Abstract
Aims
Metabolic profiling is a top-down method of analysis looking at metabolites, which are the intermediate or end products of various cellular pathways. Our primary objective was to perform a systematic review of the published literature to identify metabolites in human synovial fluid (HSF), which have been categorized by metabolic profiling techniques. A secondary objective was to identify any metabolites that may represent potential biomarkers of orthopaedic disease processes.
Methods
A systematic review was conducted in accordance with Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) guidelines using the MEDLINE, Embase, PubMed, and Cochrane databases. Studies included were case series, case control series, and cohort studies looking specifically at HSF.
Results
The primary analysis, which pooled the results from 17 published studies and four meeting abstracts, identified over 200 metabolites. Seven of these studies (six published studies, one meeting abstract) had asymptomatic control groups and collectively suggested 26 putative biomarkers in osteoarthritis, inflammatory arthropathies, and trauma. These can broadly be categorized into amino acids plus related metabolites, fatty acids, ketones, and sugars.
Conclusion
The role of metabolic profiling in orthopaedics is fast evolving with many metabolites already identified in a variety of pathologies. However, these results need to be interpreted with caution due to the presence of multiple confounding factors in many of the studies. Future research should include largescale epidemiological metabolic profiling studies incorporating various confounding factors with appropriate statistical analysis to account for multiple testing of the data.
Cite this article: Bone Joint Res. 2020;9(3):108–119.
Keywords: Metabonomics, Metabolic profiling, Osteoarthritis, Rheumatoid arthritis, Inflammatory arthropathies
Article focus
To identify all metabolites in human synovial fluid (HSF), which have been categorized by metabolic profiling techniques.
To recognize any metabolites that may represent potential biomarkers of orthopaedic disease processes.
Key messages
Over 200 metabolites have been identified in HSF from the published literature.
A total of 26 putative biomarkers have been demonstrated in osteoarthritis, inflammatory arthropathies, and trauma.
The results should be interpreted with caution due to the presence of multiple confounding factors.
Strengths and limitations
The study methodology was robust.
The search criteria were broad to ensure all relevant articles were captured.
There was notable heterogeneity between studies.
Introduction
Osteoarthritis (OA) is one of the most disabling conditions in the western world, affecting approximately 10% of the UK population and presenting a major healthcare burden. It is a heterogenous disease, which manifests in a number of different phenotypes due to various pathogenic factors, ultimately leading to an alteration of the whole joint structure.1 It results in progressive degradation of ligaments, cartilage and menisci, synovial inflammation, and changes to the subchondral bone with common clinical and radiological manifestations.2
The risk factors for OA are multifactorial and involve a complex interplay between biochemical, cellular, and mechanical factors that ultimately lead to the same endpoint. Consequently, the risk factors for OA can vary among individuals.3
Rheumatoid arthritis (RA) is a chronic autoimmune disease characterized by autoantibodies, systemic inflammation, and synovitis leading to damage of the affected joints.4 Early diagnosis is important to delay disease progression by starting early intervention. A well-known biomarker of RA is rheumatoid factor (RF). However, this is non-specific and detected in other rheumatic and non-rheumatic conditions such as malignancy, infection, and even in some normal individuals.5 Anticitrullinated protein antibodies (ACPAs) are other biomarkers that have been suggested as a useful tool to differentiate RA from other types of arthritis in the 2010 American College of Rheumatology/European League Against Rheumatism (ACR/EULAR) classification criteria.6 However, as not all RA patients are seropositive for ACPA more reliable diagnostic biomarkers are still required.
Various ‘-omics’ technologies including proteomics, transcriptomics, and genomics have been increasingly utilized for the identification of disease biomarkers including those for RA. Transcriptomics has helped discover defence-related and immunity genes in RA patients and to predict the effectiveness of infliximab, the anti-tumour necrosis factor-α (TNF-α) biological agent, in RA patients.7,8 Furthermore, genomics has demonstrated differences between ACPA-positive and ACPA-negative diseases.9
Metabolic profiling (also known as metabolic phenotyping, metabolomics, and metabonomics) is an increasingly used approach, which studies the low-molecular-weight metabolites within a cell, tissue, or biofluid. These terms have been used interchangeably, leading to some confusion. Therefore, in this article, the term ‘metabolic profiling’ will be used, which is defined as “an individual’s metabolic pattern that would be reflected in the constituents of their biological fluids.”10
Metabolic profiling is a top-down method of analysis as it is looking at the metabolites, which are the intermediate or end products of various cellular pathways.11 Analyzing their concentrations provides a useful avenue to understanding the relationship of their cellular processes and biological reactions.12 As well as genetic factors, this process accounts for various environmental factors such as diet, medication, smoking, and disease. Typically, it is conducted with biofluids, the most common of which are blood serum/plasma and urine. It can lead to the formation of a ‘metabolic fingerprint’, which is unique to a particular biochemical perturbation, characteristic of a particular disease process, or toxic stimulus among other things.13
Metabolic profiling has the ability to detect and potentially quantify hundreds or even thousands of small molecules simultaneously. The most common techniques employed are nuclear magnetic resonance (NMR) spectroscopy and mass spectrometry (MS). NMR spectroscopy is based on the same physical principles as MRI. It uses the magnetic property of the nuclei called spin to study the interaction of nuclei of the different atoms in a molecule, being therefore useful to determine the structures of molecules. The most commonly used nuclei is the proton (1H) due to its natural abundance in nature (close to 100%).14 NMR spectroscopy is fast and non-destructive, allowing multiple samples to be measured daily, and the same sample can be analyzed multiple times.15 MS is more sensitive with greater metabolite coverage than NMR spectroscopy, but it often requires prior separation of the different types of compounds using chromatography. Liquid chromatography (LC), particularly ultra-high-performance liquid chromatography (UPLC) is being more frequently used due to its increased compound resolution and higher throughput. Other techniques include gas chromatography-mass spectrometry (GC-MS), which is more useful for volatile compounds. Regarding biofluids, LC-MS is typically employed, usually with both positive and negative ion detection modes using standard protocols.16 However, MS does involve sample consumption, thus preventing multiple testing of the same sample.15 It has been used successfully in clinical medicine, toxicology, environmental science, and plant science.17–21 It has also been employed in a number of conditions to influence clinical practice.19,22,23 The various metabolic profiling techniques are often used together to provide a wider coverage of the metabolic space.
Metabolic profiling may be well suited for the purposes of orthopaedic research due to the great heterogeneity of the different disease processes including OA and inflammatory arthropathies such as RA, with recognition that no single biomarker is capable of explaining the breadth of pathological and temporal processes associated with these conditions.24 Combining several biomarkers would also increase its discriminatory capacity.25 Furthermore, as metabolic perturbations occur in real time, they indicate the current disease state, thus providing a distinct advantage over other disease monitoring and diagnostic techniques such as radiography.
More recently, metabolic profiling has been used to identify metabolites within the urine, blood, and synovial fluid (SF) of both animal models and patients with OA.26–28 Changes in joint metabolism may be a contributing factor to the pathogenesis of OA.29 Previous metabolic analysis of SF has led to a better understanding of the metabolic processes associated with OA and to the identification of some of the biomarkers of OA.30,31
The aim of this systematic review was to identify metabolites in human SF (HSF), which have been categorized by metabolic profiling techniques. The secondary aim was to identify any metabolites that may represent potential biomarkers of orthopaedic disease processes.
The scope of this systematic review is to look at the role of metabolic profiling in identifying the small molecule metabolites in HSF and identify any that may represent putative biomarkers, specifically using the techniques associated with metabolic profiling including MS and NMR spectroscopy. Therefore, studies looking at macromolecules including cytokines and interleukins (ILs), plus studies utilizing the techniques of genomics, proteomics, and transcriptomics, were considered outside the scope of this article.32–35
Methods
A systematic review was undertaken in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) guidelines.36
Eligibility criteria
Inclusion criteria consisted of published articles and abstracts in English looking at small molecule metabolism of HSF in any disease state using metabolic profiling techniques. The exclusion criteria were articles not written in English, patients less than 18 years old, expert opinions, review articles, and studies using the same cohort of patients.
Identification of studies
A systematic literature review was conducted of the MEDLINE (Medical Literature Analysis and Retrieval System Online), Embase (Excerpta Medica Database, Amsterdam, The Netherlands), PubMed, and Cochrane databases without date restrictions on 1 August 2018. The search terms used are detailed in Supplementary Table i.
