Abstract
Puromycin is a naturally occurring aminonucleoside antibiotic that inhibits protein synthesis by ribosome-catalyzed incorporation into the C-terminus of elongating nascent chains, blocking further extension and resulting in premature termination of translation. It is most commonly known as a selection marker for cell lines genetically engineered to express a resistance transgene, but its additional uses as a probe for protein synthesis have proven invaluable across a wide variety of model systems, ranging from purified ribosomes and cell-free translation to intact cultured cells and whole animals. Puromycin is comprised of a nucleoside covalently bound to an amino acid, mimicking the 3′ end of aminoacylated tRNAs that participate in delivery of amino acids to elongating ribosomes. Both moieties can tolerate some chemical substitutions and modifications without significant loss of activity, generating a diverse toolbox of puromycin-based reagents with added functionality, such as biotin for affinity purification or fluorophores for fluorescent microscopy detection. These reagents, as well as anti-puromycin antibodies, have played a pivotal role in advancing our understanding of the regulation and dysregulation of protein synthesis in normal and pathological processes, including immune response and neurological function. This manuscript reviews the current state of puromycin-based research, including structure and mechanism of action, relevant derivatives, use in advanced methodologies and some of the major insights generated using such techniques both in the lab and the clinic.
Keywords: Puromycin, Ribosome, Translation, Protein synthesis, Nascent polypeptide chains, mRNA display, O-propargyl-puromycin (OPP), SUnSET, PUNCH-P, puro-PLA
1. Introduction
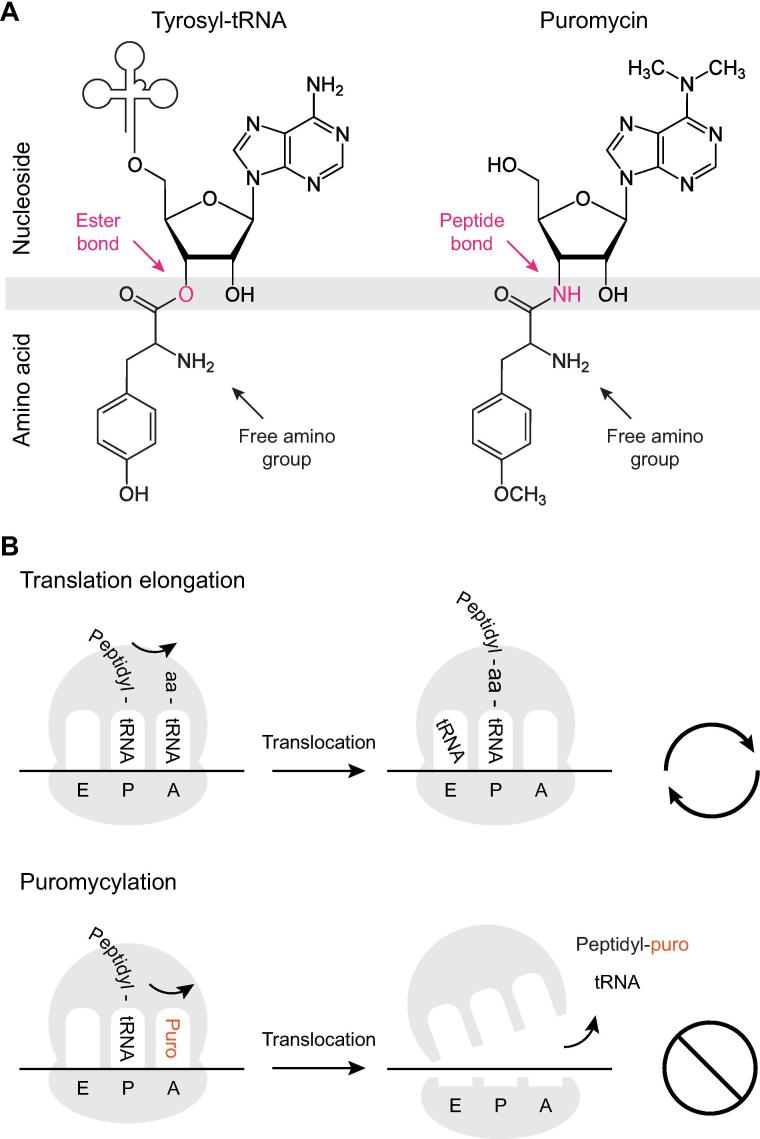
Puromycin is produced by Streptomyces alboniger, a gram-positive actinomycete, through a series of enzymatic reactions starting from adenosine triphosphate (ATP) [1]. Structurally, puromycin resembles the 3′ end of aminoacylated tRNA (aa-tRNA), with a modified adenosine base covalently linked to a tyrosine amino acid [2]. The major difference lies in the nature of the bond between the amino acid and the ribose—a labile ester in aa-tRNA versus a stable peptide bond in puromycin (Fig. 1A). Similar to aa-tRNA, puromycin can enter the ribosomal A-site, where its free amino group accepts a nascent polypeptide chain from the P-site peptidyl-tRNA, in a reaction catalyzed by the ribosome peptidyltransferase center (PTC). However, because the peptide bond between the two moieties of puromycin cannot be further cleaved by an incoming aa-tRNA, such incorporation into the C-terminus of elongating nascent chains prevents additional extension and results in irreversible premature termination of translation. This reaction, termed puromycylation, is energy independent and leads to disassembly of 80S ribosomes and release of peptidyl-puromycin products (Fig. 1B) [3], [4]. C-terminal incorporation is considered to be the major inhibitory mechanism of puromycin, although some evidence point towards additional effects on ribosome recycling or other ribosome-associated functions [5], [6]. After translation termination, truncated puromycylated polypeptides are rapidly targeted for degradation [7], [8], suggesting that these newly synthesized products are recognized as aberrant by quality control mechanisms and cleared to avoid proteotoxic stress.
Fig. 1.
Puromycin structure and mechanism of action. (A) Chemical structures of the 3′ end of an aminoacylated tyrosyl-tRNA (left) and puromycin (right). The different bonds between the nucleoside and amino acid moieties are shown in pink. (B) Basic mechanism of puromycin action. During translation elongation, aa-tRNA enters the A-site and accepts the nascent polypeptide chain from the peptidyl-tRNA in the P-site. Following translocation, the A-site becomes available to accommodate the next aa-tRNA (top). Like aa-tRNA, puromycin can enter the A-site and accept the nascent chain. This results in translation termination, ribosome disassembly and release of the nascent chain bearing a 3′ puromycin (bottom). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Despite its structural homology to tyrosyl-tRNA, puromycin incorporation is not amino acid specific. It is thought to occur at similar rates across all nascent polypeptide chains regardless of their sequence, and is therefore unaffected by the endogenous pools of amino acids or their relative content in the proteins being synthesized [9]. While the tyrosine moiety can be substituted with tryptophan without appreciable loss of activity, other naturally occurring L-amino acids significantly reduce the inhibitory effects of puromycin and some, such as glycine or proline, prevent puromycylation altogether [10]. Consistent with absence of codon-anticodon interactions, such substitutions do not increase or decrease the rate of puromycylation at positions coding for the substituted amino acids. Nevertheless, other interventions can enhance or suppress puromycylation. Pretreatment with cycloheximide, an inhibitor of ribosome translocation, can limit puromycin incorporation by blocking peptidyl-tRNA transition from the A-site [11]. In contrast, pretreatment with emetine can enhance puromycylation while preventing ribosome disassembly and peptidyl-puromycin release, likely by stabilizing a ribosome translocation intermediate that increases A-site availability [4], [12]. However, these effects may vary between cell types and experimental conditions, as some reports suggest that emetine inhibits puromycylation [13] while cycloheximide promotes it [14]. Based on these observations, as well as reports showing preferential incorporation of puromycin at specific positions during in vitro translation of model proteins, it has been proposed that puromycylation may occur more efficiently under conditions associated with reduced A-site occupancy e.g. during ribosome pausing on rare codons [5], [15].
