Abstract
Background
Atypical teratoid/rhabdoid tumors (ATRTs) are known to exhibit molecular and clinical heterogeneity even though SMARCB1 inactivation is the sole recurrent genetic event present in nearly all cases. Indeed, recent studies demonstrated 3 molecular subgroups of ATRTs that are genetically, epigenetically, and clinically distinct. As these studies included different numbers of tumors, various subgrouping techniques, and naming, an international working group sought to align previous findings and to reach a consensus on nomenclature and clinicopathological significance of ATRT subgroups.
Methods
We integrated various methods to perform a meta-analysis on published and unpublished DNA methylation and gene expression datasets of ATRTs and associated clinicopathological data.
Results
In concordance with previous studies, the analyses identified 3 main molecular subgroups of ATRTs, for which a consensus was reached to name them ATRT-TYR, ATRT-SHH, and ATRT-MYC. The ATRT-SHH subgroup exhibited further heterogeneity, segregating further into 2 subtypes associated with a predominant supratentorial (ATRT-SHH-1) or infratentorial (ATRT-SHH-2) location. For each ATRT subgroup we provide an overview of its main molecular and clinical characteristics, including SMARCB1 alterations and pathway activation.
Conclusions
The introduction of a common classification, characterization, and nomenclature of ATRT subgroups will facilitate future research and serve as a common ground for subgrouping patient samples and ATRT models, which will aid in refining subgroup-based therapies for ATRT patients.
Keywords: ATRT, consensus, meta-analysis, molecular subgroups
Key Points.
Meta-analyses confirmed the presence of 3 distinct molecular subgroups of ATRT.
Consensus was reached to name them ATRT-TYR, ATRT-SHH, and ATRT-MYC
Overview is presented of molecular and clinical characteristics of each subgroup.
Importance of the Study.
The international consensus on number and naming of ATRT molecular subgroups and their main characteristics, which we present here, will be important for the design of future clinical trials, patient stratification, and a uniform classification of patients’ tumor samples, much in line as it has been for medulloblastoma, ependymoma, and high-grade glioma. It will also be essential for a better interpretation of preclinical experiments using properly classified in vitro and in vivo ATRT models.
Atypical teratoid/rhabdoid tumors (ATRTs) arise in all compartments of the central nervous system (CNS), predominantly affect infants or young children, and display a remarkably simple cancer genome. Biallelic mutation—including partial or whole loss of chromosome 22, resulting in inactivation of SWItch/sucrose nonfermentable (SWI/SNF) related, matrix associated, actin dependent regulator of chromatin, subfamily B1 (SMARCB1)—is the main, and often only, recurrent molecular feature seen in ATRT.1 Rare cases (<5%) with an intact SMARCB1 harbor mutations in SMARCA4, both encoding components of the SWI/SNF chromatin remodeling complex.2,3 The recurrent loss of SMARCB1 in these tumors is in stark contrast to a pleomorphic histology and considerable molecular and clinical heterogeneity observed in ATRT cohorts. Therapeutic strategies in ATRT are largely influenced by the age of the patient, tumor location in the CNS, and disease stage at diagnosis.4 These factors inform extent of surgical resection and various radiological and chemotherapeutic interventions. However, there is currently no international consensus on standard therapeutic approaches, with the majority of therapeutic and survival data being published on a center-by-center basis. Numbers of patients with ATRT are small, therefore concerted international efforts to evaluate and standardize therapy are ongoing. However, it is as yet unclear how tumor biology shapes the response to treatment, outcome, and/or long-term effects in patients with ATRT. A critical step toward improving the poor outlook for these patients is therefore to define and characterize the biological heterogeneity in ATRT such that a standard subgrouping scheme is available and can be further used to investigate subgroup-specific features of ATRT and inform subgroup-specific therapies.
Earlier attempts to subgroup ATRT at the transcriptomic level had already recognized a degree of heterogeneity but were limited by a small cohort size.5 More recently, international efforts to collect and profile significantly larger cohorts of ATRTs have resulted in the identification of distinct ATRT subgroups defined by gene expression and/or DNA methylation profiling and associated with different molecular and clinicopathological features (Table 1).6–8 Since the number of subgroups and platforms used to identify these subgroups differed between studies, there is an urgent need to align findings and define the number, molecular and clinical features, and a common nomenclature for ATRT subgroups.
Table 1.
Summary of defining transcriptional features of ATRT subgroups. Data derived from publications8–10
| Study | Subgroups | Methods/Platforms Used (n = # cases) | ||
|---|---|---|---|---|
| Torchia et al, 2015 | Group 1 Overexpression of ASCL1 | Group 2 | Immunohistochemistry (ASCL1) (n = 170) Gene expression array profiling (IIlumina HT12) (n = 43) | |
| Torchia et al, 2016 | Group 1 Overexpression of Notch pathway genes ASCL1, CBL, HES1 | Group 2A Overexpression of neuronal and mesenchymal genes OTX2, PDGFRB, BMP4 | Group 2B Overexpression of HOX cluster genes | Methylation array profiling (Illumina 450K) (n = 162) Gene expression array profiling (IIlumina HT12) (n = 90) |
| Johann et al, 2016 | ATRT-SHH Overexpression of SHH pathway genes GLI2, BOC, PTCHD2, MYCN | ATRT-TYR Overexpression of melanosomal genes TYR, TYRP, MITF, OTX2 | ATRT-MYC Overexpression of MYC and HOX cluster genes | Methylation array profiling (Illumina 450K) (n = 150) Gene expression array profiling (Affymetrix U133 Plus 2.0) (n = 69) |
| Han et al, 2016 | hIC2 Overexpression of ASCL1, BOC, SOX2, GLI2, FABP7 | hIC1 Overexpression of BMP4, OTX2, SMAD7 | hIC3 Overexpression of ACTL6A, FABP7, GFAP | Gene expression array profiling (Affymetrix U133 Plus 2.0) (n = 30) |
To this end, we performed a meta-analysis of previously published and additional ATRT DNA methylation profiles with parallel transcriptomic and clinicopathological data in order to generate a consensus definition and naming for ATRT subgroups and to define their main molecular and clinicopathological characteristics.
