Abstract
Pancreatic ductal adenocarcinoma (PDAC) is an aggressive cancer with an extremely poor prognosis. There is an urgent need to identify new therapeutic targets and also understand the mechanism of PDAC progression that leads to aggressiveness of the disease. To find therapeutic targets, we analyzed data related to PDAC transcriptome sequencing and found overexpression of the de novo purine metabolic enzyme phosphoribosylaminoimidazole succinocarboxamide synthetase (PAICS). Immunohistochemical analysis of PDAC tissues showed high expression of the PAICS protein. To assess the biological roles of PAICS, we used RNA interference and knock down of its expression in PDAC cell lines that caused a reduction in PDAC cell proliferation and invasion. Furthermore, results of chorioallantoic membrane assays and pancreatic cancer xenografts demonstrated that PAICS regulated pancreatic tumor growth. Our data also showed that, in PDAC cells, microRNA-128 regulates and targets PAICS. PAICS depletion in PDAC cells caused upregulation in E-cadherin, a marker of the epithelial-mesenchymal transition. In PDAC cells, a BET inhibitor, JQ1, reduced PAICS expression. Thus, our investigations show that PAICS is a therapeutic target for PDAC and, as an enzyme, is amenable to targeting by small molecules.
Introduction
Pancreatic ductal adenocarcinoma (PDAC), an aggressive cancer, is the fourth leading cause of cancer-related deaths in the United States. By 2020, it is expected to be the second deadliest malignancy [1]. Surgical resection remains the best treatment for the 20% of PDAC patients diagnosed with early stages of the disease [2,3]. About 40% of patients have metastasis at the time of diagnosis, and their overall survival is 6 months to 1 year due to the limited availability of therapeutic strategies, disease relapse, and drug toxicity [4]. Standard chemotherapy for metastatic PDAC involves administration of gemcitabine alone or combination therapy including gemcitabine, which produces a response rate of 5%-10% [5]. New therapeutic regimens, targeted therapies, and combinational treatment options are required for improvement in survival of patients suffering from PDAC. For this to occur, a more sophisticated understanding of the biology of this cancer is needed.
Since PDACs have extensive and poorly vascularized desmoplastic stroma and adapt to metabolically challenging survival conditions, targeting of specific metabolic pathways could lead to the development of effective therapies [6]. Metabolic reprogramming and altered cellular metabolism, characteristics of cancer, are related to cancer cell proliferation, growth, and survival [7]. Since an imbalance of de novo purine metabolism is linked with progression of cancer cells, targeting of enzymes or protein-protein interactions in this pathway is a promising strategy to combat tumor growth and metastasis [8]. Most of the enzymes of the de novo pathway for purine synthesis are associated with malignancies [[9], [10], [11]]. Targeting of guanosine monophosphate synthase, an enzyme required for de novo biosynthesis of GMP, suppresses melanoma cell invasion and tumorigenicity [12]. In the de novo purine biosynthetic pathway, N-succinyl-5-aminoimidazole-4-carboxamide-1-ribose-5′-phosphate (SAICAR) is an intermediary metabolite of phosphoribosylaminoimidazole succinocarboxamide synthetase (PAICS), and in glucose-deprived conditions, SAICAR activates pyruvate kinase M2 (PKM2) [13]. In a hypoxic tumor environment, PKM2 is necessary for proliferation of PDACs and regulates VEGF-A secretion and angiogenesis through NF-κB and HIF-1α [14]. The SAICAR-PKM2 interaction is associated with survival of cancer cells [13]. The bifunctional enzyme aminoimidazole carboxamide ribonucleotide transformylase catalyzes the last two steps of de novo purine biosynthesis. A small molecule inhibitor of aminoimidazole carboxamide ribonucleotide transformylase reduces its activity, leading to a decrease in the proliferation of breast cancer cells [15].
MicroRNAs (miRNAs) are short noncoding RNAs that bind to the 3′-UTR regions of target genes and regulate gene expression post transcriptionally. miR-128 is a regulator of oncogenic properties. As a tumor suppressor, miR-128 decreases cancer cell growth by targeting ZEB1 in prostate cancer [16] and esophageal squamous cell cancer [17]; TERT in HeLa cells [18]; Bmi-1 in gastric cancer [19] and glioblastoma [20]; c-met in lung cancer stem cells, enhancing the cancer cell sensitivity to gefitinib [21]; PAICS in bladder cancer [10]; and MDM4 in pancreatic cancer [22]. Cisplatin combined with miR-128 reduces expression of cisplatin-resistant proteins ABCC5 and Bmi-1, resulting in reduced ovarian tumor growth [23].
Our earlier studies show an association of PAICS with cellular proliferation, colony formation, and invasion in lung [24], prostate [11], and bladder cancers [10]. PAICS is also involved in breast cancer growth [25,26]. In the present study, we demonstrated a role of PAICS in PDAC. The results showed overexpression of PAICS in PDACs compared with associated normal pancreatic tissue. The involvement of PAICS in various malignant properties of PDAC was assessed by employing lentiviral shRNA- or siRNA-based gene silencing. The involvement of PAICS in cellular proliferation, colony formation, invasion, migration, formation of spheroids, and tumor growth of PDAC cells was documented. Our investigation also showed that, in PDAC cells, ectopic expression of miR-128 targets PAICS expression. In cells with PAICS knockdown, expression of E-cadherin was higher. These findings provide evidence that PAICS is involved in growth of PDACs through regulation of E-cadherin, supporting our earlier results for bladder cancer [10]. Identification of PAICS as a target will allow a better understanding of the consequences of PAICS inhibition with small molecule inhibitors and could lead to the development of new therapeutics that block metabolic reprogramming in PAICS addicted-PDACs.
Materials and Methods
Cell Lines
PDAC cell lines PANC-1 and S2VP10 were obtained from Prof. Donald J Buchsbaum, University of Alabama at Birmingham, Birmingham, AL. PANC-1 cells were cultured in DMEM (high glucose) containing 10% fetal bovine serum and 1% penicillin/streptomycin; S2VP10 cells were cultured in RPMI media containing 10% fetal bovine serum and 1% penicillin/streptomycin. Cells were maintained in a humidified 5% CO2 atmosphere at 37°C.
Immunohistochemical Analysis
Measurement of PAICS protein expression was assessed by immunohistochemical analysis of paraffin-embedded PDAC sections as described [[27], [28], [29]]. Tissues were obtained with approval from the Institutional Review Board (The University of Michigan at Ann Arbor). Briefly, antigen retrieval was performed by heating paraffin-embedded tissue sections at 100°C for 10 minutes in citrate buffer, pH 6 (Cat# C9999, Sigma-Aldrich, St. Louis, MO), and rehydration. Endogenous peroxidase activity was removed by incubating specimens with BLOXALL endogenous peroxidase and alkaline phosphatase blocking solution (Cat# SP-6000, Vector Laboratories, Burlingame, CA), followed by blocking with normal horse serum (Cat# S-2000, Vector Laboratories). Specimens were incubated with a mouse monoclonal antibody against PAICS (Cat# GTX83950, GeneTex, Irvine, CA, 1:2500 dilution) followed by incubation with ImmPRESS HRP anti-mouse IgG (Cat# MP-7402, Vector Laboratories) as a secondary antibody. The immunoreactivity was visualized with 3,3-diaminobenzidine (Cat#SK-4100, Vector Laboratories), followed by counterstaining with Vector Hematoxylin QS (Cat#H-3404 Vector Laboratories). VectaMount Permanent mounting medium (Cat#H-5000 Vector Laboratories) was used to mount the specimens.
