Abstract
Aims
Osteoarthritis (OA) is the most prevalent joint disease. However, the specific and definitive genetic mechanisms of OA are still unclear.
Methods
Tissue-related transcriptome-wide association studies (TWAS) of hip OA and knee OA were performed utilizing the genome-wide association study (GWAS) data of hip OA and knee OA (including 2,396 hospital-diagnosed hip OA patients versus 9,593 controls, and 4,462 hospital-diagnosed knee OA patients versus 17,885 controls) and gene expression reference to skeletal muscle and blood. The OA-associated genes identified by TWAS were further compared with the differentially expressed genes detected by the messenger RNA (mRNA) expression profiles of hip OA and knee OA. Functional enrichment and annotation analysis of identified genes was performed by the DAVID and FUMAGWAS tools.
Results
We detected 33 common genes, eight common gene ontology (GO) terms, and one common pathway for hip OA, such as calcium and integrin-binding protein 1 (CIB1) (PTWAS = 0.025, FCmRNA = -1.575 for skeletal muscle), adrenomedullin (ADM) (PTWAS = 0.022, FCmRNA = -4.644 for blood), Golgi apparatus (PTWAS <0.001, PmRNA = 0.012 for blood), and phosphatidylinositol 3' -kinase-protein kinase B (PI3K-Akt) signalling pathway (PTWAS = 0.033, PmRNA = 0.005 for blood). For knee OA, we detected 24 common genes, eight common GO terms, and two common pathways, such as histocompatibility complex, class II, DR beta 1 (HLA-DRB1) (PTWAS = 0.040, FCmRNA = 4.062 for skeletal muscle), Follistatin-like 1 (FSTL1) (PTWAS = 0.048, FCmRNA = 3.000 for blood), cytoplasm (PTWAS < 0.001, PmRNA = 0.005 for blood), and complement and coagulation cascades (PTWAS = 0.017, PmRNA = 0.001 for skeletal muscle).
Conclusion
We identified a group of OA-associated genes and pathways, providing novel clues for understanding the genetic mechanism of OA.
Cite this article: Bone Joint Res. 2020;9(3):130–138.
Keywords: Hip osteoarthritis, Knee osteoarthritis, Transcriptome-wide association study
Article focus
To access the genetic mechanism of osteoarthritis (OA) using the transcriptome-wide association study (TWAS) analysis.
We performed functional enrichment and annotation analysis of the candidate genes associated with OA.
Key messages
A total of 33 candidate genes were identified for hip OA, such as calcium and integrin-binding protein 1 (CIB1) and adrenomedullin (ADM).
For knee OA, we detected 24 candidate genes, such as histocompatibility complex, class II, DR beta 1 (HLA-DRB1), and Follistatin-like 1 (FSTL1).
Strengths and limitations of this study
TWAS has a boosting power to detect novel disease genes.
One limitation is that TWAS may be too low power to detect the causal loci without cis-expression effects on target disease.
Introduction
Osteoarthritis (OA) is mainly characterized by the degeneration of articular cartilage in the knee and hip,1 and knee and hip OA occur in approximately 6% and 3% of Americans aged 30 years or older, respectively.2 The clinical symptoms of OA include joint pain, swelling, stiffness, and limited motion. The burden of OA is not only on healthcare and society costs of the patients and families, but also includes the patients’ quality of life, mental health, and emotional relationship.3,4
A review study has shown that the heritability of genetic factors in the development of OA was estimated at approximately 50% in twin studies and familial studies.5 Through using data across 16.5 million variants from the UK Biobank, a group of OA-associated genes have been identified by genome-wide association studies (GWAS), such as MAP2K6 and ZNF345.6 However, GWAS usually focus on a few genome-wide significant loci with large genetic effects, while they are likely to miss many biological true positive loci with moderate or weak genetic effects. In addition, a previous study found that a large section of genetic variants identified by GWAS was enriched in non-coding regulatory genomic regions, which leads to difficulty in interpreting the genetic effects.7
It has been demonstrated that the messenger RNA (mRNA) expression levels were under genetic control.8 Expression quantitative trait loci (eQTLs) are the genetic variants, which could explain the variations in expression levels of mRNAs.9 eQTLs provide a novel way to uncover the biological mechanism of identified genetic variants underlying diseases.10 Integrating the GWAS and eQTL data could boost the power to discover novel disease-associated genes. In the previous research, the transcriptome-wide association study (TWAS) was developed to explore gene-trait relationships, using publicly available GWAS results and eQTL reference datasets.11 The expression levels of thousands of genes were predicted by TWAS, and subsequently were used to evaluate the associations between gene expression levels and target diseases. Different from GWAS testing millions of single nucleotide polymorphisms (SNPs), TWAS can greatly reduce the burden of multiple comparisons in statistical analysis and enhance the ability of GWAS to detect novel disease genes.12 As a supplement to traditional GWAS analyses, five novel susceptibility loci associated with cutaneous squamous cell carcinoma (cSCC) were validated by TWAS analysis.13
To identify novel OA-associated genes, we performed a large-scale integrative analysis of TWAS and mRNA expression profiles for hip OA and knee OA. Using previous large-scale GWAS data and eQTL reference data of skeletal muscle and blood, TWAS was performed to detect novel candidate genes, the predicted expression levels of which were associated with OA. The genes identified by TWAS were further subjected to gene ontology (GO) and pathway enrichment analysis. To further confirm the functional relevance of identified genes, the TWAS results were compared with the mRNA expression profiles of OA to detect common genes, GO terms, and pathways shared by TWAS, and mRNA expression profiles of OA.
