Abstract
Gene repression using the endonucleolytically deactivated dCas9 protein and sgRNAs (CRISPR interference or CRISPRi) is a useful approach to study gene functions. Here, we established CRISPRi in Paenibacillus sonchi genomovar Riograndensis SBR5, a plant growth promoting bacterium. CRISPRi system with sgRNAs targeting SBR5 endogenous genes spo0A, yaaT and ydjJ and plasmid-borne gfpUV was constructed and analyzed. Flow cytometry analysis revealed a significant decrease of reporter protein GFPUV signal in P. sonchi strains expressing gfpUV sgRNA in comparison with non-targeting controls. CRISPRi-based repression of chromosomal genes for regulation of sporulation spo0A and yaaT decreased sporulation and increased biofilm formation in SBR5. Repression of the sorbitol catabolic gene ydjJ revealed decreased specific activity of YdjJ in crude cell extracts and reduced biomass formation from sorbitol in growth experiments. Our work on CRISPRi-based gene repression serves as basis for gene function studies of the plant growth promoter P. sonchi SBR5. To our knowledge, the present study presents the first tool for gene repression established in Paenibacillus species.
|
Key points • CRISPRi toward gene repression was applied for the first time in Paenibacillus. • CRISPRi of spo0A and yaaT depleted spores and increased biofilms in SBR5. • CRISPRi-based ydjJ repression decreased specific activity of sorbitol dehydrogenase. |
Keywords: Paenibacillus sonchi, CRISPR interference, dCas9, Sorbitol catabolism, Sporulation, Biofilms
Introduction
CRISPR (clustered regularly interspaced short palindromic repeats) associated with the type II protein Cas9 is a well-known and well-established tool applied for genome editing of diverse organisms. Furthermore, the endonucleolytically deactivated mutant of Cas9 protein, named dCas9, still maintains sgRNA-guided DNA binding, which can interfere with RNA polymerase (Qi et al. 2013). Therefore, dCas9-based CRISPR interference (CRISPRi) has been applied for gene repression (Xu and Qi 2019).
CRISPRi-based gene repression has been applied in bacteria for many purposes, for instance, to redirect metabolic fluxes toward industrially relevant products (Cleto et al. 2016; Kim et al. 2017; Schultenkämper et al. 2020). Moreover, multiplexed CRISPRi was established in Bacillus licheniformis by targeting genes involved in byproduct synthesis and l-valine degradation pathway, resulting in up to 80% increase in l-valine titers (Zhan et al. 2020). In Bacillus subtilis, CRISPRi-mediated repression of 16 genes on the branch metabolic pathways of amino acid biosynthesis resulted in up to 0.75 g L−1 surfactin production (Wang et al. 2019). Recently, CRISPRi was established in the methylotrophic Bacillus methanolicus, controlling its sporulation process by repression of sporulation regulator Spo0A and controlling its mannitol catabolism by CRISPRi of mannitol-1-phosphate dehydrogenase mtlD gene (Schultenkämper et al. 2019).
Paenibacillus riograndensis (Beneduzi et al. 2010), that was recently reclassified as a genomovar of P. sonchi (Sant'Anna et al. 2017), is a Gram-positive, biotin auxotrophic, spore-forming bacterium. The strain SBR5 was isolated from wheat (Triticum aestivum) cultivated in fields in the south of Brazil (Beneduzi et al. 2008). This organism showed plant growth promoting characteristics, such as the ability to fix nitrogen (Fernandes et al. 2014) and to produce siderophores and indole-3-acetic acid (Beneduzi et al. 2010), and for pathogen biocontrol (Bach et al. 2016). The inoculation with SBR5 improved growth of wheat under greenhouse conditions (Campos et al. 2015). The complete genome sequence of P. sonchi genomovar Riograndensis SBR5 was determined, consisting of a single chromosome of 7,893,056 base pairs containing 6705 protein coding genes (Brito et al. 2015). Moreover, comprehensive transcriptome analyses enabled the characterization of its global transcriptional landscape (Brito et al. 2017a). The genome sequence of SBR5 has been used as basis for characterization of its nitrogen fixation system through differential RNA sequencing, revealing two nif gene clusters and one alternative nitrogenase anf gene cluster (Fernandes et al. 2014). Further, genome-based RNA sequencing under iron depletion showed expression of genes related to iron uptake and revealed signs of stress resistance displayed by increased expression of genes involved in siderophore transportation, sporulation, and DNA protection (Sperb et al. 2016).
Notwithstanding its great potential for application as plant growth promoting bacterium, P. sonchi genomovar Riograndensis SBR5 is still not characterized in depth. For characterization, the uses of genetic tools for gene expression and repression are advantageous in order to better understand a given process. Genetic tools for gene expression have been developed for P. sonchi SBR5, with a theta-replicating plasmid and a rolling circle-replicating plasmid operating in fluorescent reporter gene expression and generation of a biotin prototrophic strain (Brito et al. 2017b). However, up to now, gene knockout or repression tools have not yet been developed for this bacterium. Interestingly, the genetic tools for gene expression in B. methanolicus (Irla et al. 2016) were transferable to P. sonchi SBR5 (Brito et al. 2017b). Thus, CRISPRi-mediated gene repression tools developed for B. methanolicus (Schultenkämper et al. 2019) were chosen here as a starting point for the establishment of gene repression in SBR5.
While CRISPRi has been developed and is intensively used for model bacteria, there is a need to transfer CRISPRi technology for application in non-model bacteria. Therefore, we established CRISPRi in P. sonchi genomovar Riograndensis SBR5. As proof of concept, CRISPRi was used to repress genes controlling sporulation and biofilm formation in SBR5, besides a gene involved in sorbitol catabolism. The present study represents a valuable example of CRISPRi-based gene repression in the plant growth promoter P. sonchi SBR5 and serves as groundwork for future characterization studies.
Material and methods
Strains, plasmids, and oligonucleotides
All bacterial strains, plasmids, and oligonucleotides used in this study are listed in Table 1. Paenibacillus sonchi genomovar Riograndensis SBR5 (DSM 28159) was used as the expression host; Escherichia coli strain DH5α (Stratagene) was used as general cloning host.
Table 1.
Description of strains, plasmids, and oligonucleotides of the present study
| Strain | Characteristics | Origin |
|---|---|---|
| Paenibacillus sonchi genomovar Riograndensis SBR5 | Wild-type strain, expression host | Beneduzi et al. (2010) |
| Escherichia coli DH5α | General cloning host | Stratagene |
| Plasmid | Characteristics | Origin |
| pNW33N | pNW33N derivative in which the knt-resistance gene was inserted, CmR, KanR | Irla et al. (2016) |
| piCas | pNW33N-derived plasmid carrying dcas9, driven by the mannitol-inducible m2p promoter, CmR, KanR | Schultenkämper et al. (2019) |
| piCas-tspo0A | piCas plasmid carrying the tspo0A sgRNA, targeting the non-template strand of spo0A, CmR, KanR | This study |
| piCas-tyaaT | piCas plasmid carrying the tyaaT sgRNA, targeting the non-template strand of yaaT, CmR, KanR | This study |
| piCas-tydjJ | piCas plasmid carrying the tydjJ sgRNA, targeting the non-template strand of ydjJ, CmR, KanR | This study |
| piCas-tgfpUV | piCas plasmid carrying the tgfpUV sgRNA, targeting the non-template strand of gfpUV, CmR, KanR | This study |
| pBV2mp-gfpUV | pBVmp derivative for gfpUV expression under control of mdh promoter, AmpR, KanR | Irla et al. (2016) |
| Oligonucleotides annealed and inserted in piCas XbaI and AvaI site | Sequence* (5′-3′) | |
| spo0A fw | gggatataaacgttttatgataaatatCAACGCCATCTCGTGTAGAG | |
| spo0A rv | taacttgctatttctagctctaaaacCTCTACACGAGATGGCGTTG | |
| yaaT fw | gggatataaacgttttatgataaatatCTGTCTCCACAATTACACAC | |
| yaaT rv | taacttgctatttctagctctaaaacGTGTGTAATTGTGGAGACAG | |
| ydjJ fw | gggatataaacgttttatgataaatatATCAGGCAGCCGTGGTAGGG | |
| ydjJ rv | taacttgctatttctagctctaaaacCCCTACCACGGCTGCCTGAT | |
| gfpUV fw | gggatataaacgttttatgataaatatCATCTAATTCAACAAGAATT | |
| gfpUV rv | taacttgctatttctagctctaaaacAATTCTTGTTGAATTAGATG | |
*Letters in lowercase represent plasmid overlaps
Media and cultivation conditions
E. coli strains were routinely cultivated at 37 °C in lysogeny broth (LB; Bertani 1951) or on LB agar plates (1% w/v) supplemented with antibiotics (chloramphenicol 25 μg mL−1, kanamycin 50 μg mL−1) when needed.