Materials and Methods
Integrated Analyses of ATRT Profiling Data
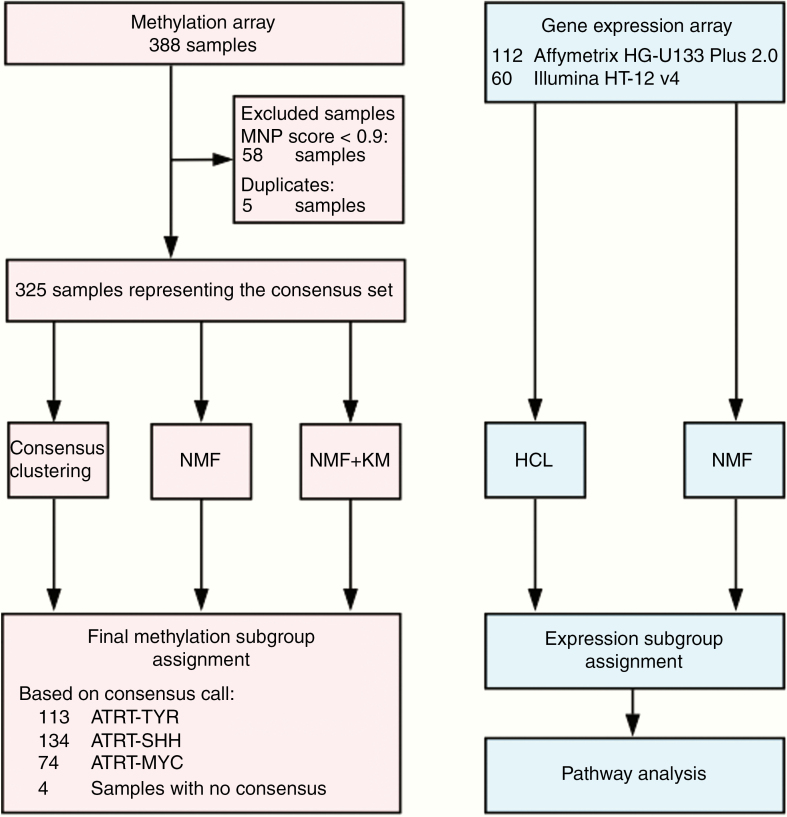
Due to the variety of data types and platforms used previously to subgroup ATRT, we first created a composite dataset of all cases (n = 388), profiled using the Illumina Infinium HumanMethylation 450K or EPIC array. We excluded all samples (n = 5) that were either duplicates or relapse cases. To exclude cases with a low tumor content or outliers for which a high-confidence classification of ATRT could not be achieved, we removed all samples (n = 58) with a calibrated score <0.9 using the Heidelberg brain tumor classifier published by Capper et al9 (www.molecularneuropathology.org). This filtering step aimed to identify potential outlier samples and generated a high-quality reference dataset for classification of subgroups. A number of factors could contribute to a sample failing to be classified as ATRT with high confidence, including high nonneoplastic cell content and low-quality tumor material from archival samples.
Of the remaining 325 samples, 137 had been published by Johann et al8 and 96 by Torchia et al,7 and 92 are newly added unpublished samples from the Northern Institute of Cancer Research (Newcastle University) (Gene Expression Omnibus [GEO] accession no. GSE141363) and the EURHAB study (GEO accession no. GSE141039) (Supplementary Table 1). Informed consent was obtained for all cases. In order to determine consensus subgroups, methylation array data were subjected to 3 different clustering methods, including consensus nonnegative matrix factorization (NMF),10 regular NMF,6,7 and unsupervised consensus clustering8 (see Supplementary Methods for full technical details). Algorithms chosen had either been previously applied to discover ATRT subgroups or used in consensus subgrouping studies for other CNS tumors (ie, medulloblastoma [MB]11).
Consensus calls were established by comparison of calls from the 3 different methods, and consensus subgrouping was based on at least equivalent calls from 2 of the 3 methods. A “no consensus call” was assigned in 4 cases. As an additional validation step, we corroborated consensus calls using t-distributed stochastic neighbor embedding (t-SNE) analysis of the consensus dataset.
To corroborate DNA methylation–based classification, we also reanalyzed published Affymetrix HG-U133 Plus 2.0 expression profiles (n = 97)8,12 and Illumina HT12 v4 gene expression array data (n = 60)6 (see Supplementary Methods for further information).
Results
DNA Methylation and Gene Expression Profiling Identify 3 Main Subgroups of ATRT
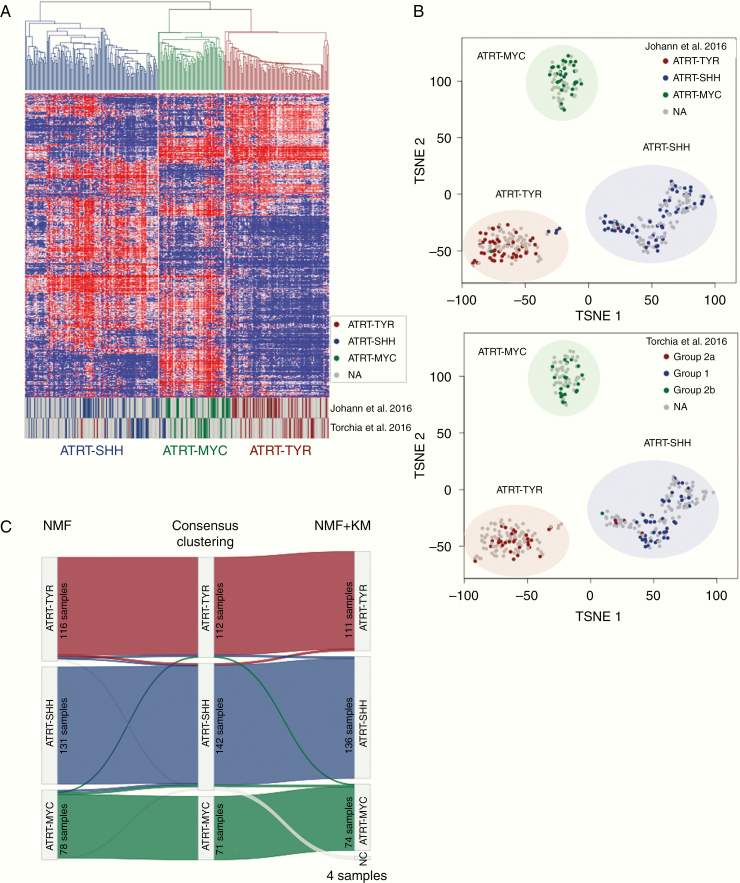
Robust DNA methylation data from 325 unique ATRT cases (Fig. 1) were classified using 3 independent clustering algorithms (consensus NMF, regular NMF, ConsensusClusterPlus) to define consensus subgroups of ATRTs. Each method was applied considering a number of possible subgroups between 2 and 8. Clustering metrics (including but not limited to cophenetic coefficients, change in area under the cumulative distribution function curve, dispersion, kappa, silhouette score; Supplementary Fig. 1) indicated that the most consistently robust clustering solution was 3 subgroups with possible further subclusters identified, as previously discussed by Johann et al,8 but supported less robustly here (Fig. 2, Supplementary Fig. 1). There was very high concordance between subgrouping based on consensus clustering (97%, 316 cases correctly classified), NMF (98%, 318 cases correctly classified), and consensus NMF (99%, 321 cases correctly classified), as shown in the Sankey plot in Fig. 2C. Notably, adding back samples excluded for low classification calls did not alter number of subgroups, as indicated in the t-SNE analysis and cluster metrics for the methylation array analyses (Supplementary Fig. 2).