Methods
GWAS summary data of hip and knee OA
The GWAS summary data of hip and knee OA were derived from the published studies.6 Briefly, 2,396 hospital-diagnosed hip OA patients and 9,593 controls, and 4,462 hospital-diagnosed knee OA patients and 17,885 controls were all derived from the UK Biobank. The diagnose standard of hospital-diagnosed OA coding in the UK Biobank comes from the International Classification of Diseases (ICD)-10 code captured from Hospital Episode Statistics (HES) data. Finally, 16,122,076 SNPs for hip OA and 16,309,199 SNPs for knee OA were applied for GWAS analysis. Association testing was conducted by SNPTEST v2.5.2 (University of Oxford, Oxford, UK).14 A detailed description of the sample characteristics, experimental design, and statistical analysis can be found in the published study.6
mRNA expression profiles of hip cartilage with OA
The mRNA expression profiles data of articular cartilage from hip OA and traumatic femoral neck fracture patients were obtained from a previous study.15 Briefly, articular cartilage specimens of hip were collected from nine OA patients and ten traumatic femoral neck fracture patients undergoing joint replacement surgery. OA status was confirmed using clinical examination and joint score.16 The subjects with hip scoring ≤ 1 were considered healthy samples, while those scoring ≥ 5 were classified as case samples. Differentially expressed genes were identified significantly with a fold change (FC) ≥ 1.5 and p < 0.05. The detailed sample characteristics and experimental design can be found in the previous study.15
mRNA expression profile of knee cartilage with OA
The mRNA expression profile data of knee OA were obtained from a previously published study.17 Briefly, normal human knee cartilage tissues of 18 people without history of joint disease or trauma were procured from tissue banks. OA-affected cartilage specimens were harvested from 20 donors accepting knee arthroplasty surgery. Differentially expressed genes were identified significantly with an adjusted p-value of < 0.05 and log2FC ≥ 1. Detailed descriptions of sample characteristics, experiment design, and statistical analysis of this dataset are available in the study by Fisch et al.17
TWAS of hip OA and knee OA
TWAS of hip OA and knee OA were performed by the FUSION software (Harvard T.H. Chan School of Public Health, Boston, Massachusetts, USA) through integrating the OA GWAS summary data and precomputed gene expression weights of skeletal muscle and blood (including peripheral blood and whole blood).18 A previous study performed TWAS analysis and identified the causal genes to pancreatic cancer (PC).19 The gene expression weights reference of skeletal muscle and blood were obtained from the FUSION website.18,20 Peripheral blood and skeletal muscle were also used in previous biological studies of OA.21,22 The gene expression weights of skeletal muscle were derived from the Genotype-Tissue Expression (GTEx) Project (version 7; National Disease Research Interchange, Philadelphia, Pennsylvania, USA; n = 361).23 The gene expression weights of peripheral blood and whole blood were collected from 1,245 unrelated control individuals from the Netherlands Twin Registry study and 1,264 subjects from the Young Finns Study.24–27 For eQTL data, a total of 3,637,328 SNPs (corresponding to 7,408 genes), 1,120,437 SNPs (corresponding to 2,454 genes), and 2,044,474 SNPs (corresponding to 4,700 genes) were used for the TWAS of OA in skeletal muscle, peripheral blood, and whole blood tissues, respectively.
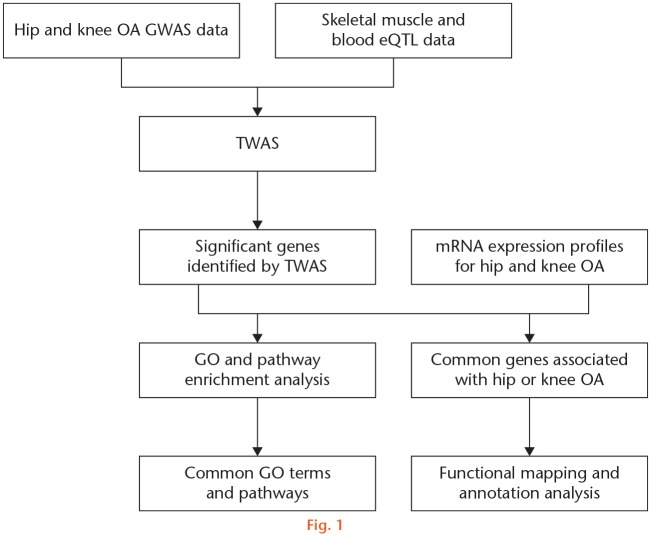
Firstly, the gene expression weights were calculated using the prediction models of FUSION. Then, the calculated expression weights were combined with GWAS results to impute association statistics between gene expression levels and target diseases. The SNP-expression weights in the 1 Mb cis loci of the gene for a given gene were computed using Bayesian sparse linear mixed model (BSLMM).28 The association testing statistics between predicted gene expression and target diseases was calculated as ZTWAS = w'Z/(w'Lw)1/2. ‘Z’ denotes the scores of OA and ‘w’ denotes the weights. ‘L’ denotes the SNP-correlation linkage disequilibrium (LD) matrix. In this analysis, we accounted for LD among SNPs and viewed the imputed gene expression data as a linear model of genotypes with weights. A TWAS p-value was calculated for each gene within skeletal muscle and blood. The genes with p < 0.05 were considered as significant. Then, the hip and knee OA-associated genes identified by TWAS were further compared with the differentially expressed genes detected by the mRNA expression profiles of hip and knee OA, respectively. ‘FCmRNA’ and ‘PmRNA’ represent the FCs and p-value in mRNA expression profiles. A flowchart illustrating the study design is shown in Figure 1.
Fig. 1.
Detailed flowchart of the study design. eQTL, expression quantitative trait locus; GO, gene ontology; GWAS, genome-wide association study; mRNA, messenger RNA; OA, osteoarthritis; TWAS, transcriptome-wide association study.