Screening and assessment of eligibility
Two independent reviewers (PA and UK) looked at the titles of the articles identified in the preliminary literature search. Any disagreement resulted in the article proceeding to the next stage of review. The same authors then read the abstracts of the remaining articles. Any disagreement resulted in the articles proceeding to full-text review. The full-text articles were then reviewed by the same authors and any conflict was discussed to achieve consensus.
Risk of bias (quality) assessment
The articles were evaluated for relevance, sample numbers, the underlying disease process, statistical power, analytical validity, quality of evidence, and conclusions. The Newcastle-Ottawa Scale was used to evaluate the study design. Relevant metabolites were highlighted and where statistical testing was performed, significance was quoted. The metabolites themselves were identified using various commercial software packages such as Chenomx NMR Suite (Chenomx, Edmonton, Canada), as well as by identifying them from published databases including the Human Metabolome Database,37 the Biological Magnetic Resonance Bank,38 and various in-house databases.
Results
Literature search
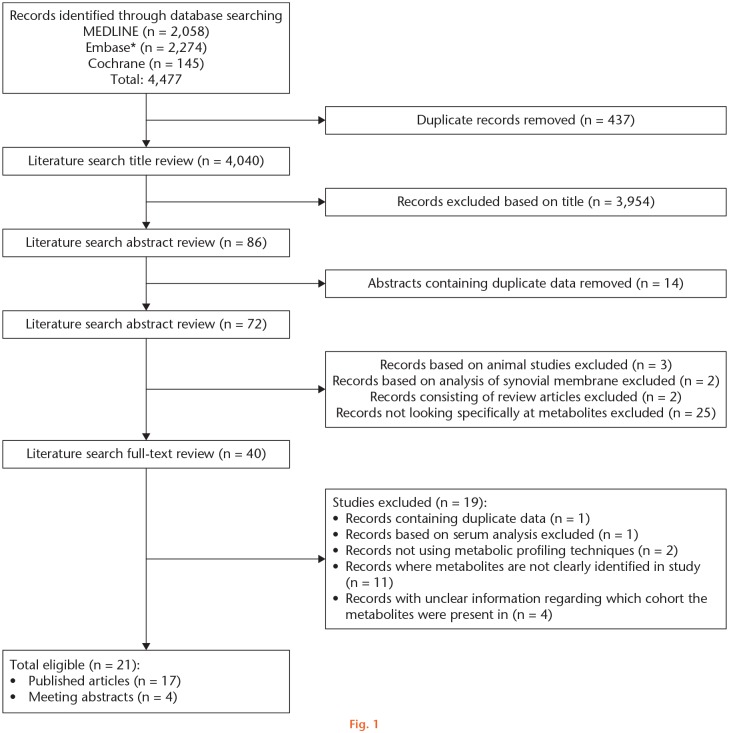
The electronic database searches identified 4,477 articles. Following exclusion of any duplicates and reviewing the titles, 4,391 articles were excluded. The abstracts of the remaining 86 articles were reviewed and a further 14 were excluded as they contained duplicate data. Of the remaining 72 articles, 25 were excluded for not meeting the entry criteria, two were removed as they contained data from the same cohort, three were excluded because they did not look at HSF, and two were removed as they looked specifically at synovial membranes and not SF. Of the remaining 40 articles, one article was excluded as it had duplicate data, one was excluded as it only looked at serum and not SF, two were excluded for not using metabolic profiling techniques, 11 were removed because the metabolites were not clearly identified or only a portion of them were presented, and four were excluded as it was unclear in these articles which cohort the metabolites were found in greater quantities. As a result, 21 studies were eventually used (17 articles and four abstracts) (Figure 1).
Fig. 1.
Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) search and screening flowchart for the role of metabolic profiling in human synovial fluid research. *Excerpta Medica Database, Amsterdam, The Netherlands.
Study characteristics and quality
The methodology of the published studies was assessed using the Cochrane criteria for bias and the Newcastle-Ottawa Scale. The studies included had similar designs and metabolic profiling techniques. Patient selection was not random, as all studies were looking at specific disease processes. Furthermore, blinding was not possible at sample collection for either the researcher or the patient. Multivariate analysis was performed to detect patterns of changes in the metabolites detected, which did not necessarily involve significance testing. These types of analyses were performed mostly using supervised methods, which required information about the sample class, and therefore the data could not be blinded. As p-values were not consistently reported in all the studies, reporting bias may exist towards those that do so. Furthermore, studies that involve assaying hundreds of metabolites may have overestimated the significance of the p-values, unless false discovery rate (FDR) or validation datasets were utilized. All the identified studies are listed in Table I.39–61
Table I.
Baseline description of all the studies included in this systematic review
| First author/year | Study design | Country of origin | Joint | Diagnosis | Disease staging | Sample size | Type of analysis | Validated analysis | Controls | Statistical validity | NOS |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Adams et al, 201439 | Case control study | USA | Ankle | Radiological | Takakura grading | n = 20; c = 20 | UHPLC-MS/MS | Weak | Healthy asymptomatic patients | Adequate | 7 |
| Ahn et al, 201540 | Case series | South Korea | N/A | Clinical | N/A | n = 24 | GC-TOF-MS | Strong | None | Adequate | 3 |
| Anderson et al, 201841 | Cohort study | UK | Knee | N/A | N/A | n = 10 (OA); n = 14 (RA) | 1H-NMR | N/A | None | Adequate | 0 |
| Carlson et al, 201842 | Case control study | USA | N/A | N/A | N/A | n = 5 (OA); n = 3 (RA); c = 5 | LC-MS | Weak | Post-mortem samples | Adequate | 3 |
| Chen et al, 201843 | Case control study | China | Knee | Clinical/radiological | KL | n = 32; c = 35 | UHPLC-TQ-MS | Weak | Healthy asymptomatic patients | Adequate | 8 |
| Dubey et al, 201944 | Case series | India | Knee | N/A | N/A | n = 8 | 1H-NMR | N/A | None | Adequate | 0 |
| Dubey et al, 201745 | Cohort study | India | Knee | Clinical | N/A | n = 19 (ReA); n = 13 (USpA) | 1H-NMR | N/A | None | Adequate | |
| Dubey et al, 201946 | Case control study | India | Knee | Clinical | Braun's, ASAS, and ACR criteria | n = 52 (SSA); n = 29 (RA); c = 82 | 1H-NMR | Weak | Healthy asymptomatic patients | Adequate | 6 |
| Furman et al, 201747 | Case control study | USA | Knee | Clinical | Not applicable | n = 8; c = 8 | UHPLC-MS/MS | N/A | Contralateral non-injured knee | Adequate | 7 |
| Hwang et al, 201348 | Cohort study | South Korea | N/A | N/A | N/A | n = 18 (RA); n = 11 (OA) | GC-TOF-MS | N/A | None | Adequate | 6 |
| Kang et al, 201549 | Case series | South Korea | Knee | Clinical/radiological | KL (OA); ACR (RA) | n = 10 (OA); n = 10 (RA) | UPLC-QTOF-MS | Weak | None | Adequate | 5 |
| Khatib et al, 201850 | Case series | UK | Knee | N/A | N/A | n = 13 | 1H-NMR | N/A | None | Adequate | 3 |
| Kim et al, 201751 | Case series | South Korea | Knee | Clinical/radiological | KL | n = 8 (KL1 to 2); n = 7 (KL3 to 4) | GC-TOF-MS | Strong | None | Adequate | 4 |
| Kim et al, 201452 | Case series | South Korea | N/A | Clinical/radiological | ACR for RA; ASAS for AS; criteria of the 1990 ISG for BD; MSU crystals in joint fluid for gout. | n = 13 (RA); n = 7 (AS); n = 5 (BD); n = 13 (gout) | GC-TOF-MS | Adequate | None | Adequate | 4 |
| Leimer et al, 201753 | Cohort study | USA | Ankle | Radiological | N/A | n = 19; c = 19 | UHPLC-MS/MS | Adequate | Contralateral non-injured ankle | Adequate | 8 |
| Meshitsuka et al, 199954 | Case series | Japan | Knee | Clinical/radiological | ACR | n = 14 (RA); n = 16 (OA) | 1H-NMR | Adequate | None | Adequate | 2 |
| Mickiewicz et al, 201555 | Cohort study | Canada | Knee | Clinical/radiological | N/A | n = 55; c = 13 (cadaveric - 6 bilateral/1 unilateral sample) | 1H-NMR; GC-MS | Strong | Cadaveric controls | Adequate | 6 |
| Naughton et al, 199356 | Cohort study | UK | Knee | N/A | N/A | n = 22 (RA); c = 6 | 1H-NMR | Adequate | Healthy asymptomatic patients | Adequate | 5 |
| Yang et al, 201557 | Case control study | China | Knee | ACR | N/A | n = 25 (RA); c = 10 | GC-TOF-MS | Adequate | Above knee amputated patients | Adequate | 6 |
| Zhang et al, 201458 | Case series | Canada | Hip/knee | ACR | ESOA | n = 80 | LC-MS | Adequate | None | Adequate | 5 |
| Zhang et al, 201559 | Case series | Canada | Knee | N/A | ESOA | n = 69 | LC-MS | Adequate | None | Adequate | 5 |
| Zhang et al, 201660 | Case control study | Canada | Knee | ACR criteria and clinical judgement | ESOA | n = 97 | LC-MS | Adequate | No SF sample controls (only serum) | Adequate | 6 |
| Zheng et al, 201761 | Cohort study | China | Knee | KL | KL2 and KL4 | n = 49, c = 21 | GC-TOF-MS and LC-MS/MS | Adequate | Asymptomatic patients | Adequate | 7 |
1H-NMR, nuclear magnetic resonance spectroscopy; ACR, American College of Rheumatology; AS, ankylosing spondylitis; ASAS, Assessment of SpondyloArthritis international Society; BD, Behçet’s disease; C, control group; ESOA, end-stage osteoarthritis; GC-MS, gas chromatograph-mass spectrometry; GC-TOF-MS, gas chromatography/time-of-flight mass spectrometry; ISG, International Study Group; KL, Kellgren and Lawrence; LC-MS, liquid-chromatography mass spectrometry; MSU, monosodium urate; N/A, not available; NOS, Newcastle-Ottawa Scale; OA, osteoarthritis; RA, rheumatoid arthritis; ReA, reactive arthritis; SSA, seronegative spondyloarthropathy; TQ MS, triple quadrupole mass spectrometry; UHPLC-MS/MS, ultra-high performance liquid chromatography/tandem mass spectrometry; UPLC-QTOF-MS, ultraperformance liquid chromatography quadruple time-of-flight mass spectrometer; UHPLC-TQ-MS, ultra-high performance liquid chromatography triple quadrupole mass spectrometry; USpA, undifferentiated spondyloarthropathy.
