Abstract
Multiple myeloma (MM) is an uncontrolled proliferation of plasma cells and these cells play an important role in the immune system. In this research, we retrospectively analyzed cytogenetic abnormalities in 381 patients with MM. Conventional cytogenetic analysis was successful in 354 patients (92.9%). Chromosomal abnormalities were detected in 31.9% (113/354) and 45.8% (116/253) of patients screened with conventional cytogenetics and FISH, respectively. Of 113 patients with chromosomal abnormalities, 31 patients (27.4%) had hyperdiploid and 26 of 31 patients with hyperdiploidy had both numerical and structural anomalies. On the other hand, non-hyperdiploidy was observed in 62 patients (54.8%). The most common gains of chromosomes were 15, 9, 19 followed by 3, 5, 11, and 21. Whole chromosome losses were also frequent involving Y, 13 and 22 chromosomes. In our patients, 1q gain was determined in a total of 25 patients (22%), including 7 structural abnormalities and 19 unbalanced translocations causing complete or partial duplication of the long arm of chromosome 1. Although the breakpoints were different, t(1;5) balanced translocation and unbalanced translocations of t(1;2), t(1;3), t(1;7), t(1;16) and t(1;19) were observed twice. The most common structural abnormality was the deletion of the short arm of chromosome 13 (13q) or monosomy of chromosome 13 (-13) (24.1%, 61/253) in patients evaluated by FISH. Deletion involving chromosome 17p (del 17p) or monosomy of chromosome 17 (-17) were found in 31 (12.3%) patients. Translocations involving IgH regions were as follows: t(11;14)(q13;q32.33) in 22 (8.7%), t(4;14)(p16.3;q32.33) in 22 (8.7%) and t(14;16)(q32.33;q23.1) in 2 (0.8%) patients. In addition, t(14;17)(q32;q21) translocation was detected in a multiple myeloma patient for the first time in this study. There are a limited number of large study groups including both cytogenetic and FISH findings in MM patients. As the number of these studies increases, it is thought that new cytogenetic data can be guiding especially in clinical risk determination.
Electronic supplementary material
The online version of this article (10.1007/s12288-019-01215-5) contains supplementary material, which is available to authorized users.
Keywords: Multiple myeloma, Cytogenetics, Chromosomal abnormalities, FISH
Introduction
Multiple myeloma (MM) is a clonal bone marrow disease resulting in differentiation of B cells by neoplastic transformations [1]. This hematological malignancy, which accounts approximately 1% of all cancer cases, is the second most common type of cancer in the United States [2]. MM seen most commonly in older people, the median age of patients is about 65 years at the time of diagnosis [2, 3]. MM causes accumulation of plasma cells in bone marrow, excessive increase in monoclonal immunoglobulins in urine or serum, and destructive lytic bone lesions. Typical symptoms are; bone pain related to lytic bone lesions, anemia, kidney failure, infections and neurological problems in the patients with MM [4, 5]. Nowadays, the treatment strategies have been changed in the era of novel agents. Autologous stem cell transplantation and novel drugs such as proteasome inhibitors (bortezomib) and immunomodulatory drugs (thalidomide and lenalidomide) have become routine therapies in MM and the median survival of patients with MM has increased from 3 to 6 years [6, 7]. However, there is still reduced survival of patients with newly diagnosed MM treated by autologous stem cell transplantation due to malignant plasma cells having complex karyotype including multiple numerical and structural chromosomal abnormalities [8–12]. Currently, detection of cytogenetic abnormalities is accepted as one of the most important tools for estimated outcome in newly diagnosed MM patients. In previous studies, abnormal karyotypes were reported in approximately 30–50% in patients with MM by conventional cytogenetics, more often in advanced stage than newly diagnosed patients [13, 14]. While conventional cytogenetic analyses can provide important diagnostic and prognostic information, low mitotic index of tumor plasma cells is considered to be a significant limiting factor [15]. Interpretation can be very difficult because some anomalies are cryptic and chromosomal morphologies can be in poor quality. It is possible to overcome these limitations by using FISH probes targeting frequently observed anomalies in multiple myeloma.
In this study, we aimed to determine the frequency of chromosomal anomalies in Turkish patients with newly diagnosed MM by analyzing both conventional cytogenetics and FISH results.
Materials and Methods
Patients
In this study, 381 patients who fulfilled the inclusion criteria with a diagnosis of Multiple Myeloma were included. Our study group consisted of patients who were sent from Hematology department of Akdeniz University Hospital to Center of Genetic Diagnosis between 2009 and 2018 years to be investigated with conventional cytogenetic and FISH analysis. Symptomatic Multiple Myeloma was diagnosed according to the International Myeloma Working Group Criteria defined in 2003 and updated in 2014 [16, 17]. The cytogenetic risk stratification which was based on Mayo Clinic Risk Stratification of Myeloma and Risk-Adapted Therapy (mSMART) [18]. Furthermore, patients who had complex karyotype without positive FISH findings were considered as high risk cytogenetics.