Immunoblot Analyses
Protein samples were resolved on NuPAGE 4%-12% Bis-Tris Midi Protein Gels (Invitrogen, Carlsbad, CA) and transferred onto Immobilon-P PVDF membranes (EMD Millipore, Billerica, MA) as described previously [30]. Immunoreactivity was evaluated by probing the membranes with mouse primary antibody followed by incubation with secondary IgG horseradish peroxidase antibody (Table S1). Immunoreactivity against PAICS protein was evaluated by use of Luminata Crescendo chemiluminescence Western blotting substrate according to the manufacturer's protocol (EMD Millipore), and images were captured with an Amersham Imager 600RGB (GE Healthcare Life Sciences, Pittsburgh, PA).
Immunofluorescence
The Lab-Tek II CC2 Chamber slide System (Cat#154917, Nunc, Rochester, NY) was used for growing 2000 PDAC cells overnight as described previously [10]. One hundred percent chilled methanol for 10 minutes was used for cell fixation, followed by blocking with horse serum for 30 minutes. Further incubation was accomplished with primary antibody diluted in horse serum for 1 hour, followed by incubation with a secondary antibody for 1 hour. Prolong Gold antifade reagent with DAPI (Cat#P36931, Life technologies, Eugene, OR) was used for nuclear staining and mounting, and image acquisition was with a ZEISS LSM 710 laser confocal microscope using a 60× lens (Carl Zeiss, Thornwood, NY) by keeping detector gains constant for all acquisitions. Images were acquired at UAB High-Resolution Imaging Facility.
RNA Interference
As described previously [10], PAICS knockdowns in PDAC cells were performed by treatment with lentivirus shRNA or siRNA. Lentiviruses for stable PAICS knockdowns were generated by the UAB Neuroscience NINDS Protein Core (P30 NS47466) using pGreenPuro shRNA expression lentivectors (Systembio, Palo Alto, CA) (Table S2). pGreenPuro shRNA expression lentivectors have GFP and puromycin resistance markers for selection of positive populations. Lentivirus infection of PDAC cells was accomplished, and selection was made with 1 μg/ml puromycin (Life Technologies, Carlsbad, CA). The siRNA duplexes (Table S3), purchased from GE Healthcare Dharmacon, Inc. (Lafayette, CO), were used as described previously [11]. For siRNA-based PAICS knockdown, reverse transfection was performed with Lipofectamine RNAiMAX (Life Technologies). For this, transfection was performed after seeding 1 × 106 PDAC cells per well.
Cell Proliferation Assay
Cell proliferation was assessed by seeding 5000 PAICS knockdown and control PDAC cells in 12-well plates and counting cells at 2, 4, and 6 days with a Z2 Coulter particle counter (Beckman Coulter, Brea, CA).
Colonogenic Assay
Colony formation assays were performed by seeding 1000 PAICS knockdown PDAC cells per well in six-well plates in triplicates, as described previously [29]. Colonies were allowed to grow for 10 days, followed by fixation with 5% glutaraldehyde in phosphate-buffered saline and staining with crystal violet (Sigma Aldrich, St. Louis, MO). Images were captured with an Amersham Imager 600RGB (GE Healthcare Life Sciences, Chicago, IL).
Invasion Assay
Corning BioCoat Matrigel matrix (Corning, NY) was used to determine the involvement of PAICS in invasion of PDAC cells, as described previously [31]. Briefly, in triplicate wells of 24-well plates, 5 × 104 PAICS knockdown PDAC cells were seeded in serum-free media onto 8-μm pore inserts, with serum-containing media as a chemoattractant in the lower chamber. After 48 hours, cells that invaded to the lower side of the chamber were fixed with 5% glutaraldehyde and stained with crystal violet. Invaded cells were photographed using bright field microscopy.
Wound Healing Assay
Wound healing assays were accomplished to ascertain changes in cell motility, as described previously [32]. PAICS knockdown and control PDAC cells (1 × 106) were seeded on 35-mm Petri dishes in triplicates. After overnight incubation, a wound was created on the confluent cell monolayer by use of an Aerosol P200 pipette tip, and photomicrographs were taken at 0 and 24 hours with an inverted phase-contrast microscope under a 4× objective.
Spheroid Model
Spheroid cultures resemble the biological features of cancer cells in animals. To evaluate the effect of PAICS knockdown in PDAC cells, spheroids were generated by Cultrex 3D spheroid BME cell invasion assays (Cat# 3500-096-K, Trevigen, Gaithersburg, MD), as described previously [10]. Following the manufacturer's guidelines, 10,000 cells (45 μl) were seeded in triplicate with 5 μl of Spheroid Formation ECM. After centrifugation at 200 ×g for 3 minutes, cells were incubated at 37°C under 5% CO2 for 72 hours. Invasion matrix (50 μl) was added to the wells, followed by centrifugation at 200 ×g for 3 minutes at 4°C. After 1 hour, warm medium (100 μl) containing chemoattractant and invasion-modulating compounds was added, followed by incubation for 4 days. Images were taken with an inverted phase-contrast microscope under a 4× objective.
Chick Chorioallantoic Membrane (CAM) Assay
To assess tumor growth, CAM assays were performed as described previously [11,29]. Fertilized chicken eggs, specific pathogen-free, were purchased from Charles River Laboratories (North Franklin, CT) and incubated for 10 days. S2VP10 cells (1 × 106) with nontargeting (NT) shRNA, PAICS shRNA1, or PAICS shRNA2 in 50 μl of culture medium were applied to the CAMs on the 11th day of embryonic growth. On the 18th day of growth, tumors were harvested and weighed. The experiment was started with 10 eggs per group, but, in each group, two eggs were nonviable at the end of the experiment. For the eight eggs remaining per group, tumors were weighed, followed by data analysis.
Pancreatic Tumor Xenografts
As described [33,34], mice were maintained with regulatory standards under a protocol approved by the University of Alabama at Birmingham Institutional Animal Care and Use Committee with Animal Project Number as IACUC-21501. S2VP10 xenograft tumors were developed for investigation of the function of PAICS on tumor growth. S2VP10 cells deficient in PAICS or control cells with transfected with NT shRNA (1 × 106) were implanted subcutaneously into the right dorsal flanks of 6-week-old NOD/SCID/IL2γ-receptor null (NSG) mice. Mice were euthanized at the end of the experiment (n = 7 for each group), tumors were excised and weighed, and the results were plotted.
Statistical Analyses
For studies with cell lines, statistical comparisons of mean values were made using Student's t test. P values < .05 were considered statistically significant. All cell culture assays were performed in triplicate, and data were expressed as means ± standard deviation.
Results
PDACs Overexpress PAICS, Which Predicts a Poor Prognosis for Patients
Studies of gene expression profiling acquired using the Oncomine [35] cancer microarray database (Oncomine Platform) (Life Technologies, Ann Arbor, MI) from two different datasets, Segara et al., 2005 (PDAC, n = 6 and pancreas, n = 11) [36] and Pei et al., 2009 (PDAC, n = 16 and pancreas, n = 36) [37], indicated dysregulated expression of PAICS in PDACs (Figure 1, A and B). Badea et al., 2008 [38] performed gene expression analysis and showed upregulation of enzymes of the de novo purine nucleotide pathway in PDAC (n = 39) compared with matched normal tissue (n = 39) (Supplementary Figure S1). As determined with UALCAN (http://ualcan.path.uab.edu), an interactive web resource for analyzing cancer transcriptome data [39], microarray profiling studies also indicated overexpression of PAICS in PDACs (Figure 1C). Transcriptome sequencing confirmed PAICS overexpression in various stages of PDAC relative to normal pancreas (Figure 1D). Kaplan-Meier survival analysis using transcript data predicted poor patient survival with higher expression of PAICS in PDACs (Figure 1E). Immunhistochemical analysis confirmed PAICS protein overexpression, as there was strong cytoplasmic, chocolate-brown immunostaining in PDAC tissue sections (Figure 1F). Immunofluorescence analysis in PANC-1 and S2VP10 cells confirmed overexpression of PAICS in PDAC cell lines (Supplementary Figure S2). These results confirmed the elevated expression of PAICS in PDACs.