2. Puromycin-based reagents
Since the discovery of its chemical structure, puromycin has been modified using both nucleotide and amino acid chemistries, generating a wide range of radiolabeled, fluorescent, biotinylated and photoactivatable derivatives (summarized in Table 1). It quickly became clear that the amino acid moiety of puromycin could only tolerate minor substitutions without a significant loss of activity, and that the free amino group is essential for inhibition of protein synthesis [5], [10]. However, modifications of the nucleoside were generally better tolerated. Conjugation of puromycin to the 3′ end of a cytidine nucleotide or dinucleotide through phosphodiester bonds, mimicking the conserved CCA tail found in tRNAs, resulted in compounds that retained a relatively strong inhibitory effect; longer oligonucleotide extensions, however, were associated with drastic loss of function [5]. A cytidine nucleotide was used as a linker to introduce other functional groups without substantially affecting the inhibitory potential of the product, generating biotin- or fluorophore-dC-puromycin conjugates that mediate efficient puromycylation in cell-free systems. Blocking the free amino group yields products that cannot inhibit protein synthesis but may be otherwise useful, as some still bind ribosomes with high affinity [16]. When blocked with photolabile protecting groups e.g. O-Nitroveratryloxycarbonyl (NVOC) or 7-Diethylamino-4-methylcoumarin (DEACM), the resulting compounds show minimal toxicity but expose their amino group and undergo puromycylation upon UV irradiation, serving as a photoactivatable puromycin [17], [18]. Some small modifications of the O-methyl-phenyl ring can also be tolerated without significant loss of function, such as the introduction of an alkyne group in O-propargyl-puromycin (OPP), which allows subsequent manipulation of puromycylated proteins by copper-catalyzed alkyne-azide cycloaddition of a “clickable” biotin or fluorophore [8]. More recently, clickable puromycin reagents were expanded to include various alkyne or azide substitutions of either the nucleoside or amino acid moiety [19]. Another related reagent in the puromycin toolbox is the anti-puromycin antibody, first developed as a rabbit polyclonal [20], [21] and later replaced by a commercial mouse monoclonal (clone 12D10) [22]. Both antibodies were raised by covalently attaching puromycin to a carrier protein via peptide bond formation, resembling the product formed by ribosome-catalyzed incorporation.
Table 1.
Major puromycin derivatives.
| Compound | Description | Uses |
|---|---|---|
| Nucleoside substitutions | ||
| 5′ Fluorophore-dC-puromycin | Cy3, Cy5, or fluorescein attached via deoxycytidine linker | Imaging protein synthesis in cultured cells [23]; In vitro C-terminal labeling of full-length proteins [24] |
| 5′ Biotin-dC-puromycin | Biotin attached via deoxycytidine linker | Labeling newly synthesized proteins under cell free conditions for subsequent affinity purification and proteomic analysis [25], [26] |
| 5′ alkyne/azide puromycin (5Y/5Z/5N/5T) | Alkyne/azide substitution of the 5′ hydroxyl group | Labeling newly synthesized proteins in cultured cells for visualization or affinity purification using click chemistry cycloaddition of fluorophore/biotin [19] |
| Amino acid substitutions | ||
| O-propargyl-puromycin (OPP) | Alkyne substitution of the O-methyl-phenyl ring | Labeling newly synthesized proteins in cultured cells, tissues and whole animals for visualization or affinity purification using click chemistry cycloaddition of fluorophore/biotin [8], [27] |
| Photocleavable N-blocked (NVOC/DEACM) puromycin | Photocleavable group attached to the free amino terminus | Labeling newly synthesized proteins in cultured cells with improved spatiotemporal resolution using laser excitation [17], [18] |
| Enzyme labile N-blocked puromycin (PhAc-puro) | Enzyme labile phenylacetyl group attached to the free amino terminus | Selective labeling of newly synthesized proteins in cultured cells engineered to express an E. coli enzyme [28] |
| N-blocked biotin puromycin (3P) | Biotin attached to the free amino terminus via double Jeffamine linker | Affinity purification of ribosomes under cell free conditions (without puromycylation) [16] |
3. Uses of puromycin and puromycin-based techniques
Puromycin inhibits protein synthesis in organisms across all kingdoms of life, including its producer bacteria. However, S. alboniger also expresses an enzyme called puromycin N-acetyltransferase (pac), which acetylates and blocks the reactive amino group in puromycin, thus preventing peptide bond formation and conferring resistance. Puromycin resistance also occurs naturally in other organisms e.g. Saccharomyces cerevisiae due to limited permeability or presence of drug resistance factors, which can be reversed by genetic manipulations [29]. In the 1980s, pac was first cloned and expressed in mammalian cells to render them resistant to puromycin [30]. It has since become a popular method for selection of stably transformed or genetically engineered cells, seemingly without major perturbations to normal cellular function, although the expression of pac and the presence of N-acetylated puromycin may have some effects on the cellular transcriptome [31], [32]. Due to its non-selectivity and high systemic toxicity, puromycin never became a clinically relevant antibiotic, but has nevertheless been extensively used to study mechanisms of protein synthesis, particularly peptide bond formation by the PTC and ribosome translocation (reviewed in [33]). The next few sections describe the uses of puromycin beyond selection e.g. to measure protein synthesis rates, monitor subcellular localization of translating ribosomes, isolate nascent polypeptide chains and generate mRNA-protein fusions. A comparison of puromycin-based and alternative methods for probing translation is provided in Table 2.
Table 2.
Comparison of methods for probing translation.
| Puromycin reagents | Non-puromycin alternatives | Method of detection | Comments | |
|---|---|---|---|---|
| Translation rate measurement | Radioactive puromycin [35] | Radioactive amino acids (AAs) [55] | Scintillation or autoradiography | AA analogs do not terminate translation, but puromycin can be used without predepletion of endogenous AAs |
| Puromycin/OPP (SUnSET) [8], [22] | Clickable AAs [56] | Immunoblot or FACS | ||
| Visualization of newly synthesized proteins | Puromycin/OPP (RPM, Puro-PLA) [8], [14], [42] | Clickable AAs (FUNCAT, FUNCAT-PLA) [42], [57] | Immunofluorescence | AA analogues do not terminate translation, but only puromycin can be used to label sites of active translation when combined with inhibitors of elongation |
| Translatome analysis | N-blocked biotinylated puromycin (RiboLace) [16] | Deep sequencing of nuclease-resistant ribosome-protected mRNA (ribo-seq) [58] | Next generation sequencing | RiboLace can be used to affinity purify translating ribosomes prior to ribo-seq analysis |
| Biotinylated puromycin/OPP (PUNCH-P/OPP-ID) [44], [47] | AA isotopes (pSILAC) [59] Clickable AAs (BONCAT/QuaNCAT) [56], [60] | Mass spectrometry | AA analogs do not terminate translation, but puromycylation is rapid, AA nonspecific and can occur on isolated ribosomes |
3.1. Measurement of global protein synthesis
Regulation of protein synthesis is intimately linked to development and homeostasis at both the cellular and organismal level, and dysregulation has been implicated in many pathological states [34], underlining the importance of monitoring translation. Soon after the initial characterization of puromycin as an inhibitor of protein synthesis, levels of puromycylated peptides were found to be proportional to the overall rate of translation, rendering the puromycylation reaction a valuable tool for measuring changes in translation. The first probe, tritiated [3H]puromycin, was used both in vitro [35] and in vivo [36] to demonstrate that translation is inhibited in response to starvation and stimulated in response to insulin administration. However, radioactive puromycin was discarded in favor of metabolic labeling using radioactive amino acids e.g. [3H]leucine and [35S]methionine/cysteine. These became the gold standard for translation rate measurements as they more directly reflect the normal physiological process, and because multiple radioactive labels can be incorporated into a single nascent chain without causing termination, allowing sequential rounds of translation and increasing final signal intensity. Nevertheless, radioactive and other amino acid analogs are significantly limited by competition with the endogenous pools, often restricting their use to conditions that allow predepletion and rendering them inapplicable for in vivo labeling.