Functional enrichment and annotation analysis
Gene ontology and pathway enrichment analysis of the genes identified by TWAS and mRNA expression profiles were performed by the DAVID tool.29–31 We compared the analysis results of TWAS and mRNA expression profiles to screen out the common genes, GO terms, and pathways, which were shared by TWAS and mRNA expression profiles. Functional Mapping and Annotation of Genome-wide Association Studies (FUMAGWAS)32 was used to annotate, prioritize, visualize, and interpret the function of the common genes shared by TWAS and mRNA expression profiles.33
Results
TWAS results of hip and knee OA
For hip OA, TWAS identified 182 genes for skeletal muscle and 390 genes for blood with p < 0.05, such as ArfGAP with SH3 domain, ankyrin repeat and PH domain 3 (ASAP3) (PTWAS < 0.001 for skeletal muscle), high-density lipoprotein binding protein (HDLBP) (PTWAS < 0.001 for skeletal muscle), transcription elongation factor A3 (TCEA3) (PTWAS < 0.001 for blood), and serine/threonine kinase 25 (STK25) (PTWAS < 0.001 for blood) (Supplementary Table i). For knee OA, TWAS identified 180 genes for skeletal muscle and 410 genes for blood with p < 0.05, such as RP11-347C12.1 (PTWAS < 0.001 for skeletal muscle), centrosomal protein 250 (CEP250) (PTWAS < 0.001 for skeletal muscle), RWD domain containing 2B (RWDD2B) (PTWAS < 0.001 for blood), and ubiquinol-cytochrome c reductase complex assembly factor 1 (UQCC) (PTWAS < 0.001 for blood) (Supplementary Table ii). The top ten significant genes of hip and knee OA identified by TWAS are shown in Table I.
Table I.
List of the top ten significant genes identified by transcriptome-wide association studies for hip and knee osteoarthritis (p < 0.05).
| OA | Tissue | Gene | CHR | ZTWAS | PTWAS* |
|---|---|---|---|---|---|
| Hip | Skeletal muscle | ASAP3 | 1 | −4.9331 | < 0.001 |
| Hip | Blood | TCEA3 | 1 | −4.5970 | < 0.001 |
| Hip | Skeletal muscle | HDLBP | 2 | −4.3684 | < 0.001 |
| Hip | Blood | STK25 | 2 | −4.0764 | < 0.001 |
| Hip | Skeletal muscle | ITIH4-AS1 | 3 | 3.8985 | < 0.001 |
| Hip | Blood | ST3GAL3 | 1 | −3.8807 | < 0.001 |
| Hip | Blood | PNO1 | 2 | −3.8675 | < 0.001 |
| Hip | Skeletal muscle | GRINA | 8 | 3.8140 | < 0.001 |
| Hip | Skeletal muscle | RP5-966M1.6 | 3 | 3.7270 | < 0.001 |
| Hip | Blood | PMM2 | 16 | −3.7253 | < 0.001 |
| Knee | Blood | RWDD2B | 21 | −4.1737 | < 0.001 |
| Knee | Blood | UQCC | 20 | 3.9040 | < 0.001 |
| Knee | Blood | N6AMT1 | 21 | −3.8769 | < 0.001 |
| Knee | Blood | MAPK3 | 16 | −3.8468 | < 0.001 |
| Knee | Skeletal muscle | RP11-347C12.1 | 16 | 3.8161 | < 0.001 |
| Knee | Blood | SSH1 | 12 | −3.7411 | < 0.001 |
| Knee | Blood | GPX4 | 19 | 3.6084 | < 0.001 |
| Knee | Blood | PLIN2 | 9 | −3.5411 | < 0.001 |
| Knee | Blood | CDK7 | 5 | 3.4976 | < 0.001 |
| Knee | Blood | MTHFSD | 16 | −3.4959 | < 0.001 |
Each PTWAS value was calculated by transcriptome-wide association study analysis.
CHR, chromosome; OA, osteoarthritis; TWAS, transcriptome-wide association study.
Gene ontology and pathway enrichment analysis
For hip OA, GO and pathway enrichment analysis results of the genes identified by TWAS are shown in Supplementary Table iii. DAVID detected 11 GO terms for skeletal muscle and 31 GO terms for blood with p < 0.05, such as UFM1 hydrolase activity (PTWAS = 0.016 for skeletal muscle), DNA damage checkpoint (PTWAS = 0.019 for skeletal muscle), and membrane (PTWAS < 0.001 for blood). Pathway enrichment analysis detected four pathways for blood, such as bile secretion (PTWAS = 0.010) and glycosaminoglycan biosynthesis-chondroitin sulfate/dermatan sulfate (PTWAS = 0.006).
For knee OA, ten GO terms for skeletal muscle and 58 GO terms for blood were detected with p < 0.05, such as protein binding (PTWAS < 0.001), poly(A) RNA binding (PTWAS = 0.013), and cytosol (PTWAS < 0.001) (Supplementary Table iv). Pathway enrichment analysis of the significant genes identified one pathway for skeletal muscle and 14 pathways for blood (PTWAS < 0.05), such as complement and coagulation cascades (PTWAS = 0.017), influenza A (PTWAS = 0.006), and viral carcinogenesis (PTWAS = 0.009) (Supplementary Table iv).