Figure 1.
Overexpression of PAICS in PDAC. Oncomine gene expression profiling showing PAICS expression from PDAC tissues as reported by (A) Segara et al., 2005 [36] and (B) Pei et al., 2009 [37]. (C) PAICS expression, determined by use of UALCAN [39], for PDACs compared to normal pancreatic tissue. (D) Stage-wise PAICS expression in pancreatic tumors. (E) Kaplan-Meier analysis of survival time for PDACs determined by use of UALCAN. (F) Representative images showing PAICS expression, assessed by immunohistochemistry, in PDACs. Hematoxylin was used for nuclear staining.
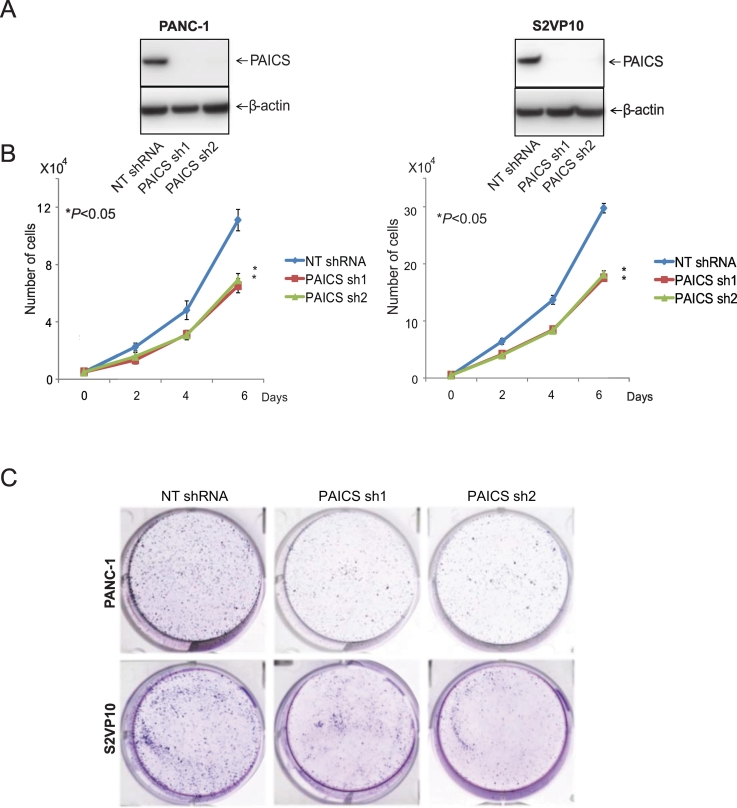
PAICS Reduction Leads to Reduced Cellular Proliferation and Colonogenicity of PDAC cells
To establish the functional relevance of PAICS to oncogenic properties of cancer cells, we employed a stable knockdown strategy by using lentivirus shRNAs and transient knockdowns by siRNAs to reduce PAICS in the PDAC cells PANC-1 and S2VP10. Western blot analyses showed knockdown of PAICS protein with either of two shRNAs for PAICS compared with a control NT shRNA (Figure 2A) as well as with two independent PAICS siRNAs (Supplementary Figure S3A). Subsequently, assays were performed with these knockdown cells to determine the role of PAICS in PDAC cell growth. Proliferation studies with PANC-1 cells revealed reductions in cell counts by PAICS shRNA1 of 41% (P < .05) and PAICS shRNA2 by 37% (P < .05) (Figure 2B). Similarly, counts of S2VP10 cells were reduced 40% (P < .05) by PAICS shRNA1 and 39% (P < .05) by PAICS shRNA2 (Figure 2B). There were also reductions in PDAC cells transfected with PAICS siRNA1 or 2 (Supplementary Figure S3B). PAICS knockdown also resulted in a reduction in the colony-forming capacity of PANC-1 and S2VP10 cells with PAICS shRNA1 and 2 (Figure 2C). The colony-forming capacity of PDAC cells was also decreased by PAICS siRNA1 and 2 (Supplementary Figure S3C). Thus, these results indicated a role of PAICS in the proliferation and clonogenicity of PDAC cells.
Figure 2.
PAICS knockdown reduced PDAC cell proliferation and colony formation. (A) Immunoblot analyses of PAICS protein levels in PANC-1 and S2VP10 cells treated with PAICS shRNA compared to levels in cells treated with NT shRNA. β-Actin was used as a loading control. (B) PAICS knockdown reduced cell proliferation at 2, 4, and 6 days. (C) Colony formation was suppressed at 10 days after seeding of 1000 shRNA-treated cells in six-well plates. Statistically significant differences (*P < .05) are indicated by an asterisk (*).
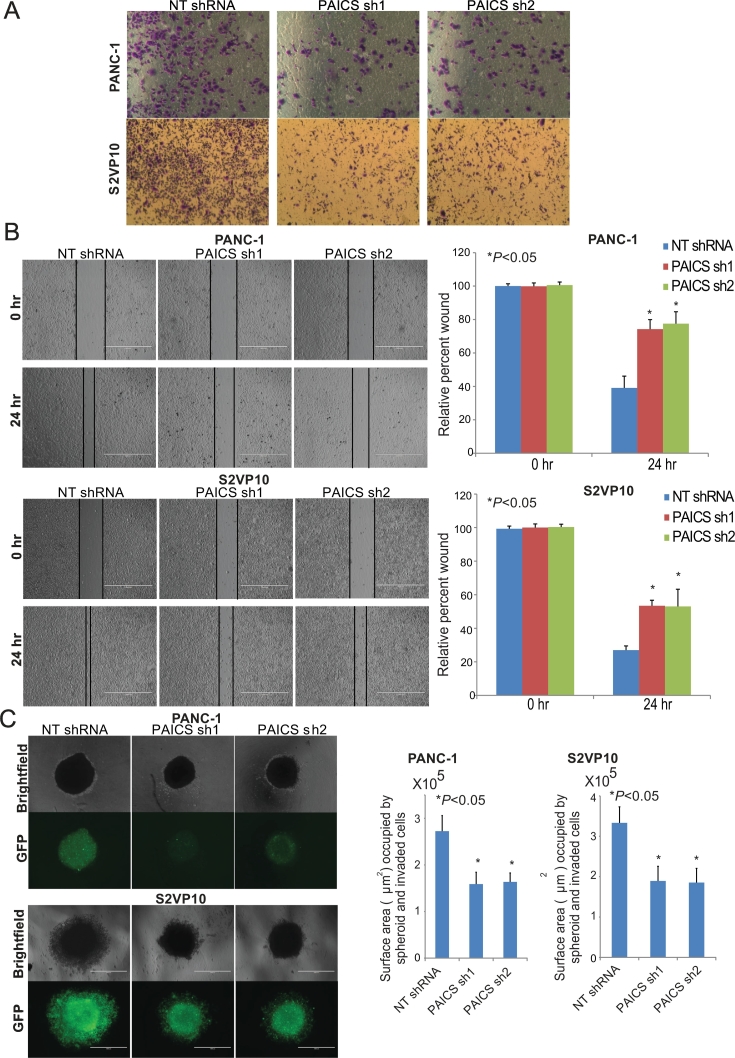
Function of PAICS in Invasion, Motility, and Spheroid Formation of PDAC Cells
To investigate the role of PAICS in invasion and motility of PDAC cells, Transwell invasion and wound healing assays were accomplished. Repression of PAICS protein by PAICS shRNA1 and 2 resulted in reductions in the invasive capacity of PANC-1 and S2VP10 cells (Figure 3A). Reductions in invasive capacity of PANC-1 and S2VP10 cells were also observed when they were transfected with PAICS siRNA1 or 2 (Supplementary Figure S4A). For wound-healing assays, an artificial wound was made in a confluent population of PDAC cells at 0 hour, and motility was assessed after 24 hours. The motility of PANC-1 and S2VP10 cells was significantly (*P < .05) retarded after transfection with PAICS shRNA1 or 2 (Figure 3B). Wound healing was also reduced when PANC-1 and S2VP10 cells were transfected with PAICS siRNA1 or 2 (Supplementary Figure S4B).