All the identified metabolites have been listed in Supplementary Table ii. The identified studies have been subdivided into those with healthy controls that have identified putative biomarkers and those looking at specific disease processes.
Studies with a healthy control group
Adams et al39 examined the cytokine and metabolic differences between healthy and end-stage post-traumatic arthritic ankle (PTAA) joint SF. They identified 29 metabolites in significantly different concentrations between the PTAA and control groups, the most important of which is glutamate. Their findings suggest a mainly oxidative and proinflammatory environment with an imbalance in amino acid (AA) and lipid metabolism among other factors. However, there are no p-values stated in the paper and no FDR or other analysis was performed to account for multiple testing.
The metabolic changes in the physiological responses of early knee OA were performed by Chen et al.43 They identified 22 significant metabolic differences between the two groups. Most serum AA levels were found to be altered in the OA group, suggesting that OA is accompanied and precipitated by changes in AA metabolism. They identified three potential biomarkers: alanine, γ-aminobutyric acid (GABA), and 4-hydroxy-L-proline (Hyp). Alanine and Hyp were increased in the OA group and GABA was reduced in the OA group.
Dubey et al46 explored whether metabolic profiling would identify a distinctive metabolic signature of seronegative spondyloarthropathy (SSA) that is not influenced by age and sex. Their control group consisted of two subgroups of healthy patients, who were stratified by age, creating a young and older control group. There were a number of patient cohorts consisting of those with reactive arthritis (ReA), SSA, and RA. They suggested low-density lipoprotein (LDL), very low-density lipoprotein (VLDL), leucine, lysine/arginine, acetone, glycine, glucose, creatine, polyunsaturated fatty acids (PUFAs), and phenylalanine as putative biomarkers for ReA when compared to an age-matched control group. Conversely, leucine, lysine/arginine, phenylalanine, and valine were suggested as putative biomarkers for discriminating between ReA and RA.
An abstract published by Furman et al47 analyzed healthy and injured knees to identify any metabolic pathways affected by the knee injury and identify any discriminatory SF biomarkers. They demonstrated significantly increased sphingomyelin (SPM) and 2-hydroxy-fatty acids in the injury group. They suggested that these may be potential SF biomarkers of knee injury and may be prognostic indicators of the risk of post-traumatic arthritis.
Leimer et al53 attempted to characterize the global metabolic profile of SF after intra-articular ankle fractures with an emphasis on changes in the lipid profile. They identified 16 lipid-based metabolites found in significantly greater quantities following an intra-articular ankle fracture, which subsequently decreased six months post-surgery. Most long-chain fatty acids (FAs) and PUFAs were acutely elevated in the fractured ankles at baseline compared to the control group. However, none of these were suggested as potential biomarkers. They suggest the distinctive lipid signature identified is reflective of injury, fracture, and early changes associated with OA.
The metabolic status of normal and RA SF was assessed qualitatively by Naughton et al.56 They demonstrated increased levels of LDL in RA compared to the control group and suggested this to be secondary to inflammation and increased synovial membrane permeability. However, the controls were not age-matched (25 to 42 years old) and were younger than the RA group (40 to 67 years old), which may result in important metabolic differences.
Zheng et al61 explored the metabolites of OA. Six metabolites were significantly different between the two groups. Three were found in significantly greater concentrations in the OA group and three in the control group. Gluconic lactone, threonine, and 1,5-Anhydroglucitol (1,5-AG) were in significantly greater concentrations in OA SF compared to the control group. Glutamine, tyramine, and 8-aminocaprylic acid were in significantly lower concentrations in OA SF compared to the control group. The authors concluded that a new diagnostic model combining two metabolites provides greater sensitivity in diagnosing OA than a single metabolite alone. Furthermore, gluconic lactone may prove to be a novel benchmark for the differential diagnosis of OA from RA due to the significant differences in the concentration of this metabolite between these conditions with a high level of sensitivity and specificity between them.
None of these studies performed FDR or other analysis to account for multiple testing of the data. Consequently, the results must be reviewed with caution. The metabolites identified in this section, which have been proposed to serve as putative biomarkers, are listed in Table II.43,46,47,56,61
Table II.
Putative biomarkers identified from studies with an asymptomatic control group. All metabolites were identified from human knee synovial fluid.
| Underlying pathology | Metabolite | Change | Multivariate analysis |
|---|---|---|---|
| OA | Alanine43 | Increased in OA | VIP 3.31, p < 0.001 |
| Hyp43 | VIP 1.75, p < 0.001 | ||
| Gluconic lactone61 | FC 1.54, p < 0.05 | ||
| Threonine61 | FC 2.71, p < 0.05 | ||
| 1,5-AG61 | FC 1.67, p < 0.05 | ||
| GABA43 | Decreased in OA | VIP 2.61, p < 0.001 | |
| Glutamine61 | FC 0.28, p < 0.05 | ||
| Tyramine61 | FC 0.30, p < 0.05 | ||
| 8-Aminocaprylic acid61 | FC 0.27, p < 0.05 | ||
| Inflammatory arthropathies | Acetone46 | Increased in ReA | FC 1.54, p < 0.006 |
| Creatine46 | FC 0.63, p < 0.001 | ||
| VLDL46 | N/A | ||
| Glucose46 | FC 1.12, p < 0.367 | ||
| Glycine46 | FC 1.03, p < 0.02 | ||
| LDL46 | N/A | ||
| Leucine46 | FC 0.83, p < 0.051 | ||
| Lysine/arginine46 | FC 0.78/1.21, p < 0.002/p < 0.46 | ||
| Phenylalanine46 | FC 1.33, p < 0.122 | ||
| PUFA46 | N/A | ||
| Leucine46 | Increased in ReA vs RA | FC 1.88, p < 0.001 | |
| Lysine/arginine46 | FC 1.46/2.07, p < 0.005/p < 0.001 | ||
| Phenylalanine46 | FC 2.56, p < 0.001 | ||
| Valine46 | FC 1.57, p < 0.001 | ||
| RA | LDL56 | Increased in RA | N/A |
| Knee injury | SPM47 | Increased in knee trauma | p < 0.0065 following FDR |
| 2-hydroxy-fatty acids47 | p < 0.0065 following FDR |
1-5 AG, 1,5-Anhydroglucitol; FC, fold change; FDR, false discovery rate; GABA, γ-aminobutyric acid; Hyp, 4-hydroxy-L-proline; LDL, low-density lipoprotein; N/A, not available; OA, osteoarthritis; PUFA, polyunsaturated fatty acid; RA, rheumatoid arthritis; ReA, reactive arthritis; SPM, sphingomyelin; VIP, variable importance on projection score; VLDL, very low-density lipoprotein.