Comparative analysis of TWAS and mRNA expression profiles
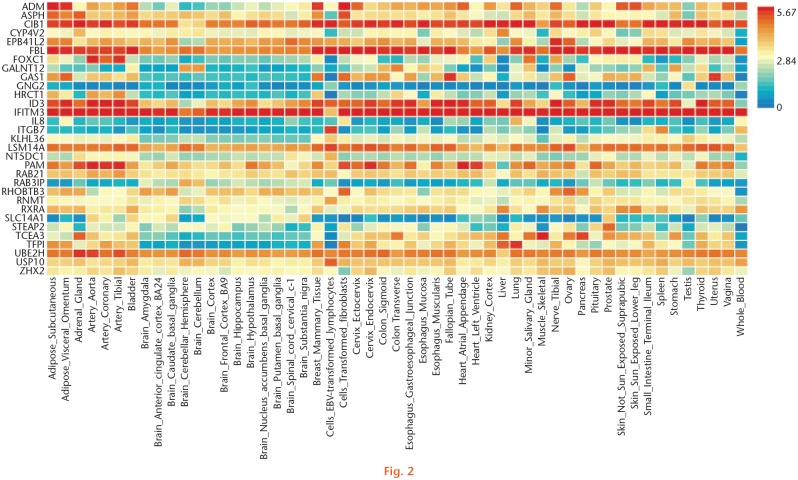
We further compared the analysis results of TWAS and mRNA expression profiles. For hip OA, we detected 33 common genes shared by the TWAS and mRNA expression profiles, such as calcium and integrin-binding protein 1 (CIB1) (PTWAS = 0.025, FCmRNA = -1.575 for skeletal muscle), adrenomedullin (ADM) (PTWAS = 0.022, FCmRNA = -4.644 for blood), and forkhead box C1 (FOXC1) (PTWAS = 0.029, FCmRNA = 1.527 for blood) (Table II). In addition, we detected eight common GO terms and one common pathway, such as cell-cell adherens junction (PTWAS = 0.037, PmRNA = 0.016 for skeletal muscle), Golgi apparatus (PTWAS < 0.001, PmRNA = 0.012 for blood), and PI3K-Akt signalling pathway (PTWAS = 0.033, PmRNA = 0.005 for blood) (Table III). The heat map of those common genes of hip OA is shown in Figure 2.
Table II.
Common genes between the significant genes identified by transcriptome-wide association studies and the differentially expressed genes identified by messenger RNA expression profiles for hip osteoarthritis.
| Tissue | Gene | PTWAS* | PmRNA† | FCmRNA |
|---|---|---|---|---|
| Skeletal muscle | CIB1 | 0.025 | 0.006 | −1.575 |
| CYP4V2 | 0.006 | 0.006 | 1.782 | |
| HRCT1 | 0.028 | 0.007 | 1.636 | |
| SLC14A1 | 0.027 | 0.009 | 2.633 | |
| STEAP2 | 0.050 | 0.006 | −1.838 | |
| TFPI | 0.033 | 0.006 | −2.637 | |
| Blood | ADM | 0.022 | 0.006 | −4.644 |
| ASPH | 0.007 | 0.006 | −1.569 | |
| EPB41L2 | 0.038 | 0.006 | 2.295 | |
| GLT25D2 | 0.024 | 0.006 | 2.816 | |
| GNG2 | 0.003 | 0.008 | 1.564 | |
| IFITM3 | 0.001 | 0.006 | −2.309 | |
| IL8 | 0.037 | 0.006 | −2.774 | |
| LSM14A | 0.031 | 0.006 | 1.527 | |
| RAB3IP | 0.044 | 0.006 | −1.952 | |
| SLC14A1 | 0.015 | 0.009 | 2.633 | |
| ZHX2 | 0.041 | 0.009 | −1.603 | |
| FOXC1 | 0.029 | 0.007 | 1.527 | |
| GALNT12 | 0.025 | 0.006 | −1.548 | |
| GAS1 | 0.025 | 0.006 | 4.096 | |
| ID3 | 0.001 | 0.006 | 2.267 | |
| ITGB7 | 0.042 | 0.006 | 1.684 | |
| KLHL36 | 0.046 | 0.006 | −1.598 | |
| NT5DC1 | 0.038 | 0.006 | 1.609 | |
| PAM | 0.032 | 0.006 | 1.557 | |
| RAB21 | 0.009 | 0.006 | −1.650 | |
| RNMT | 0.042 | 0.006 | −1.836 | |
| RXRA | 0.011 | 0.006 | 1.889 | |
| TCEA3 | < 0.001 | 0.006 | 1.843 | |
| UBE2H | 0.007 | 0.006 | −1.693 | |
| USP10 | 0.004 | 0.006 | −1.699 | |
| FBL | 0.003 | 0.009 | −1.635 | |
| RHOBTB3 | 0.038 | 0.006 | −1.696 |
Each PTWAS value was calculated by transcriptome-wide association study analysis.
Each PmRNA value was derived from the published studies.
FC, fold change; mRNA, messenger RNA; TWAS, transcriptome-wide association study.
Table III.
Common gene ontology terms and pathways between the significant genes identified by transcriptome-wide association studies and the differentially expressed genes identified by messenger RNA expression profiles for hip osteoarthritis.
| Category | Tissue | Name | PTWAS* | PmRNA† |
|---|---|---|---|---|
| GO | Skeletal muscle | GO:0005913~cell-cell adherens junction | 0.037 | 0.016 |
| Blood | GO:0000139~Golgi membrane | < 0.001 | 0.012 | |
| GO:0005794~Golgi apparatus | < 0.001 | < 0.001 | ||
| GO:0005515~protein binding | 0.008 | < 0.001 | ||
| GO:0005925~focal adhesion | 0.018 | < 0.001 | ||
| GO:0070062~extracellular exosome | 0.019 | < 0.001 | ||
| GO:0005789~endoplasmic reticulum membrane | 0.024 | 0.002 | ||
| GO:0005654~nucleoplasm | 0.031 | 0.033 | ||
| Pathway | Blood | hsa04151:PI3K-Akt signalling pathway | 0.033 | 0.005 |
Each PTWAS value was calculated by gene ontology and pathway analysis using DAVID for the significant genes identified by transcriptome-wide association study analysis.
Each PmRNA value was calculated by gene ontology and pathway analysis using DAVID for the significant genes identified by messenger RNA expression profiles.
GO, gene ontology; mRNA, messenger RNA; PI3K-Akt, phosphatidylinositol 3’ -kinase-protein kinase B; TWAS, ranscriptome-wide association study.
Fig. 2.
Gene expression heat map of the identified common genes shared by transcriptome-wide association studies (TWAS) and messenger RNA (mRNA) expression data of hip osteoarthritis (OA).