In all experiments, P. sonchi strains were cultured in 500-mL shaking flasks containing 50 mL of CASO broth (DMSZ 220) or in 24-square deep well plates (Duetz et al. 2000) containing 3 mL of PbMM (Paenibacillus minimal medium; Brito et al. 2017a), shaking at 120 rpm and at temperature of 30 °C. For each condition tested, six biological replicates were used: three replications of bacterial cells were harvested for total RNA isolation, and three for further determination of growth characteristics. The optical density (OD) at 600 nm of the cultivated cells was measured throughout growth. Recombinant P. sonchi strains were routinely plated on 1% agar (w/v) CASO broth. When appropriate, culture media were supplemented with 2.5 μg mL−1 chloramphenicol and 50 mM mannitol as dcas9 expression inducer. Glucose (100 mM) was used as standard carbon source for cultivation of P. sonchi in minimal medium, but 98.9 mM sorbitol was used as additional carbon source when appropriate.
Plasmid construction and preparation of recombinant strains
Molecular cloning was performed as described by Sambrook and Russel (2001). Chemically competent cells of E. coli DH5α were prepared for cloning (Hanahan 1983). All the information about plasmid construction and oligonucleotide sequences are described in Table 1. The NucleoSpin® Gel and PCR clean-up kit (Machery-Nagel, Düren, Germany) were used for PCR clean-up, and plasmids were isolated using the GeneJET plasmid miniprep kit (Thermo Fisher Scientific, Waltham, USA). The pNW33N derivative plasmid piCas (Schultenkämper et al. 2019) was double digested with XbaI and AvaI restriction enzymes (New England Biolabs, Ipswich, USA). Because CRISPRi targeting template DNA strand shows little repression effect (Qi et al. 2013; Cleto et al. 2016), we designed sgRNAs targeting non-template strands of P. sonchi genes. sgRNAs were selected by using CRISPy-web tool (Blin et al. 2016) and introduced to piCas plasmid XbaI and AvaI cloning site (Fig. 1). In order to build sgRNAs, oligonucleotides (Table 1) were annealed at 95 °C for 5 min and cooled to room temperature for 1 h. Later, their overlapping regions were joined by Gibson assembly (Gibson et al. 2009). For colony PCR, Taq polymerase (New England Biolabs) was used. The correctness of inserted DNA sequences was confirmed by sequencing in the sequencing facility of the Center for Biotechnology (University of Bielefeld, Germany). The constructed plasmids were transformed to P. sonchi SBR5 by means of magnesium-aminoclay method, as described by Brito et al. (2017b).
Fig. 1.

Schematic representation of plasmid piCas and sgRNA sequences used in this study. Vector pNW33N was used as backbone for contruction of piCas. Bacillus methanolicus m2p promoter has lacI operator sequence (lacO;yellow) inserted between the transcriptional start site and the dcas9 coding sequence (red) to avoid expression in the cloning host E. coli (Schultenkämper et al. 2019). sgRNA transcription is driven by m2p promoter lacking its 5′UTR (′m2p). XbaI and AvaI sites are positioned upstream dCas9 handle (blue) and Streptococcus pyogenes terminator (orange) sequence (Schultenkämper et al. 2019). Those sites were used to linearize piCas vector for insertion of gene targeting sequence (black). The PAM regions and spacer sequences were selected from SBR5 genome sequence using the CRISPy-web tool (Blin et al. 2016)
Quantification of sporulation
Paenibacillus sonchi strains were cultivated in CASO broth as described above. After 6 h, when the cells were in exponential growth phase, the cells were transferred to fresh CASO broth containing sporulation salts (15 mg L−1 MnCl2, 1 g L−1 MgCl2, and 0.8 g L−1 CaCl2; Malvar et al. 1994). The endospores, middle stage spores, and non-spore cells were counted in technical triplicates. More than 100 cells were counted with a hemocytometer per condition and sample. If needed, suitable dilutions were used. The cells were examined by bright field microscopy (Zeiss® Axio Lab.A1).
Colony phenotyping of recombinant P. sonchi strains
Iris software, an image processing software that automatically quantifies several features of microbial colonies in high throughput (Kritikos et al. 2017), was used to determine the colony phenotype of P. sonchi strains. Therefore, the recombinant P. sonchi strains were cultivated in 50-mL CASO broth until reaching exponential phase (OD = 0.6), and 24 drops were stamped with a replicator on CASO agar plates containing or not mannitol for dcas9 gene expression. Plates were imaged and analyzed with the IRIS software, in which colony opacity and size were determined.
Quantification of biofilms
Biofilm formation was determined according to O'Toole (2011) microtiter dish biofilm formation assay. Recombinant P. sonchi strains were inoculated in round-bottomed microtiter plates (Greiner Cellstar®, 96-well cell culture plate) to an OD of 1, containing 200 μL CASO broth supplemented with expression inducer mannitol, when needed, in each well. The plates were incubated for 2 days in a 30 °C incubator. After incubation, the cells were dumped out by turning the microtiter plate over. Then, the plate was gently submerged in water. After shaking out the water, 200 μL of a 0.1% (w/v) crystal violet solution was added in each well. The microtiter plate was incubated at room temperature for 15 min. The solution was then rinsed out by washing the plate gently in water for several times, and the plate was turned upside down and dried overnight. To quantify the biofilm formation, 200 μL of 30% acetic acid was added to the wells to solubilize the crystal violet. The plate was incubated at room temperature for 15 min, and 200 μL of the solubilized solutions was transferred to a new flat-bottomed microtiter dish (Thermo Fisher Scientific, 96 wells). The absorbance was measured at 560 nm in a plate reader.
GFPUV fluorescence measurement by flow cytometry
In order to quantify green fluorescent protein (GFPUV) mean fluorescence intensities (MFI) of P. sonchi SBR5 pBV2mp-gfpUV strains harboring CRISPRi plasmids and proper controls, cell cultures were analyzed by fluorescence activated cell scanning (FACS). Therefore, P. sonchi cells were cultured until reaching exponential phase and centrifuged for 10 min at 4000 rpm (5810R centrifuge, Eppendorf, Hamburg, Germany). The pellets were washed two times in NaCl 0.89%, and the OD of the cultures was adjusted to 0.5. The fluorescence of the cell suspension was measured using flow cytometer (Beckman Coulter, Brea, USA) and the data analyzed in the Beckman Coulter Kaluza® Flow Analysis Software. As described previously, the settings for the emission signal and filters within the flow cytometer for detection of GFPUV fluorescence were based on a 550/525 bandpass FL9 filter (Brito et al. 2017b).