Osteoarthritis studies
Mickiewicz et al62 identified two metabolites found in significantly greater concentrations in OA SF (fructose and citrate) and nine metabolites (O-acetylcarnitine, hexanoylcarnitine, N-phenylacetylglycine, ethanol, ethanolamine, methionine, malate, creatine, and 3-hydroxybutyrate) found in lower concentrations in OA SF compared to cadaveric controls.
Kim et al51 characterized the metabolite differences between early- and late-stage OA. They identified 28 metabolites as being significantly different between the groups, with all 28 increased significantly in late-stage OA.
Zhang et al58 examined the metabolic markers in SF that can be used to classify patients with OA into distinct subgroups. Broadly speaking, they identified numerous metabolites included 40 acylcarnitines (one free carnitine), 20 AAs, nine biogenic amines, 87 glycerophospholipids, 11 sphingolipids, and one hexose (> 90% was glucose). Following multivariate analysis, they identified subgroups of OA, which differed in acylcarnitine levels and fat metabolism. They observed distinctions in the glycophospholipids and SPMs. However, as no age-matching and no correlation to clinical factors took place, it is difficult to draw definitive conclusions.
Using a similar methodology, Zhang et al63 later investigated the differences between OA and type II diabetes mellitus. Of note, leucine and phosphatidylcholine (PC) metabolism were influenced by both diabetes mellitus and OA. Phosphatidylcholine is involved in many membrane-related phenomena including forming the essential lipid bilayer of all biological membranes, regulation of membrane trafficking, and signal transduction.64
An abstract by Khatib et al50 investigated whether the mechanical loading of the joint during pivot shift will reveal a profile of mechanically regulated metabolic biomarkers in patients with ACL deficient knees. Using nuclear magnetic resonance spectroscopy (1H-NMR), they identified a significant difference in alanine and choline between pre- and post-pivot shift testing of ACL deficient knees. These metabolites remained significant when accounting for multiple testing and the authors suggest they might be useful for rehabilitation or surgical intervention in patients with knee injuries who may be at risk of post-traumatic OA.
Inflammatory arthropathy studies
A recent study explored RA-related biochemical abnormalities by analyzing the metabolic profile of knee SF from RA patients and a control group.57 These controls were patients who had a ‘high-level’ amputation. However, the paper does not state the reason for the amputation nor whether the sample was taken before or after amputation, which may have important metabolic consequences. Following multivariate analysis and using a variable projection of importance score (VIP) > 1 plus p < 0.05, 13 of these metabolites were significant between the two groups. Glucose was decreased and lactic acid was increased in RA SF. Levels of glucose-1-phosphate and D-mannose were also decreased.
Ahn et al40 evaluated the metabolomic profile of SF in patients with Behçet’s disease (BD) with arthritis compared to those with seronegative arthritis (SNA). They identified 11 metabolites as being significantly increased in BD with arthritis compared to SNA. These include branched-chain AAs (BCAA: valine, leucine, and isoleucine), citramalate, glutamate, and methionine sulfoxide.
In an earlier study, Kim et al52 also evaluated potential biomarkers for RA. Their study consisted of patients with RA (n = 13), ankylosing spondylitis (AS) (n = 7), BD (n = 5), and gout (n = 13). These patients were then combined into two groups, which were RA and non-RA. They identified 20 metabolites that remained significantly different between the two groups following robust statistical analysis, which they proposed could be putative biomarkers. Of these, 14 were in significantly greater concentrations in the RA group and six were in greater concentrations in the non-RA group.
Osteoarthritis versus rheumatoid arthritis studies
Carlson et al42 evaluated global liquid chromatography coupled to mass spectrometry (LC-MS) based metabolic profiles as a tool for quantifying biomarkers within SF. Their control group consisted of five purchased post-mortem samples. It is unclear how long after death these samples were harvested and the death to post-mortem interval is likely to be a major confounding variable, so this uncertainty might have important metabolic consequences.65 They identified five metabolites (citric acid, D-lactic acid methyl ester, hydroxyl-L-proline, L-isoleucine, and L-methionine) found in significantly lower concentrations in OA and RA SF, compared to controls and one metabolite (L-citrulline) found in greater concentrations in OA compared to RA and controls. The authors also performed FDR analysis to account for multiple testing.
A recently published abstract explored the role of NMR spectroscopy in producing analyzable spectra from a low volume of SF taken in a clinical environment.41 They identified 11 metabolites found in significantly different concentrations between OA and RA SF. Seven were more abundant in OA and six were more abundant in RA SF. Their analysis suggested the metabolic pathways most impacted were: aminoacyl-transfer RNA (tRNA); biosynthesis; nitrogen metabolism; valine, leucine, and isoleucine biosynthesis; glycine, serine, and threonine metabolism; and taurine and hypotaurine metabolism. The authors allude to their methodology being useful for analyzing low-volume SF. However, in their methodology they state that each sample consists of approximately 100 ml. Furthermore, although the authors mention the term “FDR < 0.05”, it is unclear exactly what analysis took place.
Another abstract investigated the metabolites of SF in patients with RA and OA to identify the characteristic metabolites differentiating the two diseases.48 Using gas chromatography/time-of-flight mass spectrometry (GC/TOF MS) and following multivariate analysis, they identified 17 metabolites as being found in significantly different concentrations between the two groups. Six were upregulated in RA (maltose, lignoceric acid, uracil, mannitol, pyrophosphate, and phosphoric acid) and 11 in OA (lysine, tyrosine, valine, glyceric acid, alanine, asparagine, hydroxylamine, tryptophan, glycerol, glutamine, and citrulline).
Kang et al49 identified 21 metabolites as being in significantly different concentrations between RA and OA SF. Concentrations of lipid metabolites were typically higher in RA than OA SF, which has previously been demonstrated.66 Concentrations of tryptophan metabolites also differed significantly between the two groups.
The ratios of lactate and alanine have also been shown to be significantly greater in RA than in OA SF.54
Discussion
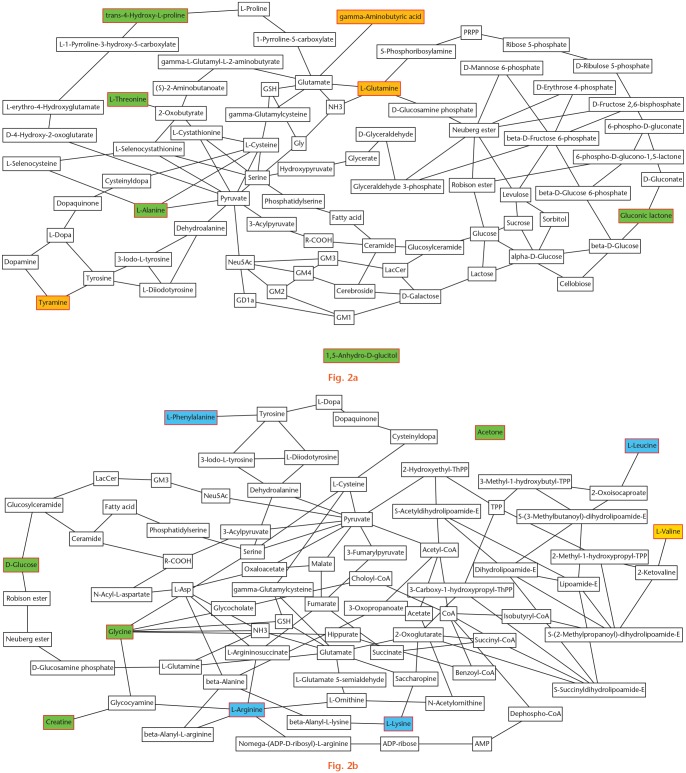
Metabolic profiling is an invaluable method of differentiating patients with various pathologies from healthy individuals in the clinical setting. This systematic review has identified numerous metabolites in different pathologies. Specifically, a few putative biomarkers have been illustrated. The studies identified have further demonstrated the important role of lipid mediators and metabolism in both OA and RA. However, its role in RA and other inflammatory arthropathies is more prominent. Furthermore, this article has summarized some of the putative biomarkers identified in the literature, although further studies are required to confirm and determine their significance (Table II). Looking specifically at these metabolites, collation of these results illustrates how metabolic changes may be interlinked in OA and inflammatory arthropathies, while postulating the potential metabolic pathways that may be affected (Figure 2).