Conventional Cytogenetics
When culturing bone marrow cells, chromosome kit M (EuroClone, Milan, Italy) was used and attention was paid to the manufacturer’s protocols. The Synchroset (EuroClone, Milan, Italy) was used in order to acquire a high band level of chromosomes and a high number of metaphases. Bone marrow cell cultures were incubated for 24 or 48 h at 37 °C. Colcemid (0.1 μg/ml) (Gibco, USA) was added 30 min before culture termination and then harvested. The cells were harvested in accordance with standard cytogenetic preparations and metaphase chromosomes were analyzed by using standard Giemsa banding. At least 20 metaphases were analysed as far as possible. Karyotypes were described according to the International System for Human Cytogenetic Nomenclature (ISCN 2016). Numerical chromosomal abnormalities were assigned to the two categories: hyperdiploid (47–57 chromosomes) and non-hyperdiploid (pesudodiploid for those with 46 chromosomes and structural or numerical abnormalities, hypodiploid if they had 45 or less chromosomes, and near-triploid if they had 58–80 chromosomes) [19].
Fluorescence In Situ Hybridization (FISH)
On slides which had been prepared from cultured bone marrow cells, the FISH studies was carried out using standardized protocols, according to the manufacturer’s instructions (Cytocell, Cambridge, UK). The following probes were used; IGH/FGFR3 Dual Fusion (DF) Probe (Cytocell, UK) for detection of t(4;14)(p16.3;q32.33), IGH/MAF DF Probe (Cytocell, UK) for detection of t(14;16)(q32.33;q23.1), IGH/CCND1/MYEOV DF Probe (Cytocell, UK) for detection of t(11;14)(q13;q32.33), 13q14.3 Dual Color Probe (Cytocell, UK) for detection of del(13q14), P53 (TP53) Dual Color Probe (Cytocell, UK) for detection of del(17p13) and ATM Dual Color Probe (Cytocell, UK) for detection of del(11q22.3). The evaluation of FISH signals was performed using a fluorescence microscope (Axio Imager. M1. Carl Zeiss, Germany) with the software Metafer 4 (version 3.9.0 MetaSystems GmbH, Altlussheim, Germany). At least 200 interphase nuclei were analyzed for each slide.
Results
Three hundred eighty-one patients with MM were studied. Conventional cytogenetic analysis failed in only 27 (7.1%) of 381 patients. One hundred thirteen (31.9%) of the rest 354 patients were found to have an abnormal karyotype. The cytogenetic results of 113 patients with MM were presented in supplementary table. In our series, of 113 patients with abnormal karyotype, 35 patients were female and 78 were male (sex ratio 1:2.2). The age of patients ranged from 42 to 84 years at diagnosis, with median age of 63. Additionally, clinical follow-up of 24 patients was available and the medications used in treatment, drug response and patient follow-up of these 24 patients were given in supplementary table.
The frequency of cytogenetic abnormalities in patients with abnormal karyotype was summarized in Table 1. The patients with abnormal karyotpe were separated into two groups according to the number of the chromosomes. There were 27.4% (31/113) hyperdiploid cases and 54.8% (62/113) non-hyperdiploid cases (31 hypodiploid, 24 pseudodiploid and 7 near-triploid). Also, Table 1 shows the cytogenetic risk stratification of MM patients. We determined the risk stratification of each patient based on the combined results of FISH and cytogenetics. The number of patients in standard, intermediate and high risk groups were 36, 25 and 52, respectively.
Table 1.
Frequencies of cytogenetic abnormalities in patients who were evaluated with conventional cytogenetic and/or FISH and risk stratification
| Cytogenetic abnormalities | N (%) |
|---|---|
| Hyperdiploidy | 31 (27.4) |
| Non-hyperdiploidy | |
| Hypodiploidy | 31 (27.4) |
| Pseudodiploidy | 24 (21.4) |
| Near-triploidy | 7 (6.2) |
| Loss of Y chromosome | 20 (17.7) |
| 1q gain | 25 (21.2) |
| 1p del | 5 (4.2) |
| FISH abnormalities | |
| -13/del(13q) | 61 (24.1) |
| -17/del(17p) | 31 (12.3) |
| t(11;14)(q13;q32)/IGH-CCND1 | 22 (8.7) |
| t(4;14)(p16;q32)/IGH-FGFR3 | 22 (8.7) |
| t(14;16)(p32;q23)/IGH-MAF | 2 (0.8) |
| Risk stratification | |
| Standard risk | 36 (31.8) |
| Intermediate risk | 25 (22.1) |
| High risk | 52 (46.0) |
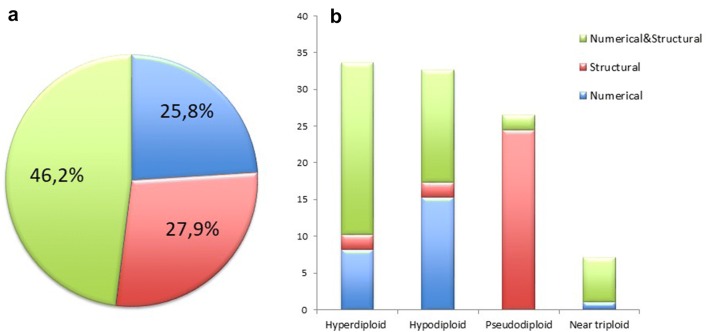
Besides, twenty patients (17.7%) with Y chromosome loss were classified separately. Of the 93 patients, 24 (25.8%) had only numerical chromosomal abnormalities, 26 (27.9%) had structural abnormalities, and 43 (46.2%) had both type of abnormalities (Fig. 1a). Patients with loss of Y chromosome had not been included in this group. The distribution of numerical and structural abnormalites in the four ploidy groups were given in Fig. 1b.