RNA isolation and qRT-PCR
For total RNA isolation, P. sonchi strains were cultivated in CASO broth containing 100 mM sorbitol and 50 mM mannitol to exponential phase (OD = 1). The cultivation was carried out in biological triplicates. Bacterial cells were harvested by 15 min centrifugation at 4 °C and 4000 rpm (5810R centrifuge, Eppendorf). The supernatant was discarded, and the pellets were immediately frozen in liquid nitrogen and stored at − 80 °C. In order to isolate RNA, the cell pellets were thawed in ice and the samples were homogenized by resuspending the cells in 100 μL TE buffer (10 mM Tris-HCl, 1 mM EDTA; pH 8) containing 5 mg mL−1 lysozyme. After 30 min incubation at 37 °C, total RNA was extracted using NucleoSpin® RNA kit (Macherey-Nagel) according to manufacturer’s instruction. Thereafter, RNA samples were treated with DNase digestion using RNase-free DNase Set and “RNeasy MinElute” kits (Qiagen, Hilden, Germany) to eliminate possible genomic DNA contamination. Furthermore, quality control with Taq polymerase (New England Biolabs) was performed in order to determine purity and integrity of isolated RNA. Additionally, total RNA concentration was measured using a spectrophotometer (NanoDrop®, ND-1000). Equal amounts of 50 ng of each RNA sample were used to perform quantitative real-time PCR (qRT-PCR). All qRT-PCRs were performed according to manufacturer’s instruction using the SensiFAST™ SYBR® No-ROX One-Step Kit (Bioline, London, UK) and the CFX96 cycler system (Bio-Rad, Hercules, USA). The temperature profile employed in all qRT-PCRs was 45 °C for 10 min (reverse transcription); 95 °C for 2 min; and 40 cycles of 95 °C for 5 s, 55 °C for 10 s, and 72 °C for 5 s; with melt curve analysis with measurements between 65 °C and 95 °C. The ΔCq method was used for calculations (Higuchi et al. 1992; Bustin et al. 2009). For each sample, three independent qRT-PCR experiments were performed.
Determination of YdjJ specific activity
For determination of YdjJ activity, P. sonchi strains harboring the plasmids pNW33N, piCas or piCas-tydjJ strains were cultivated in flasks containing 50 mL CASO broth with 100 mM sorbitol and 50 mM mannitol, to circumvent growth deficits, until they reached an OD600 of approximately 1. Then, the cells were disrupted according to Brautaset et al. (2004). Crude extracts were used to determine YdiJ specific activity in an enzymatic assay using the Sorbitol Dehydrogenase Kit (MAK317, Sigma-Aldrich, St. Louis, USA). Sorbitol dehydrogenase activity was assayed according to manufacturer’s instructions at room temperature and 565 nm using a spectrophotometer (Shimadzu, UV-1800). One unit (U) of YdjJ catalyzed the conversion of 1 mol of d-sorbitol to fructose per minute at pH 8.2.
Results
CRISPRi-mediated repression of plasmid-borne GFPUV
In order to analyze CRISPRi efficiency in P. sonchi SBR5 cells, we chose to express reporter gene gfpUV sgRNA in SBR5 using the plasmid pBV2mp-gfpUV. sgRNA-guided dCas9 activity resulted in a significant decrease of GFPUV medium fluorescence intensity (MFI). GFPUV MFI in P. sonchi SBR5(pBV2mp-gfpUV) cells expressing dCas9 and gfpUV sgRNA was about 3 times lower than in the same strain expressing dCas9 alone (Table 2). Thus, repression of a highly expressed gene on a plasmid by CRISPRi could be demonstrated.
Table 2.
CRISPRi-based repression of plasmid-borne gfpUV gene in P. sonchi SBR5. Paenibacillus sonchi SBR5(pBV2mp-gfpUV) carried gfpUV coding sequence targeted by sgRNA-guided dCas9. GfpUV mean fluorescence intensity (MFI) of recombinant cells was determined by means of flow cytometry. Data represent means and standard deviations of technical triplicates. Different numbers represent significant differences by Scott-Knott test (p < 0.05)
| GfpUV MFI | ||||
|---|---|---|---|---|
| SBR5(pBV2mp-gfpUV)(pNW33N) | 1.49 | ± | 0.33 | (a2) |
| SBR5(pBV2mp-gfpUV)(piCas) | 1.33 | ± | 0.23 | (a2) |
| SBR5(pBV2mp-gfpUV)(piCas-tgfpUV) | 0.41 | ± | 0.07 | (a1) |
Effect of ydjJ gene repression on P. sonchi SBR5 grown in d-sorbitol
After having shown that CRISPRi worked to repress a gene on a plasmid, we chose to repress a chromosomal gene. Based on the complete genome sequence of SBR5 (Brito et al. 2015), we selected a catabolic gene, namely the d-sorbitol dehydrogenase gene ydjJ, as CRISPRi target. Paenibacillus sonchi SBR5 carrying the plasmid piCas9-tydjJ and the strains carrying empty vector and non-targeting control plasmids were cultivated in minimal medium with d-sorbitol as sole carbon source and for comparison in glucose minimal medium. As expected, P. sonchi SBR5(pNW33N) was able to grow in glucose and d-sorbitol either as sole or as combined carbon sources (Fig. 2). However, expressing dCas9 alone in the non-targeting SBR5(piCas) strain surprisingly resulted in significantly decreased biomass formation in comparison with the empty vector control, regardless of the provided carbon source. Moreover, the biomass formation of SBR5(piCas-tydjJ) on glucose was compromised, but comparable with the non-targeting control (Fig. 2). In minimal media with d-sorbitol as sole or combined carbon source, SBR5 expressing ydjJ sgRNA resulted in 3 and 2 times less biomass formation in comparison with the empty vector control and non-targeting control, respectively (Fig. 2). This result indicated that YdjJ may play a crucial role in d-sorbitol catabolism in P. sonchi SBR5.
Fig. 2.
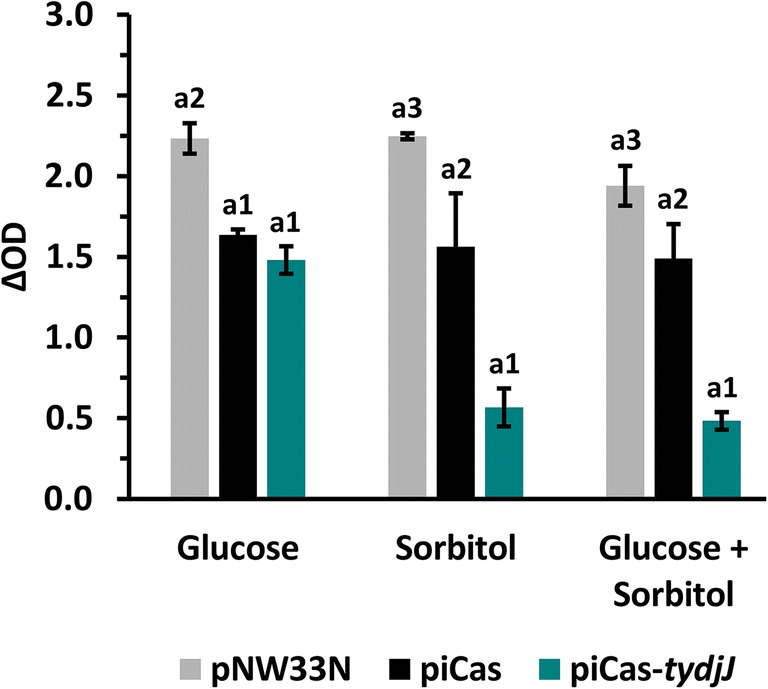
dCas9 targeting sorbitol dehydrogenase coding gene ydjJ reduced biomass formation of P. sonchi SBR5 in growth medium containing sorbitol. Final growth (ΔOD) of P. sonchi SBR5 cultivated in deep well plates (Duetz et al. 2000). SBR5 cells carried pNW33N, piCas9, or piCas9-tydjJ plasmids. Glucose (100 mM), sorbitol (98.9 mM) or glucose (50 mM), and sorbitol (49.45 mM) were used as sole or combined carbon sources in PbMM medium. ΔOD values represent the difference between the OD of cell cultures at the end of exponential phase and the OD at inoculation. The error bars represent standard deviation of technical triplicates. Different numbers represent significant differences by Scott-Knott test (p < 0.001)
Pursuing more evidence of YdjJ CRISPRi in P. sonchi SBR5, we determined the YdjJ specific enzyme activity in crude extracts of SBR5(piCas-tydjJ) in comparison with proper controls. YdjJ specific activity in SBR5(piCas-tydjJ) was 0.2 ± 0.0 U mg total protein−1, thus less than half of the activity observed in the non-targeting control strain (0.5 ± 0.1 U mg total protein−1). Surprisingly, while the highest YdjJ specific activity was reached by the non-targeting SBR5(piCas) strain, the empty vector control strain SBR5(pNW33N) presented YdjJ specific activity of 0.3 ± 0.0 U mg total protein−1 (Fig. 3). Nevertheless, targeting ydjJ led to about 33% less enzyme activity in comparison with the empty vector control and this difference was statistically significant (p < 0.05; Fig. 3). Even though CRISPRi-based attenuation of ydjJ led to only partial depletion of YdjJ activity, these results illustrate that repression of a chromosomally encoded catabolic gene by CRISPRi could successfully be demonstrated, and the functional role of d-sorbitol dehydrogenase YdjJ for utilization of d-sorbitol could be revealed.
