Abstract
Soil-borne wheat mosaic virus (SBWMV) causes a serious viral disease that can significantly reduce grain yield in winter wheat worldwide. Using resistant cultivars is the only feasible strategy to reduce the losses caused by SBWMV. To fine map the resistance gene Sbwm1, 205 wheat accessions was genotyped using wheat Infinium iSelect Beadchips with 90 K SNPs. Association analysis identified 35 SNPs in 12 wheat genes and one intergenic SNP in the Sbwm1 region that showed a significant association with SBWMV resistance. Those SNPs were converted into Kompetitive Allele-Specific Polymerase assays (KASP) and analyzed in two F6-derived recombinant inbred line (RIL) populations derived from the crosses between two resistant cultivars ‘Wesley’ and ‘Deliver’ and a susceptible line ‘OK03825-5403-6’. Linkage analysis mapped this gene on chromosome 5D at intervals of 5.1 cM and 3.4 cM in the two populations, respectively. The two flanking markers in both populations delimited the gene to a 620 kb region where 19 genes were annotated. Comparative analysis identified a syntenic region of 660 kb in Ae. tauschii with 18 annotated genes and a syntenic region in chromosome 1 of B. distachyon. The candidate region includes several disease resistance related genes and we identified a PTI1-like tyrosine-protein kinase 1 gene as a putative candidate gene for Sbwm1. The two flanking SNPs for Sbwm1 can effectively separate the resistant and susceptible lines in a new diversity panel of 159 wheat germplasm. The results from this study lay a solid foundation for the cloning, functional characterization and marker-assisted selection of Sbwm1.
Subject terms: Plant breeding, Plant breeding
Introduction
Soil-borne wheat mosaic (SBWM) disease, caused by Soil-borne wheat mosaic virus (SBWMV) was first appeared in the Great Plains of the U.S.A. in 19191 and has become an important disease in most winter wheat growing regions globally2–7. SBWMV infects wheat roots in wet soil during germination at 15 °C to 20 °C with an optimum temperature at 17 °C. Soil water is critical for the swimming zoospore to reach host roots8,9. Virus movement from the site of infection on roots to the leaves occurs at temperatures lower than 20 °C. Higher temperatures limit virus movement into leaves and further infection in developing leaves will not occur if temperatures rise to more than 20 °C after some leaves are infected9.
The SBWMV symptom usually first appears in early spring when the crop begins to green up. Low spring temperatures is usually conducive to the occurrence of SBWMV and could promote the development of the symptoms. Irregular chlorotic patches in a field indicate SBWMV infection. In dry environments, patches of symptomatic SBWMV-infected plants usually present in low-lying, wet regions of a field that are favorable for infection by the swimming zoospores of Polymyxa graminis, the vector of SBWMV10. In wet soil climates, these patches may present anywhere in an infected field. SBWMV symptoms stop developing on leaves when the average temperature exceeds 20 °C, but can be reinitiated on newly emerged leaves given conductive environmental conditions10. Reported yield losses caused by SBWMV ranged from 10% to 30% and can reach up to 80% in SBWMV severely occured fields in the U.S.A. and 50% in Brazil11–13.
Polymyxa graminis is one of the endoparasitic slime molds (Plasmodiophoromycota), which can produce resting spores that harbor viral RNA and movement protein, and can be easily distributed by wind, water and machinery. Those resting virus-containing spores can remain dormant and invasive in soil for up to 30 years, and then germinate in an environment favorable for infection when host are available. Thus, it is very difficult to control this disease once it happened in a field. The most practical SBWM control strategy is to grow resistant cultivars2,14,15.
Inheritance studies have shown that resistance to SBWMV is controlled by major genes. Winter wheat cultivars such as Shawnee, Centurk and KS73256 were reported to carry a dominant gene for SBWMV resistance. Newton might contain a gene for resistance different from that in ‘Arthur 71’, ‘Homestead’, and ‘Tascosa’14. However, a cultivar ‘Embrapa 16’ from Brazil was reported to carry two genes for SBWMV resistance2. With the rapid development and wide use of molecular markers in wheat genetic studies, quantitative trait locus (QTL) mapping approach has been used to localize SBWMV resistance genes using linkage maps. The QTL on 5DL has been reported by several independent studies on winter wheat ‘Karl 92’16, ‘Pioneer 26R61’, ‘AGS 2020’ and ‘Heyne’12,17. A breeding line KS96WGRC40 derived from Aegilops tauschii also carries this gene18. This gene was designated as Sbwm117. Interestingly, a gene conferring resistance to Soil-borne cereal mosaic virus (SBCMV) in ‘Cadenza’, ‘Tremie’ and ‘Claire’, designated as Sbm1, was also mapped in the region of Sbwm119,20. Because they tightly linked to the marker Xgwm469-5D, Sbm1 for SBCMV resistance is either a tightly linked or the same gene as Sbwm1 for SBWMV resistance.
Single nucleotide polymorphisms (SNPs) are the most widely distributed DNA sequence variations in a genome. During the past decade, next-generation sequencing (NGS) technologies have become the cheapest and fastest technology for genome-wide SNPs discovery21. Fixed SNP chips have been released and widely used for various genetic and breeding studies including genome-wide association analysis and genomic selection in many crop species21–23. In wheat, SNP chips with various number of SNPs including Wheat 9 K iSelect, Wheat 90 K iSelect and Wheat 660 K Axiom SNP chips have been developed and widely used to map genes or QTLs for different traits24–28.
In a former study, a SSR marker Xgwm469, that was associated with the SBWMV resistance on chromosome 5DL has been identified29. Further association mapping analysis using Wheat 9 K iSelect chip identified six SNPs from two genes that were highly associated with SBWMV resistance, and the identified SNPs were further mapped on 5DL using a F6 RIL population ‘Heyne’/‘Trego’17. However, those SNPs were mapped on one side of Sbwm1 and the marker flanking on the other side of Sbwm1 is about 20 cM a way. Therefore, a tightly linked flanking marker is urgently needed for effective marker-assisted transfer of Sbwm1 to new wheat cultivars and cloning Sbwm1.
In this study, the panel of 205 wheat accessions previously genotyped with 9 K SNP chips were further genotyped using Infinium 90 K wheat SNP chips to identify more flanking SNPs for Sbwm1. Two new RIL populations were used to map the SBWMV resistance genes in the two resistant cultivars Wesley and Deliver and to verify the linkage among the newly identified SNP markers Sbwm1. We also pinpointed the candidate region for Sbwm1 using the Chinese Spring reference genome, developed KASP assays for the linked SNPs to Sbwm1 for marker-assisted selection (MAS), and identified a putative candidate gene for Sbwm1 by gene expression analysis.