Figure 3.
PAICS knockdown suppressed invasion and motility of PDAC cells. (A) Representative images of PDAC cells with PAICS knockdown showing less invasion through Transwell Matrigel membranes compared to cells treated with NT shRNA. (B) Representative images showing wound healing assays of PANC-1 and S2VP10 cells lacking PAICS. Images were taken at 0 and 24 hours. Scale bar- 1000 μm. Comparative analysis of wound healing after PAICS knockdown. Asterisk (*) represents statistically significant difference. (C) PDAC cells with reduced PAICS formed smaller spheroids compared with cells treated with NT shRNA as shown by phase-contrast microscopy and GFP (due to the presence of pGreenPuro shRNA expression lentivector for GFP) images. Scale bar- 1000 μm. Histographs showed the spheroid formation capacity of PAICS shRNA relative to NT shRNA (*P < .05).
As growth of cancer cells in 3D recapitulates the morphology of tumors growing in animals, spheroid 3D assays were performed to elucidate the function of PAICS in spheroid formation by PDAC cells. Depletion of PAICS resulted in inhibition of spheroid formation. As represented in a photomicrograph and graph, targeting PAICS by shRNA resulted in significant reductions (*P < .05) in spheroid-forming capacity of PANC-1 and S2VP10 cells (Figure 3C). These data point to a role for PAICS in the invasion, motility, and spheroid formation of PDAC cells.
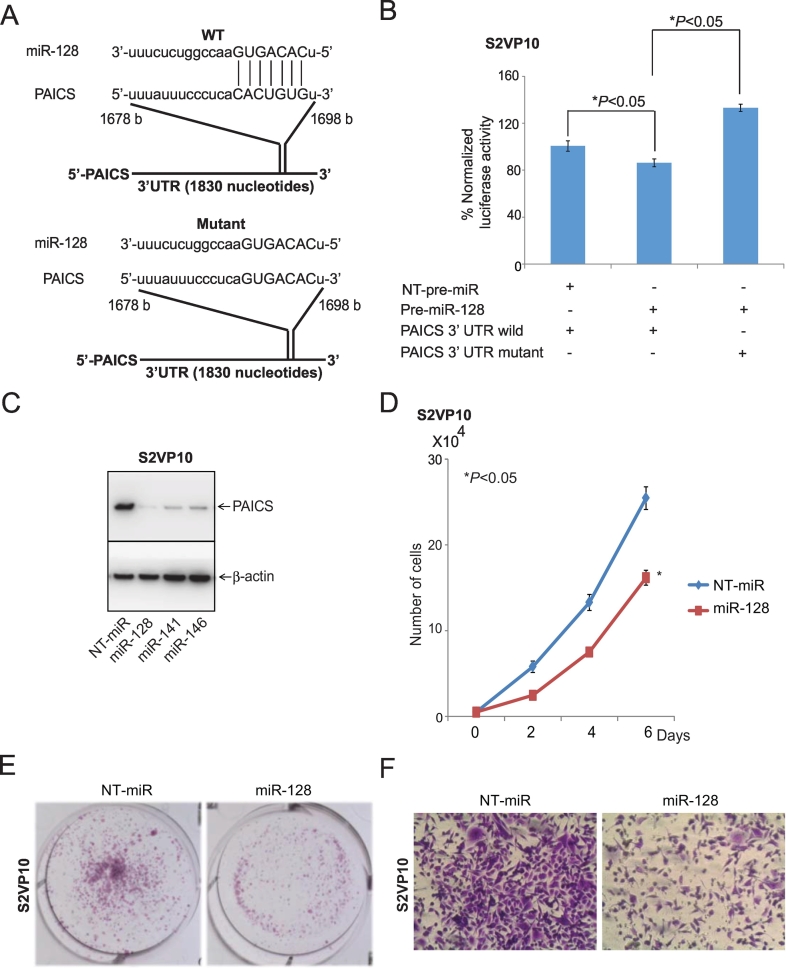
miR-128 Regulates and Targets PAICS in PDAC Cells
The microRNA target prediction bioinformatics tool TargetScan (http://www.targetscan.org/vert_61/) predicted that miR-128 could target PAICS. Wild-type and mutant pMir-REPORT-PAICS 3′-UTR plasmids were constructed as shown in Figure 4A. A luciferase assay was performed to determine if PAICS is a direct target of miR-128. Co-transfection of miR-128 and wild-type PAICS 3′UTR decreased luciferase expression compared with co-transfection of nontargeting (NT-miR) and wild-type PAICS 3′UTR, whereas co-transfection of miR-128 and the mutant PAICS 3′UTR did not (Figure 4B). Bioinformatics analysis predicted miR-128, miR-141, or miR-146 as the microRNAs supposed to target PAICS. We ectopically expressed miR-128, miR-141, or miR-146 in S2VP10 cells and tested the expression of PAICS. PAICS levels were lower after transfection of miR-128 as compared to NT-miR, miR-141, or miR-146 as determined by immunoblot analysis (Figure 4C). Additionally, we assessed the effect of miR-128 on the phenotype of S2VP10 cells after ectopic expression of miR-128. Forced miR-128 expression reduced PDAC cell proliferation (Figure 4D); colony formation (Figure 4E) and invasion (Figure 4F) were also inhibited. Collectively, these results indicated that PAICS is regulated and targeted by miR-128 in PDAC cells.
Figure 4.
miR-128 targets and regulates PAICS in PDAC. (A) Pictorial diagram of predicted binding sites of miR-128 with the 3′-UTR of PAICS as shown by Targetscan. (B) In S2VP10 PDAC cells, NT-pre-miR or miR-128 was co-transfected with luciferase constructs of either PAICS-3′UTR wild type or mutant. (C) NT-pre-miR, pre-miR-128, 141, or 146 was transfected into S2VP10 cells, and immunoblot analyses were performed for PAICS protein expression. β-Actin was used as an internal control. Ectopic expression of pre-miR-128 compared to NT-pre-miR largely canceled S2VP10 expansion in cell proliferation (D), colony formation (E) and invasion (F) assays.
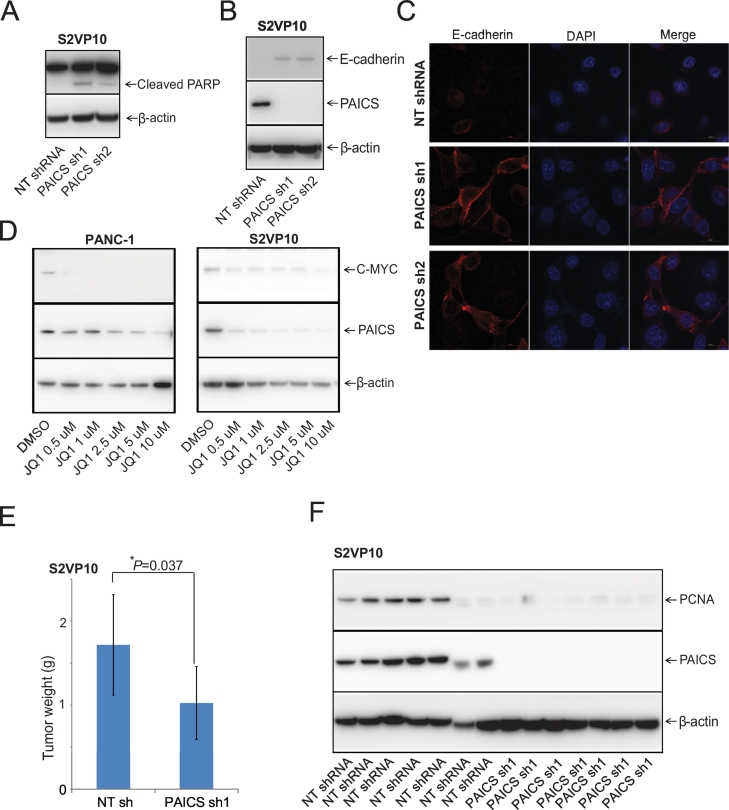
PAICS Knockdown Induces Apoptosis in PDAC Cells
For cancer cells, cleavage of poly(ADP-ribose)polymerase 1 (PARP1) by caspases is a characteristic of apoptosis [40]. PAICS depletion in S2VP10 PDAC cells showed endogenous, full-length PARP1 (116 kDa) and a fragment of PARP1 (89-kDa) resulting from caspase cleavage (Figure 5A). The PARP1 fragment of 89 kDa is an indicator of caspase activation, which signals the apoptosis process in PDAC cells with PAICS knockdown.