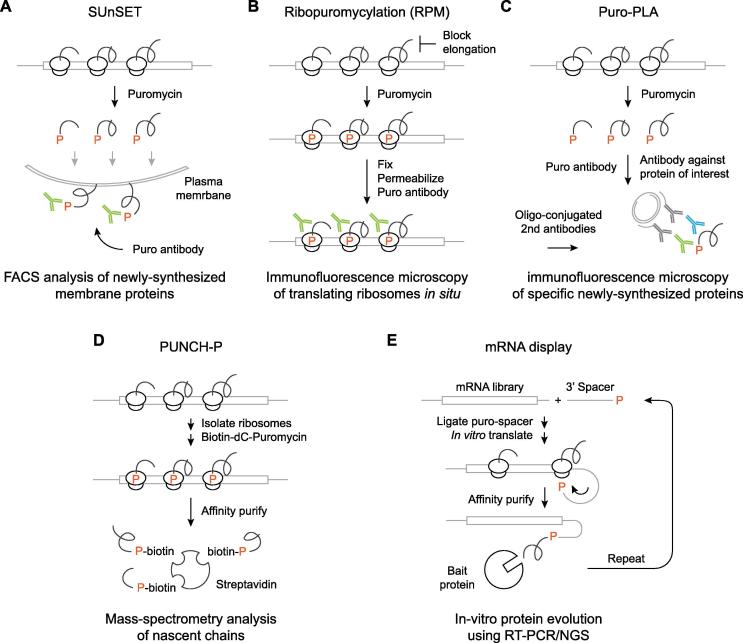
More recently, antibody-coupled detection of puromycylated peptides was introduced as a nonradioactive alternative to traditional metabolic labeling. In its simplest application, low concentrations of puromycin are added to cultured cells or administered to animals, and puromycylated peptides are detected by standard immunoblotting using anti-puromycin antibodies, generating a complex laddering pattern that reflects the broad repertoire of newly synthesized proteins [22]. Other than immunoblotting, detection can take place by fluorescence activated cell sorting (FACS) using fluorophore-conjugated anti-puromycin antibodies. After addition of puromycin, puromycylated membrane proteins traffic from the ER to the plasma membrane, where they can be detected without disrupting the cells. In SUrface SEnsing of Translation (SUnSET) (Fig. 2A), a 10-min pulse of puromycin followed by a 50-min chase resulted in detectable cell surface puromycylation in mouse embryonic fibroblasts (MEF). This was blocked by pretreatment with either cycloheximide or Brefeldin A, which inhibits egress of membrane proteins from the ER. The signal generated was proportional to immunoblot detection and comparable to metabolic labeling using radioactive amino acids, supporting the use of SUnSET as a proxy for measuring global translation rates [22]. A unique advantage of this approach is that translation can be measured at a single-cell resolution in parallel with other parameters e.g. mTOR activity, phosphorylation of translation factors and cell cycle phase. This allowed for the detection of cell-to-cell variability in steady-state inhibitory phosphorylation of elongation factor 2 (eEF2) and revealed an unexpectedly weak correlation between baseline eEF2 phosphorylation and global translation rates [37]. However, SUnSET may not be readily applicable to all cell types, as puromycylated peptides were found to be absent from the surface of mouse bone marrow cells [27], suggesting that differences in plasma membrane composition or trafficking of membrane proteins may confound comparisons of SUnSET between different cells or organisms.
Fig. 2.
Major applications of puromycin and its derivatives. (A) In SUrface SEnsing of Translation (SUnSET), global translation rates are estimated based on incorporation of puromycin into membrane proteins. After a pulse of puromycin in cultured cells or whole animals, puromycylated membrane proteins are trafficked to the plasma membrane, where they can be detected by FACS using anti-puromycin antibodies. (B) In Ribopuromycylation (RPM), cells are plated on cover slips and emetine is used to arrest elongating ribosomes. Subsequent addition of puromycin leads to incorporation into nascent chains without release from ribosomes. Cells are then fixed, permeabilized and stained with anti-puromycin antibodies to visualize translating ribosomes in situ. (C) In puro-PLA, puromycylation of cells on cover slips is followed by incubation with two primary antibodies: one against puromycin and another against a protein of interest. Oligonucleotide conjugated secondary antibodies are then added, and ligation takes place wherever the two primary antibodies bind closely to each other on the same protein molecule. After rolling circle amplification, the products are visualized by hybridization to a fluorescent oligonucleotide probe. This detects the subcellular localization of specific newly synthesized proteins. (D) In PUromycin-associated Nascent CHain Proteomics (PUNCH-P), translating ribosomes are extracted from cells or tissues and incubated with biotin-dC-puromycin. Puromycylated nascent chains are then isolated by streptavidin affinity purification and subjected to mass-spectrometry analysis, to generate a snapshot of the entire nascent proteome. (E) In mRNA display, a cDNA library is in vitro transcribed and an oligonucleotide spacer modified with a 3′ puromycin is covalently attached to each transcript. Using cell-free translation, incorporation of puromycin by ribosomes that reach the stop codon links the nascent polypeptide to its cognate transcript. These mRNA-protein fusion products are then selected by binding to bait proteins or nucleic acids and amplified by error-prone PCR for additional rounds of selection prior to detection by next generation sequencing.
The method described above has several variations. In one, OPP and fluorophores can replace puromycin and anti-puromycin antibodies. For example, OPP-labeled proteins were detected in mice as early as 1 h post-injection by fixing and permeabilizing extracted cells and coupling OPP to fluorescent azide. Incorporation of OPP can be similarly detected by either immunoblot or FACS, and may have some advantages over antibody detection in terms of signal strength [27], [38]. In another variation, ribosome runoff is induced by pretreatment with inhibitors of translation initiation. In SUnSET-based Ribosome Speed of Elongation (SunRiSE) [37], a short pulse of harringtonine, which blocks initiation, leads to partial termination of elongating ribosomes prior to addition of puromycin. The difference in the levels of puromycylated peptides between pretreated and control cells is directly proportional to the extent of ribosome runoff and therefore provides an estimate of elongation rates. In MEFs, addition of puromycin at regular intervals from 30 s to 10 min after harringtonine was associated with a gradual reduction in the overall levels of puromycylated peptides detected by either immunoblotting or FACS. Signal decay was proportional to peptide length, with rapid loss of smaller puromycylated peptides, consistent with quicker termination of ribosomes translating shorter transcripts [37].
3.2. Subcellular localization of translating ribosomes and newly synthesized proteins
Puromycin can also be used to visualize translating ribosomes or translation products by immunofluorescence microscopy. In an early report, mouse dendritic cells (DC) were treated with puromycin for 30 min and then fixed, permeabilized and stained with an anti-puromycin antibody. This revealed that, during DC maturation in response to inflammatory challenge, newly synthesized proteins are sequestered in aggregates called DC aggresome-like-induced structures (DALIS) where they are protected from degradation [39]. Ribopuromycylation (RPM) microscopy [14] went one step further, adding an emetine pretreatment step to retain puromycylated peptides on ribosomes thus allowing in situ visualization of active translation sites (Fig. 2B). Puromycylation in the presence of emetine had the same effect on SUnSET as brefeldin A, consistent with emetine preventing the release of nascent chains from ribosomes and subsequent trafficking to the cell surface. Colocalization of puromycylated peptides and ribosomal proteins was found to be high but incomplete, likely due to the presence of idle ribosomes. When applied to cells infected with vaccinia virus, which renders most ribosomes idle except for a subset that is recruited to sites of viral biosynthesis, RPM revealed that ribosomal proteins are scattered throughout the cytoplasm but only colocalize with puromycylated peptides in these so-called viral factories [14]. Nevertheless, the subcellular localization of newly synthesized proteins detected by puromycin microscopy may be different than that obtained by amino acid analogs [8], advising some caution in interpreting the results of such experiments. Similar to other techniques, puromycin derivatives can be substituted for antibodies in RPM. While bulky biotin or fluorophore derivatives cannot promote rapid and efficient puromycylation in cultured cells or whole animals due to either low permeability or reduced affinity to the ribosome A-site [23], uptake and incorporation of the smaller OPP, followed by fixation and fluorophore cycloaddition, was successfully employed to visualize protein synthesis in situ [8], [40], [41].