Materials and Methods
Plant materials
The association mapping panel includes 205 wheat accessions with 137 hard winter wheat (HWW) and 68 soft winter wheat (SWW) from six 2008 HWW and SWW nurseries (Supplementary Table S1). Seeds for DNA isolation and disease evaluation were originated from a single plant of each accession increased in a greenhouse to minimize within-line heterogeneity29. All these accessions were genotyped using Infinium iSelect 90 K wheat SNP chips25. A second panel of 159 wheat cultivars and breeding lines were used to validate the mapped KASP assays (Supplementary Table S2).
Two F6 RIL populations developed by single-seed descent from the crosses ‘Wesley’ x OK03825-5403-6 (180 RILs) and ‘Deliver’ x OK03825-5403-6 (260 RILs) were used to map the SBWMV resistance gene. ‘Wesley’ and ‘Deliver’ are SBWMV-resistant HWW cultivars, whereas OK03825-5403-6 is a highly SBWMV-susceptible HWW breeding line.
Disease evaluation
The association panel of 205 wheat accessions were evaluated for SBWMV resistance in the SBWMV-infested field at the Rocky Ford Research Farm of Kansas State University, Manhattan, KS in 2009–2010 and 2010–2011 wheat growing seasons. The nursery has shown consistent and severe SBWMV infection on susceptible wheat cultivars since 2006. Field experimental design, planting, and disease damage evaluation followed Zhang et al.30. An additional panel of 159 accessions were evaluated for SBWMV resistance in 2010–2011 and 2011–2012 wheat growing seasons in the same field nursery for marker validation. The two RIL populations were evaluated for SBWMV resistance in the same field in 2013–2014 and 2014–2015 growing seasons using the same rating scale described for association mapping. All experiments used random complete block design with two replicates. In all experiments, each accession or RIL was sowed at 1.5 m -long rows with planting density of 40 seeds. After planting, the nursery was sprinkler irrigated everyday for 7 days till the seedling emergence to maintain high moisture in the soil to promote SBWMV infection.
DNA extraction and 90 K SNP assay
The 205 accessions were genotyped using the 90 K SNP chips at USDA-ARS Cereal Crops Research Unit, Fargo, ND as described by Wang et al.25. SNPs with less than 5% minor allele frequency (MAF) or with more than 15% missing data were removed31. A total of 21,600 SNPs were scored and used in the final analysis. Sequences that harbored significant SNPs were blasted against the Chinese Spring Wheat SeqVer1.0 reference sequence32 to assign their putative chromosome positions and were converted to Kompetitive Allele Specific Polymerase Chain Reaction (KASP) assays for screening the two RIL populations (Table 1). KASP analysis following the methods described by Liu et al.17.
Table 1.
Primers for KASP-SNPs that are highly associated with SBWMV resistance in the association mapping population.
| SNP Name | Left Primer | Right Primers |
|---|---|---|
| BS00067308_51-FAM | ATGCTGCATCCACGTCCTt | CACGTCTTCACGGATGTTTTT |
| BS00067308_51-HEX | ATGCTGCATCCACGTCCTg | |
| CAP12_c5949_104-FAM | GCATCAGGAAGAGGAACAGCa | GCCGTCGTAGTAGGTGAAGG |
| CAP12_c5949_104-HEX | GCATCAGGAAGAGGAACAGCc | |
| BS00079676_51-FAM | GGGTACTCTCGTCTTCCTGCATa | CAGTACAAAGCGCAACCTCA |
| BS00079676_51-HEX | GGGTACTCTCGTCTTCCTGCATg | |
| Kukri_c5528_603-FAM | CCAGTTAAAGGCTACATGGAGAt | AGGCCACATGAATGAGATCC |
| Kukri_c5528_603-HEX | CCAGTTAAAGGCTACATGGAGAc | |
| BS00011469_51-FAM | TGACTGGAAGGAAACCGGTt | GCCCAAGTGACTAAACTTTGC |
| BS00011469_51-HEX | TGACTGGAAGGAAACCGGTc | |
| BS00073116_51-FAM | CCGTGTGATAAGGATTCTGGt | GTTGTGTGGAGCACAGCACT |
| BS00073116_51-HEX | CCGTGTGATAAGGATTCTGGg | |
| BS00105939_51-FAM | GGTTGAGCAGGAAGCAGGa | GCAAGCAGACCAGTCTCTCA |
| BS00105939_51-HEX | GGTTGAGCAGGAAGCAGGg | |
| Contig08110-FAM | CCACCAATGGACACAAGGc | TCATCGCTCCAGGAATTACA |
| Contig08110-HEX | CCACCAATGGACACAAGGt | |
| Excalibur_c22724_85-FAM | ACCATTCACAACAATTGCGt | GATAATTTGTGCCGCGAGTT |
| Excalibur_c22724_85-HEX | ACCATTCACAACAATTGCGc | |
| Excalibur_c2795_1518-FAM | ATATACAAGGTATATTAGGt | TAACCCACCACCTGGCTTCT |
| Excalibur_c2795_1518-HEX | ATATACAAGGTATATTAGGc | |
| BobWhite_c13030_406-FAM | ATGCTCCCGGGCTTGATa | CCCATCACGCTCGACTTC |
| BobWhite_c13030_406-HEX | ATGCTCCCGGGCTTGATg |
Development of SNP markers using Ae. tauschii reference genome
To identify more markers in the SBWMV resistance QTL region, the probe sequences harboring the identified SNPs for SBWMV resistance were used to blast the draft Ae. tauschii reference genome33. Fourteen mapped SNPs (listed in Supplementary Table S4) in Ae. tauschii physical map in the QTL region were identified and converted to KASP assays to screen the three parents, Wesley, Deliver and OK03825-5403-6, of the two RIL populations, and the polymorphic SNP (Contig08110) was screened across the two RIL populations (Table 1).
Statistical analysis
Data for SBWMV phenotypes were analyzed using two-way GLM in SAS for Windows v9 (SAS Institute, Cary, NC) to determine the effects of genotypes (g), environments (e), and G x E interactions. Heritability (H2) was estimated using the formula
where бg2 is the variance among RILs, бge2 is the variance for G x E; бe2 is the variance of environments, n is the number of environments, and r is the number of replicates34.
Population structure, kinship, linkage disequilibrium and association analysis of the AM Panel
Population structure was assessed by Structure 2.3.435 using a set of 1,500 SNPs that are evenly distributed on all the 21 wheat chromosomes31. The admixture model was used for structure analysis and the number of sub-populations (K) was set as 1 to 10 with variable length of burn-in period and number of iterations at 20,000–250,000. For each trait, Bayesian information criterion (BIC) was applied to determine the optimum number of subpopulations31. Kinship was calculated using the same set of 1,500 SNPs used for structure analysis and SPAGeDi package36.
Genome-wide association analysis used the generalized linear model (GLM) with the Q matrix as fixed effects and the mixed linear model (MLM) with a Q matrix as fixed effects and a kinship matrix as random effects. The models were applied to each experiment for SBWMV resistance, and model fitness was determined based on the Bayesian information criterion value.
