Abstract
Increasing evidence of involvement of non-coding RNAs, especially long non-coding RNAs (lncRNAs), in the molecular biology of various malignancies have been recently reported. Their utilization as markers for diagnosis, prognosis and evaluation of treatment response was widely investigated. As the impact of lncRNA HOTAIR on multiple myeloma (MM) was not properly highlighted, we aimed to explore the expression levels of HOTAIR in three groups of MM patients and to analyze its relationship to different patients’ characteristics. Plasma samples were withdrawn from 24 newly diagnosed MM patients, 23 post-therapy patients in complete response (CR) or very good partial response (VGPR) and 15 patients who had either progressive disease (PD) or relapse. The expression of lncRNA HOTAIR in MM patients and 20 healthy controls was analyzed by quantitative reverse transcription polymerase chain reactions. HOTAIR was significantly upregulated in newly diagnosed and PD/relapse categories in comparison with controls and MM patients who had achieved CR or VGPR (P < 0.001). Furthermore; HOTAIR expression levels correlated with the percentage of malignant plasma cells in bone marrow (P = 0.006) and disease stage (ISS stage) (P = 0.031). HOTAIR may be employed as prognostic molecular marker and novel therapeutic tool for newly diagnosed MM patients.
Keywords: HOTAIR long untranslated RNA, Multiple myeloma, Plasma cells, Polymerase chain reaction, Prognosis
Introduction
Multiple myeloma (MM), an incurable plasma cell neoplasm, is characterized by abnormal proliferation of clonal plasma cells within the bone marrow microenvironment, the secretion of monoclonal immunoglobulin in serum or urine and the frequently present lytic bone lesions [1].
An enhanced understanding of MM pathogenesis and the development of novel therapeutic strategies resulted in improved response rates and survival of MM patients. Despite these advances, the major challenge of MM being incurable illness has not been overcome and most patients experience relapse [2].
Being heterogeneous disease, MM patients have various outcomes and responses to therapy. Proper and accurate evaluation of the prognosis of MM patients is of positive impact on the selection of reasonable treatment options adjusted to each patient. With the continuous progress of technology, the early detection, prognosis and treatment response evaluation of MM patients are no longer limited to ordinary techniques [3].
Long non-coding RNAs (lncRNAs), substantial players of critical cellular functions, are RNA molecules longer than 200 nucleotides of no protein-synthesis ability [4]. The expressions and roles of lncRNAs in various begin and malignant diseases have been widely investigated [5–8].
Aberrant expression of lncRNAs has been observed in many types of hematological malignancies, suggesting that lncRNAs own either oncogenic or tumor suppressive properties [9–12]. In addition to crucial role in tumor development and progression, lncRNAs showed many interesting features as diagnostic and predictive biomarkers and also as promising therapeutic targets [13].
The identification of deregulated lncRNAs in MM has progressively raised questions concerning the link between lncRNAs and MM biology [14]. Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) was reported as the first lncRNA to be overexpressed in bone marrow samples of newly diagnosed and relapsed MM patients compared to successively treated patients and healthy individuals [15].
HOX transcript antisense intergenic RNA (HOTAIR), lncRNA with 2158 nucleotides length, is suggested to play an oncogenic role by enhancing cancer progression, migration and invasion [16]. HOTAIR interacts with pivotal epigenetic regulators such as polycomb repressive complex 2 and Lysine-specific histone demethylase 1A, regulates histone methylation and eventually results in gene silencing [17].
HOTAIR was reported in many studies as a predictor of poor patient outcome when highly expressed [18–20]. Nevertheless, the impact of HOTAIR on MM has not yet been satisfactorily investigated. To understand the potential role of HOTAIR in MM, we tested the expression level of HOTAIR in three groups of MM patients and analyzed its relationship to different patients’ characteristics.