Puromycin-based microscopy can also be coupled with proximity ligation (PLA) to detect the distribution and turnover of specific newly synthesized proteins, based on spatial proximity between an antibody against puromycin and another antibody against a specific protein of interest [42]. In puro-PLA, cultured cells are puromycylated, fixed and incubated with two primary antibodies raised in different species. Next, a pair of oligonucleotide-labeled secondary antibodies are added, and hybridization occurs wherever both antibodies bind to the same protein molecule. Ligation forms a closed oligonucleotide loop for rolling circle amplification, and the amplified product is detected by a complementary fluorescent probe (Fig. 2C). This technique has been used to visualize sites of translation of specific proteins in neuronal cells [42], [43]. PLA can also be combined with clickable amino acids, but puro-PLA may have some advantages e.g. shorter incubation times and higher signal [37].
3.3. Isolation of nascent chains and untagged ribosomes
Cell-free puromycylation enables the labeling of nascent chains on ribosomes isolated from cells or tissues, thus avoiding potential stress-related effects triggered by the presence of truncated peptides. This principle guided the development of PUromycin-associated Nascent CHain Proteomics (PUNCH-P) [44], in which translating ribosomes are first isolated by ultracentrifugation, and nascent chains are then labeled in vitro with biotin-dC-puromycin, captured on streptavidin beads and analyzed by mass-spectrometry (MS) (Fig. 2D). This technique capitalizes on the high affinity between biotin and streptavidin to generate a snapshot of the nascent proteome at the time of lysis. PUNCH-P detected over 5000 nascent chains in HeLa cells and about 2000 in mouse brains, and was used to identify rapid cell cycle variations in translation and degradation rates, revealing a regulated lag between transcription, translation and protein accumulation [25], [45]. PUNCH-P was also combined with quantitative isotope labeling proteomics to show that a large proportion of nascent chains are cotranslationally degraded by the proteasome, including proteins involved in ribosome biogenesis and translation, nuclear transport and amino acid metabolism [46]. Co-translational degradation rates were found to correlate with translation efficiency and protein length, but not half-life of the mature protein counterparts [46]. As an alternative to biotin-dC-puromycin, nascent chains can be isolated from OPP-treated cells or animals by coupling to biotin prior to streptavidin capture and MS (OPP-ID). This minimizes potential biases introduced by the ribosome isolation procedure but is also susceptible to stress-related effects. Furthermore, this and other techniques based on incorporation of amino acid analogs have so far exhibited lower sensitivity, with about 300 newly synthesized proteins measured after a 2 h pulse of OPP in MEFs [47] and 400 proteins detected in brain tissue after a 2 day feeding of mice with clickable amino acids [48]. Finally, puromycin can be used to stably capture ribosomes without tagging ribosomal proteins; when modified with a biotin linker on its amino group, puromycin binds the ribosome but is unable to form a peptide bond. As such, it is a poor inhibitor of translation but a high affinity ribosome ligand. By immobilizing this biotinylated derivative of puromycin on streptavidin beads, translating ribosomes can be affinity purified from cell lysates together with the mRNA templates they harbor, for further processing or analysis such as by ribosome profiling (ribo-seq) [16].
3.4. Protein labeling and formation of mRNA-peptide fusions
At low non-inhibitory concentrations, about two orders of magnitude less than those commonly used for selection, puromycin cannot effectively compete with aa-tRNA and therefore only becomes incorporated when ribosomes reach a stop codon, generating C-terminal labeling of full-length proteins. This was revealed through cell-free translation of a transcript bearing multiple methionines immediately upstream to a stop codon. Escherichia coli translation reactions were supplemented with radioactive methionine, and products were digested with a carboxypeptidase that cleaves after terminal amino acids but not puromycin. Adding low concentrations of puromycin to the reaction mix did not affect the overall amount of protein product, but protected it from cleavage of the terminal methionines [49]. Furthermore, depletion of translation termination factors and addition of ribosome recycling factors increased labeling efficiency [50], further supporting the proposed mechanism of action. This approach was used to generate proteins labeled on their C-terminus with fluorescent or biotinylated puromycin, later to be used as probes for protein–protein and protein-nucleic acid interaction studies [51], [52].
Another technique based on incorporation of puromycin into full-length proteins is called mRNA display or in vitro virus (IVV) [53]. This is an in vitro display technique for protein selection and evolution that establishes a physical link between genotype and phenotype. mRNA is transcribed from a cDNA library, then conjugated to a noncoding oligonucleotide spacer with a 3′ puromycin through enzymatic ligation or chemical crosslinking. The resulting transcripts are translated in a cell-free system e.g. rabbit reticulocyte lysate, where the incorporation of puromycin by ribosomes that pause on the 3′ end of coding mRNA fuses nascent polypeptides to their cognate transcripts (Fig. 2E). For directed evolution experiments, the resulting products are affinity purified by binding to immobilized bait proteins or nucleic acids, reverse transcribed and then amplified using error prone PCR or saturation mutagenesis to introduce additional variation, thereby increasing library diversity for the next round of selection. A similar principle can also be used to fabricate protein chips by in situ translation of mRNA-puromycin fusions immobilized to streptavidin-coated slides [54]. Puromycin-based mRNA display has some advantages over the more commonly used phage or yeast display techniques. Libraries can be larger, with up to trillions of mRNAs, and avidity effects are avoided because each ribosome can only display a single protein copy. The cell-free conditions also allow for incorporation of unnatural amino acids to improve affinity, permeability or stability, and the mutational landscape can be better explored. However, mRNA display is limited by the instability of RNA under various selection conditions, as well as the low efficiency of fusion formation, which can be improved by introduction of rare codons in the 3′ end of coding regions [15].
4. Biological insights generated using puromycin-based techniques
4.1. Immune response and nuclear translation
During an immune response, environmental changes can cause stress that in most cell types results in near-complete shutoff of all major biosynthetic functions. However, immune cells have different resistance mechanisms to maintain their functions under stress. Activation of such cells, as measured by in vivo puromycin labeling, is associated with several fold induction of translation rates [61]. To alert the immune system to potential pathogens, proteins are rapidly degraded and presented by class I major histocompatibility complex (MHC). Puromycin-based microscopy revealed that a major source of such peptides is defective ribosomal products (DRiPs)—nascent polypeptides that fail to fold or complete their synthesis and are targeted for proteasomal degradation. During maturation, antigen-presenting DCs rapidly sort DRiPs into cytosolic granules for storage, to delay peptide presentation until migration to lymph nodes or the spleen [39]. These are then targeted for degradation with the help of molecular chaperones [62]. Using OPP, DRiPs were also found to diffuse into the nucleus where they form nuclear aggregates that, if not cleared by proteasomes, lead to depletion of ubiquitin and defects in DNA repair [63], [64].
Similar immunofluorescent puromycylation experiments also provided compelling evidence in support of nuclear translation. It was long known that all components of the translation apparatus can be found in the nucleus, including ribosomal proteins, translation factors and tRNAs, but whether or not these individual components actively engage in protein synthesis is still controversial. Puromycylation was detected in the nucleoplasm and nucleolus of multiple cell types, including HeLa cells, primary monocytes [14] and drosophila S2 cells, where the signal accumulated around chromosomes at transcriptionally active sites [13]. Puromycin signal in the nucleus was eliminated by harringtonine and enhanced by emetine, suggesting that it depends on ribosome catalysis and is not secondary to import of proteins synthesized in the cytoplasm. Nuclear staining was also eliminated upon infection with vaccinia virus, which confines translating ribosomes to perinuclear viral factories [14]. Nuclear translation was proposed to serve as a quality control mechanism, similar to the cytoplasmic pioneer round of translation that surveys and aborts transcripts with premature stop codons [65]. Indeed, nuclear translation was shown to be a source of antigenic peptides presented by MHC I [66]. Antigenic peptides were found to be produced even when their coding sequences were introduced into introns that are spliced out prior to nuclear export, suggesting that translation of prespliced mRNAs occurs in the nucleus and contributes to immunological surveillance [67].