Fig. 2.
Metabolic network analysis of all the putative biomarkers identified in this systematic review demonstrating the associated metabolic pathways. All metabolites with a red outline were putative biomarkers. a) Putative biomarkers identified in osteoarthritic synovial fluid (SF). Those in green were raised and those in orange were reduced in osteoarthritic SF compared to an asymptomatic control group. b) Putative biomarkers identified in inflammatory arthropathies. Those in green and blue were raised in reactive arthritis (ReA) compared to an asymptomatic control group; those in blue were also raised in ReA compared to rheumatoid arthritis (RA); valine (in yellow) was raised in ReA compared to RA. ADP, adenosine 5'-diphosphate; AMP, adenosine 5'-monophosphate; CoA, coenzyme A; GD1a, N-acetylneuraminyl-D-galactosyl-N-acetyl-D-galactosaminyl-(N-acetylneuraminyl)-D-galactosyl-D-glucosylceramide; Gly, glycine; GM1, D-galactosyl-N-acetyl-D-galactosaminyl-(N-acetylneuraminyl)-D-galactosyl-D-glucosylceramide; GM2, N-acetyl-D-galactosaminyl-(N-acetylneuraminyl)-D-galactosyl-D-glucosylceramide; GM3, (N-acetylneuraminyl)-D-galactosyl-D-glucosylceramide; GM4, N-acetylneuraminyl-galactosylceramide; GSH, reduced glutathione; LacCer, lactosylceramide; L-Asp, L-aspartic acid; Neu5Ac, N-acetylneuraminic acid; NH3, ammonia; PRPP, 5-phosphoribosyl 1-pyrophosphate; R-COOH, carboxylic acid; ThPP, thiamin pyrophosphate; TPP, thiamin pyrophosphate.
The metabolic homeostasis within a joint is often disturbed in the disease state leading to an anaerobic state secondary to stress and inflammation. Whether these changes differ between the diseased joint and the normal joint is an important question when considering the importance of diagnostic or prognostic putative biomarkers.
Role of the identified putative biomarkers
Broadly speaking, the putative biomarkers in OA can be classified into two main groups: AAs plus related metabolites (alanine, Hyp, threonine, GABA, glutamine, tyramine); and sugars plus related metabolites (gluconic lactone, 1,5-AG). Alanine, Hyp, and threonine are found in articular cartilage.67 Their increase in OA SF could be associated with increased catabolism of the articular cartilage. This may also represent increased energy consumption to account for the increased bone turnover and subchondral sclerosis seen in OA. 1,5-AG is a monosaccharide occasionally used as a short-term marker of glycaemia.68 Elevation of this metabolite in SF is consistent with the reduced glucose concentration in OA SF,69 secondary to increased energy expenditure. Increased gluconic lactone in OA SF may be due to auto-oxidation induced by increased levels of reactive oxygen species (ROS). ROS are able to directly induce cartilage degradation by cleaving aggrecan and collagen plus activating matrix metalloproteinases (MMPs).70
GABA arises from glutamic acid,71 which regulates glucose, also suggesting increased energy consumption in the diseased joint due to less residual glucose. Glutamine has a role in oxidative metabolism, and reduced levels suggest altered oxidative metabolism in diseased joints secondary to increased energy expenditure.72 Glutamine has been shown to supress inflammatory cytokines73 and protect chondrocytes from heat stress and nitrous oxide (NO)-induced apoptosis.74 These effects may protect chondrocytes from various types of stress and prevent progressive cartilage degeneration in OA. Tyramine is derived from the AA tyrosine, which is thought to have a role in promoting osteophyte formation. Increased levels have been seen in subchondral bone.55,75
The putative biomarkers increased in inflammatory arthropathies can be classified into four main groups: AAs and related metabolites (creatine, glycine, leucine, lysine, arginine, phenylalanine, valine); lipids and lipoproteins (LDL, VLDL, PUFA); sugars (glucose); and ketone bodies (acetone). They identified that AAs are all constituents of articular cartilage with leucine, proline, glutamic acid, and glycine specifically being constituents of proteoglycans.67 Their increase suggests breakdown of the articular cartilage, likely related to the underlying inflammatory process. Low concentrations of FAs have been demonstrated in HSF. The increased levels of LDL, VLDL, and PUFA identified here are secondary to increased synovial membrane permeability and inflammation associated with underlying inflammatory arthropathies.56
Metabolic changes seen in osteoarthritis
Fructose elevation suggests a hypoxic condition of the diseased and inflamed knee joint. Hypoxia has been shown to result in the upregulation of glucose phosphate isomerase, which catalyzes the conversion of glucose-6-phosphate (G6P) into fructose-6-phosphate (F6P) in inflammatory arthritis.76 Lower concentrations of O-acetylcarnitine, hexanoylcarnitine, N-phenylacetylglycine, and ethanolamine indicate protracted FA and lipid metabolism in the SF of OA patients compared to controls.37 Decreased methionine concentrations indicate its use, where it is likely converted to S-adenosylmethionine (SAM), a proposed factor for cartilage damage repair and inflammatory reduction.77
Kim et al51 identified three unique pathways in their study, which corresponded to the metabolic differences they identified. These were FA metabolism, glycolipid metabolism, and the tricarboxylic acid (TCA) cycle. These pathways may be associated with an increasing degree in the severity of OA. Glycerol and various FA concentrations were more prominent in the late-stage OA group. Their findings suggest that FA biosynthesis is predominantly responsible for energy generation in late-stage OA. Furthermore, increased concentrations of malate in the late-stage OA group compared to the early-stage group suggest a possible difference in the energy level between the two groups.
Furthermore, alterations in the concentration and composition of phospholipids covering articular cartilage has been shown to be associated with the development of OA.78
Metabolic changes seen in inflammatory arthropathies
Glucose was decreased and lactic acid was increased in RA SF.57 Levels of glucose-1-phosphate and D-mannose were also decreased. These decreases may be explained by the increased energy demands caused by inflammation in RA. Furthermore, the increased consumption of glucose can lead to increased lactic acid production. Levels of citric acid were decreased in RA SF. Citric acid is an important component of the TCA cycle, which provides the complete oxidation of acetyl-coenzyme A (CoA) derived from AAs, carbohydrates, and fats. Consequently, this leads to decreased adenosine triphosphate (ATP) production from the aerobic oxidation process. Yang et al57 suggest that low glucose and high lactic acid concentrations in RA SF may represent potential biomarkers of RA.
The increased levels of BCAAs, identified in the study by Ahn et al,40 result in increased production of IL-1 and/or TNF-α, which are typically increased in RA and SNA.79 Elevated expression of citramalate has been suggested to indicate disturbed metabolism of glutamate in the setting of active inflammation.80 Elevated expression of citrulline and methionine sulfoxide were also identified in this study. This may reflect neutrophil hyperactivity documented in BD.81 Ahn et al40 suggest that these metabolites may act as potential biomarkers for discriminating BD with arthritis from SNA. However, there are no normal controls in their study.
The metabolites identified in a study by Kim et al52 are major intermediates of FA and AA metabolism, the TCA cycle, and the urea cycle. The authors suggest that AA metabolism, the TCA cycle, and the urea cycle were more activated in the RA group compared with those in the non-RA group. Although the authors suggest these metabolites may be potential biomarkers, there are no normal controls and several different disease processes were compared. Consequently, it is difficult to say with any certainty whether any of these metabolites are indeed potential biomarkers.