Association analysis of SNP data was conducted using the genome association and prediction integrated tool (GAPIT) implemented in a R package37. A threshold of p < 10−5 (1/number of markers) was set to claim significant marker-trait associations. Linkage disequilibrium and haplotype analyses of the significant SNPs were performed using HAPLOVIEW v.4.2 (http://www.broadinstitute.org/scientific-community/science/programs/medical-and-population-genetics/haploview/haploview).
Linkage mapping
Genotypes with disease rating of 1–2 were classified as resistant and 3–4 as susceptible for SBWMV resistance in the two RIL populations. SNP linkage maps were constructed for the two RIL populations using JoinMap ver. 4.038. Recombination fractions were converted into centiMorgans (cM) using the Kosambi function39. The threshold of logarithm of odd (LOD) score was set at 3.0 to claim linkage among markers with 0.4 as a maximum fraction of recombination. The goodness-of-fit between observed and expected segregation ratios between two alleles was analyzed for each marker locus using a chi-square-test.
Marker sequence analysis and candidate gene annotation
Sequences that contain significant SNPs associated with SBWMV resistance were used as queries to blast the IWGSC Chinese Spring reference genome sequence RefSeqv1.0, Ae. tauschii reference genome sequence and B. distachyon genome sequence using BLASTN32,40,41. A significant match was declared when the queried sequences showed at least 99% nucleotide identity with an e-value lower than e−20.
RNA extraction and gene expression analysis
Leaf tissues were collected from seedlings of Wesley and OK03825-5403-6 grown in the SBWMV field nursery for RNA extraction in fall 2015 and spring 2016 when the SBWMV symptom appeared. RNA was extracted and purified using the RNeasy Plant Kit with on-column DNaseI treatment (Qiagen, Valencia, CA). Complementary DNA from reverse-transcription reaction using a SuperScriptII kit (Invitrogen, Grand Island, NY) was amplified by conventional reverse transcriptase-PCR (RT-PCR). The annotated genes in the target region were tested for the gene expression using semi-quantitative PCR analysis. Each PCR reaction mixture consisted of 15.5 μL H2O, 2.4 μL 10 × buffer, 2.4 μL 50 mM MgCl2, 1 μL of 10 mM dNTPs, 1 μL each primer at 20 μM, 2.5 U Taq polymerase, and 1 μL cDNA. Cycle conditions were 95 °C for 1 min, annealing at 55–60 °C for 1 min for different primers, and extension at 72 °C for 1 min. A final extension at 72 °C was performed for 10 min. Annealing temperatures and cycle conditions for each primer pair were optimized such that each PCR was in the linear range of amplification. PCR primers were listed in Supplementary Table S5.
The identified candidate gene TraesCS5D01G531200 was further quantified with three replications by quantitative real-time PCR performed on an ABI7900HT instrument using SYBR Green (Thermo Fisher Scientific). Gene expression level was analyzed using the 2−ΔΔCt method. Actin gene was used as the endogenous control with ACCTTCAGTTGCCCAGCAAT as the forward primer, CAGAGTCGAGCACAATACCAGTTG as the reverse primer.
Results
SBWMV resistance in the association and linkage mapping populations
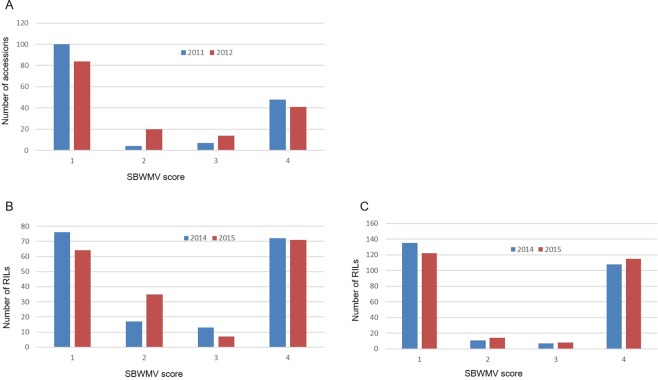
High correlation coefficients were observed for SBWMV resistance between the two years of phenotypic data from the marker validation population (r = 0.92, p < 0.01), the two RIL populations (r = 0.81 and 0.94, p < 0.01), and the association mapping population (r = 0.85, p < 0.01)17, suggesting a high repeatability of SBWMV resistance data between the field experiments. Analysis of variance showed highly significant genotypic effects and very high heritabilities (from 84.9% to 98.2%) in both the RIL populations and the validation population (Supplementary Table S3). In the two biparental populations, the distribution of SBWMV disease scores deviated significantly from a normal distribution and showed an obvious bimodal distribution with two peaks towards the resistant and susceptible parents (Fig. 1), suggesting that a major gene is responsible for SBWMV resistance in Wesley and Deliver.
Figure 1.
Frequency distribution of Soil-borne wheat mosaic virus disease scores in wheat accessions and recombinant inbred lines (RILs) evaluated in the field experiments at Manhattan, KS. (A) Accessions used for the KASP validation were phenotyped in the spring 2011 and 2012 field experiments; (B) RILs from Wesley x OK03825-5403-6 F6 population and (C) RILs from Deliver x OK03825-5403-6 population were phenotyped in the spring 2014 and 2015 field experiments. 1 = resistant, 2 = moderately resistant, 3 = moderately susceptible and 4 = susceptible.
Association analysis identified SNPs for the SBWMV resistance
Genotyping the association mapping population with the 90 K SNP chips identified 21,600 polymorphic SNPs. Using the MLM model, thirty-five SNPs were identified significantly associated with SBWMV resistance in both years (P < 10−5) (Fig. 2, Table 2), with 33 of them from 11 annotated high confidence genes (from TraesCS5D01G529700 to TraesCS5D01G532100) on chromosome 5D. SNP Excalibur_c22724_85 was from an annotated low confidence gene TraesCS5D01G625100LC, and BS00013935_51 was from an intergenic region. Using the Chinese Spring reference genome (IWGSC RefSeq v1.0), those SNPs can be located in a 69 Mb interval on 5D. However, when SNP BS00013935_51, the SNP with the lowest association among those SNPs (P = 1.44E-07) was excluded, the interval can be delimited to a 1.18 Mb region from 546,086,597 to 547,273,657 bp on the chromosome 5D.
Figure 2.