Fig. 3.

CRISPR interference-based gene repression decreased specific activity of sorbitol dehydrogenase YdjJ in crude extracts of P. sonchi SBR5. Specific activities (U mg total protein−1) of SBR5 strains harboring plasmids pNW33N, piCas, or piCas-tydjJ. One unit (U) of YdjJ catalyzed the conversion of 1 mol of d-sorbitol to fructose per minute at pH 8.2. Crude extracts were prepared after growth in CASO broth supplemented with 100 mM sorbitol and 50 mM mannitol. Error bars represent standard deviations of technical triplicates. Different numbers represent significant differences by Scott-Knott test (p < 0.05)
CRISPRi targeting sporulation regulatory gene spo0A decreased its mRNA level
In order to test if CRISPRi can be applied to repress regulatory genes of P. sonchi, we chose to target the sporulation regulatory gene spo0A. The extent of gene repression of the recombinant P. sonchi SBR5(piCas-tspo0A) generated in the present study was quantified by qRT-PCR. The expression of SBR5 chromosomal gene spo0A, here targeted by sgRNA-guided dCas9, was quantified (Cq) and normalized by the Cq expression of dcas9 gene (ΔCq). The relative qRT-PCR data of the knockdown strain revealed significantly reduced spo0A mRNA level (p < 0.05) in comparison with RNA samples from SBR5 control strain piCas (Fig. 4).
Fig. 4.

qRT-PCR revealed CRISPRi spo0A gene repression in P. sonchi SBR5. ΔCq expression of spo0A gene in the SBR5 strain piCas-tspo0A in comparison with non-targeting piCas control. Cq values obtained from dcas9 gene were used as normalization factor. ΔCq expression means of biological triplicates are depicted, and error bars represent standard deviations of samples. Different numbers represent significant differences by Scott-Knott test (p < 0.05)
Hemocytometer spore counts of P. sonchi cells targeting spo0A and yaaT
Next, we quantified the effect of CRISPRi on sporulation of P. sonchi SBR5 in media containing sporulation salts, by hemocytometer counts. In addition to targeting spo0A, yaaT was chosen as related regulatory sporulation gene. The percentages of viable SBR5 cells, cells in middle sporulation stage, and spores were calculated from the microscopic analysis. In the first quantification (t = 0, i.e., before transfer to medium with sporulation salts), SBR5 cells carrying the empty vector control plasmid pNW33N, the non-targeting piCas plasmid or piCas-tspo0A and piCas-tyaaT plasmids were all viable (Fig. 5a). Initial spore formation could be seen at 24 h of cultivation in CASO broth for SBR5 strains pNW33N (10% ± 7%) and piCas (8% ± 1%) (Fig. 5b). However, when the cultivation broth was supplemented with dCas9 expression inducer mannitol, the percentage of spores slightly increased for SBR5 strains pNW33N and piCas (20% ± 9% and 24% ± 10% of spores from total cells, respectively) (Fig. 5b). Even though some spores were observed in mannitol-induced SBR5(piCas-tyaaT) at 24 h (2% ± 1%) (Fig. 5b), no spores were observed in the analyzed cells of the same strain in later time points (Fig. 5c and d). Cells in middle sporulation stage were observed at 24 h, counting 9% ± 6%, 5% ± 2%, and 1% ± 2% of induced pNW33N, piCas, and piCas-tyaaT cells, respectively (Fig. 5b). In contrast to SBR5 control strains pNW33N and piCas and the CRISPRi targeting strain piCas-tyaaT, SBR5(piCas-tspo0A) did not proceed to the middle stage of sporulation nor fully developed sporulation at 24 h that occurred independently of mannitol addition to the cultivation medium (Fig. 5b). Although CRISPRi repression of spo0A and yaaT genes resulted in total suppression of formed spores, SBR5(piCas-tspo0A) and SBR5(piCas-tyaaT) cultures, induced with mannitol or not, presented approximately 10% of cells in middle sporulation stage at 48 and 72 h (Fig. 5c and d). For the negative control strains pNW33N and piCas9, the percentage of cells in middle stage sporulation and formed spores increased dramatically in the late stationary phase, from 48 to 72 h (Fig. 5c and d). During this final stage of cultivation, sporulation increased further, as in the end 35% ± 4% and 35% ± 1% of the induced pNW33N, and piCas cells, respectively, were fully sporulated (Fig. 5d). Furthermore, non-induced cells of pNW33N and piCas presented approximately 20% sporulation (Fig. 5d). Hence, even though middle sporulation stage cells were observed along the growth, when dCas9 targeted the sporulation genes spo0A and yaaT, no formed spores were observed in up to 72 h (Fig. 5). Thus, targeting sporulation regulatory genes spo0A and yaaT slowed progress of sporulation dramatically (Fig. 5).
Fig. 5.
CRISPR interference of spo0A and yaaT genes reduced hemocytometer spore counts of P. sonchi SBR5 cells. SBR5 cells carrying plasmids pNW33N, piCas, piCas-tspo0A, or piCas-tyaaT were cultivated in CASO broth (DSMZ 220). Spores were counted by means of hemocytometer method at 0 (a), 24 (b), 48 (c), and 72 (d) hours after transfer to CASO broth containing sporulation salts (Malvar et al. 1994). CASO broth was supplemented (I) or not (NI) with 50 mM mannitol to induce dcas9 expression. Percentage of viable cells, middle stage sporulation, and spores is given as means and standard deviations of technical triplicates
Colony phenotyping analysis
CRISPRi targeting spo0A and yaaT genes in P. sonchi SBR5 resulted in strong reduction of spore formation (Fig. 5). To independently assess sporulation, we analyzed colony opacity since colonies of sporulating SBR5 present an opaque aspect in comparison with non-sporulating colonies. Therefore, colonies from P. sonchi SBR5 carrying the plasmids pNW33N, piCas, piCas-tspo0A, or piCas-tyaaT were imaged, and their size and opacity were scored using the IRIS software. The average colony size was similar for the sporulating colonies from SBR5(pNW33N) and SBR5(piCas) and the non-sporulating colonies from SBR5(piCas-tspo0A) and SBR5(piCas-tyaaT) (Fig. 6). Furthermore, while colonies of SBR5 expressing spo0A and yaaT sgRNAs remained translucent, sporulating SBR5(pNW33N) and SBR5(piCas) colonies presented high opacity scores (Fig. 6). Opacity score values for SBR5 strains carrying plasmids pNW33N and piCas were about 2-fold and 4-fold higher, respectively, in comparison with that in the strains expressing sgRNAs (Fig. 6). Taken together, targeting spo0A and yaaT revealed reduced sporulation not only by hemocytometer analysis of cells in liquid medium (Fig. 5), but also by colony opacity analysis of cells grown an agar plates (Fig. 6).