Fig. 1.
Overview flow charts on all analyses and samples.
Fig. 2.
Methylation array analysis of the consensus dataset. (A) Unsupervised hierarchical clustering using the top 5000 most variable cytosine-guanine (CG) sites confirms the presence of 3 subgroups in the consensus dataset (325 samples). (B) t-SNE visualization of the analyzed dataset based on the 5000 most variable CG sites reproduces segregation into 3 main ATRT subgroups. Coloring of data points in the t-SNE plots displays the subgrouping as published by Johann et al8 (upper plot) or by Torchia et al7 (lower plot). Half transparent circles show the consensus subgroups used in this paper. (C) Sankey plot displaying the concordance between the subgroup calls using different methods (NMF, consensus clustering, and NMF + k-means based subgrouping). Numbers in each subgroup show the number of samples which have been assigned to the subgroup with the respective method.
Given that each clustering method consistently identified 3 main subgroups of ATRTs, with a high degree (>90%) of concordance between the different methods applied (Fig. 2), we chose this as the basis of our consensus subgrouping. As shown in Fig. 2A, B, Group 1 annotated cases from the Torchia et al study7 formed one group with the ATRT-SHH cases from the Johann et al study,8 while Group 2A or Group 2B annotated cases form groups with either ATRT-TYR or ATRT-MYC cases, respectively, indicating that our previous studies identified largely the same 3 subgroups. We have designated the 3 subgroups as ATRT-SHH, ATRT-TYR, and ATRT-MYC, based on the nomenclature proposed by Johann et al.8
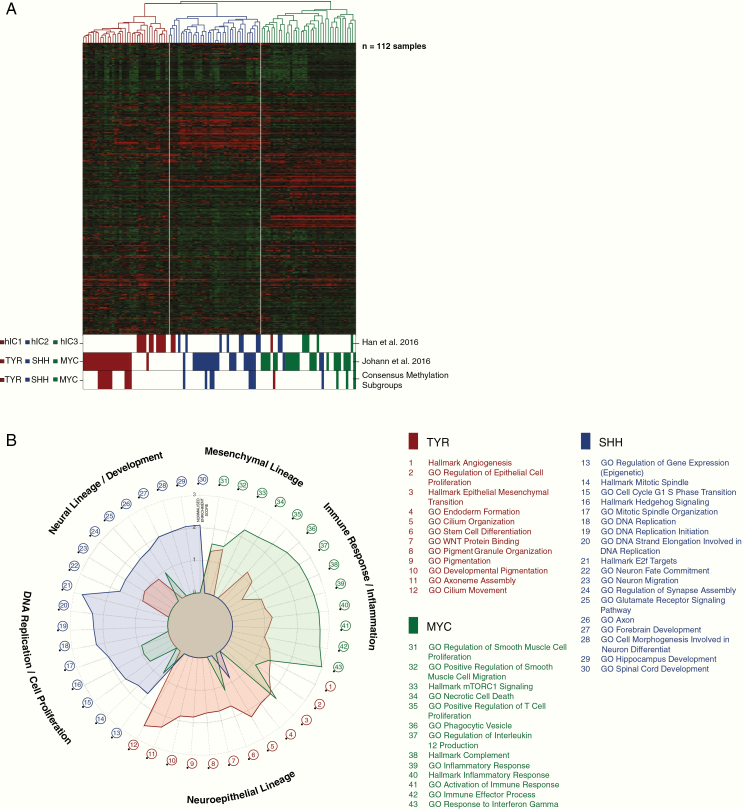
We next analyzed ATRT gene expression profiles available for a total of 172 cases, profiled on Affymetrix HG-U133 Plus 2.0 arrays (112, including 15 new cases) or Illumina HT-12 v4 arrays (60 cases).6 For 21 Affymetrix cases and for 48 Illumina profiled tumors there were matching DNA methylation data. Unsupervised hierarchical clustering analysis of the Affymetrix data using expression of the 1500 most variable genes confirmed the presence of 3 major molecular subgroups of ATRTs in line with DNA methylation analyses (Fig. 3A). These results remained stable across different numbers of differentially expressed genes (data not shown). Subgroup annotations were also highly concordant with prior publications where subsets of these data have been analyzed8,12; annotated cases of human intracranial 1 (hIC1) largely overlapped with the ATRT-SHH cases (8/10), while hIC2 and hIC3 annotated cases largely overlapped with ATRT-TYR cases (10/12) and ATRT-MYC cases (5/5), respectively (Fig. 3A). For the 21 cases with matched Affymetrix gene expression and DNA methylation data there was also good concordance, with only 2 samples annotated to different subgroups. We assigned final annotation of these 2 cases based on the DNA methylation data. Similarly, unsupervised hierarchical clustering of the Illumina gene expression data also revealed 3 molecular subgroups of ATRTs with a good concordance (96%) between DNA methylation array and Illumina based subgrouping (Supplementary Fig. 3). It is worth noting that the clustering metrics derived from the Affymetrix data also supported 3 main and not additional subgroups (Supplementary Fig. 4B).
Fig. 3.
Cluster analysis based on Affymetrix array gene expression data. (A) Unsupervised hierarchical clustering using the top 1500 most variable genes in the consensus gene expression dataset. Annotations in the lower bar show the grouping as presented by Han et al12 and by Johann et al8 and the current methylation consensus calls. (B) Visualization of gene set enrichment analysis results as a radar plot. Axis displays the normalized enrichment values. Each ATRT subgroup is represented in the respective subgroup color.
Finally, in order to gain further insights into the biology of the subgroups, we performed ingenuity pathway and gene set enrichment analyses, for which the normalized enrichment scores are shown in Fig. 3B as a radar plot. Overall, the ATRT-SHH subgroup displayed a low overlap of enriched gene sets with ATRT-TYR and ATRT-MYC, but there was some overlap between ATRT-TYR and ATRT-MYC—in particular for gene sets related to immune response. The specific gene enrichment features as well as comments on known published genes for each subgroup are described below.