Fig. 1.
Distribution of chromosome abnormalities in MM patients. a A pie chart showing the percentage of numerical and structural abnormalities in the total of 113 MM patients. b A bar graph showing the comparison of numerical and structural abnormalities in the four ploidy groups
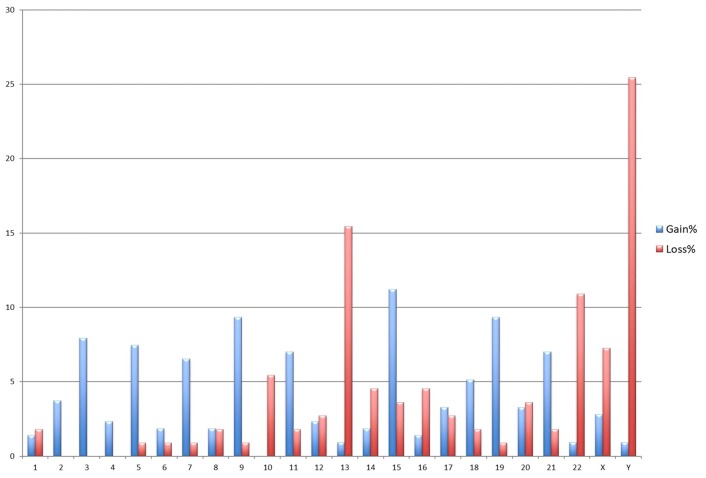
The most frequently occurring numerical chromosome abnormalities were trisomies. In 113 cases with cytogenetic abnormalities, gain of whole chromosome 15 (11.2%) was the most frequently observed, and that was followed by gain of chromosomes 9 (9.34%), 19 (9.34%), 3 (7.94%), 5 (7.47%), 11 (7%) and 21 (7%). Additionally, the most common monosomies were detected in chromosome 13 (15.4%), 22 (10.9%) and the loss of Y chromosome was present in 25.4% of patients (Fig. 2).
Fig. 2.
Frequency of chromosomal gains and losses in 113 multiple myeloma patients
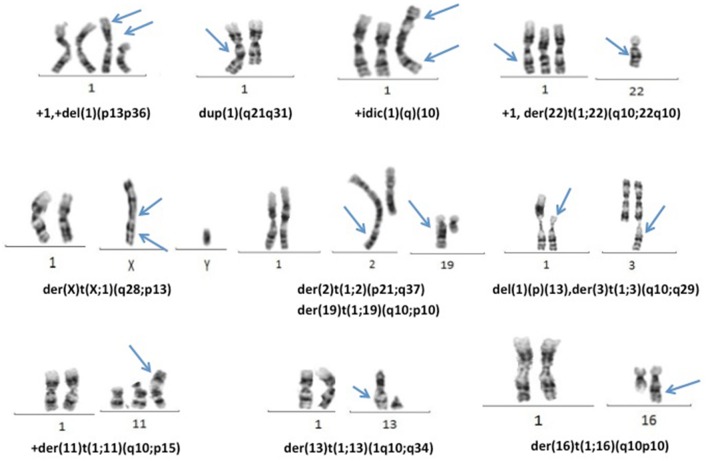
Structural chromosomal abnormalities were determined in 69 cases. The structural abnormalities of chromosome 1 were the most common chromosomal abnormality in MM patients and were determined in 33.3% (31/93). 1q gain was determined in a total of 25 patients (22%), including seven structural abnormalities such as duplication and isochromosome and also nineteen unbalanced translocations causing complete or partial duplication of the long arm of chromosome 1 (Fig. 3). In six cases deletions of chromosome 1p arm were determined. Additionally, t(1;5) balanced translocation and unbalanced translocations of t(1;2), t(1;3), t(1;7), t(1;16) and t(1;19) were observed twice with different breakpoints.
Fig. 3.
Representative examples of 1q gains
Similarly, numerical or structural abnormalities of chromosome 3 were observed in 31.2% patients (29/93). Other common chromosomal abnormality, -13/del (13q), was determined in 23.7% patients (22/93). Translocations involving the 14q32 region was found: 6 cases had t(11;14) translocation, 1 case had t(8;14) and 1 case had t(14;17).