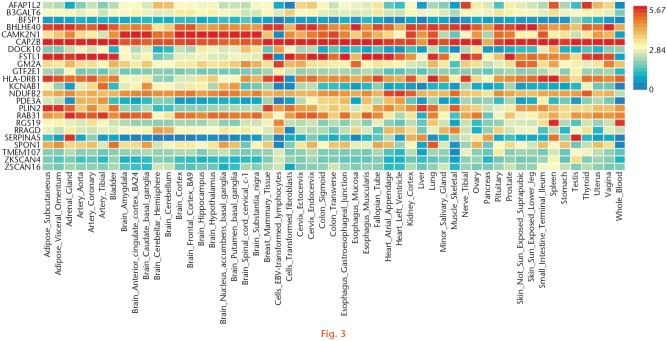
For knee OA, 24 common genes were identified, such as major histocompatibility complex, class II, DR beta 1 (HLA-DRB1) (PTWAS = 0.040, FCmRNA = 4.062 for skeletal muscle), general transcription factor IIE subunit 1 (GTF2E1) (PTWAS = 0.043, FCmRNA = 2.368 for skeletal muscle), Follistatin-like 1 (FSTL1) (PTWAS = 0.048, FCmRNA = 3.000 for blood), and beta-1,3-galactosyltransferase 6 (B3GALT6) (PTWAS < 0.001, FCmRNA = 2.221 for blood) (Table IV). In addition, we detected eight common GO terms and two common pathways, such as protein binding (PTWAS < 0.001, PmRNA < 0.001 for skeletal muscle), cytoplasm (PTWAS < 0.001, PmRNA = 0.005 for blood), complement and coagulation cascades (PTWAS = 0.017, PmRNA = 0.001 for skeletal muscle), and viral myocarditis (PTWAS = 0.009, PmRNA = 0.050 for blood) (Table V). A heat map showing the expression of common genes of knee OA is shown in Figure 3.
Table IV.
Common genes identified by comparing the gene ontology and pathway enrichment analysis results of transcriptome-wide association studies and messenger RNA expression profiles for knee osteoarthritis.
| Tissue | Gene | PTWAS* | PmRNA† | FCmRNA |
|---|---|---|---|---|
| Skeletal muscle | GTF2E1 | 0.043 | < 0.001 | 2.368 |
| HLA-DRB1 | 0.040 | 0.045 | 4.062 | |
| RRAGD | 0.001 | < 0.001 | 0.403 | |
| PDE3A | 0.048 | 0.003 | 2.826 | |
| SERPINA5 | 0.002 | < 0.001 | 3.078 | |
| RGS19 | 0.002 | 0.011 | 3.001 | |
| Blood | B3GALT6 | 0.010 | < 0.001 | 2.221 |
| BHLHE40 | 0.045 | < 0.001 | 0.196 | |
| KCNAB1 | 0.021 | < 0.001 | 0.496 | |
| NDUFB2 | 0.029 | 0.043 | 2.270 | |
| SPON1 | 0.007 | 0.025 | 2.074 | |
| BFSP1 | 0.005 | 0.003 | 2.605 | |
| CAMK2N1 | 0.018 | 0.035 | 2.012 | |
| CAPZB | 0.007 | 0.003 | 2.201 | |
| DOCK10 | 0.006 | 0.002 | 4.971 | |
| FSTL1 | 0.048 | < 0.001 | 3.000 | |
| GM2A | 0.005 | < 0.001 | 2.019 | |
| HLA-DRB1 | 0.014 | 0.048 | 4.062 | |
| ZKSCAN4 | 0.004 | < 0.001 | 2.318 | |
| ZSCAN16 | 0.008 | 0.001 | 2.324 | |
| PLIN2 | < 0.001 | < 0.001 | 0.499 | |
| AFAP1L2 | 0.019 | 0.006 | 0.486 | |
| TMEM107 | 0.037 | < 0.001 | 0.402 | |
| RAB31 | 0.042 | < 0.001 | 3.123 |
Each PTWAS value was calculated by transcriptome-wide association study analysis.
Each PmRNA value was derived from the published studies.
FC, fold change; mRNA, messenger RNA; TWAS, transcriptome-wide association study.
Table V.
Common gene ontology terms and pathways identified by comparing the gene ontology and pathway enrichment analysis results of transcriptome-wide association studies and messenger RNA expression profiles for knee osteoarthritis.
| Category | Tissue | Name | PTWAS* | PmRNA† |
|---|---|---|---|---|
| GO | Skeletal muscle | GO:0005515~protein binding | < 0.001 | < 0.001 |
| Blood | GO:0005515~protein binding | < 0.001 | < 0.001 | |
| GO:0005737~cytoplasm | < 0.001 | 0.005 | ||
| GO:0070062~extracellular exosome | 0.002 | < 0.001 | ||
| GO:0015629~actin cytoskeleton | 0.014 | 0.026 | ||
| GO:0006468~protein phosphorylation | 0.017 | 0.033 | ||
| GO:0060333~interferon-gamma-mediated signalling pathway | 0.017 | 0.047 | ||
| GO:0046982~protein heterodimerization activity | 0.033 | 0.025 | ||
| Pathway | Skeletal muscle | hsa04610:Complement and coagulation cascades | 0.017 | 0.001 |
| Blood | hsa05416:Viral myocarditis | 0.009 | 0.050 |
Each PTWAS value was calculated by gene ontology and pathway analysis using DAVID for the significant genes identified by transcriptome-wide association study analysis.
Each PmRNA value was calculated by gene ontology and pathway analysis using DAVID for the significant genes identified by messenger RNA expression profiles.
GO, gene ontology; mRNA, messenger RNA; TWAS, transcriptome-wide association study.
Fig. 3.
Gene expression heat map of the identified common genes shared by transcriptome-wide association studies (TWAS) and messenger RNA (mRNA) expression data of knee osteoarthritis (OA).