Molecular and Clinical Features of the 3 Subgroups
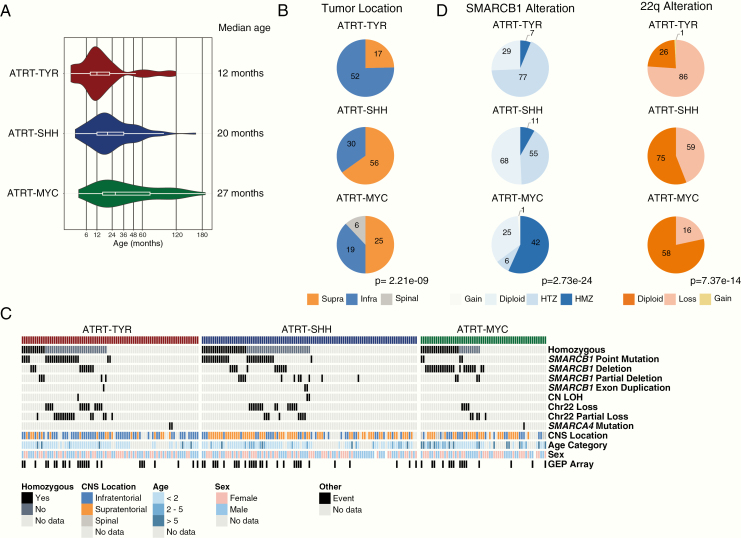
Having identified and confirmed the presence of 3 main molecular subgroups of ATRTs, we examined available pooled molecular and clinicopathological data of all cases to define the main characteristics of each group as described below and shown in Fig. 4 (and an overview of cytogenetic aberrations is in Supplementary Fig. 5).
Fig. 4.
Clinical and genetic associations of ATRT subgroups. (A) Violin plots of age distribution in ATRT subgroups. (B) Frequency of CNS tumor location by consensus subgroup represented by pie charts. (C) Oncoprint Fig. displays the distribution of various types of SMARCB1 mutations among the consensus set. (D) Distribution of SMARCB1 alterations and chr22 changes in the consensus set as determined by methylation array analyses, represented by pie charts. Significance between subgroups was calculated with chi-square test.
Correlations of the 3 Subgroups with Published ATRT Models
In the last years, a number of cell lines and genetically engineered mouse models have been established to model ATRT tumorigenesis. In order to see how these match our consensus human ATRT subgrouping, we collected RNAseq and gene expression data from previously published studies12–14 and performed a multidimensional scaling (MDS) analysis, sample-wise correlation between cell lines and mouse models against human ATRT samples (Supplementary Fig. 6A, C), and unsupervised hierarchical clustering analysis on the combined human and mouse datasets using orthologous genes (Supplementary Fig. 6B). Results of this preclinical model characterization are discussed below in the respective sections.
ATRT-TYR
The ATRT-TYR subgroup was named after the enzyme tyrosinase, which is highly overexpressed in most ATRT-TYR cases, but not in the other ATRT subgroups or other brain tumors, indicating it may be a good diagnostic marker for ATRT-TYR cases.15 The protein physiologically catalyzes the synthesis of melanin in melanocytes and is an important protagonist in neural tube development.16 Although the role of TYR in ATRT tumorigenesis remains to be established, it is notable that several other components of the melanosomal pathway, including the tyrosinase-related protein TYRP and the melanoma-oncogene MITF, are also upregulated in this subgroup, potentially reflecting restricted neuroectodermal origins.17
Other pathways and genes upregulated in ATRT-TYR tumors include the bone morphogenetic protein (BMP) pathway (eg, BMP4) and developmentally related transcription factors such as orthodenticle homeobox 2 (OTX2) (Supplementary Fig. 4A). Gene set enrichment analysis performed on the differentially overexpressed genes confirms the melanosomal pathway and tyrosine metabolism (Fig. 3B, “GO [developmental] Pigmentation,” “GO Pigment granule organization”), as well as epithelial proliferation as being enriched in ATRT-TYR (Fig. 3B). Although comprehensive histopathological studies of ATRT molecular subtypes remain pending, it is notable that cribriform neuroectodermal tumors (CRINETs), which also all express TYR, have DNA methylation profiles that are highly similar to ATRT-TYR tumors,18 suggesting that CRINET and ATRT-TYR tumors may represent 2 histological variants with a common cell of origin. Whether the favorable outcomes of CRINET patients18 also apply to patients with ATRT-TYR tumors remains to be investigated.
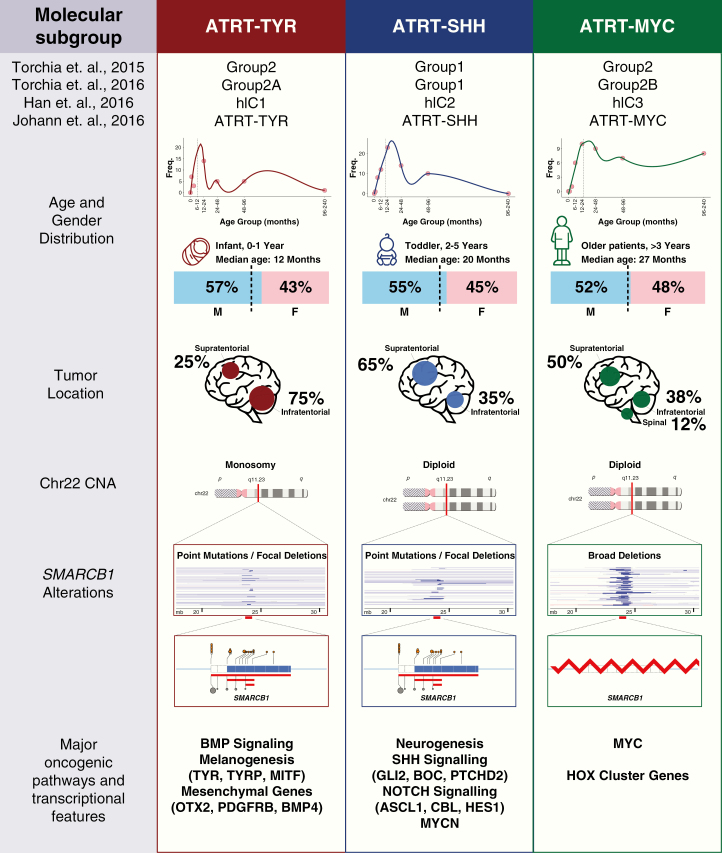
Genetically, the prototypic type of biallelic SMARCB1 inactivation in the ATRT-TYR group is whole or partial loss of one copy of chromosome 22 accompanied by an inactivating (eg, point) mutation in SMARCB1 on the other allele (Fig. 4C, D). The loss of chromosome 22 was more prevalent in ATRT-TYR (86 vs 59 cases in ATRT-SHH and 16 cases in ATRT-MYC; P = 0.053, chi-square test). Investigations using assay for transposase-accessible chromatin sequencing revealed that this subgroup harbors a more open chromatin, suggestive of a more primitive epigenetic landscape compared with the other ATRT subgroups.7
Clinically, ATRT-TYR patients represent the youngest patient group, with median age at diagnosis of 12 months (range, 0–108 mo). This subgroup also contained the highest proportion of patients under 3 years of age at time of diagnosis (90% in ATRT-TYR vs 74.6% in ATRT-SHH vs 52.3% in ATRT-MYC; Fig. 4A, Supplementary Table 1). Most (75%, 52 of 69 with location data) ATRT-TYR tumors have infratentorial location, and only 25% (17 cases) are located supratentorially (Fig. 4B). This differs significantly from ATRT-SHH and ATRT-MYC, which are more often localized supratentorially (P = 2.21e-09, chi-square test). A recent radiogenomics study of ATRT molecular subgroups suggested that ATRT-TYR tumors may have MRI appearance characterized by a bandlike enhancement of contrast media.19
For preclinical studies, the number of available in vitro and in vivo models for ATRT remains very limited, and the number of models with molecular subgroup information is even more limited. In a recently published study by Brabetz et al,20 3 ATRT patient xenograft models from the SHH and MYC subgroups were included, but none of the analyzed xenografts exhibited the profile of ATRT-TYR. The study by Torchia et al7 classified 8 ATRT cell lines as 3 representing Group 1 (equivalent to ATRT-SHH) and 5 representing Group 2 (equivalent to ATRT-TYR/ATRT-MYC). However, so far it has remained unclear how many of the Group 2 cell lines represent ATRT-TYR (Group 2A) tumors and how many represent ATRT-MYC (Group 2B). Aiming to answer this question, we have analyzed the available transcriptomic data for these cell lines. For CHLA02, CHLA04, and CHLA05 the allocation to the ATRT-SHH subgroup is confirmed as expected by unsupervised hierarchical clustering and visualized in the MDS plot (Supplementary Fig. 6). For the remaining cell lines (BT12, BT16, SH, CHLA266, CHLA06), allocation to either ATRT-TYR or ATRT-MYC has so far not been performed. Projection from the MDS plot, correlation analysis, and unsupervised clustering (Supplementary Fig. 6A–C) indicate that BT12, BT16, CHLA06, CHL266, and SH can clearly be allocated to ATRT-MYC. These 5 cell lines, previously characterized as Group 2, all exhibited high expression of MYC and elevated HOX gene expression in some, but all lacked the TYR group signature (see Supplementary Fig. 6D). Taken together, these data suggest that these cell lines are likely all derived from ATRT-MYC tumors.
