Summary
Regulatory T cells are important regulators of the immune system and have versatile functions for the homeostasis and repair of tissues. They express the forkhead box transcription factor Foxp3 as a lineage-defining protein. Negative regulators of Foxp3 expression are not well understood. Here, we generated double-stranded DNA probes complementary to the Foxp3 promoter sequence and performed a pull-down with nuclear protein in vitro, followed by elution of bound proteins and quantitative mass spectrometry. Of the Foxp3-promoter-binding transcription factors identified with this approach, one was T cell factor 1 (TCF1). Using viral over-expression, we identified TCF1 as a repressor of Foxp3 expression. In TCF1-deficient animals, increased levels of Foxp3intermediateCD25negative T cells were identified. CRISPR-Cas9 knockout studies in primary human and mouse conventional CD4 T (Tconv) cells revealed that TCF1 protects Tconv cells from inadvertent Foxp3 expression. Our data implicate a role of TCF1 in suppressing Foxp3 expression in activated T cells.
Subject Areas: Molecular Biology, Molecular Mechanism of Gene Regulation, Immunology, Proteomics
Graphical Abstract
Highlights
-
•
Quantitative proteomics identifies proteins bound to the Foxp3 gene promoter
-
•
Promoter-binding proteins are suppressing Foxp3 expression
-
•
TCF1-deficient animals have more Foxp3-expressing CTLA4−CD25−CD4+ T cells
-
•
TCF1 suppresses Foxp3 expression in activated non-Treg cells
Molecular Biology; Molecular Mechanism of Gene Regulation; Immunology; Proteomics
Introduction
Foxp3 is the master transcription factor (TF) for regulatory T (Treg) cells, and its absence leads to catastrophic autoimmune events in mice (Scurfy phenotype; Brunkow et al., 2001, Fontenot et al., 2003, Wildin et al., 2001) and humans (IPEX or immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome; Bennett et al., 2001). Foxp3 expression in concert with a specific epigenetic landscape induced in the thymus defines Treg cells (Ohkura et al., 2012). A subset of Treg cells can acquire epigenetic and transcriptional profiles defining their tissue adaptation (Delacher et al., 2017, Delacher et al., 2019, Delacher et al., 2020, Schmidl et al., 2018).
The Foxp3 gene is located on the X chromosome. It contains 14 exons, three of which are not translated (-2a, -2b, and −1). The promoter with TATA box, GC box, and CAAT box is located just upstream of the transcription start site (TSS). When comparing the sequence homology between mouse and human Foxp3 genetic code, a high degree of conservation can be appreciated for the coding exons, three regions in non-coding introns, and the promoter itself (Andersen et al., 2012, Janson et al., 2008, Sadlon et al., 2010, Xie et al., 2015). The three distinct conserved regions within the intronic sequences of the Foxp3 gene were determined as conserved non-coding sequences 1, 2, and 3 (CNS1-3). Each CNS region has a distinct function in the initiation or stabilization of Foxp3 gene expression, just like the core Foxp3 promoter (Delacher et al., 2014, Rudensky, 2011). A fourth conserved region outside the Foxp3 gene, named CNS0, has recently been described (Kitagawa et al., 2017).
CNS0 contains Treg-specific super-enhancers crucial for Treg cell lineage specification in the thymus (Kitagawa et al., 2017). CNS1 is an important transforming growth factor (TGF)-β-sensitive enhancer region for the induction of peripherally induced Treg (pTreg) from Foxp3- conventional CD4 T (Tconv) cells in vivo and for the in vitro conversion of Treg cells from Tconv. CNS1 is not relevant for thymic Treg cell generation (Josefowicz et al., 2012, Schlenner et al., 2012, Tone et al., 2008). The CNS2 region contains a high number of CpG sites, becomes demethylated in the thymus, and has an important role to stabilize Foxp3 expression (Delacher et al., 2017, Floess et al., 2007, Zheng et al., 2010). In addition, some factors bind this region to stabilize the demethylated phenotype (Kim and Leonard, 2007, Mouly et al., 2010). The CNS3 is a pioneer element required for efficient induction of Foxp3 transcription (Schuster et al., 2012, Zheng et al., 2010).