4.2. Neurological function and axonal translation
Early studies into the acquisition of long-term memory showed that intracranial injections of puromycin in goldfish [68] and mice [69] were associated with amnestic effects following shock aversion training, establishing a role for de novo protein synthesis in consolidation of learning-induced synaptic changes. More recently, in vivo puromycin labeling revealed that global translation rates are reduced in Down Syndrome due to constitutive activation of the integrated stress response [70], but elevated in a mouse model of autism due to hyperactivation of translation initiation factor 4E (eIF4E) [71], expanding the link between translation dysregulation and neurological dysfunction. eIF4E hyperactivation was also reported in Huntington’s disease, leading to increased translation of oxidative phosphorylation proteins and decreased translation of neuron-specific factors, as measured by PUNCH-P [26]. Furthermore, in vivo and in situ puromycylation showed that a specific splice isoform of initiation factor 4G (eIF4G) is associated with ribosome stalling due to recruitment of factors that promote RNA granule nucleation, and its depletion results in increased protein synthesis and cognitive impairment in mice [72]. Similar methods were used to show that expression of pathological Tau, implicated in Alzheimer’s disease, reduces translation of ribosomal proteins and translation factors in patient cells and mouse brains, leading to shortage of ribosomes [73], [74], whereas deletion of Tau increases protein synthesis and enhances long-term memory in drosophila neurons [75].
At the subcellular level, localized protein synthesis is a common feature of pre- and post-synaptic neuronal compartments [76]. Puromycin microscopy detected a large proportion of the neuronal transcriptome in granules containing stalled ribosomes, which travel from the soma to the synapse, allowing regulated reactivation of translationally-paused mRNAs. This ensures rapid synaptic synthesis in response to stimulus, even under conditions that inhibit translation initiation [77], and is critical for both axonal development and homeostasis [78]. Proteomic analysis of OPP-treated mouse neurons revealed that, in response to nerve injury, mammalian target of rapamycin (mTOR) activates local translation of its own mRNA as well as that of other genes involved in injury signaling [79]. Even without pretreatment to block elongation, puromycylated peptides remain associated with ribosomes in axons but become released from ribosomes in the cell soma, confirming the axonal-specific presence of stalled ribosomes [80]. These stalled ribosomes can associate with late endosomes, which serve as a moving platform for localized translation, particularly that of pro-survival mitochondrial proteins [81]. Localized synaptic translation was further detected by electron microscopy: cultured hippocampal neurons were pulsed with puromycin, fixed and analyzed using an immunogold labeled anti-puromycin antibody, showing discrete puromycin-positive puncta in both dendrites and synapses [43]. Strikingly, these were recently found to consist mostly of elongating 80S monosomes rather than polysomes, possibly allowing more precise control of protein copies in the synapse [82].
4.3. Protein evolution and interactome studies
mRNA display has been used to screen for antibodies and protein ligands, and some of the resulting products have been evaluated not only in the lab but also in clinical settings [83]. One unique advantages of this approach is the ability to screen for cyclic peptides, which offer benefits such as locking the peptide in its biologically active conformation, restricting proteolytic cleavage and improving membrane permeability. Natural cyclic peptides are most commonly synthesized by enzymatic complexes called nonribosomal peptide synthetases, but mRNA display can incorporate unnatural amino acids that enable cyclization, providing a platform for their ribosomal synthesis [84], [85]. This was used to develop a high-affinity protease-resistant cyclic peptide inhibitor against a disease-related G protein. A randomized library was first screened for high affinity binders, and these were then evolved for resistance to proteolytic cleavage by preincubation with an immobilized protease [86]. mRNA display was also used to screen an antibody mimetic library for pathogen receptor binders that, when fused to an antigenic peptide, triggered presentation by DCs and stimulated an immune response [87]. The same library was combined with continuous flow magnetic separation for single-round selection of immunoglobulin binders, resulting in identification of candidates with comparable affinity to that of protein A and protein G, for potential research or industrial use [88]. In another application, a library constructed from mouse brain poly-A mRNA was selected using bait DNA, generating large scale interaction data for multimeric complexes of transcription factors or other DNA-binding proteins [89].
4.4. Clinical diagnosis and treatment
Measurement of protein synthesis rate in cancer patients can guide treatment decisions and is important for long term survival. Radiolabeled amino acids showed unsatisfactory results as tracers of protein synthesis, but puromycin labeled with radioisotopes e.g. Gallium-68 [90], Scandium-44 [91] or Fluorine-18 [92] holds promise as a radiopharmaceutical for positron emission tomography (PET). Tumor uptake of radiolabeled puromycin was not only significant and rapid, but also, and unlike amino acids, proportional to and dependent on protein synthesis. In the body, puromycin is metabolized into a nephrotoxic metabolite called puromycin aminonucleoside; this metabolite is used to induce kidney damage in an animal model called puromycin aminonucleoside nephrosis (PAN), which mimics nephrotic syndrome in humans [93]. Puromycin also inhibits several aminopeptidases, including puromycin-sensitive aminopeptidase (PSA), which is involved in the proteolytic clearance of aggregation-prone substrates [94]. Substitutions of the methyl phenyl group in puromycin yield potent aminopeptidase inhibitors with minimal effects on protein synthesis, and these compound are being evaluated as candidates for the treatment of hematologic malignancies associated with elevated aminopeptidase activity [95]. Puromycin may also hypersensitize cells to chemotherapy by enhancing p53-dependent apoptosis [96]. Finally, blocked puromycin derivatives are evaluated as prodrugs for selective cancer therapy. In one strategy, blocking the free amino group by an acetylated lysine generates a masked cytotoxic agent, which is specifically activated by histone deacetylase and cathepsin L, two enzymes known to be overexpressed in some cancers [97], [98].
5. Summary and outlook
Since its discovery over half a century ago, puromycin has helped shape our understanding of ribosome function and protein synthesis kinetics. It laid the groundwork for establishing a link between translation regulation and fundamental biological processes across a diverse range of normal development and disease models, including cancer, viral infection and neurodegeneration. The toolbox of puromycin-based reagents has grown considerably over the past decade, with antibodies and derivatives opening up new avenues of research and biotechnological applications. While amino acid analogs may offer a more direct approach to measuring and visualizing translation, puromycin-based techniques have some unique advantages: they are robust, highly sensitive, amino acid nonspecific and applicable for cell-free, intact cells and whole animal systems without requiring pretreatment or predepletion of endogenous amino acids.
Nevertheless, additional work is required to better characterize potential biases associated with puromycylation. Truncated puromycylated peptides are recognized by quality control mechanisms as aberrant [7], [8], and this could introduce confounding variables e.g. increased aggregation and degradation or altered gene expression. Therefore, puromycin-based techniques are generally considered inapplicable for studying protein synthesis over extended periods of time. At high concentrations, puromycin was suggested to inhibit protein synthesis via peptide bond-independent mechanisms [5], which could further complicate data interpretation. Differences in puromycin uptake or rate of puromycylation and trafficking of membrane proteins may confound comparisons of cell surface puromycylation between cell types [27]. Furthermore, structural variations between endogenous amino acids, amino acid analogs and puromycin-based reagents may affect A-site affinity and therefore incorporation specificity. Indeed, discrepancies in labeling pattern, including molecular weight distribution and subcellular localization, have been observed between different puromycin and amino acid analogs [8], [19]. One study showed that such discrepancies are exacerbated under specific cellular conditions e.g. glucose starvation [99]. While some of these differences may be attributed to the amino acid nonspecific nature of puromycylation, they could also reflect preferential incorporation of puromycin at sites of ribosome pausing [5], [15] due to e.g. rare codons, RNA secondary structures, or limited availability of tRNAs and translation factors.
Each of the methods discussed above has its strengths and weaknesses. Therefore, combining complementary puromycin-based and alternative techniques for monitoring translation could offer a broader, more robust perspective. Regardless of the above caveats, puromycin still holds great promise for improving our understanding of ribosome function and global as well as transcript-specific translation regulation in health and disease.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Funding to pay the Open Access publication charge was provided by NIH grant GM056433.