Fig. 6.

CRISPR interference of spo0A and yaaT genes altered colony phenotypes of P. sonchi SBR5. Equal volumes of SBR5 cell suspension carrying plasmids pNW33N, piCas, piCas-tspo0A, or piCas-tyaaT were transferred to agar CASO broth (DSMZ 220) plates. Plates were imaged for IRIS software analysis of colony size and opacity. Size S-score values represent the colony area in pixels; opacity S-scores represent the sum of the brightness values for all the pixels in the colony (Kritikos et al. 2017). High S-score values of opacity and colony size represent cell sporulation. Bars represent IRIS S-score means and error bars represent standard deviations of 24 equally spaced colonies. Different numbers represent significant differences by Scott-Knott test (p < 0.001)
Effect of CRISPRi on biofilm formation by P. sonchi
Spo0A regulates biofilm development in B. subtilis and B. methanolicus (Hamon and Lazazzera 2001; Bustin et al. 2009; Schultenkämper et al. 2019). To investigate biofilm formation in P. sonchi strains expressing spo0A and yaaT sgRNAs, biofilm formation was measured in a crystal violet assay. CRISPRi-based repression of both spo0A and yaaT genes led to approximately 3-fold increase in crystal violet signal (560 nm) in comparison with empty vector and non-targeting vector controls (Fig. 7). This result reflects the negative impact of Spo0A and YaaT on biofilm formation by SBR5. Thus, as expected, significant enhancements of biofilm formation were observed upon targeting spo0A and yaaT, although less dominant in the latter case (Fig. 7).
Fig. 7.
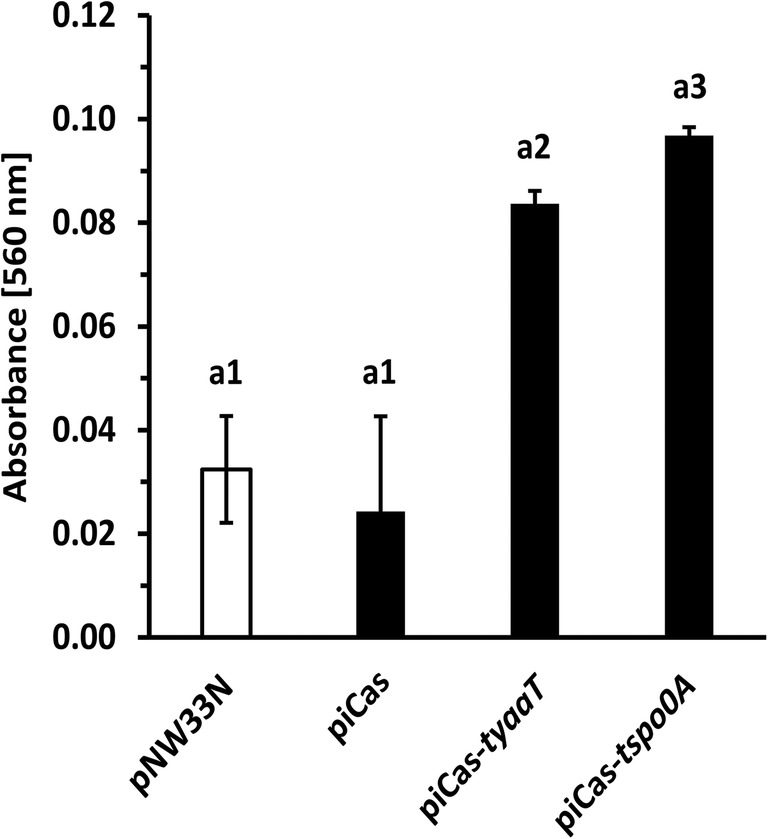
Crystal violet quantification revealed increased biofilm formation upon CRISPRi-mediated repression of spo0A and yaaT genes in P. sonchi SBR5. SBR5 cells carrying plasmids pNW33N, piCas, piCas-tspo0A, or piCas-tyaaT were cultivated in CASO broth (DSMZ 220) and transferred to 96 microwell plates. Three-day-old biofilms attached to the bottom of the individual wells in a 96-well plate were stained with crystal violet for absorbance readings (560 nm) derived from methanol elution of the crystal violet stain. Crystal violet absorbance means are depicted, and error bars represent standard deviation of 96 individual wells. Different numbers represent significant differences by Scott-Knott test (p < 0.001)
Discussion
We developed CRISPRi of plasmid and chromosomal genes in the plant growth promoter P. sonchi SBR5 for the first time. Based on the CRISPRi-mediated gene repression system developed for the methylotrophic B. methanolicus (Schultenkämper et al. 2019), we showed repression of gfpUV expressed from a plasmid in a fluorescence assay. Evaluation of CRISPRi-based repression of fluorescent proteins is a reliable and often used method to test CRISPRi activity in bacteria. dCas9 and sgRNA targeting chromosome integrated EYFP (enhanced yellow fluorescent protein) gene conferred effective and stable EYFP suppression in Synechococcus elongatus (Huang et al. 2016). Moreover, targeting a genome-integrated gene coding for a red fluorescent protein (rfp) in C. glutamicum resulted in reduced RFP fluorescence (Cleto et al. 2016). In both cases, CRISPRi was highly efficient, with less than 1% remaining reporter protein signal. Similarly, targeting plasmid GFP reporter gene in Pseudomonas putida decreased fluorescence intensity to background levels (Kim et al. 2020). In this study, flow cytometry analysis revealed substantial drop of plasmid-borne GFPUV fluorescence (~ 70%) in P. sonchi SBR5 expressing dCas9 and gfpUV targeting sgRNA, but about 30% of the reporter signal remained (Table 2). Comparable results have been reported for Klebsiella pneumoniae, where plasmid-derived enhanced green fluorescent protein (EGFP) expression level was reduced to 10–15%, depending on the different targeted regions of promoter upstream of EGFP gene (Wang et al. 2018). Variations in CRISPRi activities might be due to the fact that CRISPRi regulation levels are ruled by sgRNA design, being highly dependent on PAM site variants and gene target locations (Tan et al. 2019). Even though our study did not compare the effect of various sgRNAs on gfpUV repression, the efficiency of the system could be confirmed by the repression of chromosomal genes, which resulted in 3-fold reduction of spo0A relative expression (Fig. 4) and clear phenotypic changes when genes ydjJ, spo0A, and yaaT were targeted.