The precise location of the Foxp3 promoter and the true TSS were identified in a study utilizing rapid amplification of 5′ ends, proving that the core promoter is indeed the area where DNA-dependent RNA transcription of Foxp3 pre-mRNA begins (Tone et al., 2008).
Several studies identified Nfat (nuclear factor of activated T cells) binding to the Foxp3 promoter, and mutations in the Nfat-binding sites or deficiency in calcium sensing disrupted its activity (Mantel et al., 2006, Oh-Hora et al., 2013, Tone et al., 2008). In addition, a set of Forkhead Box proteins (Foxo1 and Foxo3a) bind the Foxp3 promoter as part of the PI3K-Akt-mTOR pathway, and their specific deletion caused multifocal inflammatory disorder (Harada et al., 2010, Ouyang et al., 2010). Stat5 (signal transducer of activated T cells 5) has also been detected at the Foxp3 gene promoter, and its selective deletion prevents Treg cell development (Burchill et al., 2007, Yao et al., 2007). Another example of direct Foxp3 promoter regulation is the study of nuclear receptor subfamily members: mice devoid of all three subfamily members (Nr4a1, Nr4a2, Nr4a3) cannot produce Treg cells and die of systemic autoimmunity (Sekiya et al., 2011, Sekiya et al., 2013). Several studies identified the c-Rel enhanceosome complex (Ruan et al., 2009) as well as Runx proteins (Bruno et al., 2009, Klunker et al., 2009) at the Foxp3 promoter. Finally, Foxp3-promoter-binding partners have been identified in the E2A-Id3 signaling axis and have been shown to influence Foxp3 expression (Wang et al., 2011, Wohlfert et al., 2011).
These studies have identified an impressive set of Foxp3-inducing factors. However, much less is known about Foxp3-repressive factors required to protect Foxp3-negative Tconv cells from unwanted Foxp3 expression, e.g., as a by-product of T cell activation.