Metabolic changes seen between osteoarthritis and rheumatoid arthritis
Kang et al49 identified 21 metabolites as being in significantly different concentrations between RA and OA SF. Concentrations of lipid metabolites were typically higher in RA than in OA SF, which has previously been demonstrated.66 Furthermore, regulation of inflammation includes the roles of lipid mediators and prostaglandins (PGs). Leukotrienes and PGs are crucial in the development of arthritic diseases.82 Concentrations of tryptophan metabolites also differed significantly between the OA and RA groups. This is an exogenous AA that must be provided in the diet. Tryptophan and its metabolites are involved in inflammation. One of the metabolites, kynurenine, has well known anti-inflammatory effects that are toxic to T cells and which cause apoptosis.83
Although this systematic review has identified many metabolites present in different disease states, including some putative biomarkers, there are some important limitations. There were only seven studies identified in the literature with healthy controls.39,43,46,47,53,56,61 Furthermore, only two studies performed an analysis to account for multiple testing of their dataset and neither of these studies had healthy controls.41,42 The presence of multiple confounding factors in many of the studies was another important limitation. Not all studies accounted for age or sex and certainly very few considered medical comorbidities. Consequently, the results must be viewed with caution. The solution to these limitations would be to conduct a largescale epidemiological metabolic profiling study incorporating multiple confounding factors such as age, sex, medical comorbidities, and medications with a view to addressing the correlations between clinical features of disease, inflammation, and metabolism. It should be noted that the majority of the reported works are untargeted metabolic profiling studies, where the identity of any putative biomarker is unknown at the outset. In those cases where metabolites were identified or annotated, no parameter of identification certainty, such as the Metabolomics Standards Initiative (MSI) level of identification, was reported.84 Therefore, the occurrence of incorrect identifications is possible hence affecting further metabolic interpretation and biomarker validation. Another important limitation of some of the studies is that they do not provide quantitative percentage or fold change, but only the direction of change.
In conclusion, metabolic profiling is proving to be an invaluable method of identifying putative biomarkers in the field of orthopaedics unique to different pathologies. Although numerous studies have been performed using these techniques in human SF, larger studies are required with healthy controls accounting for multiple confounding factors and using robust statistical analysis to identify putative biomarkers. This may lead to the development of new diagnostic techniques and possible treatment strategies. Recent advances in both proteomic and genetic studies have demonstrated the importance of these techniques to improve disease understanding and identify biomarkers.85,86 Future studies integrating genomic, proteomic, and metabolic profiling techniques may provide the greatest hope for the advancement of biomarker discovery.
Acknowledgments
The authors would like to acknowledge Rebecca Jones for her help with performing the article search. The authors would also like to thank Dr Claire L. Boulangé for her help setting up the metabolic profiling projects conducted at Imperial College, London and for the useful discussions that took place around the topics covered in this systematic review.
The authors also acknowledge the support of the Imperial NIHR Biomedical Research Centre (BRC).
Footnotes
Author Contributions: P. Akhbari: Assisted in designing the article search strategy, Screened articles for final inclusion, Wrote the manuscript.
U. Karamchandani: Assisted in designing the article search strategy, Performed the article searches according to the PRISMA guidelines, Screened the articles for final inclusion.
M. K. J. Jaggard: Assisted in writing the manuscript.
G. Graça: Assisted in writing the manuscript.
R. Bhattacharya: Reviewed the manuscript prior to submission.
J. C. Lindon: Assisted in writing the manuscript.
H. R. T. Williams: Reviewed the manuscript prior to submission.
C. M. Gupte: Assisted in writing the manuscript, Reviewed the final manuscript prior to submission.
ICMJE COI statement: None declared
Ethical review statement: This study did not require ethical approval.
Follow us @BoneJointRes
Supplementary Material
Tables showing search terms used for this systematic review on the role of metabolic profiling in human synovial fluid (HSF) (Supplementary Table i), and a list of all identified metabolites by article in HSF (Supplementary Table ii).
Funding statement
No benefits in any form have been received or will be received from a commercial party related directly or indirectly to the subject of this article.
References
- 1. Castañeda S, Roman-Blas JA, Largo R, Herrero-Beaumont G. Osteoarthritis: a progressive disease with changing phenotypes. Rheumatology (Oxford). 2014;53(1):1-3. [DOI] [PubMed] [Google Scholar]
- 2. Poole AR. Osteoarthritis as a whole joint disease. HSS J. 2012;8(1):4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Johnson VL, Hunter DJ. The epidemiology of osteoarthritis. Best Pract Res Clin Rheumatol. 2014;28(1):5-15. [DOI] [PubMed] [Google Scholar]
- 4. Scott DL, Wolfe F, Huizinga TW. Rheumatoid arthritis. Lancet. 2010;376(9746):1094-1108. [DOI] [PubMed] [Google Scholar]
- 5. Cammarata RJ, Rodnan GP, Fennell RH. Serum anti-γ-globulin and antinuclear factors in the aged. JAMA. 1967;199(7):455-458. [PubMed] [Google Scholar]
- 6. Humphreys JH, Symmons DP. Postpublication validation of the 2010 American College of Rheumatology/European League Against Rheumatism classification criteria for rheumatoid arthritis: where do we stand? Curr Opin Rheumatol. 2013;25(2):157-163. [DOI] [PubMed] [Google Scholar]
- 7. Tanino M, Matoba R, Nakamura S, et al. Prediction of efficacy of anti-TNF biologic agent, infliximab, for rheumatoid arthritis patients using a comprehensive transcriptome analysis of white blood cells. Biochem Biophys Res Commun. 2009;387(2):261-265. [DOI] [PubMed] [Google Scholar]
- 8. Teixeira VH, Olaso R, Martin-Magniette ML, et al. Transcriptome analysis describing new immunity and defense genes in peripheral blood mononuclear cells of rheumatoid arthritis patients. PLoS One. 2009;4(8):e6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kallberg H, Padyukov L, Plenge RM, et al. ; Epidemiological Investigation of Rheumatoid Arthritis study group. Gene-gene and gene-environment interactions involving HLA-DRB1, PTPN22, and smoking in two subsets of rheumatoid arthritis. Am J Hum Genet. 2007;80(5):867-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Horning EC, Horning MG. Metabolic profiles: gas-phase methods for analysis of metabolites. Clin Chem. 1971;17(8):802-809. [PubMed] [Google Scholar]
- 11. Lee SJ, Trostel A, Adhya S. Metabolite changes signal genetic regulatory mechanisms for robust cell behavior. MBio. 2014;5(1):e00972-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hu T, Zhang W, Fan Z, et al. METABOLOMICS DIFFERENTIAL CORRELATION NETWORK ANALYSIS OF OSTEOARTHRITIS. Pac Symp Biocomput. 2016;21:120-131. [PubMed] [Google Scholar]
- 13. Lindon JC, Holmes E, Nicholson JK. So what’s the deal with metabonomics? Anal Chem. 2003;75(17):384A-391A. [DOI] [PubMed] [Google Scholar]
- 14. Harwood LM, Claridge TDW. Introduction to Organic Spectroscopy. Oxford: Oxford University Press; 1999. [Google Scholar]
- 15. Lindon JC, Nicholson JK. Spectroscopic and statistical techniques for information recovery in metabonomics and metabolomics. Annu Rev Anal Chem (Palo Alto Calif). 2008;1:45-69. [DOI] [PubMed] [Google Scholar]