CRISPRi of the chromosomally encoded P. sonchi gene coding for putative d-sorbitol dehydrogenase YdjJ revealed that a catabolic gene can be repressed by CRISPRi and corroborated its hypothetical function in sugar alcohol utilization. Upon targeting ydjJ biomass formation from d-sorbitol (Fig. 3) and NAD-dependent d-sorbitol dehydrogenase (EC 1.1.1.14) activity in crude extracts was reduced (Fig. 2). In the genome sequence of P. sonchi, ydjJ is annotated to code for NADP dependent alcohol dehydrogenase (Brito et al. 2015), and it shares 43.82% protein identity with d-sorbitol dehydrogenase from B. subtilis (Ng et al. 1992) and 42.94% with that enzyme from Bacillus fructosus (Uwajima 1999). The B. subtilis enzyme is active with other sugar alcohols, but possesses higher affinity for d-sorbitol (KM = 11 mM, Vmax = 6.25 μM min−1) than for other sugar alcohol substrates (Ng et al. 1992). Crude extracts of B. fructosus, efficient in producing d-fructose from d-sorbitol, reach a specific activity of d-sorbitol dehydrogenase of 0.0021 U mg total protein−1, yielding 100% of d-sorbitol conversion (Uwajima 1999). In our study, CRISPRi of ydjJ decreased YdjJ activity and biomass formation from d-sorbitol significantly, but not completely (Figs. 2 and 3). Incomplete repression of gene coding for catabolic proteins has been reported previously: CRISPRi of B. methanolicus mannitol-1-phosphate 5-dehydrogenase gene mtlD resulted in substantial, but not complete decrease of biomass formation from mannitol as well as of MtlD specific activity (Schultenkämper et al. 2019). As an alternative explanation, it is possible that the genome of P. sonchi encodes other proteins with side activity as d-sorbitol dehydrogenase. For example, xylitol dehydrogenase from Gluconobacter oxydans (EC 1.1.1.-) catalyzes the conversion of d-sorbitol to d-fructose (Liu et al. 2019). However, it remains to be studied if such enzymes exist in P. sonchi and/or whether ydjJ gene repression was incomplete.
It is known that the entry to sporulation in bacilli is closely regulated by a cascade of genetic events started by the transcriptional master regulator Spo0A (Tan and Ramamurthi 2014). Upon histidine kinase phosphotransfer, B. subtilis Spo0A initiates response processes such as biofilm formation, cannibalism, and sporulation (Hamon and Lazazzera 2001; Ellermeier et al. 2006; Veening et al. 2009). CRISPRi-mediated gene repression led to down regulation of P. sonchi spo0A (Fig. 4), which resulted in strong reduction of sporulated cells (Fig. 5). Likewise, CRISPRi system was successfully used to repress Spo0A in B. methanolicus, resulting in substantial decrease of sporulated cells (Schultenkämper et al. 2019). The decrease of sporulation ability could also be observed in Clostridium, when CRISPRi system was applied to target spo0A gene in Clostridium beijerinckii (Li et al. 2016). Moreover, colony opacity increases with the level of sporulation (White et al. 2006). Our colony phenotyping analysis confirmed that and showed high opacity scores of colonies from non-targeting control strains, whereas colonies from strains with sgRNA targeting spo0A gene remained translucent (Fig. 6). Thus, the crucial role of Spo0A in P. sonchi sporulation was confirmed. Furthermore, CRISPRi of spo0A impacted biofilm production of P. sonchi (Fig. 7). Previously, CRISPRi of quorum sensing gene luxS was used to inhibit biofilm formation in E. coli (Zuberi et al. 2017). Later, biofilm formation and swarming motility were strongly inhibited by CRISPRi of environmental sensor histidine kinase gene gacS in Pseudomonas fluorescens (Noirot-Gros et al. 2019). In B. subtilis, mutations in spo0A caused a defect in biofilm formation (Hamon and Lazazzera 2001). In contrast, targeting spo0A with CRISPRi increased formation of biofilms in P. sonchi (Fig. 7). As in the present study, Schultenkämper et al. (2019) reported increased biofilm formation of B. methanolicus when its spo0A gene was downregulated by means of CRISPRi. That might be explained by the fact that bacterial cells form biofilms in opposition to spores in face of low level of Spo0A phosphorylation (Hamon and Lazazzera 2001). Here, few spore counts were observed in P. sonchi cultures expressing spo0A sgRNA (Fig. 5), indicating some spo0A expression. Hence, a low level of spo0A expression in P. sonchi should increase its biofilm formation.
CRISPRi in P. sonchi showed that its sporulation process is driven by Spo0A protein and that targeting spo0A gene impacted biofilm formation. In addition, an extension of sporulation and biofilm regulation could be demonstrated. The B. subtilis protein YaaT interacts with the proteins YmcA and YlbF forming a stable ternary complex that acts upstream of Spo0A, accelerating its production (Carabetta et al. 2013). In accordance with the results obtained by downregulating spo0A, targeting yaaT alone with CRISPRi system resulted in a strong decrease of sporulation (Figs. 5 and 6) and an increase of biofilm formation (Fig. 7). These results are in accordance with the findings in B. subtilis in which YaaT directly influenced Spo0A (Carabetta et al. 2013; Hosoya et al. 2002).
Our study demonstrated CRISPRi employing type II Cas9 modified dCas9 protein as an efficient gene repression tool for P. sonchi. However, a toxic effect of dCas9 protein was observed in our experiments, for example, since growth in sorbitol/glucose of the non-targeting control strain SBR5(piCas) was significantly affected as compared with the control strain bearing the parental plasmid pNW33N lacking the dcas9 gene (Fig. 2). It is widely observed that Cas9 and its mutant dCas9 may exhibit off-target activity and/or sequence-specific toxicity in target organisms such as E. coli (Cui et al. 2018; Hamilton et al. 2019) and clostridia (Xu et al. 2015). Previously, we observed cloning difficulties with plasmid piCas in E. coli DH5α which could be overcome by cloning of Lac repressor operator sequence (lacO) at the transcriptional starting site of the employed m2p promoter (Schultenkämper et al. 2019). In the target organism P. sonchi, the issue of toxicity might be circumvented by exchanging the promoter that drives the expression of dcas9 gene. The m2p promoter used in the present study is functional and inducible by 40 mM mannitol in P. sonchi SBR5 (Brito et al. 2017b). Mannitol is not a gratuitous inducer since it is a carbon source utilized by P. sonchi SBR5 (Beneduzi et al. 2010). Moreover, the m2p promoter leads to significant background gene expression (Brito et al. 2017b). This leaky expression of dCas9 might be sufficient to cause cellular toxicity in P. sonchi. A tight regulation of dcas9 is pivotal to overcome the background expression of this gene and consequently alleviate toxicity (Cho et al. 2018; Cui et al. 2018; Hofmann et al. 2019). dCas9 toxicity could also be mitigated in E. coli by mutating dCas9 PAM binding site and fusing it to TetR-family PhlF repressor in order to enhance DNA binding (Zhang and Voigt 2018). Moreover, the use of truncated sgRNA regions can decrease undesired Cas9 off-target double strand breaks (Fu et al. 2014). Altogether, our results of sporulation control and sorbitol catabolism control indicated strong on-target dCas9 repression. Thus, we conclude that despite the disadvantage of some levels of toxicity, the CRISPRi system established here is favorable for gene function studies in the plant growth promoting P. sonchi SBR5.
Our study opens doors for further gene function research in P. sonchi SBR5. More advanced tests may be amenable in the future including comparison of multiple sgRNAs pursuing higher CRISPRi efficiency levels and multiplexed repression allowing to target whole metabolic pathways rather than single genes. This has been shown in microbes in previous studies, e.g., an array of four sgRNAs was successfully applied for elimination of n-butanol byproducts in E. coli (Kim et al. 2017) and the simultaneous CRISPRi-mediated repression of seven genes competitive for β-amyrin synthesis in yeast (Ni et al. 2019). Furthermore, based on our demonstration of CRISPRi with dCas9 in P. sonchi SBR5, the technology could be adapted to genome editing using the intact Cas9 protein. In the related Paenibacillus polymyxa DSM 365, CRISPR-Cas9-based genome editing was developed and used to improve its exopolysaccharide production (Rütering et al. 2017). Our study is the first demonstration of CRISPRi-mediated gene repression in Paenibacilli. Thus, we developed a new tool for genetic manipulation in Paenibacilli such as the plant growth promoter P. sonchi SBR5, enabling its study as a strong candidate for crops inoculation in the future.
Contributions
KS and LFB carried out the experimental procedures and analyzed data of the present study. LFB prepared a draft of the manuscript. KS, LFB, LMPP, and VFW finalized the manuscript. VFW coordinated the study. All authors read and approved the manuscript.