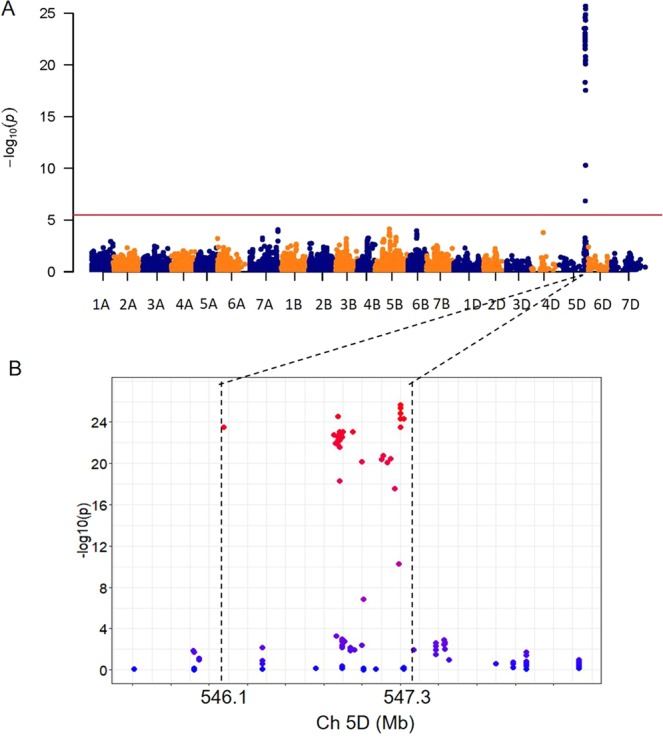
Manhattan plots for wheat resistance to Soil-borne wheat mosaic virus (SBWMV) in an association mapping population (A) and the physical positions of the significant SNPs on chromosome 5D (B). Red line represents the threshold to claim significant SNPs. Two-seasons of SBWMV ratings were collected from two field experiments plus the mean SBWMV ratings over both experiments.
Table 2.
Significant SNPs associated with Soil-borne wheat mosaic virus resistance and their corresponding annotated genes.
| SNP Name | SNP Position | MAFa | Orthologous gene | FDRb |
|---|---|---|---|---|
| BS00013935_51 | 5D.480829545 | 0.35 | Intergenic region | 8.55E-5 |
| BobWhite_c13030_406 | 5D.549670218 | 0.26 | TraesCS5D01G529700 | 7.89E-21 |
| CAP12_c5949_104 | 5D.550233279 | 0.31 | TraesCS5D01G530200 | 2.16E-20 |
| BS00079676_51 | 5D.550234521 | 0.33 | TraesCS5D01G530300 | 1.05E-19 |
| D_GA8KES401AL4GG_122 | 5D.550235948 | 0.31 | TraesCS5D01G530300 | 2.93E-20 |
| RAC875_c13169_459 | 5D.550235521 | 0.31 | TraesCS5D01G530300 | 5.32E-20 |
| BS00105939_51 | 5D.550235127 | 0.30 | TraesCS5D01G530400 | 1.28E-19 |
| BobWhite_c4438_162 | 5D.550272236 | 0.32 | TraesCS5D01G530600 | 4.72E-20 |
| BS00088587_51 | 5D.550272456 | 0.31 | TraesCS5D01G530600 | 2.34-20 |
| BS00088592_51 | 5D.550272362 | 0.37 | TraesCS5D01G530600 | 3.15E-16 |
| Excalibur_c687_886 | 5D.550271002 | 0.29 | TraesCS5D01G530600 | 1.40E-21 |
| Excalibur_c687_907 | 5D.550271023 | 0.31 | TraesCS5D01G530600 | 2.00E-20 |
| Excalibur_c687_961 | 5D.550271077 | 0.31 | TraesCS5D01G530600 | 3.91E-20 |
| IACX10520 | 5D.550272436 | 0.31 | TraesCS5D01G530600 | 2.00E-20 |
| Kukri_c1073_91 | 5D.550271228 | 0.31 | TraesCS5D01G530600 | 1.74E-20 |
| Kukri_c7786_81 | 5D.550241671 | 0.33 | TraesCS5D01G530600 | 2.22E-19 |
| wsnp_JD_c4438_5567834 | 5D.550272167 | 0.31 | TraesCS5D01G530600 | 2.00E-20 |
| wsnp_JD_c4438_5567972 | 5D.550272029 | 0.31 | TraesCS5D01G530600 | 2.39E-20 |
| wsnp_JD_c4438_5568170 | 5D.550271831 | 0.31 | TraesCS5D01G530600 | 5.22E-20 |
| D_GDS7LZN01CBWNE_99 | 5D.550282662 | 0.31 | TraesCS5D01G530700 | 2.92E-20 |
| Kukri_c5528_603 | 5D.550285051 | 0.31 | TraesCS5D01G530700 | 1.74E-20 |
| BS00073116_51 | 5D.550445864 | 0.35 | TraesCS5D01G531100 | 4.36E-18 |
| BS00011469_51 | 5D.550488971 | 0.34 | TraesCS5D01G531200 | 1.24E-18 |
| Excalibur_c14043_548 | 5D.550491920 | 0.38 | TraesCS5D01G531200 | 1.82E-15 |
| Excalibur_c28592_173 | 5D.550491839 | 0.36 | TraesCS5D01G531200 | 2.35E-18 |
| Excalibur_c28592_377 | 5D.550491410 | 0.36 | TraesCS5D01G531200 | 5.53E-18 |
| Excalibur_c42190_383 | 5D.550487798 | 0.36 | TraesCS5D01G531200 | 2.96E-18 |
| Excalibur_c2795_1518 | 5D.550503097 | 0.20 | TraesCS5D01G531300 | 3.15E-08 |
| BS00067308_51 | 5D.550837863 | 0.32 | TraesCS5D01G531900 | 9.53E-22 |
| BS00011794_51 | 5D.550854024 | 0.29 | TraesCS5D01G532100 | 4.09E-22 |
| BS00022036_51 | 5D.550854228 | 0.32 | TraesCS5D01G532100 | 1.66E-21 |
| CAP7_rep_c12715_390 | 5D.550853989 | 0.32 | TraesCS5D01G532100 | 7.89E-21 |
| CAP8_c145_89 | 5D.550893835 | 0.30 | TraesCS5D01G532100 | 4.09E-22 |
| wsnp_CAP11_c209_198671 | 5D.550854078 | 0.32 | TraesCS5D01G532100 | 1.66E-21 |
| Excalibur_c22724_85 | 5D.550362064 | 0.31 | TraesCS5D01G625100LC | 1.74E-20 |
aMinor allele frequency. badjusted P value.
Among the 11 high confidence genes that carry significantly associated SNPs for SBWMV resistance, TraesCS5D01G530600 harbors 12 SNPs including nine new SNPs and three previously reported SNPs (wsnp_JD_c4438_5567972, wsnp_JD_ c4438_5568170 and wsnp_JD_c4438_5567834)17. TraesCS5D01G532100 carries five SNPs with four new SNPs. All the remaining SNPs were distributed in nine other genes.
Linkage mapping
To validate the marker-trait association, one SNP was selected from each of the 12 genes harboring the identified SNPs in the association mapping study for KASP assay conversion. The SNPs wsnp_JD_c4438_5568170 from the gene TraesCS5D01G530600 and wsnp_CAP11_c209_198467 from the gene TraesCS5D01G532100 (Table 1) were used to represent the two genes identified in the assays of 90 K wheat SNP chips. The 12 newly designed KASP-SNPs were used to screen the three parents, Deliver, Wesley, and OK03825-5403-6 for polymorphism, and ten of the newly designed KASP-SNPs were polymorphic between Wesley and OK03825-5403-6 and eight were polymorphic between Deliver and OK03825-5403-6, respectively. Those polymorphic SNPs also segregated in the two RIL populations.