Additional models have been employed in several other publications investigating drug targets in ATRT xenografts, but the subgroup identity of these models remains unknown. In the cell line BT37, for instance, the role of high-mobility group adenine thymine–hook 2 (HMGA2) as an oncogene has been highlighted.21 This protein is overexpressed specifically in ATRT-TYR (Supplementary Fig. 4), suggesting that BT37 may represent an ATRT-TYR model. While numerous studies have examined drug targets in ATRT irrespective of their subgroup, knowledge of subgroup-specific vulnerabilities is sparse. Platelet derived growth factor receptor B (PDGFRB), for example, has been shown to be a drug target in Group 2 cell lines BT12, BT16, CHLA266, CHLA06, and SH, and they all displayed a higher susceptibility to the PDGFRB inhibitors nilotinib and dasatinib than did Group 1 (ATRT-SHH) cell lines. Again, whether this means that both ATRT-TYR and ATRT-MYC tumors can be targeted by these inhibitors remains to be seen, as PDGFRB expression levels in ATRT-TYR tumors are much higher than in ATRT-MYC tumors (Supplementary Fig. 4). Transcriptome analyses further suggest other promising drug targets that have or have not been tested already in rhabdoid tumors. For instance, fibroblast growth factor receptor 2 (FGFR2) is specifically upregulated in ATRT-TYR, and FGFR signaling (together with PDGFR inhibition) has been described as a vulnerability in rhabdoid tumors.22,23 Another possible candidate is Janus kinase 1 (JAK1), a protein tyrosine kinase overexpressed in ATRT-TYR that regulates the signaling cascade of JAK–signal transducers and activators of transcription (STAT). Approved inhibitors such as ruxolitinib are available and hold the promise of a possible targeted therapy. Further drug screening using robustly subgrouped cell lines will be important to determine which of the prior preclinically tested substances have subgroup specificity.
ATRT-SHH
The ATRT-SHH subgroup, in the Torchia et al publication also referred to as Group 1,7 displays an overexpression of both sonic hedgehog (SHH) and Notch pathway members, such as GLI2, PTCH1, and BOC (all SHH pathway) or ASCL1, HES1, DTX1 (all regulators of the Notch pathway; Supplementary Fig. 4A). Protein expression of achaete-scute homolog 1 (ASCL1), a neuronal differentiation transcription factor, has been suggested as an immunohistochemical marker for this subgroup.6 Moreover, Torchia et al6 showed that ASCL1 protein expression could be a biomarker for improved survival, suggesting that ASCL1-positive ATRT-SHH (Group 1) cases have a better overall survival than ASCL1-negative (Group 2) ATRTs.6 However, as ASCL1 is not expressed in all samples of the SHH subgroup and can also be expressed in some cases of the other subgroups, it remains to be seen whether patients with ATRT-SHH tumors have a better outcome than other ATRT patients. More analyses on prospective cohorts of ATRT patients are needed to see whether there are survival differences between the 3 molecular subgroups defined by DNA methylation.
Beyond the oncogenic signaling pathways, gene set enrichment analyses confirmed previous observations that ATRT-SHH is mainly a neuronally differentiated subgroup with enrichment of genes involved in “axon guidance pathways” and “neuronal system” pathways compared with other subgroups (Fig. 3B).
Torchia et al showed using small interfering RNA and gamma secretase inhibitors that the Group 1/SHH cell lines depended on Notch signaling for growth.7 However, therapeutic significance of SHH signaling for ATRT-SHH remains to be tested, as unlike SHH-activated MBs in the MB-SHH subgroup, genomic aberrations of SHH pathway members including PTCH1, SMO, and SUFU have to date not been found in any ATRT-SHH. All SHH pathway marker genes overexpressed in this subgroup (such as GLI2) are thus most likely directly or indirectly activated by the SMARCB1 loss in these tumors as reported previously.24 Why SMARCB1 loss does not activate the SHH pathway to the same extent in the other subgroups remains unknown but may be related to different cellular origins for these subgroups. Thus, whereas clear therapeutic indication for SHH pathway inhibitors has been established for SHH MB, the role of vismodegib and other Smoothened inhibitors in ATRTs is unclear and remains to be further investigated.25
Genetically, ATRT-SHH cases differ from the other 2 subgroups regarding the type of SMARCB1 alterations. Most ATRT-SHH cases display compound heterozygous point mutations (P < 0.00025, chi-square test) compared with the other groups (Fig. 4C), while homo- or heterozygous SMARCB1 deletions are less frequently found in this subgroup compared with the other groups (66 from 134 samples, 44% in ATRT-SHH vs 77% in ATRT-TYR and 64% in ATRT-MYC) (Fig. 4D). With regard to age, ATRT-SHH represent a more intermediate subgroup (median age 20 mo, range 0–96), with patients on average younger than ATRT-MYC patients and older than ATRT-TYR patients.