Funding information
Open Access funding provided by Projekt DEAL. This work was supported in part by the ERA CoBioTech project C1Pro (No. 722361).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval and consent to participate
This article does not contain any studies with human participants or animal experiments performed by any of the authors.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Bach E, Seger GDS, Fernandes GC, Lisboa BB, Passaglia LMP. Evaluation of biological control and rhizosphere competence of plant growth promoting bacteria. Appl Soil Ecol. 2016;99:141–149. doi: 10.1016/j.apsoil.2015.11.002. [DOI] [Google Scholar]
- Beneduzi A, Peres D, da Costa PB, Bodanese Zanettini MH, Passaglia LM. Genetic and phenotypic diversity of plant-growth-promoting bacilli isolated from wheat fields in southern Brazil. Res Microbiol. 2008;159(4):244–250. doi: 10.1016/j.resmic.2008.03.003. [DOI] [PubMed] [Google Scholar]
- Beneduzi A, Costa PB, Parma M, Melo IS, Bodanese-Zanettini MH, Passaglia LM. Paenibacillus riograndensis sp. nov., a nitrogen-fixing species isolated from the rhizosphere of Triticum aestivum. Int J Syst Evol Microbiol. 2010;60(Pt 1):128–133. doi: 10.1099/ijs.0.011973-0. [DOI] [PubMed] [Google Scholar]
- Bertani G. Studies on lysogenesis. I The mode of phage liberation by lysogenic Escherichia coli1. J Bacteriol. 1951;62(3):293–300. doi: 10.1128/JB.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blin K, Pedersen LE, Weber T, Lee SY. CRISPy-web: an online resource to design sgRNAs for CRISPR applications. Synth Syst Biotechnol. 2016;1(2):118–121. doi: 10.1016/j.synbio.2016.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brautaset T, Jakobsen M ØM, Flickinger MC, Valla S, Ellingsen TE (2004) Plasmid-dependent methylotrophy in thermotolerant Bacillus methanolicus. J Bacteriol 186(5):1229–1238 [DOI] [PMC free article] [PubMed]
- Brito LF, Bach E, Kalinowski J, Ruckert C, Wibberg D, Passaglia LM, Wendisch VF. Complete genome sequence of Paenibacillus riograndensis SBR5(T), a gram-positive diazotrophic rhizobacterium. J Biotechnol. 2015;207:30–31. doi: 10.1016/j.jbiotec.2015.04.025. [DOI] [PubMed] [Google Scholar]
- Brito LF, Irla M, Kalinowski J, Wendisch VF (2017a) Detailed transcriptome analysis of the plant growth promoting Paenibacillus riograndensis SBR5 by using RNA-seq technology. BMC Genomics 18(1):846. 10.1186/s12864-017-4235-z [DOI] [PMC free article] [PubMed]
- Brito LF, Irla M, Walter T, Wendisch VF. Magnesium aminoclay-based transformation of Paenibacillus riograndensis and Paenibacillus polymyxa and development of tools for gene expression. Appl Microbiol Biotechnol. 2017;101(2):735–747. doi: 10.1007/s00253-016-7999-1. [DOI] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55(4):611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Campos SB, Beneduzi A, Fernandes GC, Passaglia LMP. Biological nitrogen fixation. 2015. Genetic and functional characterization of Paenibacillus riograndensis: a novel plant growth-promoting bacterium isolated from wheat; pp. 941–948. [Google Scholar]
- Carabetta VJ, Tanner AW, Greco TM, Defrancesco M, Cristea IM, Dubnau D. A complex of YlbF, YmcA and YaaT regulates sporulation, competence and biofilm formation by accelerating the phosphorylation of Spo0A. Mol Microbiol. 2013;88(2):283–300. doi: 10.1111/mmi.12186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S, Choe D, Lee E, Kim SC, Palsson B, Cho BK. High-level dCas9 expression induces abnormal cell morphology in Escherichia coli. ACS Synth Biol. 2018;7(4):1085–1094. doi: 10.1021/acssynbio.7b00462. [DOI] [PubMed] [Google Scholar]
- Cleto S, Jensen JV, Wendisch VF, Lu TK. Corynebacterium glutamicum metabolic engineering with CRISPR interference (CRISPRi) ACS Synth Biol. 2016;5(5):375–385. doi: 10.1021/acssynbio.5b00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L, Vigouroux A, Rousset F, Varet H, Khanna V, Bikard D. A CRISPRi screen in E. coli reveals sequence-specific toxicity of dCas9. Nat Commun. 2018;9(1):1912. doi: 10.1038/s41467-018-04209-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duetz WA, Rüedi L, Hermann R, O'Connor K, Büchs J, Witholt B. Methods for intense aeration, growth, storage, and replication of bacterial strains in microtiter plates. Appl Environ Microbiol. 2000;66(6):2641–2646. doi: 10.1128/aem.66.6.2641-2646.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellermeier CD, Hobbs EC, Gonzalez-Pastor JE, Losick R. A three-protein signaling pathway governing immunity to a bacterial cannibalism toxin. Cell. 2006;124(3):549–559. doi: 10.1016/j.cell.2005.11.041. [DOI] [PubMed] [Google Scholar]
- Fernandes GC, Trarbach LJ, de Campos SB, Beneduzi A, Passaglia LM (2014) Alternative nitrogenase and pseudogenes: unique features of the Paenibacillus riograndensis nitrogen fixation system. Res Microbiol 165(7):571–580. 10.1016/j.resmic.2014.06.002 [DOI] [PubMed]
- Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol. 2014;32(3):279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6(5):343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- Hamilton TA, Pellegrino GM, Therrien JA, Ham DT, Bartlett PC, Karas BJ, Gloor GB, Edgell DR. Efficient inter-species conjugative transfer of a CRISPR nuclease for targeted bacterial killing. Nat Commun. 2019;10(1):4544. doi: 10.1038/s41467-019-12448-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamon MA, Lazazzera BA. The sporulation transcription factor Spo0A is required for biofilm development in Bacillus subtilis. Mol Microbiol. 2001;42:1199–1209. doi: 10.1046/j.1365-2958.2001.02709.x. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Higuchi R, Dollinger G, Walsh PS, Griffith R. Simultaneous amplification and detection of specific DNA sequences. Biotechnol Nat Publ Co. 1992;10:413–417. doi: 10.1038/nbt0492-413. [DOI] [PubMed] [Google Scholar]
- Hofmann A, Falk J, Prangemeier T, Happel D, Kober A, Christmann A, Koeppl H, Kolmar H. A tightly regulated and adjustable CRISPR-dCas9 based AND gate in yeast. Nucleic Acids Res. 2019;47(1):509–520. doi: 10.1093/nar/gky1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoya S, Asai K, Ogasawara N, Takeuchi M, Sato T. Mutation in yaaT leads to significant inhibition of phosphorelay during sporulation in Bacillus subtilis. J Bacteriol. 2002;184(20):5545–5553. doi: 10.1128/jb.184.20.5545-5553.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CH, Shen CR, Li H, Sung LY, Wu MY, Hu YC. CRISPR interference (CRISPRi) for gene regulation and succinate production in cyanobacterium S. elongatus PCC 7942. Microb Cell Factories. 2016;15(1):196. doi: 10.1186/s12934-016-0595-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irla M, Heggeset TM, Naerdal I, Paul L, Haugen T, Le SB, Brautaset T, Wendisch VF. Genome-based genetic tool development for Bacillus methanolicus: theta- and rolling circle-replicating plasmids for inducible gene expression and application to methanol-based cadaverine production. Front Microbiol. 2016;7:1481. doi: 10.3389/fmicb.2016.01481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SK, Seong W, Han GH, Lee DH, Lee SG. CRISPR interference-guided multiplex repression of endogenous competing pathway genes for redirecting metabolic flux in Escherichia coli. Microb Cell Factories. 2017;16(1):188. doi: 10.1186/s12934-017-0802-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SK, Yoon PK, Kim SJ, Woo SG, Rha E, Lee H, Yeom SJ, Kim H, Lee DH, Lee SG. CRISPR interference-mediated gene regulation in Pseudomonas putida KT2440. Microb Biotechnol. 2020;13(1):210–221. doi: 10.1111/1751-7915.13382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kritikos G, Banzhaf M, Herrera-Dominguez L, Koumoutsi A, Wartel M, Zietek M, Typas A. A tool named Iris for versatile high-throughput phenotyping in microorganisms. Nat Microbiol. 2017;2:17014. doi: 10.1038/nmicrobiol.2017.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Chen J, Minton NP, Zhang Y, Wen Z, Liu J, Yang H, Zeng Z, Ren X, Yang J, Gu Y, Jiang W, Jiang Y, Yang S. CRISPR-based genome editing and expression control systems in Clostridium acetobutylicum and Clostridium beijerinckii. Biotechnol J. 2016;11(7):961–972. doi: 10.1002/biot.201600053. [DOI] [PubMed] [Google Scholar]