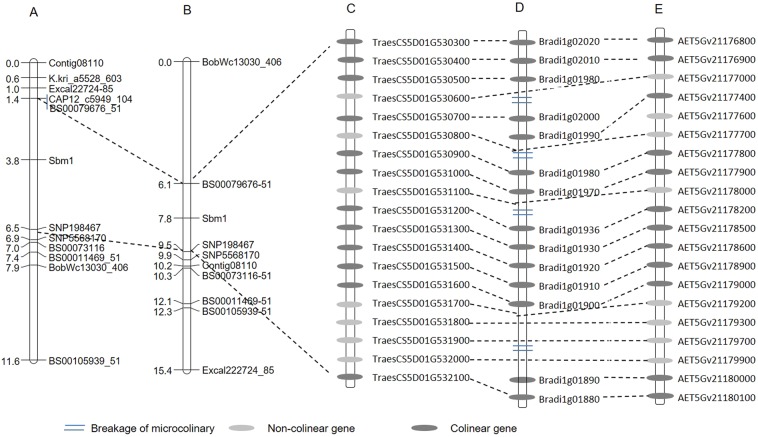
BLASTN of the 35 marker sequences harboring the significant SNPs against Ae. tauschii physical map identified 14 SNP markers mapped in the physical map (Supplementary Table S4). Ten of the 14 SNPs were successfully converted into KASP assays, but only one SNP (Contig08110_553) showed polymorphism between the two pairs of parents and segregated in the two RIL populations. Together with previously designed 10 and 8 KASP-SNPs in the two populations, a total of 11 and 9 KASP-SNPs linked to SBWMV resistance were mapped on chromosome 5D and spanned 11.6 cM and 15.4 cM in Wesley x OK03825-5403-6 and Deliver x OK03825-5403-6 populations, respectively (Fig. 3). The two flanking markers, wsnp_CAP11_c209_198467 and BS0000079676_51 delimited the gene for SBWMV resistance to 5.1 cM and 3.4 cM intervals in the populations Wesley x OK03825-5403-6 and Deliver x OK03825-5403-6, respectively. The two flanking SNPs represent the two genes that are the closest to Sbwm117 in the two populations, thus, the resistance gene in Wesley and Deliver was most likely the same as Sbwm1 (Fig. 3A).
Figure 3.
SNP maps of SBWMV-resistance gene (Sbwm1) on chromosome 5D and its syntenic regions in B. distoschyn and Ae. tauschii. (A) A SNP map of Sbwm1 developed from the Wesley x OK03825-5403-6 F6 population; (B) A SNP map of Sbwm1 developed from the Deliver x OK03825-5403-6 F6 population; (C) Annotated genes in the Sbwm1 region flanked by the two closet markers based on the Chinese Spring reference genome; (D) The syntenic region in B. distachyon with homologous genes; and (E) The syntenic region of Sbwm1 in Ae. tauschii with annotated genes.
Putative candidate genes in the genomic region of Sbwm1
BLASTN of the sequences of the two flanking markers, wsnp_CAP11_c209_198467 and BS0000079676_51, for Sbwm1 against the Chinese Spring SeqVer1.0 reference genome identified a 620 kb interval in wheat on chromosome 5D (Fig. 3). Annotation of the genes in this interval revealed 19 high confidence genes and one low confidence gene (Fig. 3C, Supplementary Table S5). Comparative analysis between wheat and Ae. tauschii, the donor of D genome of hexaploid wheat, identified the conserved collinear orthologous region of Sbwm1 with 20 high confidence orthologous genes including the two flanking genes (AET5Gv21176900–AET5Gv21180000) in a 660 kb interval on the chromosome 5D of Ae. tauschii (Supplementary Table S5). Among the 20 annotated genes of Ae. tauschii, 17 were the same to those in Chinese Spring wheat, and two genes (AET5Gv21177600, AET5Gv21180000) present in Ae. tauschii, but absent in wheat, and one gene (TraesCS5D01G530500) present in wheat, but absent in Ae. tauschii (Fig. 3E). Comparative analysis between wheat and B. distachyon identified a syntenic region (Bradi1g01880 to Bradi1g02020) on chromosome 1. However, this synteny in B. distachyon was interrupted by four non-syntenic regions (Fig. 3D). Twelve genes were common between B. distachyon and wheat or Ae. tauschii.
Besides the two flanking genes, 17 genes were in the genomic interval annotated from TraesCS5D01G530400 to TraesCS5D01G532000, which include genes encoding a sister chromatid cohesion protein DCC1 (TraesCS5D01G530400), a poly (ADP-ribose) glycohydrolase activity proteins (TraesCS5D01G530500 and TraesCS5D01G530900), a protein kinase family protein (TraesCS5D01G530600), a potassium channel activity protein (TraesCS5D01G530700), a S-adenosylmethionine-dependent methyltransferase (TraesCS5D01G530800), a pentatricopeptide repeat domain family protein (TraesCS5D01G531000), a zinc ion binding protein and zinc finger containing proteins (TraesCS5D01G531100 and TraesCS5D01G531400), a pto-interacting protein 1 (PTI1) (TraesCS5D01G531200), a protein with serine/threonine phosphatase activity (TraesCS5D01G531300), an albino-like protein (TraesCS5D01G531600), a neutral amino acid transmembrane transporter activity protein (TraesCS5D01G531700), a KDEL sequence binding protein (TraesCS5D01G531800), a major pollen allergen-like protein (TraesCS5D01G531900), and a gene encoding a protein with unknown function (TraesCS5D01G531500) (Supplementary Table S3).
Expression test of these genes in leaves showed that eight genes did not express (TraesCS5D01G530400, TraesCS5D01G530400, TraesCS5D01G531400, TraesCS5D01G531500, TraesCS5D01G531700, TraesCS5D01G531900, TraesCS5D01G531200, TraesCS5D01G625100LC), whereas nine expressed but showed the same expression levels between the resistant and susceptible parents (TraesCS5D01G530600, TraesCS5D01G530700, TraesCS5D01G530800, TraesCS5D01G530900, TraesCS5D01G531000, TraesCS5D01G531100, TraesCS5D01G531300, TraesCS5D01G531600, TraesCS5D01G531800) before and after SBWMV symptoms appeared. Only one gene, TraesCS5D01G531200 showed significant differential expression between the resistant and susceptible parents when SBWMV symptoms appeared in the leaf tissues (Fig. 4), indicating that SBWMV infection differentially induced expression of TraesCS5D01G531200. Considering that TraesCS5D01G531200 was the only one of the three genes (the other two were TraesCS5D01G530600 and TraesCS5D01G531300) in this region encoding proteins that may be involved in disease resistance, we believed TraesCS5D01G531200 might play an important role in the SBWMV resistance.
Figure 4.