Materials and Methods
The study was conducted in the period between February 2017 and December 2018 and included 62 MM patients who consecutively attended Clinical Oncology Department and refereed to Clinical Pathology Department, Menoufia University to be appropriately investigated. MM patients were grouped as 24 newly diagnosed MM patients (16 males, 8 females; median age 55 years), 23 post-therapy patients whose response criteria were matched with complete or very good partial response category (14 males, 9 females; median age 54 years) and 15 patients who experienced progressive disease (PD) or relapse (10 males, 5 females; median age 56 years). For the purpose of comparison, twenty healthy volunteers were included as a control group (12 males, 8 females; median age 55.5 years).
The updated criteria of International Myeloma Working Group (IMWG) were used as a guide for proper diagnosis of MM patients [21]. The treatment response categories were in accordance with the uniform response criteria created by IMWG [22].
The selection of therapy protocol based on whether the patient was candidate for stem cell transplantation or not. Bortezomib-based therapy was applied for transplant candidate patients while for non-transplant candidates, melphalan-based therapy was used.
Approval for this study protocol was obtained by the Ethics Committee of Menoufia University. Each patient and control provided informed consent according to the guidelines of the Declaration of Helsinki.
Methods
Plasma Collection
Whole blood was collected from each patient and control in EDTA tubes and samples were centrifuged at 2000 g for 10 min. Plasma was preserved into nuclease-free tubes at − 80 °C for further manipulation.
RNA Isolation
200 μL of plasma was adequate to extract total RNA by miRNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. Before being transcribed into cDNA; isolated RNA was assessed for concentration and purity by NanoDrop 2000 (Thermo Fisher Scientific, Wilmington, Delaware, USA).
Quantitative Reverse Transcription Polymerase Chain Reaction (RT-qPCR)
RT2 first strand Kit (Qiagen, Hilden, Germany) was used to synthesize cDNA; the genomic DNA elimination mix (4 μL RNA, 2 μL buffer GE and 4 μL nuclease-free H2O) for each RNA sample was prepared and incubated at 42 °C for 5 min. Genomic DNA elimination mix (10 μL) was mixed with the10 μL of the prepared reverse transcription mix (4 μL buffer BC3, 1 μL control P2, 2 μL RE3 reverse transcriptase mix and 3 μL nuclease-free H2O). The mixture was incubated at 37 °C for 60 min and finally the reaction was halted by incubation at 95 °C for 5 min. Real-time PCR protocol was delayed and the reactions were stored at − 20 °C.
PCR amplification mix was supplied by (Qiagen, Hilden, Germany) and included mixure of 12.5 μL RT2 SYBR Green Master Mix, 1 μL RT2 lncRNA qPCR Assay, 2 μL cDNA and 9.5 μL nuclease-free H2O to a final volume of 25 μL. Relative gene expression was quantified by ABI 7500 Real-Time PCR detection system (Applied Biosystems). Each sample was analyzed in duplicate. The sequences of primers specific for HOTAIR were (forward, 5′-CAGTGGGGAACTCTGACTCG-3′; reverse, 5′-GTGCCTGGTGCTCTCTTACC-3′). GAPDH was used as a reference gene for standardizing the expression of targeted HOTAIR LncRNA. GAPDH specific primers were (forward, 5′-AGCCACATCGCTCAGACAC-3′; and reverse, 5′-GCCCAATACGACCAAATCC-3′). QRT-PCR cycling conditions were stated at 95 °C for 10 min, followed by 45 cycles at 95 °C for 15 s and at 60 °C for 1 min. After normalization of the data according to the expression of GAPDH, the relative expression levels of HOTAIR were calculated using the 2−ΔΔCt method: in which ΔΔCT = ΔCt experimental − ΔCt control.
Statistical Analysis
Data analysis was conducted with SPSS software (version 20; IBM Corporation, USA). The relationships between the studied MM groups and healthy controls were assessed using Chi square (χ2), ANOVA and Kruskal–Wallis tests. Mann–Whitney and Kruskal–Wallis tests were used to study the relationship between the expression level of HOTAIR and characteristics of newly diagnosed MM. When P value was less than 0.05, the result was reported as statistically significant.