Figure 5.
PAICS regulates apoptosis, the EMT, and pancreatic tumor growth. Immunoblot analyses of stable PAICS knockdown S2VP10 cells showing (A) PARP-1 cleavage, (B) E-cadherin levels, and (C) immunofluorescence staining of E-cadherin (red) in these cells. Nuclear staining was accomplished with DAPI (blue). Merged images are shown. (D) Immunoblot analysis of JQ1-treated PDAC cells for PAICS and c-MYC. NSG mice were implanted in right dorsal flank with 1 million S2VP10 pancreatic cancer cells. (E) Representative histogram to show tumors weight of control NT shRNA (n = 7) and PAICS shRNA1 (n = 7) xenograft groups, mean ± SD; *P < .01. (F) Immunoblot analysis of S2VP10 xenografts treated with NT shRNA or PAICS shRNA.
PAICS Activates the Epithelial-Mesenchymal Transition (EMT) in PDAC Cells
Markers of the EMT are loss of the epithelial marker E-cadherin and gain of the mesenchymal marker vimentin, which promote PDAC cell progression and invasion [41,42]. In S2VP10 cells, there were higher levels of E-cadherin after PAICS knockdown (Figure 5B). Previously, it was reported that PANC-1 cells do not express E-cadherin [43], and our immunoblot analysis confirmed no expression (data not shown). Further, with S2VP10 cells, we examined intracellular localization of the E-cadherin protein using immunofluorescence, which showed higher levels of E-cadherin at the intercellular region upon PAICS knockdown (Figure 5C).
MYC Regulates PAICS Expression in PDACs
MYC regulates the expression of PAICS by binding at its promoter [11,25]. We treated PDAC cells with JQ1, a bromodomain and extra-terminal motif (BET) inhibitor that blocks MYC expression, and found that the expression of PAICS was reduced in PDAC cells (Figure 5D), validating previous observation that PAICS is regulated by MYC.
PAICS Depletion Induces Pancreatic Tumor Regression in the CAM and Xenograft Models
Since there was less spheroid-forming capacity of PDAC cells after PAICS reduction, we investigated the role of PAICS in the chick CAM model. Eggs were injected with control NT shRNA or stable PAICS knockdown S2VP10 cells, and tumors were excised after 7 days. A representative photograph showed less tumor growth in groups treated with PAICS shRNA compared with control NT shRNA (Supplementary Figure S5A). Treatment with PAICS shRNA1 caused a 36% (P < .05) lower tumor weight, and treatment with shRNA 2 lowered tumor weights by 38% (P < .05) (Supplementary Figure S5B). Immunoblot analysis of CAM tumor tissue showed elimination of PAICS protein in groups treated with PAICS shRNA relative to control NT shRNA (Supplementary Figure S5C).
To assess the function of PAICS in tumor growth, we established, in NSG mice, pancreatic cancer xenografts with S2VP10 cells infected with PAICS shRNA or NT shRNA. Mice were injected with S2VP10 cells with NT shRNA or stable PAICS knockdown. As shown in a representative histogram (Figure 5E), excised tumors from the PAICS knockdown group (n = 7) had lower weight compared to those of NT shRNA controls (n = 7). Immunoblot analysis of tumors procured from mice also showed lower protein levels of PAICS and proliferating cell nuclear antigen in those treated with PAICS shRNA (Figure 5F). The results obtained with the CAM and xenograft models supported those derived with cell cultures, indicating a role of PAICS in tumor growth.
Discussion
PDAC is an incurable and lethal malignancy, often diagnosed at advanced stages and fatal to most patients due to the lack of diagnostic markers and early symptoms [44]. For 2019, it is estimated that, in the United States, 56,770 new cases of PDAC will be diagnosed and that 45,750 PDAC-related deaths will occur [1]. Among tumors of the digestive system, PDAC has the worst prognosis, with a 5-year survival rate of less than 8% [45]. New therapeutic models and treatment strategies are required for improved prognosis and outcomes of PDAC patients.
Cellular biosynthetic processes, such as metabolism and protein synthesis, drive the proliferation of cancer cells. Metabolic alterations and reprogramming associated with tumorigenesis are characteristics of cancer [46]. Transcription factors and metabolic enzymes that induce alterations in cancer metabolism could be targets for cancer therapy [47]. Rapidly proliferating cancer cells often depend on the de novo purine biosynthetic pathway for synthesis of adenine and guanine, whereas normal cells generally depend on the salvage pathway. Phosphoribosyl pyrophosphate synthetase 2 catalyzes the first step of purine biosynthesis by forming 5-phosphoribosyl-1-pyrophosphate [48]. For glioblastoma patients, elevated expression of the de novo purine synthesis enzymes adenylosuccinate lyase, adenylosuccinate synthase, inosine-5′-monophosphate dehydrogenase 1, and phosphoribosyl pyrophosphate amidotransferase is associated with a poor prognosis [9]. PAICS, a bifunctional enzyme involved in de novo biosynthesis, catalyzes the conversion of aminoimidazole ribonucleotide to 4-carboxy-5-aminoimidazole ribonucleotide and 4-carboxy-5-aminoimidazole ribonucleotide to SAICAR [49]. Earlier, our group demonstrated a role of PAICS in lung [24], prostate [11], and bladder cancer growth [10]. In the present study, we showed that PAICS is involved in the proliferation of PDAC cells.
Since cell migration and invasion are steps in cancer spread, progression, and metastasis [50], we investigated the function of PAICS in cellular motility and invasion of PDAC cells by employing gene silencing. PAICS knockdown inhibited the cell motility, invasion, and spheroid-forming capacity of PDAC cells. Use of the CAM and xenograft models showed that PAICS reduction inhibited tumor growth. These results corroborate our previous studies with lung and prostate adenocarcinomas in which PAICS knockdown inhibited cell invasion and tumor growth [11,24]. PAICS-depleted PDAC cells showed a cleaved fragment of PARP-1, an indicator of apoptosis.
MicroRNAs are regulators in PDAC [51]. Our investigation employing luciferase assays showed that miR-128 binds to the 3′UTR of PAICS and regulates the expression of PAICS in PDAC. Pancreatic cancer has lower expression of miR-128 compared to adjacent noncancerous tissues, suggesting that reduced miR-128 levels drive pancreatic cancer [22]. Our study showed an inverse relation between miR-128 and PAICS, as ectopic expression of miR-128 in S2VP10 PDAC cells decreased PAICS expression and inhibited cell proliferation, colony formation, and invasion of PDAC cells. These results show that miR-128 acts as a tumor suppressor in PDAC.
A reduction in E-cadherin expression is associated with advanced tumor stages and poor survival [52]. During cancer progression, low E-cadherin expression is frequently observed in solid cancers, and loss of E-cadherin expression is associated with the EMT [53]. Together, these results support a link between PAICS and E-cadherin in PDAC.