In this research, specific probes for t(4;14), t(11;14), t(14;16), del(11q22.3), del(17p13) and del(13q14) were performed on the slides of 253 patients by FISH. Genetic abnormalities were observed in 45.8% (116/253) of patients in only FISH analysis, and this frequency was higher than that observed in conventional cytogenetics solely (31.9%, 113/354). Of the 162 patients that were found to be normal karyotype by conventional cytogenetics, 61 (37.65%) had at least one genetic aberration by FISH and these FISH results were given in Table 1. Del(13q) or -13 was observed in 61 (24.1%) of 253 patients by FISH. Del(17p) or -17 was found in 31 (12.3%), t(11;14) in 22 (8.7%), t(4;14) in 22 (8.7%) and t(14;16) in 2 (0.8%) patients. Amplification of 11q13 locus or trisomy 11 was found in 30 (11.9%) patients.
Discussion
In recent years, the identification of genetic abnormalities in MM patients has gained more importance for risk stratification and the risk-adapted treatment strategies [9, 20]. Consequently, conventional cytogenetics, FISH or microarray-based technologies are frequently used to identify genetic abnormalities in MM patients. Each of these techniques provides a diagnostic and a prognostic information, also have distinct utilities and limitations. Although conventional cytogenetics is an efficient approach providing an advantage to whole chromosome analysis with a single experiment [21], conventional cytogenetic studies on large series of MM patients have been limited to a few reports because of the low mitotic index of plasma cells. In our study group consisting of 381 patients, conventional cytogenetic analysis was successful in 92.9% (354/381) of samples. The rate of outcome with conventional cytogenetics in MM patients varies between 29.5 and 64% in the literature [22, 23]. Our success rate was higher than this range even without using no specific stimulant for plasma cells. The reason for this might be the synchronization kit which has a proven effect for both very high success rate of metaphase analysis and the best quality of chromosomal morphology.
Abnormal karyotypes were detected in 31.9% (113/354) of patients with MM. In previous studies, the ratio of abnormal karyotypes in MM by conventional cytogenetics ranged between 16.9 and 75.6% [24–32].The rate of our patients with abnormal karyotype was similar to some previously reported studies [24, 28, 33]. The differences of ratio in genetic abnormalities in multiple myeloma patients reflect the low proliferative activity and different infiltration degrees of plasma cells.
Abnormal karyotypes of our patients were classified into 4 groups according to their total chromosome number as hyperdiploid, near triploid, hypodiploid and pseudodiploid. The ratio of hyperdiploid and hypodiploid was found to be equal and the ratio was determined as 27.4%. Hyperdiploid frequency in our study was lower than reported in two large series by Avet-Loiseau et al. and Fonseca et al. [34, 35]. However, incidence of our hyperdiploid karyotype is similar to the 26% rate reported by Durak-Aras et al. [23]. Similarity of the rates in two studies may be explained by both works instantly be made in Turkey. Near triploidy was observed in 6.2% of patients whereas pseudodiploidy was determined in 21.2% of MM patients. Both numerical and structural aberrations of chromosomes were observed 46.2% (43/93) in all groups. When hyperdiploid cases were evaluated, most of them had both numerical and structural anomalies (68.7%). Li et al. reported that 80% of the cases with chromosomal abnormalities in 222 patients had both numerical and structural abnormalities. The same group stated that many of the hyperdiploid cases had structural abnormalities (44%). In our study group, it was determined that 92.3% of pseudodiploid cases had solely structural abnormalities.
Hyperdiploid karyotypes had been found with multiple gains of chromosome 3, 5, 6, 7, 9, 11, 15, 18, 19 and 21 in MM patients [24]. In our patients, the most common trisomies were observed as follows: trisomies 15 (11.2), 9 (9.34%), 19 (9.34%), 3 (7.94%), 5 (7.47%), 11 (7%) and 21 (7%). Hypodiploid karyotypes had been found by recurrent losses of chromosome 8, 13 and 14 [24]. In our hypodiploid cases, the most common monosomies were as follows; 13 (15.4%), 22 (10.9%), X (7.27%), and Y (25.4%). The chromosomal gains and losses observed in our patients were similar to those seen in previous studies with different percentages. Distinctly, loss of chromosome 8 was observed quite low in our cases [29, 36–38].
In the present study, chromosome 1 abnormalities were observed in 33.3% of patients with abnormal karyotype, and it was the one which had been observed most frequently. Previous studies showed that the presence of 1q amplifications in MM was associated with poor clinical outcome and survival [25, 39]. The incidence of the 1q amplifications was reported by several groups at ranging from 16.9 to 48% [25, 27, 39, 40]. In our patients, 1q gain was determined in a total of 25 patients (22%), including seven structural abnormalities and nineteen unbalanced translocations causing complete or partial duplication of the long arm of chromosome 1. Among 25 patients with 1q gain, having relapsed or died 21 patients supports the association between poor prognosis and presence of 1q gain.