ATRT-SHH tumors can have either a supratentorial (56/68, 75%) or infratentorial (30/68, 35%) localization (Fig. 4B). However, as reported previously,8 DNA methylation analyses suggest a further molecular heterogeneity within the ATRT-SHH subgroup. Indeed, when performing cluster analyses for ATRT-SHH profiles only, we find that the ATRT-SHH subgroup splits up in 2 subtypes associated with either a mainly supratentorial location (ATRT-SHH-1) or a mainly infratentorial (ATRT-SHH-2) location (Supplementary Fig. 7A, B). The split into 2 subtypes is supported by consensus clustering when analyzing the SHH subgroup separately (data not shown). It is important to note that both subtypes of ATRT-SHH express marker genes from the Notch and SHH pathways. More samples and analyses are needed to investigate whether there are other molecular or clinical differences between these 2 SHH subtypes. From a radiological point of view, MRI analysis of ATRT-SHH tumors revealed that this is the only subgroup containing tumors that extend both infra- and supratentorially. Moreover, there was a lower degree of contrast enhancement in these tumors compared with ATRT-TYR and ATRT-MYC.19
With respect to in vivo models, the xenograft lines ATRT-310FH and ATRT-311FH have been classified as ATRT-SHH and may thus represent a good tool for future studies.20 Interestingly, a Rosa26CreERT2;Smarcb1flox/flox mouse model of rhabdoid tumors reported by Han et al develops spontaneous brain tumors, a subset of which have gene expression profiles very similar to that of human ATRT-SHH (shown in Supplementary Fig. 6A–C).12 Similarly, a Snf5 Flox/Flox /p53 lox/lox /GFAP-Cre (as derived from the study by Ng et al13) rhabdoid model seems to be closer to SHH than to the other subgroups. In general, development of ATRT in murine models seems to require inactivation of SMARCB1 during early embryonic (E6–E7) development, suggesting very early progenitors as cells of origin. Regarding in vitro models, the CHLA02, CHLA04, and CHLA05 cell lines all represent ATRT-SHH (formerly Group 1).7 Overexpressed drug targets that merit further investigation include the discoidin domain receptor tyrosine kinase 1 (DDR1), but also enhancer of zeste homolog 2 (EZH2), which is a candidate drug target for ATRT in general.26 In vitro studies have shown that cell lines derived from ATRT-SHH (Group 1) tumors are more sensitive to EZH2 inhibitors.7 Of note, recent epigenomic characterizations of primary ATRT suggest that EZH2 overexpression in ATRT is not accompanied by global increase of repressive mark H3K27me3,27 indicating that additional non-enzymatic functions of EZH2 may be important in ATRTs.28 Although promising preclinical data have fueled phase I trials using EZH2 inhibitors (eg, with tazemetostat, NCT02601937), it remains to be seen if this therapeutic regimen will be efficacious in the clinical setting.
ATRT-MYC
The ATRT-MYC subgroup was named based on elevated expression of the MYC oncogene as opposed to the MYCN oncogene, which is enriched in the ATRT-SHH group. However, different from other MYC or MYCN-driven pediatric brain tumors like MB-Group 3, MB-SHH, and HGG-MYCN, MYC or MYCN amplifications have not been observed respectively in ATRT-MYC and ATRT-SHH tumors. In addition, one of the most striking mRNA expression patterns in these tumors is the overexpression of several HOXC cluster genes (Fig. 3B), driven by super enhancers8 (ie, very long stretched enhancers with abundant H3K27-acetylation signal). Similar to ATRT-TYR, a broad categorization into neuronal and mesenchymal subgroups would assign these tumors a more mesenchymal expression profile.7 The typical genetic pattern that leads to SMARCB1 inactivation in these tumors is a homozygous, broad loss of SMARCB1 (which is present in 42/74 cases, 57%), covering several hundred kilobases.7,8 In contrast to ATRT-TYR or ATRT-SHH tumors, point mutations are rare in ATRT-MYC tumors (Fig. 4C).
The median age of ATRT-MYC patients is significantly higher than in the 2 other subgroups (27 mo; range, 0–190.9; Fig. 4A). This is mainly due to a number of older patients and not primarily due to a lack of very young patients in this subgroup.
Although a majority of ATRT-MYC tumors arise supratentorially (25/50, 50%), all spinal tumors in our cohort (6/50, 12%) were of the ATRT-MYC subgroup (Fig. 4B). MRI studies suggest ATRT-MYC tumors are distinguished by the presence of a strong peritumoral edema.19 Of note, recent reports have highlighted similarities between extracranial malignant rhabdoid tumor (MRT) and ATRT-MYC on the DNA methylation level.29,30 As the DNA methylation profile of tumor entities is highly reflective of the cell of origin, it raises the questions of whether these entities share common cellular origins and whether the behavior of ATRT-MYC tumors may more closely resemble extracranial rhabdoid tumors. Of note, recent studies of adult ATRT suggest that these fall mostly in the ATRT-MYC group and that further clinical and molecular heterogeneity in ATRT-MYC may be revealed.28
We have shown here that several cell lines (BT12, BT16, CHLA266, CHLA06, SH) described by Torchia et al7 (Supplementary Fig. 6C, D) exhibit features more similar to the ATRT-MYC than to the ATRT-TYR subgroup. Several xenograft models of these cell lines suitable for in vivo drug testing have been reported.20,31 Our correlative analysis of genetically engineered mouse models and ATRT primary samples also revealed that as a subset of the published tumor samples generated from P0-CreC;Smarcb1 flox/flox, mice display high correlation with ATRT-MYC samples and may thus represent a model for the ATRT-MYC subgroup (Supplementary Fig. 6A, B).14
Given the previously mentioned similarities between ATRT-MYC and a subgroup of rhabdoid kidney tumors, common molecular targets between extracranial rhabdoid tumors and subgroups may exist. In fact, Oberlick et al32 found indeed a dependency of both extracranial malignant rhabdoid tumor and ATRT cell lines on a number of rhabdoid kidney tumors, thus highlighting novel drug targets in these tumors.