Results
Demographic characteristics of the four studied groups were shown in Table 1. There were no significant differences as regard age and gender distribution (P = 0.883, P = 0.952 respectively).
Table 1.
Demographic characteristics and HOTAIR expression levels in patients with multiple myeloma and healthy individuals
| Characteristics | Newly diagnosed MM (n = 24) | PD or relapsed MM (n = 15) | MM-CR or MM-VGPR (n = 23) | Healthy individuals (n = 20) | P value |
|---|---|---|---|---|---|
| Age | 55.08 ± 5.85 | 54.73 ± 6.26 | 55.26 ± 5.71 | 56.2 ± 5.43 | 0.883 |
| Gender | |||||
| Male | 16 (66.7%) | 10 (66.7%) | 14 (60.9%) | 12 (60%) | 0.952 |
| Female | 8 (33.3%) | 5 (33.3%) | 9 (39.1%) | 8 (40%) | |
| HOTAIR expression levels | 3.13 ± 1.48 | 2.83 ± 1.26 | 1.02 ± 0.45 | 0.99 ± 0.38 | < 0.001* |
| Comparison between groups according to HOTAIR expression levels | |||||
| Newly diagnosed versus CR/VGPR | < 0.001* | ||||
| Newly diagnosed versus Healthy individuals | < 0.001* | ||||
| Newly diagnosed versus PD/relapsed | 0.692 | ||||
| PD/relapsed versus CR/VGPR | < 0.001* | ||||
| CR/VGPR versus Healthy individuals | 0.921 | ||||
MM multiple myeloma, PD progressive disease, CR complete response, VGPR very good partial response
*Statistically significant at P < 0.05
Expression Levels of lncRNA HOTAIR in Multiple Myeloma Patients and Healthy Individuals
LncRNA HOTAIR expression levels were: Newly diagnosed patients (range 0.9–6.9; median 2.7), CR/VGPR MM patients (range 0.1–1.8; median 1.0), patients with PD or relapse (range 1.1–5.2; median 2.8) and healthy controls (range 0.1–1.5, median 1.1) (Fig. 1).
Fig. 1.
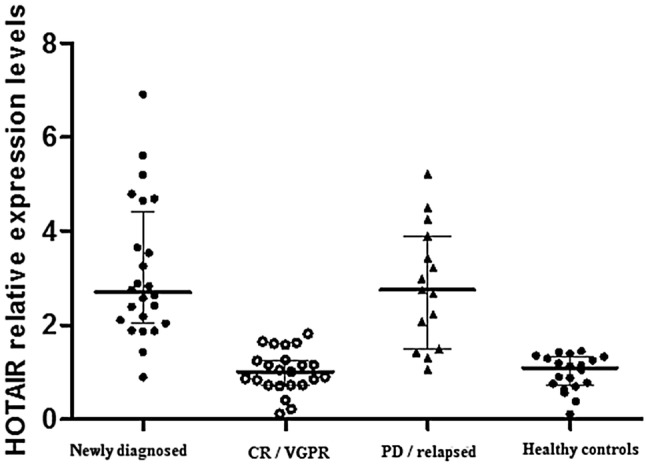
Relative lncRNA HOTAIR expression levels in MM patients and healthy controls
Clinical Importance of Dysregulated lncRNA HOTAIR in MM
Our data revealed that the lncRNA HOTAIR was significantly upregulated in newly diagnosed and MM patients with PD or relapse in comparison with controls and MM patients who got CR/VGPR (P < 0.001). There were no significant differences between newly diagnosed and progressed or relapsed MM patients (P = 0.692), or between MM patients who achieved CR/VGPR and controls (P = 0.921) (Table 1).