In the previous studies, the median number of karyotypic changes consisting of structural or numerical abnormalities have been found 8–10 in each cell of MM patients tumors [41, 42]. In our 22 of 31 hyperdiploid patients, number of chromosomal changes was found higher than eight so this finding has supported the literatures. It was remarkable that 26 of our cases with hyperdiploidy had both numerical and structural anomalies. When the clinical follow-up of these cases was evaluated, poor prognosis was determined in 15 cases. The most prominent chromosomal abnormality in these cases was the gain of the long arm of chromosome 1 which raised from unbalanced translocations. Although it was reported that the deletion of long arm of chromosome 6 was revealed in non-hyperdiploid MM patients [43], we determined the deletion of long arm of chromosome 6 as a recurrent structural abnormality in four hyperdiploidy cases with poor prognosis. On the other hand, the deletion of 6q was shown in three non-hyperdiploidy cases (two of them were relapsed and one of them died). When these findings are evaluated together, the poor prognosis in our hyperdiploid MM patients can be explained by the presence of both an increased number of the anomalies and type of the chromosomal abnormalities.
Other common chromosomal abnormality, -13/del(13q), was determined in 23.7% (22/93) patients by using conventional cytogenetics. This ratio was determined as 24.1% (61/253) for all patients with FISH analysis. Recently, monosomy 13/del(13q) was reported as 40% by Li et al., as 54% by Jung and as 52% by Mohamed et al. [25, 29, 37].
In our series, the most commonly observed translocation was t(11;14)(q13;q32) translocation determined by both conventional cytogenetics and FISH methods. While the translocation was determined as 6.5% (6/93) by cytogenetic analysis, translocation of t(11;14) was found to be in 22 (8.7%) patients by FISH. Our findings have supported that the combined use of cytogenetic studies and FISH is essential. Furthermore, other translocations involving IgH regions were as follows: in t(4;14) in 22 (8.7%) and t(14;16) in 2 (0.8%) patients by FISH. In our study, in 8 of 22 patients with t(4;14), complex karyotype involving to structural abnormalities of chromosomes 1, 13, and 17 had been found. This situation can be explained although translocations involving IgH locus (14q32) are believed to occur as an early event in MM pathogenesis, why t(4;14) translocation has been related to the poor prognosis [44, 45]. In a recent series, translocations t(11;14) and t(4;14) were found in 11–16% and 11–12% of patients with MM, respectively [46]. According to the literature, translocation t(11;14) is accompanied by monosomy 13 in 25% of patients [21]. On the contrary, this association had not been observed any of our patients. We also found rearrangements of 14q32 in 2 patients, of which t(8;14) and t(14;17) were existed. Interestingly, we described a translocation involving IgH locus that was not previously reported in multiple myeloma. The t(14;17)(q32;q21) translocation was determined in case 34 which was in the hypodiploid group. This translocation was previously described in a 16-year-old boy who was diagnosed with B acute lymphoblastic leukemia by Gu et al. In the literature, this is the first report of translocation t(14;17)(q32;q21) resulting in IGH-IGF2BP1 fusion which results in the up-regulation of IGF2BP1 [47].
In this study, three patients presented constitutional structural chromosomal changes including Robertsonian translocation; der(13;14)(q10;q10) in two cases and one case with a pericentric inversion of chromosome nine; inv(9)(p11q13). While the pericentric inversion of 9 is well known as a heteromorphism and a non-significant rearrangement, Robertsonian translocation between chromosome 13 and chromosome 14 is the most seen Robertsonian translocation in haematological malignancies [48]. Due to the observation of the patients who had hematological malignancies with acquired Robertsonian translocations [49], it could be suggested that especially Robertsonian translocation of (13;14) might have a potential role on the increase of hematological malignancy formation.
Due to loss of sex chromosome in females and males has been a phenomena associated with aging [50] loss of Y chromosome was evaluated as an independent group of classification based on chromosomal ploidy in our study. The mean age of male patients with chromosomal abnormality was detected as 62.9, whereas the mean age with Y chromosome loss was 68.8. These data suggests an association between loss of Y chromosome and aging in males. On the other hand, it is noticeable that in cases where Y chromosome loss is accompanied by complex karyotype, the presence of Y chromosome in normal cells may indicate that Y chromosome loss may be mediating genomic instability. Also, in 4 of 6 female patients the loss of chromosome X accompanied by complex karyotype and in other two female cases loss of chromosome X was a sole anomaly suggesting that sex chromosome loses cannot be an innocent in cancer cells.