We wanted to identify Foxp3-promoter-binding proteins in an unbiased way using quantitative mass spectrometry (Mittler et al., 2009). With this approach, we identified several binding partners to the Foxp3 promoter region with repressive effect on the Foxp3 promoter. One of those Foxp3-promoter-suppressive factors was T cell factor 1 (TCF1), which we followed up by Luciferase-based-binding studies, by overexpression and deletion studies in primary T cells, and by the analysis of a TCF1-deficient mouse strain. Our data point toward a specific role of TCF1 to suppress Foxp3 expression in activated non-Treg cells.
Results
Quantitative Proteomics Identifies Foxp3-Promoter-Binding Factors
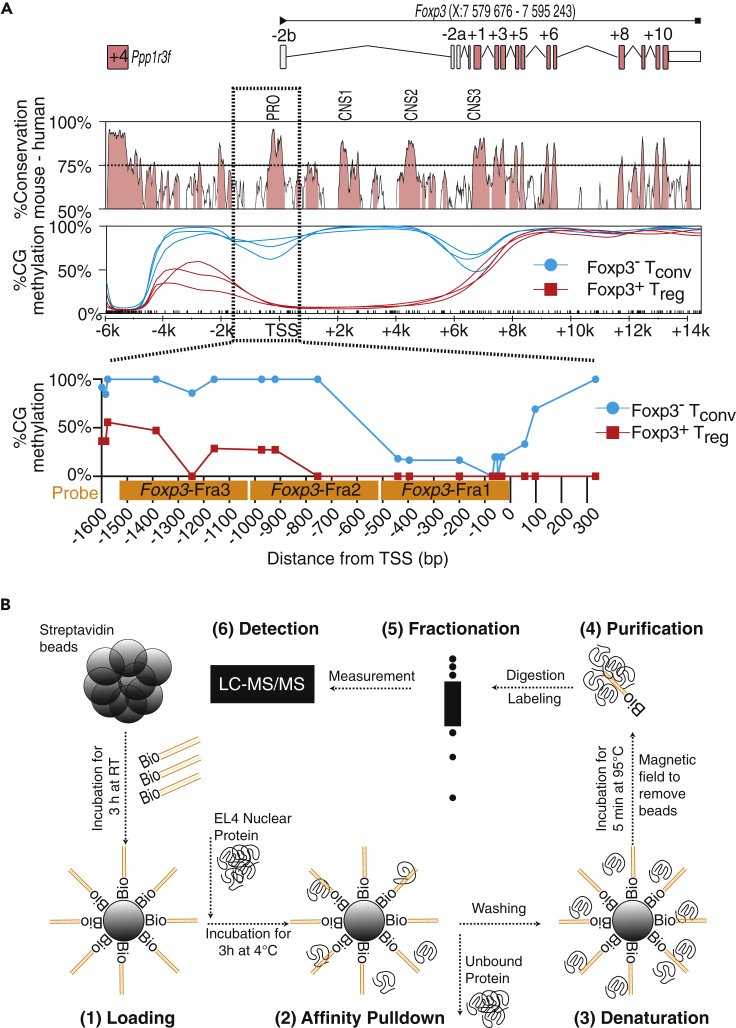
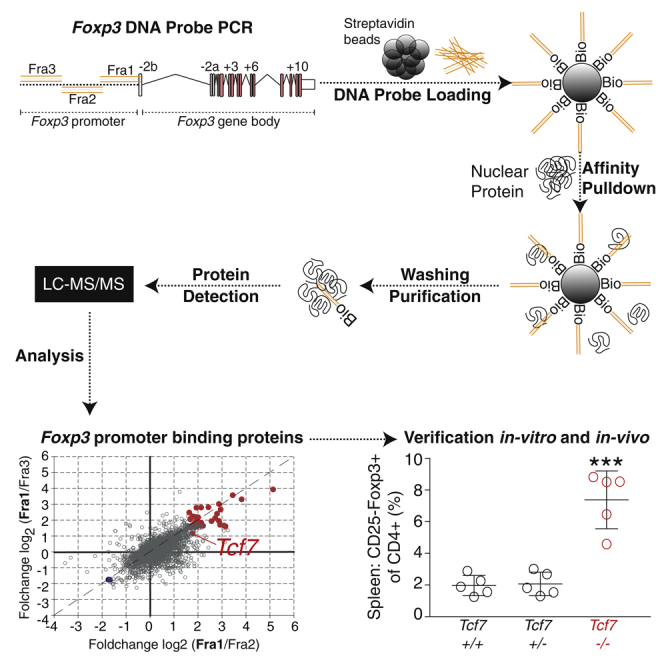
We visualized the conservation of Foxp3 genetic code between mouse and human and superimposed the Foxp3 gene structure to identify target regions for protein binding identification (Figure 1A). We could observe that the Foxp3 gene promoter, at least in its very proximal 500 bp, was highly conserved between mouse and human. In addition, the proximal promoter was demethylated in both Treg and Tconv cells, whereas intron-1 was specifically demethylated only in Treg cells. We generated three 500-bp DNA probes complementary to the Foxp3 promoter region: Foxp3-Fra1, starting from the Foxp3 TSS and extending 500 bp upstream into the promoter region (−500); Foxp3-Fra2, extending −500 bp to −1000 bp into the Foxp3 promoter; and Foxp3-Fra3, extending from −1000 bp to −1500 bp into the distal Foxp3 promoter region (Figure 1A). All three fragments were generated with biotin-labeled primers to use them as probes for an in vitro pull-down followed by mass spectrometry (Mittler et al., 2009) (Figure 1B). First, streptavidin beads were linked to biotinylated Foxp3 promoter Fra1, Fra2, or Fra3 probes, followed by incubation with nuclear proteins isolated from EL4 T cells. We used EL4 T cells as a Foxp3-negative cell line to study potential repressive elements binding to the Foxp3 promoter. Unbound protein was washed off, and beads including attached proteins were isolated in a magnetic field. Protein was eluted from the beads and purified, digested, labeled with stable isotopes, fractionated, and finally subjected to nano-liquid chromatography-mass spectrometry analysis allowing the quantitative detection of peptides bound to each DNA probe.
Figure 1.