The oncogenic transcription factor MYC is an effector in cell signaling and a driver of PDAC [54]. The present study showed that PAICS expression, which is responsible for the expansion of PDAC, is inhibited after treatment with the BET inhibitor JQ1, which reduces MYC expression. These findings are in accordance with previous studies showing that JQ1 decreases PDAC cell growth and tumor growth [55,56]. From the present experiments, we concluded that MYC also regulates PAICS expression in PDAC.
In conclusion, the present study showed overexpression of PAICS in PDAC and suggested that PAICS conferred motility and invasion to PDAC cells. Knocking down PAICS decreased cell proliferation, colony formation, invasion, cell motility, and spheroid-forming capacity. In the CAM and xenograft models, pancreatic tumors deficient in PAICS showed suppressed tumor growth. Our study also showed that miR-128 regulated and targeted PDAC cells. Further, incubating PDAC cells with the BET inhibitor JQ1 decreased PAICS levels. Thus, targeting PAICS, a de novo purine biosynthetic enzyme, is a promising therapeutic strategy for PDAC.
Acknowledgments
Acknowledgements
These studies were partly supported by R01CA157845 (S.V.) and 5U54CA118948 (U.M.) and by institutional funds (Department of Pathology and School of Medicine of the University of Alabama at Birmingham) awarded to U.M. Also, we thank Dr. Donald Hill, a Professor at the University of Alabama at Birmingham, for his editorial assistance.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.tranon.2020.100776.
Appendix A. Supplementary data
Supplementary Figure S1 Differential expression of de novo purine biosynthetic enzymes in PDAC. Gene expression profiling studies acquired from Oncomine show elevated expression of de novo purine biosynthetic enzymes in PDAC as compared to normal pancreatic tissue. The data were acquired from Badea data sets. Color scale highlights different expression as blue represents downregulation, white is no alteration, and red shows upregulation of transcripts.
Supplementary Figure S2 Cytoplasmic expression of PAICS in PDAC cells. Immunofluorescence analysis showing PAICS expression in PANC-1 and S2VP10 cells. Red color represents PAICS expression; DAPI binds to nucleus (blue color).
Supplementary Figure S3 PAICS siRNA knockdown decreased pancreatic cancer cell proliferation and colony formation. (A) Immunoblot analysis showed PAICS protein levels in PAICS siRNA-transfected pancreatic cancer cells PANC-1 and S2VP10 compared to NT siRNA. β-Actin was used as a loading control. (B) PAICS knockdown cells were seeded in 24-well plates for cell proliferation assay at 2, 4, and 6 days. (C) Colony formation was assessed by seeding 1000 cells in six-well plates. After 10 days, colonies were fixed with glutaraldehyde and stained with crystal violet. Statistically significant differences (*P < .05) are shown by an asterisk (*).
Supplementary Figure S4 PAICS siRNA knockdown impedes invasion and cellular motility of pancreatic cancer cells. (A) PAICS siRNA-treated pancreatic cancer cells showed fewer cells invading through Transwell Matrigel membranes relative to normal NT siRNA-treated cells. (B) Representative images showing wound healing for pancreatic cancer cells at 0 and 24 hours.
Supplementary Figure S5 PAICS knockdown inhibits tumor growth in the CAM model. (A) Representative images showing reduced tumor sizes of PAICS-deficient PDACs compared to tumors treated with NT shRNA. (B) Histogram showing suppressed tumor weights of PAICS-deficient S2VP10 tumors (n = 8) (*P < .05). (C) PAICS-depleted and control tumors were excised from CAMs, and immunoblot analyses were performed by probing with PAICS antibody. β-Actin was used as a loading control.
Supplementary materials
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2019. CA Cancer J. Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Kosmidis C., Sapalidis K., Kotidis E., Mixalopoulos N., Zarogoulidis P., Tsavlis D., Baka S., Man Y.G., Kanellos J. Pancreatic cancer from bench to bedside: molecular pathways and treatment options. Ann Transl Med. 2016;4:165. doi: 10.21037/atm.2016.05.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Quaranta V., Rainer C., Nielsen S.R., Raymant M.L., Ahmed M.S., Engle D.D., Taylor A., Murray T., Campbell F., Palmer D.H., Tuveson D.A., Mielgo A., Schmid M.C. Macrophage-derived granulin drives resistance to immune checkpoint inhibition in metastatic pancreatic cancer. Cancer Res. 2018;78:4253–4269. doi: 10.1158/0008-5472.CAN-17-3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Teague A., Lim K.H., Wang-Gillam A. Advanced pancreatic adenocarcinoma: a review of current treatment strategies and developing therapies. Ther Adv Med Oncol. 2015;7:68–84. doi: 10.1177/1758834014564775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang J.P., Wu C.Y., Yeh Y.C., Shyr Y.M., Wu Y.Y., Kuo C.Y., Hung Y.P., Chen M.H., Lee W.P., Luo J.C., Chao Y., Li C.P. Erlotinib is effective in pancreatic cancer with epidermal growth factor receptor mutations: a randomized, open-label, prospective trial. Oncotarget. 2015;6:18162–18173. doi: 10.18632/oncotarget.4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen R., Neuzillet C., Tijeras-Raballand A., Faivre S., de Gramont A., Raymond E. Targeting cancer cell metabolism in pancreatic adenocarcinoma. Oncotarget. 2015;6:16832–16847. doi: 10.18632/oncotarget.4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ashkavand Z., O'Flanagan C., Hennig M., Du X., Hursting S.D., Krupenko S.A. Metabolic reprogramming by folate restriction leads to a less aggressive cancer phenotype. Mol. Cancer Res. 2017;15:189–200. doi: 10.1158/1541-7786.MCR-16-0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pedley A.M., Benkovic S.J. A new view into the regulation of purine metabolism: the purinosome. Trends Biochem. Sci. 2017;42:141–154. doi: 10.1016/j.tibs.2016.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang X., Yang K., Xie Q., Wu Q., Mack S.C., Shi Y., Kim L.J.Y., Prager B.C., Flavahan W.A., Liu X., Singer M., Hubert C.G., Miller T.E. Purine synthesis promotes maintenance of brain tumor initiating cells in glioma. Nat. Neurosci. 2017;20:661–673. doi: 10.1038/nn.4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chakravarthi B., Rodriguez Pena M.D.C., Agarwal S., Chandrashekar D.S., Hodigere Balasubramanya S.A., Jabboure F.J., Matoso A., Bivalacqua T.J., Rezaei K., Chaux A., Grizzle W.E., Sonpavde G., Gordetsky J. A role for de novo purine metabolic enzyme PAICS in bladder cancer progression. Neoplasia. 2018;20:894–904. doi: 10.1016/j.neo.2018.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chakravarthi B.V., Goswami M.T., Pathi S.S., Dodson M., Chandrashekar D.S., Agarwal S., Nepal S., Hodigere Balasubramanya S.A., Siddiqui J., Lonigro R.J., Chinnaiyan A.M., Kunju L.P., Palanisamy N. Expression and role of PAICS, a de novo purine biosynthetic gene in prostate cancer. Prostate. 2017;77:10–21. doi: 10.1002/pros.23243. [DOI] [PubMed] [Google Scholar]