Discussion
To improve the understanding of the genetic aetiology of OA, we performed a TWAS of hip OA and knee OA through integrating the GWAS summary data and precomputed gene expression weights of skeletal muscle and blood. The TWAS results were further compared with mRNA expression profiles of OA cartilage, which detected multiple common genes, GO terms, and pathways shared by TWAS and mRNA expression profiles.
One of the important candidate genes identified in this study is ADM. This gene encodes preprohormone, which is cleaved into two biologically active peptides, including ADM and proadrenomedullin N-terminal 20 peptide. Previous studies have shown that ADM is expressed in bone and joint structures including cartilage and synovium.34,35 ADM is one of the top-ranking differentially expressed genes in OA bone, and plays a role in osteoblast and osteocyte differentiation and function.36 Downregulation of ADM is capable of inhibiting adipogenesis and osteoblastogenesis.37
FOXC1 belongs to the forkhead family of transcription factors (TFs), which is characterized by a distinct DNA-binding forkhead domain. A systematic analysis of six Gene Expression Omnibus (GEO) databases for synovial expression profiling identified the top ten TFs covering the most downstream differentially expressed genes and crucial TFs involved in the development of OA, including FOXC1.38 MicroRNAs have been identified in the development of OA and microRNA (miR)-138 has been reported to be involved with osteogenesis and regulation of chondrocyte phenotype.39,40 Research has also shown that miR-138-5p promotes cartilage degradation induced by interleukin (IL)-1β in human chondrocytes, possibly by targeting FOXC1.41
FSTL1/follistatin-related protein (FRP), an extracellular protein, was found in mesenchymal stem cells (MSCs) and cartilage. The FSTL1 mRNA and protein levels in the serum and synovial fluid were significantly higher in OA patients than in controls.42 Therefore, FSTL1 gene expression may be increased in OA patients.42 In addition, the findings show that FSTL1 is an important proinflammatory factor in the pathogenesis of OA through activating the canonical NF-κB pathway and enhancing synoviocyte proliferation, which may lead to the development of novel strategies for cartilage repair and be a promising target for the treatment of OA.43,44
HLA-DRB1 belongs to the major histocompatibility complex (HLA) class II beta chain paralogs. In the previous study, two SNPs (rs7775228 and rs10947262) in a region containing HLA class II/III genes associated with susceptibility to knee OA were identified through a GWAS and a replication, using a total of approximately 4,800 Japanese subjects.45 However, the rs7775228 and rs10947262 variants were not associated with risk of knee OA in European populations compared with Japanese individuals.46 The previous study showed that DR2 and DR5 were associated with OA, which hinted at LD between HLA-DRB1 genes and genes involved in the pathogenesis of OA.47
We also detected eight common GOs associated with hip and knee OA, respectively. For instance, protein binding (GO:0005515) was significant in blood of hip OA (PTWAS = 0.008, PmRNA < 0.001), skeletal muscle of knee OA (PTWAS < 0.001, PmRNA < 0.001), and blood of knee OA (PTWAS < 0.001, PmRNA < 0.001). To our knowledge, cold-inducible RNA-binding protein (CIRP) is a kind of inflammatory cytokine. In addition, research has found that the concentration of synovial fluid CIRP is closely associated with the synovial inflammation of OA and CIRP could be used as a potential marker for synovial inflammation of OA.48 The binding protein of the growth arrest and DNA damage-inducible protein 45 β (GADD45β) gene is down-regulated in ageing articular cartilage and chondrocyte clusters in OA cartilage.49 In all, these results further confirm the role of Golgi modifications and apoptosis in OA pathogenesis.
Although the heritability of OA is partly explained by GWAS, they still could ignore the genetic variants with expression-trait associations. In this study, TWAS analysis was used to identify novel genes associated with OA in the mRNA expression levels and gene expression profiles were used to verify the results. Compared with previous studies,50,51 TWAS is prone to spurious prioritization based on the expression data from OA-related tissues. In addition, TWAS was more accurate in prioritizing candidate causal genes than simple baselines. TWAS has been successful in identifying many genes and has the required boosting power to detect novel disease genes.12
There are three limitations that need to be noted in this study. Firstly, TWAS was developed to identify the genes, the regulated expression of which is associated with target diseases. TWAS may be too low power to detect the causal loci without cis-expression effects on target disease. Secondly, the objective of this study was to scan novel candidate genes related with OA. Further functional studies are needed to confirm our findings and clarify the potential biological mechanisms underlying OA, as detailed in a previous experimental study by Zhang et al.52 Thirdly, fracture may have had an impact on the hip cartilage expression profiles. However, in previous studies the individuals undergoing hip arthroplasty following femoral neck fracture were selected as the normal controls to perform the expression profiles.53,54 Therefore, the hip cartilage expression profile datasets, in which the patients with femoral neck fracture were used as the control cartilage specimens,15 could be used in this study analysis. Given these limitations, our results should be interpreted with caution and further studies are needed to confirm our findings.
In conclusion, this study combined TWAS and gene expression profiling datasets to identify the candidate gene associated with OA. We have identified 33 and 24 common genes in hip OA and knee OA, respectively, such as FOXC1, ADM, FSTL1, and HLA-DRB1. In view of these limitations, the results should be explained with caution. Therefore, we need further studies to verify our findings and reveal the potential effect of identified genes in the development of OA.
Footnotes
Author Contributions: X. Qi: Wrote the manuscript, Designed the study, Analyzed the data.
F. Yu: Wrote the manuscript.
Y. Wen: Wrote the manuscript.
P. Li: Wrote the manuscript.
B. Cheng: Wrote the manuscript.
M. Ma: Wrote the manuscript.
S. Cheng: Wrote the manuscript.
L. Zhang: Wrote the manuscript.
C. Liang: Wrote the manuscript.
L. Liu: Wrote the manuscript.
F. Zhang: Wrote the manuscript, Designed the study.
Ethical review statement: This study did not require ethical approval.