Quantitative Proteomics of the Foxp3 Promoter
(A) Conservation between mouse (CCDS29965) and human (CCDS14323) Foxp3 genetic code. The y axis indicates conservation between genetic code in %, and the x axis indicates genomic location. Histogram generated with Vista (Mayor et al., 2000). Labels PRO (promoter) and CNS (conserved non-coding sequence) on top. Below, CG methylation of the Foxp3 gene (X:7,579,676-7,595,243) with three replicate Treg and Tconv cells, data published previously (Delacher et al., 2017). Beneath histograms, magnification of the Foxp3 promoter region from −1600 bp to +300 bp relative to the Foxp3 TSS with methylation levels of individual CGs. Probes for proteomics are labeled in orange.
(B) Overview of the quantitative proteomics procedure. First, beads are loaded with biotinylated Foxp3-Fra1, Foxp3-Fra2, or Foxp3-Fra3 probes (Loading [1]). Then, loaded beads are incubated with nuclear proteins and purified in magnetic field (Affinity pull-down [2]). Proteins are denatured (Denaturation [3]) and purified (Purification [4]), followed by Fractionation (5) and Detection (6) via nano-liquid chromatography-tandem mass spectrometry (LC-MS/MS).
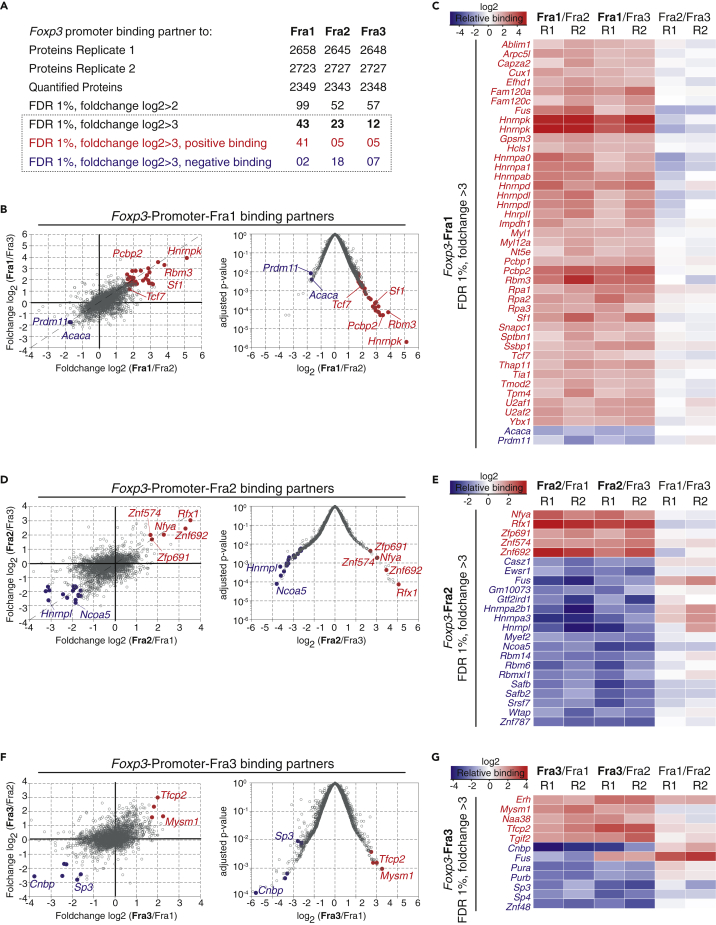
The experiment was done in two replicates, yielding more than 2,500 proteins bound to each DNA probe (Figure 2A). Of the around 2,400 proteins that could be quantified with each probe, 43, 23, and 23 were differentially bound to Fra1, Fra2, and Fra3, respectively (fold-change log2>3 compared with the other two probes, false discovery rate [FDR] <1%). All binding partners to Fra1 are displayed in a dot plot for relative binding as well as a volcano plot to visualize selection based on statistics (p < 0.01) and fold-change (log2>3, Figure 2B). Proteins with positive binding values (in the comparison Fra1 versus Fra2 and Fra1 versus Fra3) were identified and labeled in red, whereas proteins with negative binding values (e.g., not binding to Fra1, but to Fra2 or Fra3) were labeled in blue. Proteins that bound equally to both fragments were labeled in gray. A heatmap with differential binding values for all selected proteins for Fra1 clarifies selective binding patterns of all candidate factors: out of the 43 proteins that bound differentially to Fra1 versus Fra2 and Fra 1 versus Fra3, 41 were enriched on Fra1, being potential candidates for downstream testing. Two proteins bound strongly to Fra2 or Fra3, but not Fra1, and were excluded.