In conclusion, although technological advances have led to the classification of multiple myeloma at the molecular level, it is still inevitable to perform both conventional cytogenetics and FISH applications in MM patients in order to (1) reveal new recurrent specific chromosomal abnormalities, (2) to observe the frequency of the genetic defects among the different populations, (3) to explain the heterogeneous structure of the disease and (4) to determine the true clinical stage of the disease.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Compliance with Ethical Standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Palumbo A, Anderson K. Multiple myeloma. N Engl J Med. 2011;364(11):1046–1060. doi: 10.1056/NEJMra1011442. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 3.Rajkumar SV, Gahrton G, Bergsagel PL. Approach to the treatment of multiple myeloma: a clash of philosophies. Blood. 2011;118(12):3205–3211. doi: 10.1182/blood-2011-06-297853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fairfield H, Falank C, Avery L, Reagan MR. Multiple myeloma in the marrow: pathogenesis and treatments. Ann N Y Acad Sci. 2016;1364(1):32–51. doi: 10.1111/nyas.13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.San-Miguel JF, Mateos MV. Can multiple myeloma become a curable disease? Haematologica. 2011;96(9):1246–1248. doi: 10.3324/haematol.2011.051169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Avet-Loiseau H, Soulier J, Fermand JP, Yakoub-Agha I, Attal M, Hulin C, et al. Impact of high-risk cytogenetics and prior therapy on outcomes in patients with advanced relapsed or refractory multiple myeloma treated with lenalidomide plus dexamethasone. Leukemia. 2010;24(3):623–628. doi: 10.1038/leu.2009.273. [DOI] [PubMed] [Google Scholar]
- 7.Laubach JP, Schlossman RL, Mitsiades CS, Anderson KC, Richardson PG. Thalidomide, lenalidomide and bortezomib in the management of newly diagnosed multiple myeloma. Expert Rev Hematol. 2011;4(1):51–60. doi: 10.1586/ehm.10.83. [DOI] [PubMed] [Google Scholar]
- 8.Fonseca R, Bergsagel PL, Drach J, Shaughnessy J, Gutierrez N, Stewart AK, et al. International myeloma working group molecular classification of multiple myeloma: spotlight review. Leukemia. 2009;23(12):2210–2221. doi: 10.1038/leu.2009.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kapoor P, Fonseca R, Rajkumar SV, Sinha S, Gertz MA, Stewart AK, et al. Evidence for cytogenetic and fluorescence in situ hybridization risk stratification of newly diagnosed multiple myeloma in the era of novel therapies. Mayo Clin Proc. 2010;85(6):532–537. doi: 10.4065/mcp.2009.0677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Terpos E, Eleutherakis-Papaiakovou V, Dimopoulos MA. Clinical implications of chromosomal abnormalities in multiple myeloma. Leuk Lymphoma. 2006;47(5):803–814. doi: 10.1080/10428190500464104. [DOI] [PubMed] [Google Scholar]
- 11.Sonneveld P, Avet-Loiseau H, Lonial S, Usmani S, Siegel D, Anderson KC, et al. Treatment of multiple myeloma with high-risk cytogenetics: a consensus of the International Myeloma Working Group. Blood. 2016;127(24):2955–2962. doi: 10.1182/blood-2016-01-631200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nemec P, Zemanova Z, Kuglik P, Michalova K, Tajtlova J, Kaisarova P, et al. Complex karyotype and translocation t(4;14) define patients with high-risk newly diagnosed multiple myeloma: results of CMG2002 trial. Leuk Lymphoma. 2012;53(5):920–927. doi: 10.3109/10428194.2011.634042. [DOI] [PubMed] [Google Scholar]
- 13.Sawyer JR. The prognostic significance of cytogenetics and molecular profiling in multiple myeloma. Cancer Genet. 2011;204(1):3–12. doi: 10.1016/j.cancergencyto.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 14.Viguié F. Multiple myeloma. Atlas Genet Cytogenet Oncol Haematol. 2004;8:255–257. [Google Scholar]
- 15.Rajkumar SV, Fonseca R, Dewald GW, Therneau TM, Lacy MQ, Kyle RA, et al. Cytogenetic abnormalities correlate with the plasma cell labeling index and extent of bone marrow involvement in myeloma. Cancer Genet Cytogenet. 1999;113(1):73–77. doi: 10.1016/s0165-4608(99)00009-6. [DOI] [PubMed] [Google Scholar]
- 16.International Myeloma Working Group Criteria for the classification of monoclonal gammopathies, multiple myeloma and related disorders: a report of the International Myeloma Working Group. Br J Haematol. 2003;121(5):749–757. [PubMed] [Google Scholar]