Correlation Between HOTAIR Expression and Characteristics of Newly Diagnosed MM Group
The increased expression level of lncRNA HOTAIR was associated with higher percentage of malignant plasma cells in bone marrow (P = 0.006) and with advanced disease stage (ISS stage) (P = 0.031). However, no associations were found between HOTAIR expression levels and other clinical and laboratory parameters (Table 2).
Table 2.
The relationship between the expression level of lnc HOTAIR and characteristics of newly diagnosed MM
| Characteristics | Number (n = 24) | Relative expression of HOTAIR Mean ± SD | P value |
|---|---|---|---|
| Age (years) | |||
| < 65 | 22 | 3.06 ± 1.43 | 0.652 |
| ≥ 65 | 2 | 3.86 ± 2.47 | |
| Gender | |||
| Male | 16 | 3.13 ± 1.34 | 0.569 |
| Female | 8 | 3.12 ± 1.82 | |
| Ig isotype | |||
| IgG | 18 | 3.34 ± 1.49 | 0.069 |
| IgA | 3 | 1.63 ± 0.64 | |
| LC | 3 | 3.37 ± 1.41 | |
| Hemoglobin (g/L) | |||
| < 100 | 20 | 3.33 ± 1.54 | 0.08 |
| ≥ 100 | 4 | 2.13 ± 0.33 | |
| B2-Micro globulin (mg/L) | |||
| < 3.5 | 8 | 2.24 ± 0.78 | 0.087 |
| ≥ 3.5 | 16 | 3.58 ± 1.56 | |
| Albumin (g/L) | |||
| < 35 | 14 | 3.47 ± 1.46 | 0.096 |
| ≥ 35 | 10 | 2.65 ± 1.43 | |
| Creatinine (μmol/L) | |||
| < 177 | 15 | 2.93 ± 1.32 | 0.640 |
| ≥ 177 | 9 | 3.45 ± 1.74 | |
| Calcium (mmol/L) | |||
| < 2.65 | 15 | 2.73 ± 1.20 | 0.215 |
| ≥ 2.65 | 9 | 3.79 ± 1.72 | |
| Myeloma cells in BM (%)a | |||
| < 48 | 12 | 2.29 ± 0.67 | 0.006* |
| ≥ 48 | 12 | 3.97 ± 1.60 | |
| Bone disease | |||
| Absent | 9 | 2.78 ± 0.93 | 0.599 |
| Present | 15 | 3.34 ± 2.83 | |
| D–S stage | |||
| I | 3 | 2.21 ± 0.36 | 0.160 |
| II | 7 | 2.53 ± 1.19 | |
| III | 14 | 3.62 ± 1.59 | |
| ISS stage | |||
| I | 5 | 1.85 ± 0.58 | 0.031* |
| II | 10 | 3.06 ± 1.17 | |
| III | 9 | 3.91 ± 1.69 |
D-S Durie–Salmon, ISS International Staging System, BM bone marrow, LC light chain, SD standard deviation
*Statistically significant at P < 0.05
a48% is the mean value of the percentage of myeloma cells in BM of 24 MM patients
Discussion
Unfortunately, MM is incurable disease with most patients experience relapse, therapy unresponsiveness or progression [23]. Therefore, there is a crucial demand for discovery of new biomarkers of early progression and for development of novel therapeutic modalities for these patients.
LncRNAs are heterogeneous family with growing pivotal role in biological processes of normal and cancer cells. Their stability in body fluids and their tissue-specific expression support their use as potential diagnostic and predictive biomarkers in many cancer types [24]. Researches involving the link between lncRNAs and MM are still limited. However, the available data suggest important role of lncRNAs in MM [25].
Recent studies have investigated the clinical implication of various lncRNAs in MM patients. Hu et al. [26] studied 176 cancer-associated long non-coding RNAs and identified five promising lncRNA with predictive value in MM patients. Another study by Yu et al. [27] identified LINC00152 as oncogenic lncRNA with possible diagnostic and therapeutic benefits for MM patients.