Figure 2.
Proteins Identified at the Foxp3 Promoter
(A) Results of quantitative proteomics of the Foxp3 promoter with two replicates; FDR, false discovery rate. Bold numbers indicate differential binding partners to the respective fragment (FDR 1%, fold-change log2>3). Red numbers indicate positive binding partners, blue numbers negative binding partners to the respective fragment.
(B) Results for Foxp3-Fra1. Left dot plots illustrate all detected proteins with relative binding values to Fra1 versus Fra3 (y axis) and Fra1 versus Fra2 (x axis). Right graph, volcano plot with Fra1 versus Fra2 relative binding (x axis) versus adjusted p value (y axis). Key genes are highlighted. Positive and negative binding partners (FDR 1%, fold-change log2>3) are highlighted.
(C) Heatmap illustrating relative binding of Fra1 candidates to Fra1, Fra2, and Fra3 (FDR 1%, fold-change log2>3).
(D) Results for Foxp3-Fra2. Left dot plots illustrate all detected proteins with relative binding values to Fra2 versus Fra3 (y axis) and Fra2 versus Fra1 (x axis). Right graph, volcano plot with Fra2 versus Fra3 relative binding (x axis) versus adjusted p value (y axis). Key genes are highlighted. Positive and negative binding partners (FDR 1%, fold-change log2>3) are highlighted.
(E) Heatmap illustrating relative binding of Fra2 candidates to Fra1, Fra2, and Fra3 (FDR 1%, fold-change log2>3).
(F) Results for Foxp3-Fra3. Left dot plots illustrate all detected proteins with relative binding values to Fra3 versus Fra2 (y axis) and Fra3 versus Fra1 (x axis). Right graph, volcano plot with Fra3 versus Fra1 relative binding (x axis) versus adjusted p value (y axis). Key genes are highlighted. Positive and negative binding partners (FDR 1%, fold-change log2>3) are highlighted.
(G) Heatmap illustrating relative binding of Fra3 candidates to Fra1, Fra2, and Fra3 (FDR 1%, fold-change log2>3). Data are representative of two independent experiments.
To identify candidates' binding to Fra2, we again display dot plot and heatmap (Figures 2D and 2E). Of the 23 proteins that bound differentially to Fra2 versus Fra1 and Fra2 versus Fra3, five bound specifically to Fra2. All other candidates bound strongly to Fra1 or Fra3 and were excluded for downstream testing. For Fra3, 5 of 12 proteins were enriched on the Fra3-binding sites and used to select candidate factors (Figures 2F and 2G). Therefore, in summary, we identified 41 factors bound to Fra1, 5 factors bound to Fra2, and 5 factors bound to Fra3.
Foxp3-Promoter-Binding TFs Down-modulate Foxp3 Promoter Activity In Vitro
For our proteomics experiments, we used nuclear protein derived from a T cell line. To avoid cell line artifacts, we measured expression levels of some candidate factors in primary murine Treg and Tconv cells as well as in EL4 and RMA mouse T cell lines by real-time PCR. Most factors were expressed in primary Treg and Tconv cells and cell lines (Figure S1A). In addition, we isolated Treg and Tconv cells from human peripheral blood and measured the expression of some candidate factors by real-time PCR and included three T cell lines (Jurkat, CEM, and BE, Figure S1B). As seen for mouse, most factors were also expressed in primary human T cells.