Discussion
Identification of distinct molecular subgroups in an otherwise relatively genetically homogeneous disease has been a major step in further understanding the molecular heterogeneity of ATRTs (Fig. 5).6–8,12 In this study we aimed to establish a consensus regarding the number of ATRT subgroups, their main molecular and clinical characteristics, and a commonly accepted nomenclature in order to enable a better understanding of the clinical heterogeneity in ATRTs. Here, we have shown that the 3 molecular subgroups identified in previous studies based on DNA methylation and/or gene expression profiling closely match with each other and a consensus was reached to name them ATRT-TYR, ATRT-SHH, and ATRT-MYC, according to the nomenclature published by Johann et al.8 The activated genes or pathways, which were chosen for this nomenclature, emerged when performing overexpression analyses. However, their biological and therapeutic role in ATRT requires further investigation.
Fig. 5.
Consensus overview of ATRT subgroups. Schema of salient clinical and molecular characteristics of ATRT subgroups.
A consensus on number and naming of molecular subgroups has in other entities, like MB,33 ependymoma,34 and glioblastoma,35 proven to be essential for a uniform classification of patients’ tumor samples, subgroup-specific experiments using properly classified preclinical in vitro and in vivo models, and ultimately the design of clinical trials and patient stratification. A further heterogeneity within these main 3 ATRT subgroups may still exist, similar to what has been reported for MB or ependymoma subgroups, for instance.36,37 Thus far, DNA methylation profiling has identified 2 further subtypes within the ATRT-SHH subgroup, which correlates with predominant supratentorial (ATRT-SHH-1) or infratentorial (ATRT-SHH-2) locations, but larger cohort studies are needed to better define molecular differences between these subtypes and whether they are clinically relevant.
As the outcome for ATRT patients is still relatively poor, new treatment strategies are urgently needed. Identification and characterization of ATRT subgroups may help discovery of subgroup-specific treatments, but will also help to elucidate new pan-ATRT therapies. Given the reported relatedness of ATRT-MYC to extracranial rhabdoids, these investigations should also include extracranial malignant rhabdoid tumors such as those occurring in the kidney, liver, or other soft tissues.29,38 Additionally, more molecularly characterized models—including cell lines, patient-derived orthotopic xenograft models, and tumor organoid cultures—which represent the molecular spectrum of ATRTs are needed to critically advance ATRT therapeutics.
The prognostic value of the ATRT subgroups remains to be fully investigated. Torchia et al reported that ASCL1 protein expression, which is highly expressed in ATRT-SHH, was associated with a better outcome.6 However, it is not clear whether this is true for the whole SHH subgroup, as not all ATRT-SHH cases may express ASCL1. There clearly is a need for assessing the predictive power of the subgroups in well-characterized cohorts in prospective, clinical studies.
Finally, to get a better understanding of the clinical relevance of ATRT subgroups, molecular subgrouping should be included in any future clinical trial for ATRT patients. The method of choice is currently DNA methylation profiling, as this requires very little input material (tumor DNA isolated from either frozen or formalin-fixed paraffin-embedded tumor tissue) and shows little or no bias when performed at different centers. However, as DNA methylation profiling may not always be available, it would still be helpful if more readily available markers or methods to subgroup ATRTs, such as immunohistochemical staining for tyrosinase or ASCL1, or Nanostring subgrouping methods, as have been developed for MB, could also be developed for broader use in clinical labs globally.
Supplementary Material
Acknowledgments
The authors would like to thank their national and international colleagues for providing ATRT samples and associated clinical data for this study.
Funding
BH, FY, and AH are supported by the Canadian Cancer Society Research Institute (CCSRI) (705056), a Canadian Institutes of Health Research (CIHR) project grant (409302), and Tali’s Fund. PDJ and MH are supported by the Deutsche Forschungsgemeinschaft (DFG) (JO 1598/1-1 and HA 3060/8-1, respectively). YG and DW are supported by grants from the INSTINCT network, The Brain Tumor Charity, Great Ormond Street Children’s Charity, Children with Cancer UK (16/193), CRUK center core, and LoveOliver. FB is supported by 101 des Arts, Abigael, Marabout de Ficelle, Etoile de Martin, Enfants et Cancer, a St Baldrick Robert Arceci Innovation award, the French Society for the Fight against Cancer and Leukemia in Children and Adolescents (SFCE), and the Federation Enfants et Santé.
Conflict of interest statement. The authors declare no conflicts of interest.
Authorship statement. BH, PDJ, YG, discoidin domain receptor tyrosine kinase 1 (DDR1), and MDDJA performed data analyses and generated all figures. MF, MH, FB, DW, AH, and MK contributed to data, concept, and design of the paper. DW, AH, and MK supervised data analyses. All authors contributed to the writing of the paper.
References
- 1. Frühwald MC, Biegel JA, Bourdeaut F, Roberts CW, Chi SN. Atypical teratoid/rhabdoid tumors—current concepts, advances in biology, and potential future therapies. Neuro Oncol. 2016;18(6):764–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hasselblatt M, Nagel I, Oyen F, et al. SMARCA4-mutated atypical teratoid/rhabdoid tumors are associated with inherited germline alterations and poor prognosis. Acta Neuropathol. 2014;128(3):453–456. [DOI] [PubMed] [Google Scholar]
- 3. Schneppenheim R, Frühwald MC, Gesk S, et al. Germline nonsense mutation and somatic inactivation of SMARCA4/BRG1 in a family with rhabdoid tumor predisposition syndrome. Am J Hum Genet. 2010;86(2):279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ginn KF, Gajjar A. Atypical teratoid rhabdoid tumor: current therapy and future directions. Front Oncol. 2012;2:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Birks DK, Donson AM, Patel PR, et al. High expression of BMP pathway genes distinguishes a subset of atypical teratoid/rhabdoid tumors associated with shorter survival. Neuro Oncol. 2011;13(12):1296–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Torchia J, Picard D, Lafay-Cousin L, et al. Molecular subgroups of atypical teratoid rhabdoid tumours in children: an integrated genomic and clinicopathological analysis. Lancet Oncol. 2015;16(5):569–582. [DOI] [PubMed] [Google Scholar]
- 7. Torchia J, Golbourn B, Feng S, et al. Integrated (epi)-genomic analyses identify subgroup-specific therapeutic targets in CNS rhabdoid tumors. Cancer Cell. 2016;30(6):891–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Johann PD, Erkek S, Zapatka M, et al. Atypical teratoid/rhabdoid tumors are comprised of three epigenetic subgroups with distinct enhancer landscapes. Cancer Cell. 2016;29(3):379–393. [DOI] [PubMed] [Google Scholar]