- 16. Want EJ, Wilson ID, Gika H, et al. Global metabolic profiling procedures for urine using UPLC-MS. Nat Protoc. 2010;5(6):1005-1018. [DOI] [PubMed] [Google Scholar]
- 17. Odunsi K, Wollman RM, Ambrosone CB, et al. Detection of epithelial ovarian cancer using 1H-NMR-based metabonomics. Int J Cancer. 2005;113(5):782-788. [DOI] [PubMed] [Google Scholar]
- 18. Wang Z, Chen Z, Yang S, et al. (1)H NMR-based metabolomic analysis for identifying serum biomarkers to evaluate methotrexate treatment in patients with early rheumatoid arthritis. Exp Ther Med. 2012;4(1):165-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yu B, Zheng Y, Nettleton JA, Alexander D, Coresh J, Boerwinkle E. Serum metabolomic profiling and incident CKD among African Americans. Clin J Am Soc Nephrol. 2014;9(8):1410-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Athersuch TJ. The role of metabolomics in characterizing the human exposome. Bioanalysis. 2012;4(18):2207-2212. [DOI] [PubMed] [Google Scholar]
- 21. Hong J, Yang L, Zhang D, Shi J. Plant Metabolomics: An Indispensable System Biology Tool for Plant Science. Int J Mol Sci. 2016;17(6):E767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fischer K, Kettunen J, Würtz P, et al. Biomarker profiling by nuclear magnetic resonance spectroscopy for the prediction of all-cause mortality: an observational study of 17,345 persons. PLoS Med. 2014;11(2):e1001606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Holmes E, Loo RL, Stamler J, et al. Human metabolic phenotype diversity and its association with diet and blood pressure. Nature. 2008;453(7193):396-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang A, Sun H, Wang P, Han Y, Wang X. Recent and potential developments of biofluid analyses in metabolomics. J Proteomics. 2012;75(4):1079-1088. [DOI] [PubMed] [Google Scholar]
- 25. Garnero P, Rousseau JC, Delmas PD. Molecular basis and clinical use of biochemical markers of bone, cartilage, and synovium in joint diseases. Arthritis Rheum. 2000;43(5):953-968. [DOI] [PubMed] [Google Scholar]
- 26. Lamers RJ, van Nesselrooij JH, Kraus VB, et al. Identification of an urinary metabolite profile associated with osteoarthritis. Osteoarthritis Cartilage. 2005;13(9):762-768. [DOI] [PubMed] [Google Scholar]
- 27. Lamers RJ, DeGroot J, Spies-Faber EJ, et al. Identification of disease- and nutrient-related metabolic fingerprints in osteoarthritic Guinea pigs. J Nutr. 2003;133(6):1776-1780. [DOI] [PubMed] [Google Scholar]
- 28. Zhai G, Wang-Sattler R, Hart DJ, et al. Serum branched-chain amino acid to histidine ratio: a novel metabolomic biomarker of knee osteoarthritis. Ann Rheum Dis. 2010;69(6):1227-1231. [DOI] [PubMed] [Google Scholar]
- 29. Blanco FJ, Ruiz-Romero C. Osteoarthritis: metabolomic characterization of metabolic phenotypes in OA. Nat Rev Rheumatol. 2012;8(3):130-132. [DOI] [PubMed] [Google Scholar]
- 30. Damyanovich AZ, Staples JR, Chan ADM, Marshall KW. Comparative study of normal and osteoarthritic canine synovial fluid using 500 MHz 1H magnetic resonance spectroscopy. J Orthop Res. 1999;17(2):223-231. [DOI] [PubMed] [Google Scholar]
- 31. Lacitignola L, Fanizzi FP, Francioso E, Crovace A. 1H NMR investigation of normal and osteo-arthritic synovial fluid in the horse. Vet Comp Orthop Traumatol. 2008;21(1):85-88. [DOI] [PubMed] [Google Scholar]
- 32. Xie K, Dai K, Qu X, Yan M. Serum and Synovial Fluid Interleukin-6 for the Diagnosis of Periprosthetic Joint Infection. Sci Rep. 2017;7(1):1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ivy MI, Thoendel MJ, Jeraldo PR, et al. Direct Detection and Identification of Prosthetic Joint Infection Pathogens in Synovial Fluid by Metagenomic Shotgun Sequencing. J Clin Microbiol. 2018;56(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Peffers MJ, Smagul A, Anderson JR. Proteomic analysis of synovial fluid: current and potential uses to improve clinical outcomes. Expert Rev Proteomics. 2019;16(4):287-302. [DOI] [PubMed] [Google Scholar]
- 35. Griswold AJ, Perez J, Nuytemans K, Strong TA, Wang L, Vance DD, et al. Transcriptomic analysis of synovial extracellular RNA following knee trauma: A pilot study. J Orthop Res. 2018;36(6):1659-1665. [DOI] [PubMed] [Google Scholar]
- 36. Moher D, Liberati A, Tetzlaff J, Altman DG; PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Int J Surg. 2010;8(5):336-341. [DOI] [PubMed] [Google Scholar]
- 37. Wishart DS, Jewison T, Guo AC, et al. HMDB 3.0—The Human Metabolome Database in 2013. Nucleic Acids Res. 2013;41(Database issue):D801-D807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ulrich EL, Akutsu H, Doreleijers JF, et al. BioMagResBank. Nucleic Acids Res. 2008;36(Database issue):D402-D408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Adams SB, Jr, Nettles DL, Jones LC, Miller SD, Guyton GP, Schon LC. Inflammatory cytokines and cellular metabolites as synovial fluid biomarkers of posttraumatic ankle arthritis. Foot Ankle Int. 2014;35(12):1241-1249. [DOI] [PubMed] [Google Scholar]
- 40. Ahn JK, Kim S, Kim J, Hwang J, Kim KH, Cha HS. A Comparative Metabolomic Evaluation of Behcet’s Disease with Arthritis and Seronegative Arthritis Using Synovial Fluid. PLoS One. 2015;10(8):e0135856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Anderson JR, Chokesuwattanaskul S, Phelan MP, et al. Synovial fluid metabolite profiles differ between osteoarthritis and rheumatoid arthritis. 2018;57(suppl_3):iii152-iii153. [Google Scholar]
- 42. Carlson AK, Rawle RA, Adams E, Greenwood MC, Bothner B, June RK. Application of global metabolomic profiling of synovial fluid for osteoarthritis biomarkers. Biochem Biophys Res Commun. 2018;499(2):182-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chen R, Han S, Liu X, et al. Perturbations in amino acids and metabolic pathways in osteoarthritis patients determined by targeted metabolomics analysis. J Chromatogr B Analyt Technol Biomed Life Sci. 2018;1085:54-62. [DOI] [PubMed] [Google Scholar]
- 44. Dubey D, Chaurasia S, Guleria A, et al. Metabolite assignment of ultrafiltered synovial fluid extracted from knee joints of reactive arthritis patients using high resolution NMR spectroscopy. Magn Reson Chem. 2019;57(1):30-43. [DOI] [PubMed] [Google Scholar]
- 45. Dubey D, Kumar S, Ahmed S, et al. NMR based serum and synovial fluid metabolomics reveal similar metabolomic profile in patients with reactive arthritis and undifferentiated spondyloarthropathy. Poster Presentations. Indian Journal of Rheumatology. 2017;12(5):S24-25. [Google Scholar]
- 46. Dubey D, Kumar S, Chaurasia S, et al. NMR-Based Serum Metabolomics Revealed Distinctive Metabolic Patterns in Reactive Arthritis Compared with Rheumatoid Arthritis. J Proteome Res. 2019;18(1):130-146. [DOI] [PubMed] [Google Scholar]
- 47. Furman BD, Kimmerling KA, Ramamoorthy S, et al. Articular fracture upregulates lipid metabolites in human synovial fluid. Osteoarthritis Cartilage. 2017;25(Supplement 1):S381. [Google Scholar]
- 48. Hwang J, Ahn JK, Lee J, et al. Discriminative metabolite profiling of synovial fluid in rheumatoid arthritis compared to osteoarthritis. Ann Rheum Dis. 2013;72(Suppl 3):A548-A549. [Google Scholar]
- 49. Kang KY, Lee SH, Jung SM, Park SH, Jung BH, Ju JH. Downregulation of tryptophan-related metabolomic profile in rheumatoid arthritis synovial fluid. J Rheumatol. 2015;42(11):2003-2011. [DOI] [PubMed] [Google Scholar]
- 50. Khatib N, Papageorgiou A, Fairhurst S, Wilson C, Mason DJ. Identifying load responsive synovial fluid metabolic markers following pivot-shift testing in ACL injury subjects. Osteoarthritis Cartilage. 2018;26 Suppl 1:S168–S169. [Google Scholar]
- 51. Kim S, Hwang J, Kim J, Ahn JK, Cha HS, Kim KH. Metabolite profiles of synovial fluid change with the radiographic severity of knee osteoarthritis. Joint Bone Spine. 2017;84(5):605-610. [DOI] [PubMed] [Google Scholar]