To explore the function of candidate proteins with Luciferase reporter vectors, we cloned nine factors from Fra1, five factors from Fra2, and three factors from Fra3 into eukaryotic production vectors. All vectors were sequenced to confirm plasmid identity and sequence integrity, and we verified plasmid stability by gel electrophoresis (Figure S1C). In addition, we confirmed transgene expression for selected transgenes by western blot (Figure S2A) and confirmed plasmid identity (Figure S2B). Using a eukaryotic vector expression system, we transfected the respective candidate factors, a Foxp3 promoter vector with Luciferase reporter (Sekiya et al., 2011) and a beta-galactosidase transfection and normalization control vector into HEK293 cells (Figure S3A). After 2 days, we measured Luciferase and β-galactosidase enzymatic activity. To cross-validate our dataset, we used a Luciferase basic vector without promoter, a full Foxp3 promoter Luciferase vector (containing 3,500 bp of the Foxp3 promoter, location X:7,576,145- X:7,589,866 [10190 bp]), and short 500-bp Foxp3 promoter Luciferase vectors (Figure S3B). These short fragments were identical to the probes used for quantitative proteomics (Figure 1A). Using this system, we first measured light induction by GFP (negative control) and Nr4a1 (positive control). As expected, GFP expression did not induce activity together with the basic vector, the full Foxp3 promoter vector, or a vector containing only Foxp3 Fra1 (Figure S3C, left). In contrast to this, the Nr4a1 transgene induced significant activity at the full Foxp3 promoter vector, but not in the Fra1 or the basic vector (Figure S3C, right). Nr4a1 binding to the Foxp3 gene has already been described in the literature (Sekiya et al., 2011, Sekiya et al., 2013). Next, we tested the candidates identified with our screening method with this assay. None of the factors induced Foxp3 promoter Luciferase activity in the short (Fra1, Fra2, Fra3) or the full Foxp3 promoter vector. In contrast to this, some of the factors such as Sf1 (splicing factor 1), Znf574 (zinc finger protein 574), Rfx1 (regulatory factor X,1), or Naa38 (Nα-acetyltransferase 38, NatC auxiliary subunit) showed a significant down-modulation of Luciferase activity at one of the Foxp3 promoter vectors (Figures S3D–S3F). Taken together, our Luciferase screens demonstrate that some of our candidate factors showed significant repressive activity and down-modulated basic Foxp3 promoter activity.
Candidate Factors Overrule Activation-Induced Foxp3 Promoter Activity
In the previous experiments, we identified Foxp3-promoter-binding TFs and observed that they down-modulated basic Foxp3 promoter activity. Foxp3 promoter signaling can also be induced by T cell receptor (TCR) stimulation (Mantel et al., 2006, Oh-Hora et al., 2013, Tone et al., 2008). Therefore, we established a system where Jurkat T cells were electroporated with a Foxp3 promoter vector, a eukaryotic production vector carrying the candidate transgene, and a Renilla normalization and transfection control vector (Figure S4A). After 24 h, TCR signaling was mimicked by phorbol myristate acetate (PMA)/Ionomycin (PMA/IM) stimulation. After 24 h, Luciferase and Renilla activities were measured by luminescence, as before. When comparing results for the basic vector without any promoter sequence and the Foxp3 promoter vector, PMA/IM stimulation increased Foxp3 promoter activity by about 10-fold (Figure S4B). We tested several candidate transgenes with this system. No changes were observed when using the basic Luciferase vector. In contrast to this, PMA/IM stimulation induced activity with the Foxp3 promoter vector. Interestingly, some of the factors such as Pcbp1, Pcbp2, and Thap11 significantly down-regulated Luciferase activity with the Foxp3 promoter, but not with the basic vector without a promoter sequence (Figures S3C–S3E). These data indicate that certain TCR signals induced by treatment of cells with PMA/IM can be suppressed by individual candidate factors, providing additional evidence of their Foxp3-suppressive nature.