References
- 1.Tercero J.A., Espinosa J.C., Lacalle R.A., Jiménez A. The biosynthetic pathway of the aminonucleoside antibiotic puromycin, as deduced from the molecular analysis of the pur cluster of Streptomyces alboniger. J Biol Chem. 1996;271(3):1579–1590. doi: 10.1074/jbc.271.3.1579. [DOI] [PubMed] [Google Scholar]
- 2.Yarmolinsky M.B., Haba G.L. Inhibition by puromycin of amino acid incorporation into protein. Proc Natl Acad Sci USA. 1959;45(12):1721–1729. doi: 10.1073/pnas.45.12.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Semenkov Y.U., Shapkina T., Makhno V., Kirillov S. Puromycin reaction for the A site-bound peptidyl-tRNA. FEBS Lett. 1992;296(2):207–210. doi: 10.1016/0014-5793(92)80380-y. [DOI] [PubMed] [Google Scholar]
- 4.Baliga B.S., Cohen S.A., Munro H.N. Effect of cycloheximide on the reaction of puromycin with polysome-bound peptidyl-tRNA. FEBS Lett. 1970;8(5):249–252. doi: 10.1016/0014-5793(70)80278-2. [DOI] [PubMed] [Google Scholar]
- 5.Starck S.R., Roberts R.W. Puromycin oligonucleotides reveal steric restrictions for ribosome entry and multiple modes of translation inhibition. RNA. 2002;8(7):890–903. doi: 10.1017/s1355838202022069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kandala D.T., Del Piano A., Minati L., Clamer M. Targeting translation activity at the ribosome interface with UV-active small molecules. ACS Omega. 2019;4(6):10336–10345. doi: 10.1021/acsomega.9b00366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goldberg A.L. Degradation of abnormal proteins in Escherichia coli (protein breakdown-protein structure-mistranslation-amino acid analogs-puromycin) Proc Natl Acad Sci USA. 1972;69(2):422–426. doi: 10.1073/pnas.69.2.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu J., Xu Y., Stoleru D., Salic A. Imaging protein synthesis in cells and tissues with an alkyne analog of puromycin. Proc Natl Acad Sci USA. 2012;109(2):413–418. doi: 10.1073/pnas.1111561108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nathans D. Puromycin inhibition of protein synthesis: incorporation of puromycin into peptide chains. Proc Natl Acad Sci USA. 1964;51:585–592. doi: 10.1073/pnas.51.4.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nathans D., Neidle A. Structural requirements for puromycin inhibition of protein synthesis. Nature. 1963;197:1076–1077. doi: 10.1038/1971076a0. [DOI] [PubMed] [Google Scholar]
- 11.Hobden A.N., Cundliffe E. The mode of action of alpha sarcin and a novel assay of the puromycin reaction. Biochem J. 1978;170(1):57–61. doi: 10.1042/bj1700057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.David A., Bennink J.R., Yewdell J.W. Emetine optimally facilitates nascent chain puromycylation and potentiates the ribopuromycylation method (RPM) applied to inert cells. Histochem Cell Biol. 2013;139(3):501–504. doi: 10.1007/s00418-012-1063-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Al-Jubran K. Visualization of the joining of ribosomal subunits reveals the presence of 80S ribosomes in the nucleus. RNA. 2013;19(12):1669–1683. doi: 10.1261/rna.038356.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.David A. Nuclear translation visualized by ribosome-bound nascent chain puromycylation. J Cell Biol. 2012;197(1):45–57. doi: 10.1083/jcb.201112145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagumo Y., Fujiwara K., Horisawa K., Yanagawa H., Doi N. PURE mRNA display for in vitro selection of single-chain antibodies. J Biochem. 2016;159(5):519–526. doi: 10.1093/jb/mvv131. [DOI] [PubMed] [Google Scholar]
- 16.Clamer M. Active ribosome profiling with RiboLace. Cell Rep. 2018;25(4):1097–1108.e5. doi: 10.1016/j.celrep.2018.09.084. [DOI] [PubMed] [Google Scholar]
- 17.Herzig L.-M., Elamri I., Schwalbe H., Wachtveitl J. Light-induced antibiotic release from a coumarin-caged compound on the ultrafast timescale. Phys Chem Chem Phys. 2017;19(22):14835–14844. doi: 10.1039/c7cp02030a. [DOI] [PubMed] [Google Scholar]
- 18.Buhr F. Design of photocaged puromycin for nascent polypeptide release and spatiotemporal monitoring of translation. Angew Chem Int Ed Engl. 2015;54(12):3717–3721. doi: 10.1002/anie.201410940. [DOI] [PubMed] [Google Scholar]
- 19.Ge J. Puromycin analogues capable of multiplexed imaging and profiling of protein synthesis and dynamics in live cells and neurons. Angew Chem Int Ed Engl. 2016;55(16):4933–4937. doi: 10.1002/anie.201511030. [DOI] [PubMed] [Google Scholar]
- 20.Eggers D.K., Welch W.J., Hansen W.J. Complexes between nascent polypeptides and their molecular chaperones in the cytosol of mammalian cells. Mol Biol Cell. 1997;8(8):1559–1573. doi: 10.1091/mbc.8.8.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang J.X., Braakman I., Matlack K.E., Helenius A. Quality control in the secretory pathway: the role of calreticulin, calnexin and BiP in the retention of glycoproteins with C-terminal truncations. Mol Biol Cell. 1997;8(10):1943–1954. doi: 10.1091/mbc.8.10.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmidt E.K., Clavarino G., Ceppi M., Pierre P. SUnSET, a nonradioactive method to monitor protein synthesis. Nat Methods. 2009;6(4):275–277. doi: 10.1038/nmeth.1314. [DOI] [PubMed] [Google Scholar]
- 23.Starck S.R., Green H.M., Alberola-Ila J., Roberts R.W. A general approach to detect protein expression in vivo using fluorescent puromycin conjugates. Chem Biol. 2004;11(7):999–1008. doi: 10.1016/j.chembiol.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 24.Nemoto N., Miyamoto-Sato E., Yanagawa H. Fluorescence labeling of the C-terminus of proteins with a puromycin analogue in cell-free translation systems. FEBS Lett. 1999;462(1–2):43–46. doi: 10.1016/S0014-5793(99)01474-X. [DOI] [PubMed] [Google Scholar]
- 25.Aviner R., Geiger T., Elroy-Stein O. Novel proteomic approach (PUNCH-P) reveals cell cycle-specific fluctuations in mRNA translation. Genes Dev. 2013;27(16):pp. doi: 10.1101/gad.219105.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Creus-Muncunill J. Increased translation as a novel pathogenic mechanism in Huntington’s disease. Brain. 2019;142(10):3158–3175. doi: 10.1093/brain/awz230. [DOI] [PubMed] [Google Scholar]
- 27.Hidalgo San Jose L., Signer R.A.J. Cell-type-specific quantification of protein synthesis in vivo. Nat Protoc. 2019;14(2):441–460. doi: 10.1038/s41596-018-0100-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barrett R.M., Liu H.-W.W., Jin H., Goodman R.H., Cohen M.S. Cell-specific profiling of nascent proteomes using orthogonal enzyme-mediated puromycin incorporation. ACS Chem Biol. 2016;11(6):1532–1536. doi: 10.1021/acschembio.5b01076. [DOI] [PubMed] [Google Scholar]
- 29.Cary G.A. Identification and characterization of a drug-sensitive strain enables puromycin-based translational assays in Saccharomyces cerevisiae. Yeast. 2014;31(5):167–178. doi: 10.1002/yea.3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vara J.A., Portela A., Ortín J., Jiménez A. Expression in mammalian cells of a gene from Streptomyces alboniger conferring puromycin resistance. Nucl Acids Res. 1986;14(11):4617–4624. doi: 10.1093/nar/14.11.4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hernández-Damián J. Downregulation of SnoN oncoprotein induced by antibiotics anisomycin and puromycin positively regulates transforming growth factor-β signals. Biochim Biophys Acta. 2013;1830(11):5049–5058. doi: 10.1016/j.bbagen.2013.07.006. [DOI] [PubMed] [Google Scholar]
- 32.Guo R., Lee Y., Byrnes C., Miller J. Puromycin selection confounds the RNA-Seq profiles of primary human erythroblasts. Transcr Open Access. 2017;05(01):pp. doi: 10.4172/2329-8936.1000140. [DOI] [Google Scholar]