- 12.Bianchi-Smiraglia A., Wawrzyniak J.A., Bagati A., Marvin E.K., Ackroyd J., Moparthy S., Bshara W., Fink E.E., Foley C.E., Morozevich G.E., Berman A.E., Shewach D.S., Nikiforov M.A. Pharmacological targeting of guanosine monophosphate synthase suppresses melanoma cell invasion and tumorigenicity. Cell Death Differ. 2015;22:1858–1864. doi: 10.1038/cdd.2015.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keller K.E., Tan I.S., Lee Y.S. SAICAR stimulates pyruvate kinase isoform M2 and promotes cancer cell survival in glucose-limited conditions. Science. 2012;338:1069–1072. doi: 10.1126/science.1224409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Azoitei N., Becher A., Steinestel K., Rouhi A., Diepold K., Genze F., Simmet T., Seufferlein T. PKM2 promotes tumor angiogenesis by regulating HIF-1alpha through NF-kappaB activation. Mol. Cancer. 2016;15:3. doi: 10.1186/s12943-015-0490-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spurr I.B., Birts C.N., Cuda F., Benkovic S.J., Blaydes J.P., Tavassoli A. Targeting tumour proliferation with a small-molecule inhibitor of AICAR transformylase homodimerization. Chembiochem. 2012;13:1628–1634. doi: 10.1002/cbic.201200279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun X., Li Y., Yu J., Pei H., Luo P., Zhang J. miR-128 modulates chemosensitivity and invasion of prostate cancer cells through targeting ZEB1. Jpn. J. Clin. Oncol. 2015;45:474–482. doi: 10.1093/jjco/hyv027. [DOI] [PubMed] [Google Scholar]
- 17.Zhao L., Li R., Xu S., Li Y., Zhao P., Dong W., Liu Z., Zhao Q., Tan B. Tumor suppressor miR-128-3p inhibits metastasis and epithelial-mesenchymal transition by targeting ZEB1 in esophageal squamous-cell cancer. Acta Biochim. Biophys. Sin. Shanghai. 2018;50:171–180. doi: 10.1093/abbs/gmx132. [DOI] [PubMed] [Google Scholar]
- 18.Guzman H., Sanders K., Idica A., Bochnakian A., Jury D., Daugaard I., Zisoulis D.G., Pedersen I.M. miR-128 inhibits telomerase activity by targeting TERT mRNA. Oncotarget. 2018;9:13244–13253. doi: 10.18632/oncotarget.24284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu W.W., Jiang H., Zhang C.T., Peng Y. The SNAIL/miR-128 axis regulated growth, invasion, metastasis, and epithelial-to-mesenchymal transition of gastric cancer. Oncotarget. 2017;8:39280–39295. doi: 10.18632/oncotarget.16849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shan Z.N., Tian R., Zhang M., Gui Z.H., Wu J., Ding M., Zhou X.F., He J. miR128-1 inhibits the growth of glioblastoma multiforme and glioma stem-like cells via targeting BMI1 and E2F3. Oncotarget. 2016;7:78813–78826. doi: 10.18632/oncotarget.12385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang J., Feng X., Zhou W., Wu Y., Yang Y. MiR-128 reverses the gefitinib resistance of the lung cancer stem cells by inhibiting the c-met/PI3K/AKT pathway. Oncotarget. 2016;7:73188–73199. doi: 10.18632/oncotarget.12283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han H., Wang L., Xu J., Wang A. miR-128 induces pancreas cancer cell apoptosis by targeting MDM4. Exp Ther Med. 2018;15:5017–5022. doi: 10.3892/etm.2018.6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li B., Chen H., Wu N., Zhang W.J., Shang L.X. Deregulation of miR-128 in ovarian cancer promotes cisplatin resistance. Int. J. Gynecol. Cancer. 2014;24:1381–1388. doi: 10.1097/IGC.0000000000000252. [DOI] [PubMed] [Google Scholar]
- 24.Goswami M.T., Chen G., Chakravarthi B.V., Pathi S.S., Anand S.K., Carskadon S.L., Giordano T.J., Chinnaiyan A.M., Thomas D.G., Palanisamy N., Beer D.G., Varambally S. Role and regulation of coordinately expressed de novo purine biosynthetic enzymes PPAT and PAICS in lung cancer. Oncotarget. 2015;6:23445–23461. doi: 10.18632/oncotarget.4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gallenne T., Ross K.N., Visser N.L., Salony Desmet C.J., Wittner B.S., LFA Wessels, Ramaswamy S., Peeper D.S. Systematic functional perturbations uncover a prognostic genetic network driving human breast cancer. Oncotarget. 2017;8:20572–20587. doi: 10.18632/oncotarget.16244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meng M., Chen Y., Jia J., Li L., Yang S. Knockdown of PAICS inhibits malignant proliferation of human breast cancer cell lines. Biol. Res. 2018;51:24. doi: 10.1186/s40659-018-0172-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Agarwal S., Saini S., Parashar D., Verma A., Sinha A., Jagadish N., Batra A., Suri S., Gupta A., Ansari A.S., Lohiya N.K., Suri A. The novel cancer-testis antigen A-kinase anchor protein 4 (AKAP4) is a potential target for immunotherapy of ovarian serous carcinoma. Oncoimmunology. 2013;2 doi: 10.4161/onci.24270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Agarwal S., Saini S., Parashar D., Verma A., Jagadish N., Batra A., Suri S., Bhatnagar A., Gupta A., Ansari A.S., Lohiya N.K., Suri A. Expression and humoral response of A-kinase anchor protein 4 in cervical cancer. Int. J. Gynecol. Cancer. 2013;23:650–658. doi: 10.1097/IGC.0b013e31828a0698. [DOI] [PubMed] [Google Scholar]
- 29.Chakravarthi B.V., Goswami M.T., Pathi S.S., Robinson A.D., Cieslik M., Chandrashekar D.S., Agarwal S., Siddiqui J., Daignault S., Carskadon S.L., Jing X., Chinnaiyan A.M., Kunju L.P. MicroRNA-101 regulated transcriptional modulator SUB1 plays a role in prostate cancer. Oncogene. 2016;35:6330–6340. doi: 10.1038/onc.2016.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chakravarthi B., Chandrashekar D.S., Agarwal S., Balasubramanya S.A.H., Pathi S.S., Goswami M.T., Jing X., Wang R., Mehra R., Asangani I.A., Chinnaiyan A.M., Manne U., Sonpavde G. miR-34a regulates expression of the stathmin-1 oncoprotein and prostate cancer progression. Mol. Cancer Res. 2018;16:1125–1137. doi: 10.1158/1541-7786.MCR-17-0230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Agarwal S., Behring M., Kim H.G., Bajpai P., Chakravarthi B., Gupta N., Elkholy A., Al Diffalha S., Varambally S., Manne U. Targeting P4HA1 with a small molecule inhibitor in a colorectal cancer PDX model. Transl. Oncol. 2020;13:100754. doi: 10.1016/j.tranon.2020.100754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Agarwal S., Behring M., Hale K., Al Diffalha S., Wang K., Manne U., Varambally S. MTHFD1L, a folate cycle enzyme, is involved in progression of colorectal cancer. Transl. Oncol. 2019;12:1461–1467. doi: 10.1016/j.tranon.2019.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jagadish N., Agarwal S., Gupta N., Fatima R., Devi S., Kumar V., Suri V., Kumar R., Suri V., Sadasukhi T.C., Gupta A., Ansari A.S., Lohiya N.K. Heat shock protein 70-2 (HSP70-2) overexpression in breast cancer. J. Exp. Clin. Cancer Res. 2016;35:150. doi: 10.1186/s13046-016-0425-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sinha A., Agarwal S., Parashar D., Verma A., Saini S., Jagadish N., Ansari A.S., Lohiya N.K., Suri A. Down regulation of SPAG9 reduces growth and invasive potential of triple-negative breast cancer cells: possible implications in targeted therapy. J. Exp. Clin. Cancer Res. 2013;32:69. doi: 10.1186/1756-9966-32-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rhodes D.R., Kalyana-Sundaram S., Mahavisno V., Varambally R., Yu J., Briggs B.B., Barrette T.R., Anstet M.J., Kincead-Beal C., Kulkarni P., Varambally S., Ghosh D., Chinnaiyan A.M. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Segara D., Biankin A.V., Kench J.G., Langusch C.C., Dawson A.C., Skalicky D.A., Gotley D.C., Coleman M.J., Sutherland R.L., Henshall S.M. Expression of HOXB2, a retinoic acid signaling target in pancreatic cancer and pancreatic intraepithelial neoplasia. Clin. Cancer Res. 2005;11:3587–3596. doi: 10.1158/1078-0432.CCR-04-1813. [DOI] [PubMed] [Google Scholar]
