plant Cell 20, 1915–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel BN, and Harper CP (2018). The roles of auxin during interactions between bacterial plant pathogens and their hosts. J. Exp. Bot. 69, 245–254. [DOI] [PubMed] [Google Scholar]
- Liu T, Song T, Zhang X, Yuan H, Su L, Li W, Xu J, Liu S, Chen L, Chen T, et al. (2014). Unconventionally secreted effectors of two filamentous pathogens target plant salicylate biosynthesis. Nat. Commun. 5, 4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorang J, Kidarsa T, Bradford CS, Gilbert B, Curtis M, Tzeng SC, Maier CS, and Wolpert TJ (2012). Tricking the guard: exploiting plant defense for disease susceptibility. Science 338, 659–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano-Durán R, Macho AP, Boutrot F, Segonzac C, Somssich IE, and Zipfel C (2013). The transcriptional regulator BZR1 mediates trade-off between plant innate immunity and growth. ELife 2, e00983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchante C, Alonso JM, and Stepanova AN (2013). Ethylene signaling: simple ligand, complex regulation. Curr. Opin. Plant Biol. 16, 554–560. [DOI] [PubMed] [Google Scholar]
- Métraux JP, Signer H, Ryals J, Ward E, Wyss-Benz M, Gaudin J, Raschdorf K, Schmid E, Blum W, and Inverardi B (1990). Increase in salicylic acid at the onset of systemic acquired resistance in cucumber. Science 250,1004–1006. [DOI] [PubMed] [Google Scholar]
- Mitchell RE (1982). Coronatine production by some phytopathogenic pseudomonads. Physiol. Plant Pathol. 20, 83–89. [Google Scholar]
- Monte I, Ishida S, Zamarrerño AM, Hamberg M, Franco-Zorrilla JM, García-Casado G, Gouhier-Darimont C, Reymond P, Takahashi K, García-Mina JM, et al. (2018). Ligand-receptor co-evolution shaped the jasmonate pathway in land plants. Nat. Chem. Biol. 14, 480–488. [DOI] [PubMed] [Google Scholar]
- Murphy AM, Pryce-Jones E, Johnstone K, and Ashby AM (1997). Comparison of cytokinin production in vitro by Pyrenopeziza brassicae with other plant pathogens. Physiol. Mol. Plant Pathol. 50, 53–65. [Google Scholar]
- Mutka AM, Fawley S, Tsao T, and Kunkel BN (2013). Auxin promotes susceptibility to Pseudomonas syringae via a mechanism independent of suppression of salicylic acid-mediated defenses. Plant J. 74, 746–754. [DOI] [PubMed] [Google Scholar]
- Nahar K, Kyndt T, Hause B, Höfte M, and Gheysen G (2013). Brassinosteroids suppress rice defense against root-knot nematodes through antagonism with the jasmonate pathway. Mol. Plant Microbe Interact. 26, 106–115. [DOI] [PubMed] [Google Scholar]
- Návarová H, Bernsdorff F, Döring AC, and Zeier J (2012). Pipecolic acid, an endogenous mediator of defense amplification and priming, is a critical regulator of inducible plant immunity. Plant Cell 24, 5123–5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro L, Bari R,Achard P, Lisón P, Nemri A, Harberd NP, and Jones JDG (2008). DELLAs control plant immune responses by modulating the balance of jasmonic acid and salicylic acid signaling. Curr. Biol. 18, 650–655. [DOI] [PubMed] [Google Scholar]
- Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, Estelle M, Voinnet O, and Jones JDG (2006). A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312, 436–439. [DOI] [PubMed] [Google Scholar]
- Park SW, Kaimoyo E, Kumar D, Mosher S, and Klessig DF (2007). Methyl salicylate is a critical mobile signal for plant systemic acquired resistance. Science 318, 113–116. [DOI] [PubMed] [Google Scholar]
- Pertry I, Václavíková K, Depuydt S, Galuszka P, Spíchal L, Temmerman W, Stes E, Schmülling T, Kakimoto T, Van Montagu MCE, et al. (2009). Identification of Rhodococcus fascians cytokinins and their modus operandi to reshape the plant. Proc. Natl. Acad. Sci. USA 106, 929–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rekhter D, Lüdke D, Ding Y, Feussner K, Zienkiewicz K, Lipka V, Wiermer M, Zhang Y, and Feussner I (2019). Isochorismate-derived biosynthesis of the plant stress hormone salicylic acid. Science 365, 498, 10.1126/science.aaw1720. [DOI] [PubMed] [Google Scholar]
- Salt SD, Tuzun S, and Kuć J (1986). Effects of β-ionone and abscisic acid on the growth of tobacco and resistance to blue mold. Mimicry of effects of stem infection by Peronospora tabacina Adam. Physiol. Mol. Plant Pathol 28, 287–297. [Google Scholar]
- Sasaki A, Itoh H, Gomi K, Ueguchi-Tanaka M, Ishiyama K, Kobayashi M, Jeong DH, An G, Kitano H, Ashikari M, et al. (2003). Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science 299, 1896–1898. [DOI] [PubMed] [Google Scholar]
- Schellenberg B, Ramel C, and Dudler R (2010). Pseudomonas syringae virulence factor syringolin A counteracts stomatal immunity by proteasome inhibition. Mol. Plant Microbe Interact. 23, 1287–1293. [DOI] [PubMed] [Google Scholar]
- Schlaich NL (2007). Flavin-containing monooxygenases in plants: looking beyond detox. Trends Plant Sci. 12, 412–418. [DOI] [PubMed] [Google Scholar]