Follow us @BoneJointRes
Supplementary Material
Supplementary tables showing: transcriptome-wide association study results of hip osteoarthritis; transcriptome-wide association study results of knee osteoarthritis; gene ontology and pathway enrichment analysis results of the significant genes identified by transcriptome-wide association studies for hip osteoarthritis; gene ontology and pathway enrichment analysis results of the significant genes identified by transcriptome-wide association studies for knee osteoarthritis.
Funding statement
The authors report institutional grants related to this study (paid to Xi'an Jiaotong University) from the National Natural Science Foundation of China, the Key projects of international cooperation among governments in scientific and technological innovation, the Fundamental Research Funds for the Central Universities, and the Natural Science Basic Research Plan in Shaanxi Province of China.
No benefits in any form have been received or will be received from a commercial party related directly or indirectly to the subject of this article.
References
- 1. Li H, Yang HH, Sun ZG, Tang HB, Min JK. Whole-transcriptome sequencing of knee joint cartilage from osteoarthritis patients. Bone Joint Res. 2019;8(6):288-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Felson DT, Lawrence RC, Dieppe PA, et al. Osteoarthritis: new insights. Part 1: the disease and its risk factors. Ann Intern Med. 2000;133(8):635-646. [DOI] [PubMed] [Google Scholar]
- 3. Pereira D, Ramos E, Branco J. Osteoarthritis. Acta Med Port. 2015;28(1):99-106. [DOI] [PubMed] [Google Scholar]
- 4. March LM, Bachmeier CJM. Economics of osteoarthritis: a global perspective. Baillieres Clin Rheumatol. 1997;11(4):817-834. [DOI] [PubMed] [Google Scholar]
- 5. Martel-Pelletier J, Barr AJ, Cicuttini FM, et al. Osteoarthritis. Nat Rev Dis Primers. 2016;2(1):16072. [DOI] [PubMed] [Google Scholar]
- 6. Zengini E, Hatzikotoulas K, Tachmazidou I, et al. Genome-wide analyses using UK Biobank data provide insights into the genetic architecture of osteoarthritis. Nat Genet. 2018;50(4):549-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Farhat MR, Freschi L, Calderon R, et al. GWAS for quantitative resistance phenotypes in Mycobacterium tuberculosis reveals resistance genes and regulatory regions. Nat Commun. 2019;10(1):2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schwanhäusser B, Busse D, Li N, et al. Global quantification of mammalian gene expression control. Nature. 2011;473(7347):337-342. [DOI] [PubMed] [Google Scholar]
- 9. Peng S, Deyssenroth MA, Di Narzo AF, et al. Expression quantitative trait loci (eQTLs) in human placentas suggest developmental origins of complex diseases. Hum Mol Genet. 2017;26(17):3432-3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lappalainen T, Sammeth M, Friedländer MR, et al. Geuvadis Consortium. Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501(7468):506-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Veturi Y, Ritchie MD. How powerful are summary-based methods for identifying expression-trait associations under different genetic architectures? Pac Symp Biocomput. 2018;23:228-239. [PMC free article] [PubMed] [Google Scholar]
- 12. Barfield R, Feng H, Gusev A, et al. Transcriptome-wide association studies accounting for colocalization using Egger regression. Genet Epidemiol. 2018;42(5):418-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ioannidis NM, Wang W, Furlotte NA, et al. 23andMe Research Team. Gene expression imputation identifies candidate genes and susceptibility loci associated with cutaneous squamous cell carcinoma. Nat Commun. 2018;9(1):4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39(7):906-913. [DOI] [PubMed] [Google Scholar]
- 15. Xu Y, Barter MJ, Swan DC, et al. Identification of the pathogenic pathways in osteoarthritic hip cartilage: commonality and discord between hip and knee OA. Osteoarthritis Cartilage. 2012;20(9):1029-1038. [DOI] [PubMed] [Google Scholar]
- 16. Kijowski R, Blankenbaker D, Stanton P, Fine J, De Smet A. Arthroscopic validation of radiographic grading scales of osteoarthritis of the tibiofemoral joint. AJR Am J Roentgenol. 2006;187(3):794-799. [DOI] [PubMed] [Google Scholar]
- 17. Fisch KM, Gamini R, Alvarez-Garcia O, et al. Identification of transcription factors responsible for dysregulated networks in human osteoarthritis cartilage by global gene expression analysis. Osteoarthritis Cartilage. 2018;26(11):1531-1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gusev A, Ko A, Shi H, et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat Genet. 2016;48(3):245-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gong L, Zhang D, Lei Y, Qian Y, Tan X, Han S. Transcriptome-wide association study identifies multiple genes and pathways associated with pancreatic cancer. Cancer Med. 2018;7(11):5727-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. No authors listed. TWAS / FUSION. 2019. http://gusevlab.org/projects/fusion/ (date last accessed 21 January 2020).
- 21. Ponchel F, Burska AN, Hensor EMA, et al. Changes in peripheral blood immune cell composition in osteoarthritis. Osteoarthritis Cartilage. 2015;23(11):1870-1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Krishnasamy P, Hall M, Robbins SR. The role of skeletal muscle in the pathophysiology and management of knee osteoarthritis. Rheumatology (Oxford). 2018;57(suppl_4):iv22-iv33. [DOI] [PubMed] [Google Scholar]
- 23. Lonsdale J, Thomas J, Moore HF, et al. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45(6):580-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Boomsma DI, de Geus EJ, Vink JM, et al. Netherlands Twin Register: from twins to twin families. Twin Res Hum Genet. 2006;9(6):849-857. [DOI] [PubMed] [Google Scholar]
- 25. Wright FA, Sullivan PF, Brooks AI, et al. Heritability and genomics of gene expression in peripheral blood. Nat Genet. 2014;46(5):430-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nuotio J, Oikonen M, Magnussen CG, et al. Cardiovascular risk factors in 2011 and secular trends since 2007: the Cardiovascular Risk in Young Finns Study. Scand J Public Health. 2014;42(7):563-571. [DOI] [PubMed] [Google Scholar]
- 27. Raitakari OT, Juonala M, Rönnemaa T, et al. Cohort profile: the cardiovascular risk in Young Finns Study. Int J Epidemiol. 2008;37(6):1220-1226. [DOI] [PubMed] [Google Scholar]
- 28. Zhou X, Carbonetto P, Stephens M. Polygenic modeling with bayesian sparse linear mixed models. PLoS Genet. 2013;9(2):e1003264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44-57. [DOI] [PubMed] [Google Scholar]
- 30. Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. No authors listed. DAVID Bioinformatics Resources 6.8. 2020. https://david.ncifcrf.gov/ (date last accessed 7 February 2020).