- Liu L, Zeng W, Du G, Chen J, Zhou J. Identification of NAD-dependent xylitol dehydrogenase from Gluconobacter oxydans WSH-003. ACS Omega. 2019;4(12):15074–15080. doi: 10.1021/acsomega.9b01867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malvar T, Gawron-Burke C, Baum JA. Overexpression of Bacillus thuringiensis HknA, a histidine protein kinase homology, bypasses early Spo mutations that result in CryIIIA overproduction. J Bacteriol. 1994;176:4742–4749. doi: 10.1128/JB.176.15.4742-4749.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng K, Ye R, Wu XC, Wong SL. Sorbitol dehydrogenase from Bacillus subtilis. Purification, characterization, and gene cloning. J Biol Chem. 1992;267(35):24989–24994. [PubMed] [Google Scholar]
- Ni J, Zhang G, Qin L, Li J, Li C. Simultaneously down-regulation of multiplex branch pathways using CRISPRi and fermentation optimization for enhancing beta-amyrin production in Saccharomyces cerevisiae. Synth Syst Biotechnol. 2019;4(2):79–85. doi: 10.1016/j.synbio.2019.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noirot-Gros MF, Forrester S, Malato G, Larsen PE, Noirot P. CRISPR interference to interrogate genes that control biofilm formation in Pseudomonas fluorescens. Sci Rep. 2019;9(1):15954–15914. doi: 10.1038/s41598-019-52400-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole GA (2011) Microtiter dish biofilm formation assay. J Vis Exp 47. 10.3791/2437 [DOI] [PMC free article] [PubMed]
- Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152(5):1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rütering M, Cress BF, Schilling M, Rühmann B, Koffas MAG, Sieber V, Schmid J (2017) Tailor-made exopolysaccharides-CRISPR-Cas9 mediated genome editing in Paenibacillus polymyxa. Synth Biol 2(1). 10.1093/synbio/ysx007 [DOI] [PMC free article] [PubMed]
- Sambrook J, Russel DW. Molecular cloning. 3. New York: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Sant'Anna FH, Ambrosini A, de Souza R, de Carvalho FG, Bach E, Balsanelli E, Baura V, Brito LF, Wendisch VF, de Oliveira PF, de Souza EM, Passaglia LMP. Reclassification of Paenibacillus riograndensis as a genomovar of Paenibacillus sonchi: genome-based metrics improve bacterial taxonomic classification. Front Microbiol. 2017;8:1849. doi: 10.3389/fmicb.2017.01849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultenkämper K, Brito LF, Lopez MG, Brautaset T, Wendisch VF. Establishment and application of CRISPR interference to affect sporulation, hydrogen peroxide detoxification, and mannitol catabolism in the methylotrophic thermophile Bacillus methanolicus. Appl Microbiol Biotechnol. 2019;103(14):5879–5889. doi: 10.1007/s00253-019-09907-8. [DOI] [PubMed] [Google Scholar]
- Schultenkämper K, Brito LF, Wendisch VF (2020) Impact of CRISPR interference on strain development in biotechnology. Biotechnol Appl Biochem 67(1):7–21. 10.1002/bab.1901 [DOI] [PubMed]
- Sperb ER, Tadra-Sfeir MZ, Sperotto RA, Fernandes Gde C, Pedrosa Fde O, de Souza EM, Passaglia LM. Iron deficiency resistance mechanisms enlightened by gene expression analysis in Paenibacillus riograndensis SBR5. Res Microbiol. 2016;167(6):501–509. doi: 10.1016/j.resmic.2016.04.007. [DOI] [PubMed] [Google Scholar]
- Tan IS, Ramamurthi KS. Spore formation in Bacillus subtilis. Environ Microbiol Rep. 2014;6(3):212–225. doi: 10.1111/1758-2229.12130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J, Zhang F, Karcher D, Bock R. Engineering of high-precision base editors for site-specific single nucleotide replacement. Nat Commun. 2019;10(1):439. doi: 10.1038/s41467-018-08034-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uwajima T. Purification and properties of NAD(+)-dependent sorbitol dehydrogenase from Bacillus fructosus. Biosci Biotechnol Biochem. 1999;63(3):573–574. doi: 10.1271/bbb.63.573. [DOI] [PubMed] [Google Scholar]
- Veening JW, Murray H, Errington J. A mechanism for cell cycle regulation of sporulation initiation in Bacillus subtilis. Genes Dev. 2009;23(16):1959–1970. doi: 10.1101/gad.528209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zhao P, Li Y, Xu L, Tian P. Engineering CRISPR interference system in Klebsiella pneumoniae for attenuating lactic acid synthesis. Microb Cell Factories. 2018;17(1):56. doi: 10.1186/s12934-018-0903-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Cao Y, Wang Y, Sun L, Song H. Enhancing surfactin production by using systematic CRISPRi repression to screen amino acid biosynthesis genes in Bacillus subtilis. Microb Cell Factories. 2019;18(1):90. doi: 10.1186/s12934-019-1139-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White AK, Hoch JA, Grynberg M, Godzik A, Perego M. Sensor domains encoded in Bacillus anthracis virulence plasmids prevent sporulation by hijacking a sporulation sensor histidine kinase. J Bacteriol. 2006;188(17):6354–6360. doi: 10.1128/JB.00656-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Qi LS. A CRISPR-dCas toolbox for genetic engineering and synthetic biology. J Mol Biol. 2019;431(1):34–47. doi: 10.1016/j.jmb.2018.06.037. [DOI] [PubMed] [Google Scholar]
- Xu T, Li Y, Shi Z, Hemme CL, Li Y, Zhu Y, Van Nostrand JD, He Z, Zhou J. Efficient genome editing in Clostridium cellulolyticum via CRISPR-Cas9 nickase. Appl Environ Microbiol. 2015;81(13):4423–4431. doi: 10.1128/AEM.00873-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan Y, Xu Y, Zheng P, He M, Sun S, Wang D, Cai D, Ma X, Chen S. Establishment and application of multiplexed CRISPR interference system in Bacillus licheniformis. Appl Microbiol Biotechnol. 2020;104(1):391–403. doi: 10.1007/s00253-019-10230-5. [DOI] [PubMed] [Google Scholar]
- Zhang S, Voigt CA. Engineered dCas9 with reduced toxicity in bacteria: implications for genetic circuit design. Nucleic Acids Res. 2018;46(20):11115–11125. doi: 10.1093/nar/gky884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuberi A, Misba L, Khan AU. CRISPR interference (CRISPRi) inhibition of luxS gene expression in E. coli: an approach to inhibit biofilm. Front Cell Infect Microbiol. 2017;7:214. doi: 10.3389/fcimb.2017.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]