- 17.Rajkumar SV, Dimopoulos MA, Palumbo A, Blade J, Merlini G, Mateos MV, et al. International Myeloma Working Group updated criteria for the diagnosis of multiple myeloma. Lancet Oncol. 2014;15(12):e538–e548. doi: 10.1016/S1470-2045(14)70442-5. [DOI] [PubMed] [Google Scholar]
- 18.Mikhael JR, Dingli D, Roy V, Reeder CB, Buadi FK, Hayman SR, et al. Management of newly diagnosed symptomatic multiple myeloma: updated Mayo Stratification of Myeloma and Risk-Adapted Therapy (mSMART) consensus guidelines 2013. Mayo Clin Proc. 2013;88(4):360–376. doi: 10.1016/j.mayocp.2013.01.019. [DOI] [PubMed] [Google Scholar]
- 19.McGowan-Jordan J, Simons A, Schmid M (2016) ISCN 2016: an international system for human cytogenomic nomenclature (2016) cytogenetic and genome research, vol 149, no 1–2, 1st edn [DOI] [PubMed]
- 20.Munshi NC, Anderson KC, Bergsagel PL, Shaughnessy J, Palumbo A, Durie B, et al. Consensus recommendations for risk stratification in multiple myeloma: report of the International Myeloma Workshop Consensus Panel 2. Blood. 2011;117(18):4696–4700. doi: 10.1182/blood-2010-10-300970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saxe D, Seo EJ, Bergeron MB, Han JY. Recent advances in cytogenetic characterization of multiple myeloma. Int J Lab Hematol. 2018 doi: 10.1111/ijlh.12882. [DOI] [PubMed] [Google Scholar]
- 22.Kaufmann H, Krömer E, Nösslinger T, Weltermann A, Ackermann J, Reisner R, et al. Both chromosome 13 abnormalities by metaphase cytogenetics and deletion of 13q by interphase FISH only are prognostically relevant in multiple myeloma. Eur J Haematol. 2003;71(3):179–183. doi: 10.1034/j.1600-0609.2003.00111.x. [DOI] [PubMed] [Google Scholar]
- 23.Durak BA, Akay OM, Sungar G, Bademci G, Aslan V, Caferler J, et al. Conventional and molecular cytogenetic analyses in Turkish patients with multiple myeloma. Turk J Haematol. 2012;29(2):135–142. doi: 10.5152/tjh.2011.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang SY, Yao M, Tang JL, Tsay W, Lee FY, Liu MC, et al. Clinical significance of cytogenetics and interphase fluorescence in situ hybridization analysis in newly diagnosed multiple myeloma in Taiwan. Ann Oncol. 2005;16(9):1530–1538. doi: 10.1093/annonc/mdi273. [DOI] [PubMed] [Google Scholar]
- 25.Jung HA, Jang MA, Kim K, Kim SH. Clinical utility of a diagnostic approach to detect genetic abnormalities in multiple myeloma: a single institution experience. Ann Lab Med. 2018;38(3):196–203. doi: 10.3343/alm.2018.38.3.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trcić RL, Skelin IK, Sustercić D, Planinc-Peraica A, Ajduković R, Haris V, et al. Cytogenetics of multiple myeloma. Coll Antropol. 2010;34(1):41–44. [PubMed] [Google Scholar]
- 27.Jekarl DW, Min CK, Kwon A, Kim H, Chae H, Kim M, et al. Impact of genetic abnormalities on the prognoses and clinical parameters of patients with multiple myeloma. Ann Lab Med. 2013;33(4):248–254. doi: 10.3343/alm.2013.33.4.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jian Y, Chen X, Zhou H, Zhu W, Liu N, Geng C, et al. Prognostic impact of cytogenetic abnormalities in multiple myeloma: a retrospective analysis of 229 patients. Medicine (Baltimore) 2016;95(19):e3521. doi: 10.1097/MD.0000000000003521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li S, Lim HH, Woo KS, Kim SH, Han JY. A retrospective analysis of cytogenetic alterations in patients with newly diagnosed multiple myeloma: a single center study in Korea. Blood Res. 2016;51(2):122–126. doi: 10.5045/br.2016.51.2.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lim AS, Lim TH, See KH, Ng YJ, Tan YM, Choo NS, et al. Cytogenetic and molecular aberrations of multiple myeloma patients: a single-center study in Singapore. Chin Med J (Engl) 2013;126(10):1872–1877. [PubMed] [Google Scholar]
- 31.Lai YY, Huang XJ, Cai Z, Cao XS, Chen FP, Chen XQ, et al. Prognostic power of abnormal cytogenetics for multiple myeloma: a multicenter study in China. Chin Med J (Engl) 2012;125(15):2663–2670. [PubMed] [Google Scholar]
- 32.He J, Yang L, Meng X, Wei G, Wu W, Han X, et al. A retrospective analysis of cytogenetic and clinical characteristics in patients with multiple myeloma. Am J Med Sci. 2013;345(2):88–93. doi: 10.1097/MAJ.0b013e31825b32bc. [DOI] [PubMed] [Google Scholar]