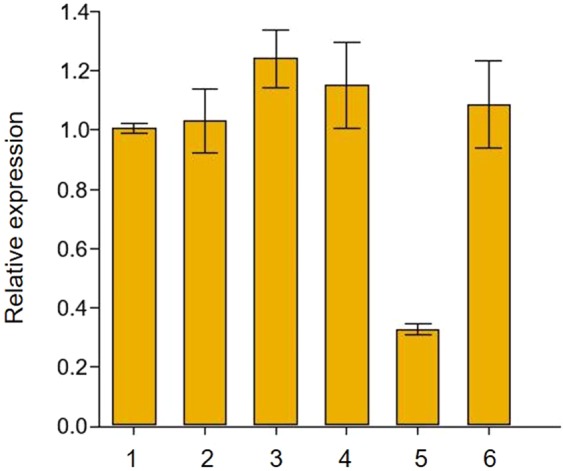
Gene expression analysis of TraesCS5D01G531200 in leaf tissue in the resistant and susceptible parents. Leaf tissue were collected from Wesley (resistant) and OK03825-5403-6 (susceptible) in early spring from uninfected field (1, 2), before winter in infected field (3,4), and in early spring from infected field with SBWMV symptom present (5, 6). Actin was used as the endogenous control.
Validation of the flanking markers in cultivars and breeding lines
To further validate the utility of markers that are tightly linked to Sbwm1 in wheat breeding, the two flanking KASP markers, wsnp_CAP11_c209_198467 and BS0000079676_51, were used to screen 159 additional wheat cultivars and breeding lines from U.S. hard winter wheat breeding programs and the validation panel was also phenotyped for SBWMV resistance in the field experiments (Table 3, Supplementary Table S2). SBWMV resistance of those accessions was consistent between two years with a high correlation of 0.91 (p < 0.001). Among these accessions, 104 were resistant and 55 were susceptible. The two SNPs identified four haplotypes across these accessions (Table 3) with the A-A haplotype associated with resistant accessions, and G-G haplotypes with susceptible accessions. For SNP wsnp_CAP11_c209_198467, A presents in 99% of the resistant accessions and G presents in 98.2% of susceptible accessions. For SNP BS00079676_51, A and G alleles present in 95.2 and 96.6% of the resistant and susceptible accessions, respectively. Two recombinant haplotypes A-G and G-A were identified in 5 and 3 accessions, respectively. All the accessions with the A-G haplotype were resistant. For G-A haplotype, however, only one G-A accession was resistant and other two accessions were susceptible, indicating there is recombination between Sbwm1 and BS00079676_51 in these two accessions. The two KASP assays together can effectively distinguish resistance and susceptibility alleles in the diverse wheat accessions and can be used for the selection of Sbwm1 in breeding programs.
Table 3.
Haplotypes and SBWMV reactions of the two flanking SNPs for Sbwm1 in 159 wheat cultivars and breeding lines.
| wsnp_CAP11_ c209_198467 | B00079676_51 | Accession num | SBWMV resistance |
|---|---|---|---|
| A | A | 99 | resistant |
| A | G | 5 | resistant |
| G | G | 52 | susceptible |
| G | A | 4 | resistant (1), susceptible (3) |
Discussion
Severity of SBWM infection is affected by environmental factors such as soil water content after planting and temperatures in early spring. In this study, highly repeatable phenotypic data was generated from the two RIL populations, the association mapping population and a validation panel of the 159 wheat accessions in repeated experiments by carefully managing the SBWMV nursery. In the two RIL populations, a slight difference in disease ratings between two years was observed for some RILs (Fig. 1). The rating changes usually occurred within the resistant or susceptible categories, not between the two categories, which can be visualized by high correlation coefficients (from 0.81 to 0.94) of SBWMV resistance between years for the four populations investigated. Those results indicated that SBWMV resistance evaluated in the field nursery in this study was highly repeatable, thus is adequate for QTL analysis.
Our previous association mapping study identified an SSR marker, Xgwm469 and two SNPs wsnp_JD_c4438_5568170, wsnp_CAP11_c209_198467 from 9 K SNP chips that closely linked with Sbwm117,30. In the current study, we used the new 90 K wheat SNP chips with high marker density to conduct a genome-wide association study on the same association mapping panel. We found 21,600 polymorphic SNPs from the 90 K SNP chips, which is three times of those identified from the 9 K chips (6,895 SNPs). Among the polymorphic SNPs, 35 SNPs were identified to be highly associated with Sbwm1. More trait associated SNPs identified in this study is mainly due to the high SNP density (10 times) in the 90 K SNP chip than the 9 K SNP chip25. Among those Sbwm1-associated SNPs, wsnp_JD_c4438_5568170, c4438_5567972, wsnp_JD_c4438_5567834 and wsnp_CAP11_c209_198671 were the same as previously identified from the 9 K SNP chips, and the other 31 were newly identified SNPs from the new 90 K chips in this study, indicating that increasing marker density facilitate identification of more trait-associated SNPs. Those common SNPs identified in both chips were owing to that the most probes in the 9 K chip were included in the 90 K chip25.
Several independent mapping studies have identified a major locus for SBWMV resistance in the similar location on the long arm of chromosome 5D from different wheat cultivars and an Ae. tauschii-derived breeding line12,16,18. Those QTLs most likely underline the same gene, Sbwm1 as reported by Liu et al.17. A gene for SBCMV resistance, designated as Sbm119, was also mapped in the similar location as Sbwm1 in wheat and they may be either tightly linked or the same gene.
In the linkage maps from Wesley/OK03825-5403-6 and Deliver/OK03825-5403-6 populations, those significant SNPs identified from the AM panel were all mapped near the SBWMV resistance gene in the same linkage group corresponding to the distal end of chromosome 5DL (Fig. 3). The SBWMV resistance gene was flanked by BS00079676_51 and wsnp_CAP11_c209_198467 with BS00079676_51 as the closest marker in the two populations, respectively. Since wsnp_CAP11_c209_198467 has been identified as the closest marker to Sbwm1 in cultivar Heyne17, the resistance gene in Wesley and Deliver is in the same region of Sbwm1 and they are most likely the same gene. Therefore, this study defined Sbwm1 region to a 3.4 cM region in the Deliver/OK03825-5403-6 population and identified closely linked flanking markers for Sbwm1, which laid solid ground for map-based cloning and marker-assisted breeding of Sbwm1. Using association mapping together with linkage mapping can effectively determine the marker-trait association.
Using the IWGSC Chinese Spring reference genome RefSeqv1.0, all the newly identified SNPs were mapped in the interval between 546,086,597 and 547,273,657 on 5D. However, the two flanking SNPs further delimited the 3.4 cM Sbwm1 region to a physical region of 620 kb (Fig. 3), suggesting Sbwm1 is in a high recombination region. In this region, only 17 high confidence genes were annotated in wheat RefSeqv1.0 reference. Using Ae. tauschii reference genome, Sbwm1 was delimited to a 660 kb region on 5D with 18 annotated genes (Fig. 3C,E). Only one predicted wheat gene, is absent in Ae. tauschii, and two Ae. tauschii genes are absent in wheat (Supplementary Table S5), indicating good microcolinearity between the two species in Sbmv1 region. In the Sbwm1 genomic region, we identified three candidate genes that are involved in disease resistance. Gene expression analysis identified only one gene showed significantly differential expression between the resistant parents and the susceptible parent (Fig. 4) in the tissues with SBWMV symptoms, which is selected as the candidate for further function analysis.