The utilization of blood-based cancer biomarkers has aroused a great attention. Previous researches have shown that lncRNAs can be detected in blood, and can be employed as non-invasive tools of diagnostic and prognostic importance in malignancies [28, 29].
HOTAIR was suggested as a modulator of cancer progression and as a novel prognostic and predictive marker in cancer patients [30, 31]. However, in MM, the HOTAIR expression status and its prognostic significance were not perfectly elucidated.
In the present study, we demonstrated a novel and valuable knowledge about the dysregulated expression of lncRNA HOTAIR in plasma of MM patients. HOTAIR was overexpressed in newly diagnosed and progressed/relapsed patients in comparison with MM-CR/MM-VGPR patients and healthy controls. These data indicate that HOTAIR may act as crucial player in biology of multiple myeloma and raise the possibility of its use as therapeutic target.
Previous study conducted by Isin et al. [32] reported that plasma HOTAIR levels were lower in MM patients in comparison with healthy controls. The authors in this study mentioned that 58 MM patients were investigated but they did not clarify whether those patient were newly diagnosed or under therapy. Actually their findings, as they mentioned, were inconsistent with previous studies that reported up-regulation of HOTAIR in multiple malignancies.
Identification of different roles of lncRNAs in MM will be essential step toward the comprehension of pathobiology and cancer-promoting pathways in MM. Furthermore; lncRNA may be utilized as excellent anticancer therapeutic tools, because of their cancer-specific expression that represent a major advantage over other therapeutic approaches [33].
We also analyzed the association between HOTAIR expression and clinical characteristics, and we found that the expression level of HOTAIR was associated with the percentage of malignant plasma cells in bone marrow and ISS stage. These findings show that HOTAIR may be useful as a novel prognostic biomarker for MM.
Some limitations still exist in our study. First: The definite role of the dysregulated lncRNA HOTAIR in pathogenesis of MM has yet to be identified. Subsequently, further experimental studies are required to determine the biological effects of lncRNA HOTAIR in MM. Second: HOTAIR expression levels were analyzed using peripheral blood samples only. Validation of results using bone marrow plasma samples should be done in subsequent studies.
To conclude, our study identified the dysregulated expression of lncRNA HOTAIR in plasma samples of MM patients. Our data may be the cornerstone for further investigation of lncRNA HOTAIR in MM. These results may raise attention to explore the efficiency of lncRNA HOTAIR as a novel therapeutic target and prognostic molecular biomarker for newly diagnosed MM patients. However, further studies are required to confirm the prognostic potential using larger groups of patients with long-term follow-up.
Financial Support and Sponsorship
None.
Compliance with Ethical Standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Gao D, Lv AE, Li HP, Han DH, Zhang YP. LncRNA MALAT-1 elevates HMGB1 to promote autophagy resulting in inhibition of tumor cell apoptosis in multiple myeloma. J Cell Biochem. 2017;118:3341–3348. doi: 10.1002/jcb.25987. [DOI] [PubMed] [Google Scholar]