- 9. Capper D, Jones DTW, Sill M, et al. DNA methylation-based classification of central nervous system tumours. Nature. 2018;555(7697):469–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schwalbe EC, Lindsey JC, Nakjang S, et al. Novel molecular subgroups for clinical classification and outcome prediction in childhood medulloblastoma: a cohort study. Lancet Oncol. 2017;18(7):958–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sharma T, Schwalbe EC, Williamson D, et al. Second-generation molecular subgrouping of medulloblastoma: an international meta-analysis of Group 3 and Group 4 subtypes. Acta Neuropathol. 2019;138(2):309–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Han ZY, Richer W, Fréneaux P, et al. The occurrence of intracranial rhabdoid tumours in mice depends on temporal control of Smarcb1 inactivation. Nat Commun. 2016;7:10421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ng JM, Martinez D, Marsh ED, et al. Generation of a mouse model of atypical teratoid/rhabdoid tumor of the central nervous system through combined deletion of Snf5 and p53. Cancer Res. 2015;75(21):4629–4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vitte J, Gao F, Coppola G, Judkins AR, Giovannini M. Timing of Smarcb1 and Nf2 inactivation determines schwannoma versus rhabdoid tumor development. Nat Commun. 2017;8(1):300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hasselblatt M, Thomas C, Nemes K, et al. Tyrosinase immunohistochemistry can be employed for the diagnosis of atypical teratoid/rhabdoid tumours of the tyrosinase subgroup (ATRT-TYR). Neuropathol Appl Neurobiol. 2019. doi: 10.1111/nan.12560. [DOI] [PubMed] [Google Scholar]
- 16. Simões-Costa M, Bronner ME. Establishing neural crest identity: a gene regulatory recipe. Development. 2015;142(2):242–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tief K, Schmidt A, Aguzzi A, Beermann F. Tyrosinase is a new marker for cell populations in the mouse neural tube. Dev Dyn. 1996;205(4):445–456. [DOI] [PubMed] [Google Scholar]
- 18. Johann PD, Hovestadt V, Thomas C, et al. Cribriform neuroepithelial tumor: molecular characterization of a SMARCB1-deficient non-rhabdoid tumor with favorable long-term outcome. Brain Pathol. 2017;27(4):411–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nowak J, Nemes K, Hohm A, et al. Magnetic resonance imaging surrogates of molecular subgroups in atypical teratoid/rhabdoid tumor. Neuro Oncol. 2018;20(12):1672–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Brabetz S, Leary SES, Gröbner SN, et al. A biobank of patient-derived pediatric brain tumor models. Nat Med. 2018;24(11):1752–1761. [DOI] [PubMed] [Google Scholar]
- 21. Kaur H, Hütt-Cabezas M, Weingart MF, et al. The chromatin-modifying protein HMGA2 promotes atypical teratoid/rhabdoid cell tumorigenicity. J Neuropathol Exp Neurol. 2015;74(2):177–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wong JP, Todd JR, Finetti MA, et al. Dual targeting of PDGFRα and FGFR1 displays synergistic efficacy in malignant rhabdoid tumors. Cell Rep. 2016;17(5):1265–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chauvin C, Leruste A, Tauziede-Espariat A, et al. High-throughput drug screening identifies pazopanib and clofilium tosylate as promising treatments for malignant rhabdoid tumors. Cell Rep. 2017;21(7):1737–1745. [DOI] [PubMed] [Google Scholar]
- 24. Jagani Z, Mora-Blanco EL, Sansam CG, et al. Loss of the tumor suppressor Snf5 leads to aberrant activation of the Hedgehog-Gli pathway. Nat Med. 2010;16(12):1429–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kerl K, Moreno N, Holsten T, et al. Arsenic trioxide inhibits tumor cell growth in malignant rhabdoid tumors in vitro and in vivo by targeting overexpressed Gli1. Int J Cancer. 2014;135(4):989–995. [DOI] [PubMed] [Google Scholar]
- 26. Knutson SK, Kawano S, Minoshima Y, et al. Selective inhibition of EZH2 by EPZ-6438 leads to potent antitumor activity in EZH2-mutant non-Hodgkin lymphoma. Mol Cancer Ther. 2014;13(4):842–854. [DOI] [PubMed] [Google Scholar]
- 27. Erkek S, Johann PD, Finetti MA, et al. Comprehensive analysis of chromatin states in atypical teratoid/rhabdoid tumor identifies diverging roles for SWI/SNF and polycomb in gene regulation. Cancer Cell. 2019;35(1):95–110.e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Alig SK, Dreyling M, Seppi B, Aulinger B, Witkowski L, Rieger CT. Severe cytokine release syndrome after the first dose of Brentuximab Vedotin in a patient with relapsed systemic anaplastic large cell lymphoma (sALCL): a case report and review of literature. Eur J Haematol. 2015;94(6):554–557. [DOI] [PubMed] [Google Scholar]
- 29. Pinto EM, Hamideh D, Bahrami A, et al. Malignant rhabdoid tumors originating within and outside the central nervous system are clinically and molecularly heterogeneous. Acta Neuropathol. 2018;136(2):315–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chun HE, Johann PD, Milne K, et al. Identification and analyses of extra-cranial and cranial rhabdoid tumor molecular subgroups reveal tumors with cytotoxic T cell infiltration. Cell Rep. 2019;29(8):2338–2354 e2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hashizume R, Zhang A, Mueller S, et al. Inhibition of DNA damage repair by the CDK4/6 inhibitor palbociclib delays irradiated intracranial atypical teratoid rhabdoid tumor and glioblastoma xenograft regrowth. Neuro Oncol. 2016;18(11):1519–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Oberlick EM, Rees MG, Seashore-Ludlow B, et al. Small-molecule and CRISPR screening converge to reveal receptor tyrosine kinase dependencies in pediatric rhabdoid tumors. Cell Rep. 2019;28(9):2331–2344 e2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Taylor MD, Northcott PA, Korshunov A, et al. Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol. 2012;123(4):465–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pajtler KW, Mack SC, Ramaswamy V, et al. The current consensus on the clinical management of intracranial ependymoma and its distinct molecular variants. Acta Neuropathol. 2017;133(1):5–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sturm D, Witt H, Hovestadt V, et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell. 2012;22(4):425–437. [DOI] [PubMed] [Google Scholar]
- 36. Cavalli FMG, Hübner JM, Sharma T, et al. Heterogeneity within the PF-EPN-B ependymoma subgroup. Acta Neuropathol. 2018;136(2):227–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cavalli FMG, Remke M, Rampasek L, et al. Intertumoral heterogeneity within medulloblastoma subgroups. Cancer Cell. 2017;31(6):737–754 e736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chun HE, Johann PD, Milne K, et al. Identification and analyses of extra-cranial and cranial rhabdoid tumor molecular subgroups reveal tumors with cytotoxic T cell infiltration. Cell Rep. 2019;29(8):2338–2354.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.