Wheat SNP chip may not be cost effective for breeding selection due to high cost per sample if only a few SNPs are interested. KASP assay is a time saving and cost-effective genotyping assay for single SNP analysis and has been successfully applied in wheat research and breeding42–44. In this study, all the identified SNPs that were highly associated with Sbwm1 were converted into KASP assays to map Sbwm1 in the two RIL populations. The KASP assays for those SNPs developed in this study will facilitate isolation and marker-assisted selection of Sbwm1 in wheat.
The effectiveness of the two flanking KASP assays wsnp_CAP11_c209_198467 and BS00079676_51 were also tested in another panel of 159 wheat cultivars and breeding lines. The result showed that when an accession carries the two SNPs in coupling phase, the accession consistently showed expected (resistant or susceptible) phenotypes. However, six accessions carry the two SNPs in repulsion phase and inconsistent phenotypes were found for each recombinant haplotype. When allele A (resistance allele) at wsnp_CAP11_c209_198467 and allele G (susceptibility allele) at BS00079676_51 were together, all accessions with this haplotype showed SBWMV resistance; whereas when accessions carry G (susceptibility allele) at wsnp_CAP11_c209_198467 and A at BS00079676_51 (resistance allele), their reactions to SBWMV infection were mixed (Table 3), indicating that Sbwm1 located between the two SNPs, and the two SNP together can separate the resistant and susceptible genotypes effectively and can be widely used in marker-assisted breeding to improve SBWMV resistance.
Supplementary information
Acknowledgements
This project is partly funded by the National Key Research and Development Programs of China (2016YFD0101802, 2016ZX08009003-001-006), National Research Initiative Competitive Grants 2017-67007-25939, and 2017-67007-25929 from the USDA National Institute of Food and Agriculture, and the Shandong Province Agricultural Fine Seeds Project (2016LZGC023). Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. USDA is an equal opportunity provider and employer.
Author contributions
S.L. and G.B. wrote the manuscript; S.L., F.J. and D.Z. performed field evaluation and genotyping; S.L., M.L. and G.B. analyzed the data; M.L. contributed to the marker development; B.T., H.N.T. and L.Y. contributed to the writing of the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Shubing Liu, Email: sbliu@sdau.edu.cn.
Guihua Bai, Email: guihua.bai@ars.usda.gov.
Supplementary information
is available for this paper at 10.1038/s41598-020-64993-3.
References
- 1.McKinney HH. Investigations on the rosette disease of wheat and its control. J. Agric. Res. 1923;23:771–800. [Google Scholar]
- 2.Barbosa M, Goulart L, Prestes A, Juliatti F. Genetic control of resistance to Soil-borne wheat mosaic virus in Brazilian cultivars of Triticum aestivum L. Thell. Euphytica. 2001;122:417–422. doi: 10.1023/A:1012937116394. [DOI] [Google Scholar]
- 3.Hariri D, Courtillot M, Zaoui P, Lapierre H. Multiplication of Soil-borne wheat mosaic virus (SBWMV) in wheat roots infected by a soil carrying SBWMV and wheat yellow mosaic virus (WYMV) Agronomie. 1987;7:789–796. doi: 10.1051/agro:19871005. [DOI] [Google Scholar]
- 4.Kapooria RG, Ndunguru J, Clover GRG. First reports of Soil-borne wheat mosaic virus and Wheat spindle streak mosaic virus in Africa. Plant Dis. 2000;84:921. doi: 10.1094/PDIS.2000.84.8.921C. [DOI] [PubMed] [Google Scholar]
- 5.Koenig R, Huth W. Soil-borne rye mosaic and European wheat mosaic virus: two names for a furovirus with variable genome properties which are widely distributed in several cereal crops in Europe. Arch. Virol. 2000;145:689–697. doi: 10.1007/s007050050663. [DOI] [PubMed] [Google Scholar]
- 6.Lebas BSM, et al. Investigation of an outbreak of Soil-borne wheat mosaic virus in New Zealand. Aust. Plant Pathol. 2009;38:85–90. doi: 10.1071/AP08082. [DOI] [Google Scholar]
- 7.Sawada E. Control of Wheat yellow mosaic virus. J. Plant Prot. 1927;14:444–449. [Google Scholar]
- 8.Driskel BA, Hunger RM, Payton ME, Verchot-Lubicz J. Response of hard red winter wheat to Soil-borne wheat mosaic virus using novel inoculation methods. Phytopathology. 2002;92:347–54. doi: 10.1094/PHYTO.2002.92.4.347. [DOI] [PubMed] [Google Scholar]
- 9.Shirako Y, Suzuki N, French RC. Similarity and divergence among viruses in the genus Furovirus. Virol. 2000;270:201–207. doi: 10.1006/viro.2000.0251. [DOI] [PubMed] [Google Scholar]
- 10.Cadle-Davidson L, Sorrells ME, Gray SM, Bergstrom GC. Identification of small grains genotypes resistant to Soil-borne mosaic virus. Plant Dis. 2006;90:1039–1044. doi: 10.1094/PD-90-1039. [DOI] [PubMed] [Google Scholar]
- 11.Bever WM, Pendleton JW. The effect of Soil-borne wheat mosaic on yield of winter wheat. Plant Dis. Rep. 1954;38:266–267. [Google Scholar]
- 12.Hao Y, et al. A conserved locus conditioning Soil-borne wheat mosaic virus resistance on the long arm of chromosome 5D in common wheat. Mol. Breed. 2012;30:1453–1464. doi: 10.1007/s11032-012-9731-x. [DOI] [Google Scholar]
- 13.Myers LD, Sherwood JL, Siegerist WC, Hunger RM. Temperature-influenced virus movement in expression of resistance to Soil-borne wheat mosaic virus in hard red winter wheat (Triticum aestivum) Phytopathology. 1993;83:548–551. doi: 10.1094/Phyto-83-548. [DOI] [Google Scholar]
- 14.Merkle OG, Smith EL. Inheritance of resistance to Soil-borne mosaic in wheat. Crop Sci. 1983;23:1075–1076. doi: 10.2135/cropsci1983.0011183X002300060012x. [DOI] [Google Scholar]
- 15.Modawi RS, Heyne EG, Brunetta D, Willis WG. Genetic studies of field reaction to wheat Soil-borne mosaic virus. Plant Dis. 1982;66:1183–1184. doi: 10.1094/PD-66-1183. [DOI] [Google Scholar]
- 16.Narasimhamoorthy B, Gill BS, Fritz AK, Nelson JC, Brown- Guedira GL. Advanced backcross QTL analysis of a hard winter wheat x synthetic wheat population. Theor. Appl. Genet. 2006;112:787–796. doi: 10.1007/s00122-005-0159-0. [DOI] [PubMed] [Google Scholar]