- 2.Lee HC, Wang H, Baladandayuthapani V, Lin H, He J, Jones RJ, et al. RNA polymerase I inhibition with CX-5461 as a novel therapeutic strategy to target MYC in multiple myeloma. Br J Haematol. 2017;177:80–94. doi: 10.1111/bjh.14525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levin A, Hari P, Dhakal B. Novel biomarkers in multiple myeloma. Transl Res. 2018;201:49–59. doi: 10.1016/j.trsl.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 4.Sun M, Kraus WL. From discovery to function: the expanding roles of long noncoding RNAs in physiology and disease. Endocr Rev. 2015;36:25–64. doi: 10.1210/er.2014-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yao X, Yan C, Zhang L, Li Y, Wan Q. LncRNA ENST00113 promotes proliferation, survival, and migration by activating PI3K/Akt/mTOR signaling pathway in atherosclerosis. Medicine (Baltimore) 2018;97:e0473. doi: 10.1097/MD.0000000000010473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fachrul M, Utomo DH, Parikesit AA. lncRNA-based study of epigenetic regulations in diabetic peripheral neuropathy. Silico Pharmacol. 2018;6:7. doi: 10.1007/s40203-018-0042-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng H, Wang JM, Li M, Tang R, Tang K, Su Y, et al. Long non-coding RNAs: new biomarkers for prognosis and diagnosis of colon cancer. Tumour Biol. 2017;39:1010428317706332. doi: 10.1177/1010428317706332. [DOI] [PubMed] [Google Scholar]
- 8.Chen H, Xu Z, Liu X, Gao Y, Wang J, Qian P, Yang B. Increased expression of Lncrna RP11-397A15.4 in gastric cancer and its clinical significance. Ann Clin Lab Sci. 2018;48:707–711. [PubMed] [Google Scholar]
- 9.Qin J, Bao H, Li H. Correlation of long non-coding RNA taurine-upregulated gene 1 with disease conditions and prognosis, as well as its effect on cell activities in acute myeloid leukemia. Cancer Biomark. 2018;23:569–577. doi: 10.3233/CBM-181834. [DOI] [PubMed] [Google Scholar]
- 10.Li ZY, Yang L, Liu XJ, Wang XZ, Pan YX, Luo JM. The long noncoding RNA MEG3 and its target miR-147 regulate JAK/STAT pathway in advanced chronic myeloid leukemia. EBioMedicine. 2018;34:61–75. doi: 10.1016/j.ebiom.2018.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chung IH, Lu PH, Lin YH, Tsai MM, Lin YW, Yeh CT, Lin KH. The long non-coding RNA LINC01013 enhances invasion of human anaplastic large-cell lymphoma. Sci Rep. 2017;7:295. doi: 10.1038/s41598-017-00382-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li QY, Chen L, Hu N, Zhao H. Long non-coding RNA FEZF1-AS1 promotes cell growth in multiple myeloma via miR-610/Akt3 axis. Biomed Pharmacother. 2018;103:1727–1732. doi: 10.1016/j.biopha.2018.04.094. [DOI] [PubMed] [Google Scholar]
- 13.Bonetti A, Carninci P. From bench to bedside: the long journey of long non-coding RNAs. Curr Opin Syst Biol. 2017;3:119–124. doi: 10.1016/j.coisb.2017.04.016. [DOI] [Google Scholar]
- 14.Ronchetti D, Manzoni M, Todoerti K, Neri A, Agnelli L. In silico characterization of miRNA and long non-coding RNA interplay in multiple myeloma. Genes (Basel) 2016;7:107. doi: 10.3390/genes7120107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cho SF, Chang YC, Chang CS, Lin SF, Liu YC, Hsiao HH, Chang JG, Liu TC. MALAT1 long non-coding RNA is overexpressed in multiple myeloma and may serve as a marker to predict disease progression. BMC Cancer. 2014;14:809. doi: 10.1186/1471-2407-14-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hajjari M, Rahnama S. HOTAIR long non-coding RNA: characterizing the locus features by the in silico approaches. Genom Inform. 2017;15:170–177. doi: 10.5808/GI.2017.15.4.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bhan A, Mandal SS. LncRNA HOTAIR: a master regulator of chromatin dynamics and cancer. Biochim Biophys Acta. 2015;1856:151–164. doi: 10.1016/j.bbcan.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu XH, Liu ZL, Sun M, Liu J, Wang ZX, De W. The long non-coding RNA HOTAIR indicates a poor prognosis and promotes metastasis in non-small cell lung cancer. BMC Cancer. 2013;13:464. doi: 10.1186/1471-2407-13-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu ZH, Wang XL, Tang HM, Jiang T, Chen J, Lu S, et al. Long non-coding RNA HOTAIR is a powerful predictor of metastasis and poor prognosis and is associated with epithelial-mesenchymal transition in colon cancer. Oncol Rep. 2014;32:395–402. doi: 10.3892/or.2014.3186. [DOI] [PubMed] [Google Scholar]