- 32. No authors listed. FUMA GWAS: Functional Mapping and Annotation of Genome-Wide Association Studies. 2020. http://fuma.ctglab.nl/ (date last accessed 7 February 2020).
- 33. Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017;8(1):1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Matsushita T, Matsui N, Yoshiya S, Fujioka H, Kurosaka M. Production of adrenomedullin from synovial cells in rheumatoid arthritis patients. Rheumatol Int. 2004;24(1):20-24. [DOI] [PubMed] [Google Scholar]
- 35. Cornish J, Naot D, Reid IR. Adrenomedullin—a regulator of bone formation. Regul Pept. 2003;112(1-3):79-86. [DOI] [PubMed] [Google Scholar]
- 36. Hopwood B, Tsykin A, Findlay DM, Fazzalari NL. Microarray gene expression profiling of osteoarthritic bone suggests altered bone remodelling, WNT and transforming growth factor-beta/bone morphogenic protein signalling. Arthritis Res Ther. 2007;9(5):R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hopwood B, Tsykin A, Findlay DM, Fazzalari NL. Gene expression profile of the bone microenvironment in human fragility fracture bone. Bone. 2009;44(1):87-101. [DOI] [PubMed] [Google Scholar]
- 38. Fei Q, Lin J, Meng H, et al. Identification of upstream regulators for synovial expression signature genes in osteoarthritis. Joint Bone Spine. 2016;83(5):545-551. [DOI] [PubMed] [Google Scholar]
- 39. Eskildsen T, Taipaleenmäki H, Stenvang J, et al. MicroRNA-138 regulates osteogenic differentiation of human stromal (mesenchymal) stem cells in vivo. Proc Natl Acad Sci U S A. 2011;108(15):6139-6144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nugent M. MicroRNAs: exploring new horizons in osteoarthritis. Osteoarthritis Cartilage. 2016;24(4):573-580. [DOI] [PubMed] [Google Scholar]
- 41. Yuan Y, Zhang GQ, Chai W, Ni M, Xu C, Chen JY. Silencing of microRNA-138-5p promotes IL-1β-induced cartilage degradation in human chondrocytes by targeting FOXC1: miR-138 promotes cartilage degradation. Bone Joint Res. 2016;5(10):523-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang Y, Li D, Xu N, et al. Follistatin-like protein 1: a serum biochemical marker reflecting the severity of joint damage in patients with osteoarthritis. Arthritis Res Ther. 2011;13(6):R193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chaly Y, Blair HC, Smith SM, et al. Follistatin-like protein 1 regulates chondrocyte proliferation and chondrogenic differentiation of mesenchymal stem cells. Ann Rheum Dis. 2015;74(7):1467-1473. [DOI] [PubMed] [Google Scholar]
- 44. Ni S, Miao K, Zhou X, et al. The involvement of follistatin-like protein 1 in osteoarthritis by elevating NF-κB-mediated inflammatory cytokines and enhancing fibroblast like synoviocyte proliferation. Arthritis Res Ther. 2015;17:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nakajima M, Takahashi A, Kou I, et al. New sequence variants in HLA class II/III region associated with susceptibility to knee osteoarthritis identified by genome-wide association study. PLoS One. 2010;5(3):e9723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Valdes AM, Styrkarsdottir U, Doherty M, et al. Large scale replication study of the association between HLA class II/BTNL2 variants and osteoarthritis of the knee in European-descent populations. PLoS One. 2011;6(8):e23371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Moos V, Menard J, Sieper J, Sparmann M, Müller B. Association of HLA-DRB1*02 with osteoarthritis in a cohort of 106 patients. Rheumatology (Oxford). 2002;41(6):666-669. [DOI] [PubMed] [Google Scholar]
- 48. Yu L, Li QH, Deng F, Yu ZW, Luo X-Z, Sun JL. Synovial fluid concentrations of cold-inducible RNA-binding protein are associated with severity in knee osteoarthritis. Clin Chim Acta. 2017;464:44-49. [DOI] [PubMed] [Google Scholar]
- 49. Ijiri K, Zerbini LF, Peng H, et al. Differential expression of GADD45β in normal and osteoarthritic cartilage: potential role in homeostasis of articular chondrocytes. Arthritis Rheum. 2008;58(7):2075-2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. He A, Ning Y, Wen Y, et al. Use of integrative epigenetic and mRNA expression analyses to identify significantly changed genes and functional pathways in osteoarthritic cartilage. Bone Joint Res. 2018;7(5):343-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhang X, Bu Y, Zhu B, et al. Global transcriptome analysis to identify critical genes involved in the pathology of osteoarthritis. Bone Joint Res. 2018;7(4):298-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhang M, Lu Q, Budden T, Wang J. NFAT1 protects articular cartilage against osteoarthritic degradation by directly regulating transcription of specific anabolic and catabolic genes. Bone Joint Res. 2019;8(2):90-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Davidson RK, Waters JG, Kevorkian L, et al. Expression profiling of metalloproteinases and their inhibitors in synovium and cartilage. Arthritis Res Ther. 2006;8(4):R124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rushton MD, Reynard LN, Barter MJ, et al. Characterization of the cartilage DNA methylome in knee and hip osteoarthritis. Arthritis Rheumatol. 2014;66(9):2450-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]