- 37.Pei H., Li L., Fridley B.L., Jenkins G.D., Kalari K.R., Lingle W., Petersen G., Lou Z., Wang L. FKBP51 affects cancer cell response to chemotherapy by negatively regulating Akt. Cancer Cell. 2009;16:259–266. doi: 10.1016/j.ccr.2009.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Badea L., Herlea V., Dima S.O., Dumitrascu T., Popescu I. Combined gene expression analysis of whole-tissue and microdissected pancreatic ductal adenocarcinoma identifies genes specifically overexpressed in tumor epithelia. Hepatogastroenterology. 2008;55:2016–2027. [PubMed] [Google Scholar]
- 39.Chandrashekar D.S., Bashel B., Balasubramanya S.A.H., Creighton C.J., Ponce-Rodriguez I., Chakravarthi B., Varambally S. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weaver A.N., Yang E.S. Beyond DNA repair: additional functions of PARP-1 in cancer. Front. Oncol. 2013;3:290. doi: 10.3389/fonc.2013.00290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xie C.G., Wei S.M., Chen J.M., Xu X.F., Cai J.T., Chen Q.Y., Jia L.T. Down-regulation of GEP100 causes increase in E-cadherin levels and inhibits pancreatic cancer cell invasion. PLoS One. 2012;7 doi: 10.1371/journal.pone.0037854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiong H., Hong J., Du W., Lin Y.W., Ren L.L., Wang Y.C., Su W.Y., Wang J.L., Cui Y., Wang Z.H., Fang J.Y. Roles of STAT3 and ZEB1 proteins in E-cadherin down-regulation and human colorectal cancer epithelial-mesenchymal transition. J. Biol. Chem. 2012;287:5819–5832. doi: 10.1074/jbc.M111.295964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gradiz R., Silva H.C., Carvalho L., Botelho M.F., Mota-Pinto A. MIA PaCa-2 and PANC-1 — pancreas ductal adenocarcinoma cell lines with neuroendocrine differentiation and somatostatin receptors. Sci. Rep. 2016;6:21648. doi: 10.1038/srep21648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang X., Luo G., Zhang K., Cao J., Huang C., Jiang T., Liu B., Su L., Qiu Z. Hypoxic tumor-derived exosomal miR-301a mediates M2 macrophage polarization via PTEN/PI3Kgamma to promote pancreatic cancer metastasis. Cancer Res. 2018;78:4586–4598. doi: 10.1158/0008-5472.CAN-17-3841. [DOI] [PubMed] [Google Scholar]
- 45.Chen P.Y., Muzumdar M.D., Dorans K.J., Robbins R., Bhutkar A., Del Rosario A., Mertins P., Qiao J., Schafer A.C., Gertler F., Carr S., Jacks T. Adaptive and reversible resistance to Kras inhibition in pancreatic cancer cells. Cancer Res. 2018;78:985–1002. doi: 10.1158/0008-5472.CAN-17-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pavlova N.N., Thompson C.B. The emerging hallmarks of cancer metabolism. Cell Metab. 2016;23:27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.DeBerardinis R.J., Chandel N.S. Fundamentals of cancer metabolism. Sci. Adv. 2016;2 doi: 10.1126/sciadv.1600200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cunningham J.T., Moreno M.V., Lodi A., Ronen S.M., Ruggero D. Protein and nucleotide biosynthesis are coupled by a single rate-limiting enzyme, PRPS2, to drive cancer. Cell. 2014;157:1088–1103. doi: 10.1016/j.cell.2014.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lane A.N., Fan T.W. Regulation of mammalian nucleotide metabolism and biosynthesis. Nucleic Acids Res. 2015;43:2466–2485. doi: 10.1093/nar/gkv047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Paul C.D., Mistriotis P., Konstantopoulos K. Cancer cell motility: lessons from migration in confined spaces. Nat. Rev. Cancer. 2017;17:131–140. doi: 10.1038/nrc.2016.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pai P., Rachagani S., Are C., Batra S.K. Prospects of miRNA-based therapy for pancreatic cancer. Curr Drug Targets. 2013;14:1101–1109. doi: 10.2174/13894501113149990181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Di Domenico M., Giordano A. Signal transduction growth factors: the effective governance of transcription and cellular adhesion in cancer invasion. Oncotarget. 2017;8:36869–36884. doi: 10.18632/oncotarget.16300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Talmadge J.E., Fidler I.J. AACR centennial series: the biology of cancer metastasis: historical perspective. Cancer Res. 2010;70:5649–5669. doi: 10.1158/0008-5472.CAN-10-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wirth M., Mahboobi S., Kramer O.H., Schneider G. Concepts to target MYC in pancreatic cancer. Mol. Cancer Ther. 2016;15:1792–1798. doi: 10.1158/1535-7163.MCT-16-0050. [DOI] [PubMed] [Google Scholar]
- 55.Leal A.S., Williams C.R., Royce D.B., Pioli P.A., Sporn M.B., Liby K.T. Bromodomain inhibitors, JQ1 and I-BET 762, as potential therapies for pancreatic cancer. Cancer Lett. 2017;394:76–87. doi: 10.1016/j.canlet.2017.02.021. [DOI] [PubMed] [Google Scholar]
- 56.Garcia P.L., Miller A.L., Kreitzburg K.M., Council LN, Gamblin T.L., Christein J.D., Heslin M.J., Arnoletti J.P., Richardson J.H., Chen D., Hanna C.A., Cramer S.L., Yang E.S. The BET bromodomain inhibitor JQ1 suppresses growth of pancreatic ductal adenocarcinoma in patient-derived xenograft models. Oncogene. 2016;35:833–845. doi: 10.1038/onc.2015.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1 Differential expression of de novo purine biosynthetic enzymes in PDAC. Gene expression profiling studies acquired from Oncomine show elevated expression of de novo purine biosynthetic enzymes in PDAC as compared to normal pancreatic tissue. The data were acquired from Badea data sets. Color scale highlights different expression as blue represents downregulation, white is no alteration, and red shows upregulation of transcripts.
Supplementary Figure S2 Cytoplasmic expression of PAICS in PDAC cells. Immunofluorescence analysis showing PAICS expression in PANC-1 and S2VP10 cells. Red color represents PAICS expression; DAPI binds to nucleus (blue color).
Supplementary Figure S3 PAICS siRNA knockdown decreased pancreatic cancer cell proliferation and colony formation. (A) Immunoblot analysis showed PAICS protein levels in PAICS siRNA-transfected pancreatic cancer cells PANC-1 and S2VP10 compared to NT siRNA. β-Actin was used as a loading control. (B) PAICS knockdown cells were seeded in 24-well plates for cell proliferation assay at 2, 4, and 6 days. (C) Colony formation was assessed by seeding 1000 cells in six-well plates. After 10 days, colonies were fixed with glutaraldehyde and stained with crystal violet. Statistically significant differences (*P < .05) are shown by an asterisk (*).
Supplementary Figure S4 PAICS siRNA knockdown impedes invasion and cellular motility of pancreatic cancer cells. (A) PAICS siRNA-treated pancreatic cancer cells showed fewer cells invading through Transwell Matrigel membranes relative to normal NT siRNA-treated cells. (B) Representative images showing wound healing for pancreatic cancer cells at 0 and 24 hours.
Supplementary Figure S5 PAICS knockdown inhibits tumor growth in the CAM model. (A) Representative images showing reduced tumor sizes of PAICS-deficient PDACs compared to tumors treated with NT shRNA. (B) Histogram showing suppressed tumor weights of PAICS-deficient S2VP10 tumors (n = 8) (*P < .05). (C) PAICS-depleted and control tumors were excised from CAMs, and immunoblot analyses were performed by probing with PAICS antibody. β-Actin was used as a loading control.
Supplementary materials