- 20.Li H, Li J, Zhang B, Zeng H. Long-chain non-coding RNA HOTAIR expression in tissue samples correlates with gastric cancer survival. Int J Clin Exp Med. 2018;11:856–862. [Google Scholar]
- 21.Rajkumar SV, Dimopoulos MA, Palumbo A, Blade J, Merlini G, Mateos MV, et al. International Myeloma Working Group updated criteria for the diagnosis of multiple myeloma. Lancet Oncol. 2014;15:e538–e548. doi: 10.1016/S1470-2045(14)70442-5. [DOI] [PubMed] [Google Scholar]
- 22.Durie BG, Harousseau JL, Miguel JS, Bladé J, Barlogie B, Anderson K, et al. International uniform response criteria for multiple myeloma. Leukemia. 2006;20:1467–1473. doi: 10.1038/sj.leu.2404284. [DOI] [PubMed] [Google Scholar]
- 23.Nijhof IS, van de Donk NWCJ, Zweegman S, Lokhorst HM. Current and new therapeutic strategies for relapsed and refractory multiple myeloma: an update. Drugs. 2018;78:19–37. doi: 10.1007/s40265-017-0841-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77:3965–3981. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ronchetti D, Agnelli L, Pietrelli A, Todoerti K, Manzoni M, Taiana E, Neri A. A compendium of long non-coding RNAs transcriptional fingerprint in multiple myeloma. Sci Rep. 2018;8:6557. doi: 10.1038/s41598-018-24701-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu AX, Huang ZY, Zhang L, Shen J. Potential prognostic long non-coding RNA identification and their validation in predicting survival of patients with multiple myeloma. Tumour Biol. 2017;39:1010428317694563. doi: 10.1177/1010428317694563. [DOI] [PubMed] [Google Scholar]
- 27.Yu T, Xu Z, Zhang X, Men L, Nie H. Long intergenic non-protein coding RNA 152 promotes multiple myeloma progression by negatively regulating microRNA-497. Oncol Rep. 2018;40:3763–3771. doi: 10.3892/or.2018.6721. [DOI] [PubMed] [Google Scholar]
- 28.Shi T, Gao G, Cao Y. Long noncoding RNAs as novel biomarkers have a promising future in cancer diagnostics. Dis Markers. 2016;2016:9085195. doi: 10.1155/2016/9085195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bolha L, Ravnik-Glavač M, Glavač D. Long noncoding RNAs as biomarkers in cancer. Dis Markers. 2017;2017:7243968. doi: 10.1155/2017/7243968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang B, Qu XL, Liu J, Lu J, Zhou ZY. HOTAIR promotes osteosarcoma development by sponging miR-217 and targeting ZEB1. J Cell Physiol. 2019;234:6173–6181. doi: 10.1002/jcp.27394. [DOI] [PubMed] [Google Scholar]
- 31.Shen J, Hodges TR, Song R, Gong Y, Calin GA, Heimberger AB, Zhao H. Serum HOTAIR and GAS5 levels as predictors of survival in patients with glioblastoma. Mol Carcinog. 2018;57:137–141. doi: 10.1002/mc.22739. [DOI] [PubMed] [Google Scholar]
- 32.Isin M, Ozgur E, Cetin G, Erten N, Aktan M, Gezer U, Dalay N. Investigation of circulating lncRNAs in B-cell neoplasms. Clin Chim Acta. 2014;431:255–259. doi: 10.1016/j.cca.2014.02.010. [DOI] [PubMed] [Google Scholar]
- 33.Nobili L, Ronchetti D, Agnelli L, Taiana E, Vinci C, Neri A. Long non-coding RNAs in multiple myeloma. Genes (Basel) 2018;9:69. doi: 10.3390/genes9020069. [DOI] [PMC free article] [PubMed] [Google Scholar]