- 52. Kim S, Hwang J, Xuan J, Jung YH, Cha HS, Kim KH. Global metabolite profiling of synovial fluid for the specific diagnosis of rheumatoid arthritis from other inflammatory arthritis. PLoS One. 2014;9(6):e97501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Leimer EM, Pappan KL, Nettles DL, et al. Lipid profile of human synovial fluid following intra-articular ankle fracture. J Orthop Res. 2017;35(3):657-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Meshitsuka S, Yamazaki E, Inoue M, Hagino H, Teshima R, Yamamoto K. Nuclear magnetic resonance studies of synovial fluids from patients with rheumatoid arthritis and osteoarthritis. Clin Chim Acta. 1999;281(1-2):163-167. [DOI] [PubMed] [Google Scholar]
- 55. Mickiewicz B, Heard BJ, Chau JK, et al. Metabolic profiling of synovial fluid in a unilateral ovine model of anterior cruciate ligament reconstruction of the knee suggests biomarkers for early osteoarthritis. J Orthop Res. 2015;33(1):71-77. [DOI] [PubMed] [Google Scholar]
- 56. Naughton DP, Haywood R, Blake DR, Edmonds S, Hawkes GE, Grootveld M. A comparative evaluation of the metabolic profiles of normal and inflammatory knee-joint synovial fluids by high resolution proton NMR spectroscopy. FEBS Lett. 1993;332(3):221-225. [DOI] [PubMed] [Google Scholar]
- 57. Yang XY, Zheng KD, Lin K, et al. Energy Metabolism Disorder as a Contributing Factor of Rheumatoid Arthritis: A Comparative Proteomic and Metabolomic Study. PLoS One. 2015;10(7):e0132695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhang W, Likhodii S, Zhang Y, et al. Classification of osteoarthritis phenotypes by metabolomics analysis. BMJ Open. 2014;4(11):e006286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhang W, Likhodii S, Aref-Eshghi E, et al. Relationship between blood plasma and synovial fluid metabolite concentrations in patients with osteoarthritis. J Rheumatol. 2015;42(5):859-865. [DOI] [PubMed] [Google Scholar]
- 60. Zhang WD, Sun G, Likhodii S, et al. Metabolomic analysis of human synovial fluid and plasma reveals that phosphatidylcholine metabolism is associated with both osteoarthritis and diabetes mellitus. Metabolomics. 2016;12(2):24. [Google Scholar]
- 61. Zheng K, Shen N, Chen H, et al. Global and targeted metabolomics of synovial fluid discovers special osteoarthritis metabolites. J Orthop Res. 2017;35(9):1973-1981. [DOI] [PubMed] [Google Scholar]
- 62. Mickiewicz B, Kelly JJ, Ludwig TE, et al. Metabolic analysis of knee synovial fluid as a potential diagnostic approach for osteoarthritis. J Orthop Res. 2015;33(11):1631-1638. [DOI] [PubMed] [Google Scholar]
- 63. Zhang W, Sun G, Likhodii S, et al. Metabolomic analysis of human synovial fluid and plasma reveals that phosphatidylcholine metabolism is associated with both osteoarthritis and diabetes mellitus. Metabolomics. 2016;12(2):1-10. [Google Scholar]
- 64. Farooqui AA, Horrocks LA, Farooqui T. Glycerophospholipids in brain: their metabolism, incorporation into membranes, functions, and involvement in neurological disorders. Chem Phys Lipids. 2000;106(1):1-29. [DOI] [PubMed] [Google Scholar]
- 65. Ghauri FYK, Nicholson JK, Sweatman BC, et al. NMR spectroscopy of human post mortem cerebrospinal fluid: distinction of Alzheimer’s disease from control using pattern recognition and statistics. NMR Biomed. 1993;6(2):163-167. [DOI] [PubMed] [Google Scholar]
- 66. Oliviero F, Lo Nigro A, Bernardi D, et al. A comparative study of serum and synovial fluid lipoprotein levels in patients with various arthritides. Clin Chim Acta. 2012;413(1-2):303-307. [DOI] [PubMed] [Google Scholar]
- 67. Miller EJ, Van der Korst JK, Sokoloff L. Collagen of human articular and costal cartilage. Arthritis Rheum. 1969;12(1):21-29. [DOI] [PubMed] [Google Scholar]
- 68. Januszewski AS, Karschimkus C, Davis KE, O’Neal D, Ward G, Jenkins AJ. Plasma 1,5 anhydroglucitol levels, a measure of short-term glycaemia: assay assessment and lower levels in diabetic vs. non-diabetic subjects. Diabetes Res Clin Pract. 2012;95(1):e17-e19. [DOI] [PubMed] [Google Scholar]
- 69. Tillmann K. Pathological aspects of osteoarthritis related to surgery. Inflammation. 1984;8(suppl):S57-S74. [DOI] [PubMed] [Google Scholar]
- 70. Davies CM, Guilak F, Weinberg JB, Fermor B. Reactive nitrogen and oxygen species in interleukin-1-mediated DNA damage associated with osteoarthritis. Osteoarthritis Cartilage. 2008;16(5):624-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Thomas-Reetz A, Hell JW, During MJ, Walch-Solimena C, Jahn R, De Camilli P. A gamma-aminobutyric acid transporter driven by a proton pump is present in synaptic-like microvesicles of pancreatic beta cells. Proc Natl Acad Sci U S A. 1993;90(11):5317-5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Handley CJ, Speight G, Leyden KM, Lowther DA. Extracellular matrix metabolism by chondrocytes. 7. Evidence that L-glutamine is an essential amino acid for chondrocytes and other connective tissue cells. Biochim Biophys Acta. 1980;627(3):324-331. [DOI] [PubMed] [Google Scholar]
- 73. Kim DS, Shin MR, Kim YS, et al. Anti-inflammatory effects of glutamine on LPS-stimulated human dental pulp cells correlate with activation of MKP-1 and attenuation of the MAPK and NF-κB pathways. Int Endod J. 2015;48(3):220-228. [DOI] [PubMed] [Google Scholar]
- 74. Tonomura H, Takahashi KA, Mazda O, et al. Glutamine protects articular chondrocytes from heat stress and NO-induced apoptosis with HSP70 expression. Osteoarthritis Cartilage. 2006;14(6):545-553. [DOI] [PubMed] [Google Scholar]
- 75. Xu Z, Chen T, Luo J, Ding S, Gao S, Zhang J. Cartilaginous Metabolomic Study Reveals Potential Mechanisms of Osteophyte Formation in Osteoarthritis. J Proteome Res. 2017;16(4):1425-1435. [DOI] [PubMed] [Google Scholar]
- 76. Chang X, Wei C. Glycolysis and rheumatoid arthritis. Int J Rheum Dis. 2011;14(3):217-222. [DOI] [PubMed] [Google Scholar]
- 77. Glass GG. Osteoarthritis. Dis Mon. 2006;52(9):343-362. [DOI] [PubMed] [Google Scholar]
- 78. Hills BA. Surface-active phospholipid: a Pandora’s box of clinical applications. Part II. Barrier and lubricating properties. Intern Med J. 2002;32(5-6):242-251. [DOI] [PubMed] [Google Scholar]
- 79. Bassit RA, Sawada LA, Bacurau RF, et al. Branched-chain amino acid supplementation and the immune response of long-distance athletes. Nutrition. 2002;18(5):376-379. [DOI] [PubMed] [Google Scholar]
- 80. Perlman S, Carr SA. Citramalic acid in cerebrospinal fluid of patients with bacterial meningitis. Clin Chem. 1984;30(7):1209-1212. [PubMed] [Google Scholar]
- 81. Pineton de Chambrun M, Wechsler B, Geri G, Cacoub P, Saadoun D. New insights into the pathogenesis of Behçet’s disease. Autoimmun Rev. 2012;11(10):687-698. [DOI] [PubMed] [Google Scholar]
- 82. Chen M, Lam BK, Kanaoka Y, et al. Neutrophil-derived leukotriene B4 is required for inflammatory arthritis. J Exp Med. 2006;203(4):837-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Fallarino F, Grohmann U, Vacca C, et al. T cell apoptosis by tryptophan catabolism. Cell Death Differ. 2002;9(10):1069-1077. [DOI] [PubMed] [Google Scholar]
- 84. Sumner LW, Amberg A, Barrett D, et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics. 2007;3(3):211-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Soul J, Dunn SL, Anand S, et al. Stratification of knee osteoarthritis: two major patient subgroups identified by genome-wide expression analysis of articular cartilage. Ann Rheum Dis. 2018;77(3):423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Kung LHW, Ravi V, Rowley L, et al. Cartilage MicroRNA Dysregulation During the Onset and Progression of Mouse Osteoarthritis Is Independent of Aggrecanolysis and Overlaps With Candidates From End-Stage Human Disease. Arthritis Rheumatol. 2018;70(3):383-395. [DOI] [PubMed] [Google Scholar]