- 17.Liu S, Yang X, Chao S, Bockus W, Bai G. Genome-wide association analysis identified a major QTL conferring wheat Soil-borne wheat mosaic virus resistance. Theor. Appl. Genet. 2014;127:1039–1047. doi: 10.1007/s00122-014-2277-z. [DOI] [PubMed] [Google Scholar]
- 18.Hall MD, Brown-Guedira G, Klatt A, Fritz AK. Genetic analysis of resistance to Soil-borne wheat mosaic virus derived from Aegilops tauschii. Euphytica. 2009;169:169–176. doi: 10.1007/s10681-009-9910-y. [DOI] [Google Scholar]
- 19.Bass C, Hendley R, Adams MJ, Hammond-Kosack KE, Kanyuka K. The Sbm1 locus conferring resistance to Soil-borne cereal mosaic virus maps to a gene-rich region on 5DL in wheat. Genome. 2006;2006(49):1140–8. doi: 10.1139/g06-064. [DOI] [PubMed] [Google Scholar]
- 20.Perovic D, et al. Mapping and diagnostic marker development for Soil-borne cereal mosaic virus resistance in bread wheat. Mol. Breed. 2009;23:641–653. doi: 10.1007/s11032-009-9262-2. [DOI] [Google Scholar]
- 21.Manish KP, et al. Development and evaluation of a high density genotyping ‘Axiom_Arachis’ array with 58 K SNPs for accelerating genetics and breeding in groundnut. Sci. Reports. 2017;7:40577. doi: 10.1038/srep40577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang, X. et al. Genome-wide association studies of 14 agronomic traits in rice landraces. Nat. Genet. 42, 961–967 (2010). [DOI] [PubMed]
- 23.Belamkar V, et al. Genomic selection in preliminary yield trials in a winter wheat breeding program. 2018;G3(8):2735–2747. doi: 10.1534/g3.118.200415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cavanagh CR, et al. Genome-wide comparative diversity uncovers multiple targets of selection for improvement in hexaploid wheat landraces and cultivars. Proc. Natl. Acad. Sci. USA. 2013;110:8057–8062. doi: 10.1073/pnas.1217133110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang S, et al. Characterization of polyploid wheat genomic diversity using a high-density 90,000 SNP array. Plant Biotech. J. 2014;12:787–96. doi: 10.1111/pbi.12183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Winfield MQ, et al. High‐density SNP genotyping array for hexaploid wheat and its secondary and tertiary gene pool. Plant Biotechnol. J. 2016;14:1195–1206. doi: 10.1111/pbi.12485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rasheed A, et al. Crop breeding chips and genotyping platforms: progress, challenges, and perspectives. Mol. Plant. 2017;10:1047–1064. doi: 10.1016/j.molp.2017.06.008. [DOI] [PubMed] [Google Scholar]
- 28.Allen AM, et al. Characterization of a Wheat Breeders’ Array suitable for high‐throughput SNP genotyping of global accessions of hexaploid bread wheat (Triticum aestivum) Plant Biotech. J. 2017;15:390–401. doi: 10.1111/pbi.12635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang D, Bai G, Zhu C, Yu J, Carver BF. Genetic diversity, population structure, and linkage disequilibrium in U.S. elite winter wheat. The Plant Genome. 2010;3:117–127. doi: 10.3835/plantgenome2010.03.0004. [DOI] [Google Scholar]
- 30.Zhang D, et al. Association study of resistance to Soil-borne wheat mosaic virus in U.S. winter wheat. Phytopathology. 2011;101:1322–9. doi: 10.1094/PHYTO-02-11-0041. [DOI] [PubMed] [Google Scholar]
- 31.Lin M, et al. Genome-wide association analysis on pre-harvest sprouting resistance and grain color in U.S. winter wheat. BMC Genomics. 2016;17:794. doi: 10.1186/s12864-016-3148-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rudi A, et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science. 2018;361:6403. doi: 10.1126/science.aar7191. [DOI] [PubMed] [Google Scholar]
- 33.Luo MC, et al. A 4-gigabase physical map unlocks the structure and evolution of the complex genome of Aegilops tauschii, the wheat D-genome progenitor. Proc. Natl. Acad. Sci. USA. 2013;110:7940–7945. doi: 10.1073/pnas.1219082110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Toojinda T, Baird E, Booth A, Broers L, Hayes P. Introgression of quantitative trait loci (QTLs) determining stripe rust resistance in barley: an example of marker assisted line development. Theor. Appl. Genet. 1998;96:123–131. doi: 10.1007/s001220050718. [DOI] [Google Scholar]
- 35.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multi-locus genotype data. Genetics. 2000;155:945–59. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hardy OJ, Vekemans X. Spagedi: a versatile computer program to analyze spatial genetic structure at the individual or population levels. Mol. Ecol. Notes. 2002;2:618–20. doi: 10.1046/j.1471-8286.2002.00305.x. [DOI] [Google Scholar]
- 37.Lipka AE, et al. GAPIT: genome association and prediction integrated tool. Bioinformatics. 2012;28:2397–9. doi: 10.1093/bioinformatics/bts444. [DOI] [PubMed] [Google Scholar]
- 38.Van Ooijen, J. W. & Voorrips, R. E. JionMap version 3.0: Software for the calculation of genetic linkage maps (2001).
- 39.Kosambi DD. The estimation of map distances from recombination values. Ann. Eugen. 1944;12:172–175. doi: 10.1111/j.1469-1809.1943.tb02321.x. [DOI] [Google Scholar]
- 40.Luo M, et al. Genome sequence of the progenitor of the wheat D genome Aegilops tauschii. Nature. 2018;551:498–502. doi: 10.1038/nature24486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.The International Brachypodium Initiative. Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature463, 63–768 (2010). [DOI] [PubMed]
- 42.Terracciano I, et al. Development of COS-SNP and HRM markers for high throughput and reliable haplotype-based detection of Lr14a in durum wheat (Triticum durum Desf.) Theor. Appl. Genet. 2013;126:1077–1101. doi: 10.1007/s00122-012-2038-9. [DOI] [PubMed] [Google Scholar]
- 43.Semagn K, Babu R, Hearne S, Olsen M. Single nucleotide polymorphism genotyping using Kompetitive Allele Specific PCR (KASP): overview of the technology and its application in crop improvement. Mol. Breed. 2014;33:1–14. doi: 10.1007/s11032-013-9917-x. [DOI] [Google Scholar]
- 44.Rasheed A, et al. Development and validation of KASP assays for genes underpinning key economic traits in bread wheat. Theor. Appl. Genet. 2016;129:1843–1860. doi: 10.1007/s00122-016-2743-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.