- 33.Steitz T.A. A structural understanding of the dynamic ribosome machine. Nat Rev Mol Cell Biol. 2008;9(3):242–253. doi: 10.1038/nrm2352. [DOI] [PubMed] [Google Scholar]
- 34.Scheper G.C., van der Knaap M.S., Proud C.G. Translation matters: protein synthesis defects in inherited disease. Nat Rev Genet. 2007;8(9):711–723. doi: 10.1038/nrg2142. [DOI] [PubMed] [Google Scholar]
- 35.Wool I.G., Kurihara K. Determination of the number of active muscle ribosomes: effect of diabetes and insulin. Proc Natl Acad Sci USA. 1967;58(6):2401–2407. doi: 10.1073/pnas.58.6.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakano K., Hara H. Measurement of the protein-synthetic activity in vivo of various tissues in rats by using [3H]Puromycin. Biochem J. 1979;184(3):663–668. doi: 10.1042/bj1840663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Argüello R.J. SunRiSE – measuring translation elongation at single-cell resolution by means of flow cytometry. J Cell Sci. 2018;131(10) doi: 10.1242/jcs.214346. [DOI] [PubMed] [Google Scholar]
- 38.Signer R.A.J., Magee J.A., Salic A., Morrison S.J. Haematopoietic stem cells require a highly regulated protein synthesis rate. Nature. 2014;509(7498):49–54. doi: 10.1038/nature13035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lelouard H. Dendritic cell aggresome-like induced structures are dedicated areas for ubiquitination and storage of newly synthesized defective proteins. J Cell Biol. 2004;164(5):667–675. doi: 10.1083/jcb.200312073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chau K.F. Downregulation of ribosome biogenesis during early forebrain development. Elife. 2018;7 doi: 10.7554/eLife.36998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Costa R.O. Synaptogenesis stimulates a proteasome-mediated ribosome reduction in axons. Cell Rep. 2019;28(4):864–876.e6. doi: 10.1016/j.celrep.2019.06.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.tom Dieck S. Direct visualization of newly synthesized target proteins in situ. Nat Methods. 2015;12(5):411–414. doi: 10.1038/nmeth.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hafner A.-S., Donlin-Asp P.G., Leitch B., Herzog E., Schuman E.M. Local protein synthesis is a ubiquitous feature of neuronal pre- and postsynaptic compartments. Science. 2019;364(6441):pp. doi: 10.1126/science.aau3644. [DOI] [PubMed] [Google Scholar]
- 44.Aviner R., Geiger T., Elroy-Stein O. Genome-wide identification and quantification of protein synthesis in cultured cells and whole tissues by puromycin-associated nascent chain proteomics (PUNCH-P) Nat Protoc. 2014;9(4):pp. doi: 10.1038/nprot.2014.051. [DOI] [PubMed] [Google Scholar]
- 45.Aviner R., Shenoy A., Elroy-Stein O., Geiger T. Uncovering hidden layers of cell cycle regulation through integrative multi-omic analysis. PLoS Genet. 2015;11(10):pp. doi: 10.1371/journal.pgen.1005554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ha S.W., Ju D., Hao W., Xie Y. Rapidly translated polypeptides are preferred substrates for cotranslational protein degradation. J Biol Chem. 2016;291(18):9827–9834. doi: 10.1074/jbc.M116.716175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Forester C.M. Revealing nascent proteomics in signaling pathways and cell differentiation. Proc Natl Acad Sci USA. 2018;115(10):2353–2358. doi: 10.1073/pnas.1707514115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McClatchy D.B. Pulsed azidohomoalanine labeling in mammals (PALM) detects changes in liver-specific LKB1 knockout mice. J Proteome Res. 2015;14(11):4815–4822. doi: 10.1021/acs.jproteome.5b00653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miyamoto-Sato E. Specific bonding of puromycin to full-length protein at the C-terminus. Nucl Acids Res. 2000;28(5):1176–1182. doi: 10.1093/nar/28.5.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ohashi H., Ishizaka M., Hirai N., Miyamoto-Sato E. Efficiency of puromycin-based technologies mediated by release factors and a ribosome recycling factor. Protein Eng Des Sel. 2013;26(8):533–537. doi: 10.1093/protein/gzt031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kawahashi Y. High-throughput fluorescence labelling of full-length cDNA products based on a reconstituted translation system. J Biochem. 2007;141(1):19–24. doi: 10.1093/jb/mvm003. [DOI] [PubMed] [Google Scholar]
- 52.Doi N. Novel fluorescence labeling and high-throughput assay technologies for in vitro analysis of protein interactions. Genome Res. 2002;12(3):487–492. doi: 10.1101/gr.218802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nemoto N., Miyamoto-Sato E., Husimi Y., Yanagawa H. In vitro virus: bonding of mRNA bearing puromycin at the 3′-terminal end to the C-terminal end of its encoded protein on the ribosome in vitro. FEBS Lett. 1997;414(2):405–408. doi: 10.1016/S0014-5793(97)01026-0. [DOI] [PubMed] [Google Scholar]
- 54.Tao S.-C., Zhu H. Protein chip fabrication by capture of nascent polypeptides. Nat Biotechnol. 2006;24(10):1253–1254. doi: 10.1038/nbt1249. [DOI] [PubMed] [Google Scholar]
- 55.DeCaprio J., Kohl T.O. Pulse-chase labeling of protein antigens with [35S]methionine. Cold Spring Harb Protoc. 2018;2018(9) doi: 10.1101/pdb.prot098525. pdb.prot098525. [DOI] [PubMed] [Google Scholar]
- 56.Dieterich D.C., Lee J.J., Link A.J., Graumann J., Tirrell D.A., Schuman E.M. Labeling, detection and identification of newly synthesized proteomes with bioorthogonal non-canonical amino-acid tagging. Nat Protoc. 2007;2(3):532–540. doi: 10.1038/nprot.2007.52. [DOI] [PubMed] [Google Scholar]
- 57.Dieterich D.C. In situ visualization and dynamics of newly synthesized proteins in rat hippocampal neurons. Nat Neurosci. 2010;13(7):897–905. doi: 10.1038/nn.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ingolia N.T., Brar G.A., Rouskin S., McGeachy A.M., Weissman J.S. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat Protoc. 2012;7(8):1534–1550. doi: 10.1038/nprot.2012.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schwanhäusser B., Gossen M., Dittmar G., Selbach M. Global analysis of cellular protein translation by pulsed SILAC. Proteomics. 2009;9(1):205–209. doi: 10.1002/pmic.200800275. [DOI] [PubMed] [Google Scholar]
- 60.Howden A.J.M. QuaNCAT: quantitating proteome dynamics in primary cells. Nat Methods. 2013;10(4):343–346. doi: 10.1038/nmeth.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Seedhom M.O., Hickman H.D., Wei J., David A., Yewdell J.W. Protein translation activity: a new measure of host immune cell activation. J Immunol. 2016;197(4):1498–1506. doi: 10.4049/jimmunol.1600088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ganassi M. A surveillance function of the HSPB8-BAG3-HSP70 chaperone complex ensures stress granule integrity and dynamism. Mol Cell. 2016;63(5):796–810. doi: 10.1016/j.molcel.2016.07.021. [DOI] [PubMed] [Google Scholar]
- 63.Mediani L. Defective ribosomal products challenge nuclear function by impairing nuclear condensate dynamics and immobilizing ubiquitin. EMBO J. 2019;38(15):1–19. doi: 10.15252/embj.2018101341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Uozumi N., Matsumoto H., Saitoh H. Detection of O-propargyl-puromycin with SUMO and ubiquitin by click chemistry at PML-nuclear bodies during abortive proteasome activities. Biochem Biophys Res Commun. 2016;474(2):247–251. doi: 10.1016/j.bbrc.2016.03.155. [DOI] [PubMed] [Google Scholar]