- Schulze B, Mentzel T, Jehle AK, Mueller K, Beeler S, Boller T, Felix G, and Chinchilla D (2010). Rapid heteromerization and phosphorylation of ligand-activated plant transmembrane receptors and their associated kinase BAK1.J. Biol. Chem. 285, 9444–9451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahnejat-Bushehri S, Nobmann B, Devi Allu A, and Balazadeh S (2016). JUB1 suppresses Pseudomonas syringae-induced defense responses through accumulation of DELLA proteins. Plant Signal. Behav. 11, e1181245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan L, He P, Li J, Heese A, Peck SC, Nürnberger T, Martin GB, and Sheen J (2008). Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor-signaling complexes and impede plant immunity. Cell Host Microbe 4, 17–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheard LB, Tan X, Mao H, Withers J, Ben-Nissan G, Hinds TR, Kobayashi Y, Hsu FF, Sharon M, Browse J, et al. (2010). Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468, 400–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Wang H, Sun M, Tang J, Zheng B, Wang X, and Tan YW (2019). Reactive oxygen species-mediated BIN2 activity revealed by single-molecule analysis. New Phytol. 223, 692–704. [DOI] [PubMed] [Google Scholar]
- Stintzi A, Weber H, Reymond P, Browse J, and Farmer EE (2001). Plant defense in the absence of jasmonic acid: the role of cyclopentenones. Proc. Natl. Acad. Sci. USA 98, 12837–12842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada Y, Spoel SH, Pajerowska-Mukhtar K, Mou Z, Song J, Wang C, Zuo J, and Dong X (2008). Plant immunity requires conformational changes [corrected] of NPR1 via S-nitrosylation and thioredoxins. Science 321, 952–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L, Rong W, Luo H, Chen Y, and He C (2014). The Xanthomonas campestris effector protein XopDXcc8004 triggers plant disease tolerance by targeting DELLA proteins. New Phytol. 204, 595–608. [DOI] [PubMed] [Google Scholar]
- Tan X, Calderon-Villalobos LI, Sharon M, Zheng C, Robinson CV, Estelle M, and Zheng N (2007). Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446, 640–645. [DOI] [PubMed] [Google Scholar]
- Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He SY, Howe GA, and Browse J (2007). JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448, 661–665. [DOI] [PubMed] [Google Scholar]
- Tian Y, Fan M, Qin Z, Lv H, Wang M, Zhang Z, Zhou W, Zhao N, Li X, Han C, et al. (2018). Hydrogen peroxide positively regulates brassinosteroid signaling through oxidation of the brassinazole-RESISTANTI transcription factor. Nat. Commun. 9, 1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton J, Van Pelt JA, Van Loon LC, and Pieterse CMJ (2002). Differential effectiveness of salicylate-dependent and jasmonate/ethylene-dependent induced resistance in Arabidopsis. Mol. Plant Microbe Interact. 15, 27–34. [DOI] [PubMed] [Google Scholar]
- Torrens-Spence MP, Bobokalonova A, Carballo V, Glinkerman CM, Pluskal T, Shen A, and Weng J-K (2019). PBS3 and EPS1 complete salicylic acid biosynthesis from isochorismate in Arabidopsis. bioRxiv. 10.1101/601948. [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Ashikari M, Nakajima M, Itoh H, Katoh E, Kobayashi M, Chow TY, Hsing YI, Kitano H, Yamaguchi I, et al. (2005). Gibberellin INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437, 693–698. [DOI] [PubMed] [Google Scholar]
- Üstün S, Bartetzko V, and Börnke F (2013). The Xanthomonas campestris type III effector XopJ targets the host cell proteasome to suppress salicylic-acid mediated plant defence. PLoS Pathog. 9, e1003427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Pajerowska-Mukhtar K, Culler AH, and Dong X (2007). Salicylic acid inhibits pathogen growth in plants through repression of the auxin signaling pathway. Curr. Biol. 17, 1784–1790. [DOI] [PubMed] [Google Scholar]
- Wildermuth MC, Dewdney J, Wu G, and Ausubel FM (2001). Isochoris-mate synthase is required to synthesize salicylic acid for plant defence. Nature 414, 562–565. [DOI] [PubMed] [Google Scholar]
- Wu Y, Zhang D, Chu JY, Boyle P, Wang Y, Brindle ID, DeLuca V, and Despres C (2012). The Arabidopsis NPR1 protein is a receptor for the plant defense hormone salicylic acid. Cell Rep. 1, 639–647. [DOI] [PubMed] [Google Scholar]
- Yabuta T, and Sumiki T (1938). On the crystal of gibberellin, a substance to promote plant growth. J. Agric. Chem. Soc. Jpn. 14, 1526. [Google Scholar]
- Yamaguchi S (2008). Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 59,225–251. [DOI] [PubMed] [Google Scholar]
- Zheng XY, Spivey NW, Zeng W, Liu PP, Fu ZQ, Klessig DF, He SY, and Dong X (2012). Coronatine promotes Pseudomonas syringae virulence in plants by activating a signaling cascade that inhibits salicylic acid accumulation. Cell Host Microbe 11, 587–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler DM (2002). An overview of the mechanism, substrate specificities, and structure of FMOs. Drug Metab. Rev. 34, 503–511. [DOI] [PubMed] [Google Scholar]