- 33.Rack K, Vidrequin S, Dargent JL. Genomic profiling of myeloma: the best approach, a comparison of cytogenetics, FISH and array-CGH of 112 myeloma cases. J Clin Pathol. 2016;69(1):82–86. doi: 10.1136/jclinpath-2015-203054. [DOI] [PubMed] [Google Scholar]
- 34.Avet-Loiseau H, Attal M, Moreau P, Charbonnel C, Garban F, Hulin C, et al. Genetic abnormalities and survival in multiple myeloma: the experience of the Intergroupe Francophone du Myéloma. Blood. 2007;8:3489–3495. doi: 10.1182/blood-2006-08-040410. [DOI] [PubMed] [Google Scholar]
- 35.Fonseca R, Blood E, Rue M, Harrington D, Oken MM, Kyle RA, et al. Clinical and biologic implications of recurrent genomic aberrations in myeloma. Blood. 2003;11:4569–4575. doi: 10.1182/blood-2002-10-3017. [DOI] [PubMed] [Google Scholar]
- 36.Calasanz MJ, Cigudosa JC, Odero MD, Ferreira C, Ardanaz MT, Fraile A, et al. Cytogenetic analysis of 280 patients with multiple myeloma and related disorders: primary breakpoints and clinical correlations. Genes Chromosomes Cancer. 1997;18(2):84–93. [PubMed] [Google Scholar]
- 37.Mohamed AN, Bentley G, Bonnett ML, Zonder J, Al-Katib A. Chromosome aberrations in a series of 120 multiple myeloma cases with abnormal karyotypes. Am J Hematol. 2007;82(12):1080–1087. doi: 10.1002/ajh.20998. [DOI] [PubMed] [Google Scholar]
- 38.Sawyer JR, Waldron JA, Jagannath S, Barlogie B. Cytogenetic findings in 200 patients with multiple myeloma. Cancer Genet Cytogenet. 1995;82(1):41–49. doi: 10.1016/0165-4608(94)00284-i. [DOI] [PubMed] [Google Scholar]
- 39.Lee JW, Lee JK, Hong YJ, Hong SI, Chang YH. Correlation of chromosomal aberrations with prognostic markers in multiple myeloma patients—a single institution studyF. Korean J Lab Med. 2008;28(6):413–418. doi: 10.3343/kjlm.2008.28.6.413. [DOI] [PubMed] [Google Scholar]
- 40.Bang SM, Kim YR, Cho HI, Chi HS, Seo EJ, Park CJ, et al. Identification of 13q deletion, trisomy 1q, and IgH rearrangement as the most frequent chromosomal changes found in Korean patients with multiple myeloma. Cancer Genet Cytogenet. 2006;168(2):124–132. doi: 10.1016/j.cancergencyto.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 41.Chng WJ, Ketterling RP, Fonseca R. Analysis of genetic abnormalities provides insights into genetic evolution of hyperdiploid myeloma. Genes Chromosomes Cancer. 2006;45(12):1111–1120. doi: 10.1002/gcc.20375. [DOI] [PubMed] [Google Scholar]
- 42.Nilsson T, Höglund M, Lenhoff S, Rylander L, Turesson I, Westin J, et al. A pooled analysis of karyotypic patterns, breakpoints and imbalances in 783 cytogenetically abnormal multiple myelomas reveals frequently involved chromosome segments as well as significant age- and sex-related differences. Br J Haematol. 2003;120(6):960–969. doi: 10.1046/j.1365-2141.2003.04221.x. [DOI] [PubMed] [Google Scholar]
- 43.Brigaudeau C. del(6q) in multiple myeloma. Atlas Genet Cytogenet Oncol Haematol. 1999;3(1):17–18. [Google Scholar]
- 44.Rajan AM, Rajkumar SV. Interpretation of cytogenetic results in multiple myeloma for clinical practice. Blood Cancer J. 2015;5:e365. doi: 10.1038/bcj.2015.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bergsagel PL, Nardini E, Brents L, Chesi M, Kuehl WM. IgH translocations in multiple myeloma: a nearly universal event that rarely involves c-myc. Curr Top Microbiol Immunol. 1997;224:283–287. doi: 10.1007/978-3-642-60801-8_30. [DOI] [PubMed] [Google Scholar]
- 46.Kumar SK, Rajkumar V, Kyle RA, van Duin M, Sonneveld P, Mateos MV, et al. Multiple myeloma. Nat Rev Dis Primers. 2017;3:17046. doi: 10.1038/nrdp.2017.46. [DOI] [PubMed] [Google Scholar]
- 47.Gu G, Sederberg MC, Drachenberg MR, South ST. IGF2BP1: a novel IGH translocation partner in B acute lymphoblastic leukemia. Cancer Genet. 2014;207(7–8):332–334. doi: 10.1016/j.cancergen.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 48.Panani AD. Is there an association with constitutional structural chromosomal abnormalities and hematologic neoplastic process? A short review. Ann Hematol. 2009;88(4):293–299. doi: 10.1007/s00277-008-0672-8. [DOI] [PubMed] [Google Scholar]
- 49.Welborn J. Acquired Robertsonian translocations are not rare events in acute leukemia and lymphoma. Cancer Genet Cytogenet. 2004;151(1):14–35. doi: 10.1016/j.cancergencyto.2003.09.019. [DOI] [PubMed] [Google Scholar]
- 50.Stone JF, Sandberg AA. Sex chromosome aneuploidy and aging. Mutat Res. 1995;338(1–6):107–113. doi: 10.1016/0921-8734(95)00016-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.