- 65.Maquat L.E., Tarn W.-Y., Isken O. The pioneer round of translation: features and functions. Cell. 2010;142(3):368–374. doi: 10.1016/j.cell.2010.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Martins R.P. Nuclear processing of nascent transcripts determines synthesis of full-length proteins and antigenic peptides. Nucl Acids Res. 2019;47(6):3086–3100. doi: 10.1093/nar/gky1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Apcher S., Millot G., Daskalogianni C., Scherl A., Manoury B., Fåhraeus R. Translation of pre-spliced RNAs in the nuclear compartment generates peptides for the MHC class I pathway. Proc Natl Acad Sci USA. 2013;110(44):17951–17956. doi: 10.1073/pnas.1309956110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Agranoff B.W., Klinger P.D. Puromycin effect on memory fixation in the goldfish. Science. 1964;146(3646):952–953. doi: 10.1126/science.146.3646.952. [DOI] [PubMed] [Google Scholar]
- 69.Flexner J.B., Flexner L.B., Stellar E. Memory in mice as affected by intracerebral puromycin. Science. 1963;141(3575):57–59. doi: 10.1126/science.141.3575.57. [DOI] [PubMed] [Google Scholar]
- 70.Zhu P.J. Activation of the ISR mediates the behavioral and neurophysiological abnormalities in Down syndrome. Science. 2019;366(6467):843–849. doi: 10.1126/science.aaw5185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Santini E. Exaggerated translation causes synaptic and behavioural aberrations associated with autism. Nature. 2013;493(7432):411–415. doi: 10.1038/nature11782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gonatopoulos-Pournatzis T. Autism-misregulated eIF4G microexons control synaptic translation and higher order cognitive functions. Mol Cell. 2020 doi: 10.1016/j.molcel.2020.01.006. [DOI] [PubMed] [Google Scholar]
- 73.Koren S.A. Tau drives translational selectivity by interacting with ribosomal proteins. Acta Neuropathol. 2019;137(4):571–583. doi: 10.1007/s00401-019-01970-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Meier S. Pathological tau promotes neuronal damage by impairing ribosomal function and decreasing protein synthesis. J Neurosci. 2016;36(3):1001–1007. doi: 10.1523/JNEUROSCI.3029-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Papanikolopoulou K. Drosophila tau negatively regulates translation and olfactory long-term memory, but facilitates footshock habituation and cytoskeletal homeostasis. J Neurosci. 2019;39(42):8315–8329. doi: 10.1523/JNEUROSCI.0391-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Holt C.E., Martin K.C., Schuman E.M. Local translation in neurons: visualization and function. Nat Struct Mol Biol. 2019;26(7):557–566. doi: 10.1038/s41594-019-0263-5. [DOI] [PubMed] [Google Scholar]
- 77.Graber T.E. Reactivation of stalled polyribosomes in synaptic plasticity. Proc Natl Acad Sci USA. 2013;110(40):16205–16210. doi: 10.1073/pnas.1307747110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Batista A.F.R., Martínez J.C., Hengst U. Intra-axonal synthesis of SNAP25 is required for the formation of presynaptic terminals. Cell Rep. 2017;20(13):3085–3098. doi: 10.1016/j.celrep.2017.08.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Terenzio M. Locally translated mTOR controls axonal local translation in nerve injury. Science. 2018;359(6382):1416–1421. doi: 10.1126/science.aan1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Langille J.J., Ginzberg K., Sossin W.S. Polysomes identified by live imaging of nascent peptides are stalled in hippocampal and cortical neurites. Learn Mem. 2019;26(9):351–362. doi: 10.1101/lm.049965.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cioni J.M. Late endosomes act as mRNA translation platforms and sustain mitochondria in axons. Cell. 2019;176(1–2) doi: 10.1016/j.cell.2018.11.030. 56–72.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Biever A. Monosomes actively translate synaptic mRNAs in neuronal processes. Science. 2020;367(6477) doi: 10.1126/science:aay4991. [DOI] [PubMed] [Google Scholar]
- 83.Davis A.M., Plowright A.T., Valeur E. Directing evolution: the next revolution in drug discovery? Nat Rev Drug Discov. 2017;16(10):681–698. doi: 10.1038/nrd.2017.146. [DOI] [PubMed] [Google Scholar]
- 84.Bashiruddin N.K., Suga H. Construction and screening of vast libraries of natural product-like macrocyclic peptides using in vitro display technologies. Curr Opin Chem Biol. 2015;24:131–138. doi: 10.1016/j.cbpa.2014.11.011. [DOI] [PubMed] [Google Scholar]
- 85.Zorzi A., Deyle K., Heinis C. Cyclic peptide therapeutics: past, present and future. Curr Opin Chem Biol. 2017;38:24–29. doi: 10.1016/j.cbpa.2017.02.006. [DOI] [PubMed] [Google Scholar]
- 86.Howell S.M. Serum stable natural peptides designed by mRNA display. Sci Rep. 2014;4:6008. doi: 10.1038/srep06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Koide S., Koide A., Lipovšek D. Target-binding proteins based on the 10th human fibronectin type III domain (10Fn3) Methods Enzymol. 2012;503:135–156. doi: 10.1016/B978-0-12-396962-0.00006-9. [DOI] [PubMed] [Google Scholar]
- 88.Olson C.A. Single-round, multiplexed antibody mimetic design through mRNA display. Angew Chem Int Ed Engl. 2012;51(50):12449–12453. doi: 10.1002/anie.201207005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tateyama S., Horisawa K., Takashima H., Miyamoto-Sato E., Doi N., Yanagawa H. Affinity selection of DNA-binding protein complexes using mRNA display. Nucl Acids Res. 2006;34(3) doi: 10.1093/nar/gnj025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nguyen-Trung N.Q., Botta O., Terenzi S., Strazewski P. A practical route to 3′-amino-3′-deoxyadenosine derivatives and puromycin analogues. J Org Chem. 2003;68(5):2038–2041. doi: 10.1021/jo026627c. [DOI] [PubMed] [Google Scholar]
- 91.Eigner S. Imaging of protein synthesis: in vitro and in vivo evaluation of (44)Sc-DOTA-puromycin. Mol Imag Biol. 2013;15(1):79–86. doi: 10.1007/s11307-012-0561-3. [DOI] [PubMed] [Google Scholar]
- 92.Betts H.M. Synthesis, in vitro evaluation, and radiolabeling of fluorinated puromycin analogues: potential candidates for PET imaging of protein synthesis. J Med Chem. 2016;59(20):9422–9430. doi: 10.1021/acs.jmedchem.6b00968. [DOI] [PubMed] [Google Scholar]
- 93.Borowsky B., Kessner D., Recant L. Structural analogues of puromycin in production of experimental nephrosis in rats. Proc Soc Exp Biol Med. 1958;97(4):857–860. doi: 10.3181/00379727-97-23900. [DOI] [PubMed] [Google Scholar]
- 94.Bhutani N., Venkatraman P., Goldberg A.L. Puromycin-sensitive aminopeptidase is the major peptidase responsible for digesting polyglutamine sequences released by proteasomes during protein degradation. EMBO J. 2007;26(5):1385–1396. doi: 10.1038/sj.emboj.7601592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Singh R., Williams J., Vince R. Puromycin based inhibitors of aminopeptidases for the potential treatment of hematologic malignancies. Eur J Med Chem. 2017;139:325–336. doi: 10.1016/j.ejmech.2017.07.048. [DOI] [PubMed] [Google Scholar]
- 96.Jung J.H. p53-dependent apoptotic effect of puromycin via binding of ribosomal protein L5 and L11 to MDM2 and its combination effect with RITA or doxorubicin. Cancers (Basel) 2019;11(4) doi: 10.3390/cancers11040582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ueki N., Lee S., Sampson N.S., Hayman M.J. Selective cancer targeting with prodrugs activated by histone deacetylases and a tumour-associated protease. Nat Commun. 2013;4:2735. doi: 10.1038/ncomms3735. [DOI] [PubMed] [Google Scholar]
- 98.Ueki N., Wang W., Swenson C., McNaughton C., Sampson N.S., Hayman M.J. Synthesis and preclinical evaluation of a highly improved anticancer prodrug activated by histone Deacetylases and Cathepsin L. Theranostics. 2016;6(6):808–816. doi: 10.7150/thno.13826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Marciano R., Leprivier G., Rotblat B. Puromycin labeling does not allow protein synthesis to be measured in energy-starved cells. Cell Death Dis. 2018;9(2):4–6. doi: 10.1038/s41419-017-0056-x. [DOI] [PMC free article] [PubMed] [Google Scholar]