Candidate TF Expression Levels in Primary Treg and Tconv Cells Identifies TCF1
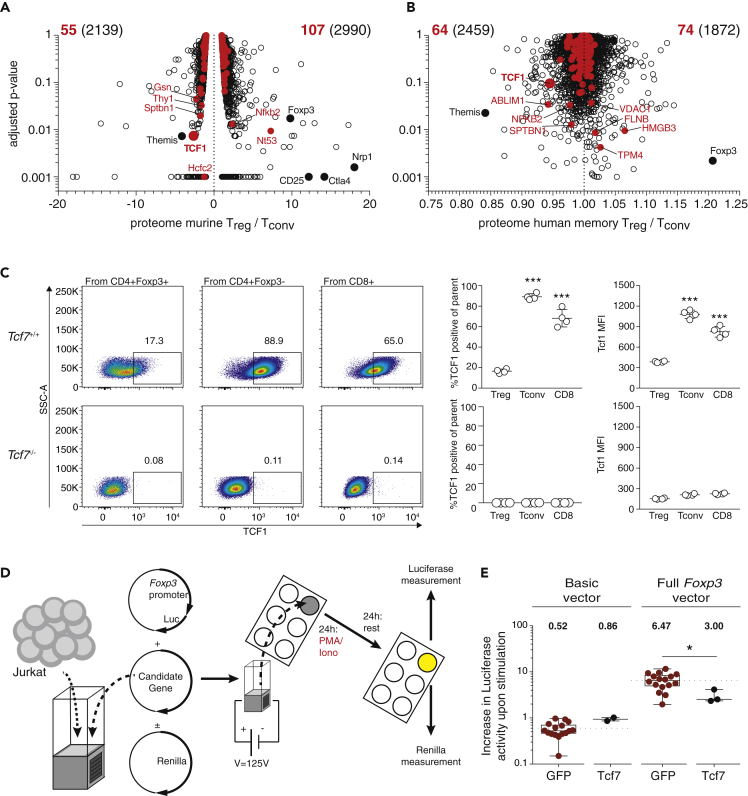
In the previous experiments, we confirmed the Foxp3-suppressive nature of multiple TFs with our proteomics approach. Now, to select a candidate TF for further evaluation, we used a funnel approach using published proteome datasets. Ideally, a candidate TF should be up-regulated in Foxp3-negative Tconv cells. Two recent studies investigated the differential proteome between murine Treg and Tconv cells (Barra et al., 2015a) and human Treg and Tconv cells (Cuadrado et al., 2018). We extracted both datasets and identified our target Foxp3-promoter-binding factors. When comparing the 5,129 proteins identified in bulk murine Foxp3+ Treg versus Foxp3- Tconv, we could map 162 of 209 Foxp3-promoter-binding factors (fold-change log2>2, FDR < 1%) to this dataset (Figure 3A). One example for a highly significant Tconv-over-expressed factor that also binds the Foxp3 promoter in our study is TCF1. When using the dataset derived from human effector Tconv versus effector Treg cells, we could map 138 proteins of 209 Foxp3-promoter-binding factors (Figure 3B). Again, TCF1 was detected as Tconv-specific over-expressed factor. Therefore, human and mouse Treg/Tconv proteomic datasets indicate that TCF1, a candidate protein identified with our Foxp3-promoter-binding screening (Figures 2B and 2C), is also a differentially expressed TF in Foxp3-positive versus Foxp3-negative T cells. To further validate this, we stained CD4+Foxp3- Tconv cells, CD4+Foxp3+ Treg cells, and CD4−CD8+ cytotoxic T cells and measured TCF1 expression levels by flow cytometry. As a control, we used Tcf7−/− animals (Tcf7−/− animals lack the TCF1 protein) (Verbeek et al., 1995). In spleen, Tconv and CD8 T cells expressed elevated TCF1 levels, whereas Treg cells had significantly lower expression values (Figure 3C). To test if TCF1 has a repressive function on the Foxp3 promoter, we used PMA/IM-stimulated Jurkat cells and electroporated a Tcf7 eukaryotic production vector (Figure 3D). Interestingly, the TCF1 overexpression showed significant capacity to down-modulate the activity of the Foxp3 promoter, but not the basic promoter (Figure 3E). In summary, we showed that TCF1 is expressed more specifically in Foxp3-negative Tconv cells and that the TCF1 protein has the capacity to down-modulate Foxp3 promoter activity, similar to other proteins identified by quantitative proteomics of the Foxp3 promoter.
Figure 3.
Funnel Approach Identifies TCF1 as Foxp3-Regulating TF
(A) 5,129 proteins comparing bulk murine Foxp3+ Treg versus Foxp3- Tconv were extracted from Barra et al. (2015a) and compared with our proteomics dataset. Data are displayed in a volcano plot with adjusted p value (y axis) versus protein expression Treg versus Tconv (x axis). 162 out of 209 Foxp3-promoter-binding factors (fold-change log2>2, FDR<1%) were mapped to this dataset and highlighted in red. Key TF labeled.
(B) 4,331 proteins comparing bulk human effector Treg versus Tconv were extracted from Cuadrado et al. (2018) and compared with our proteomics dataset. Data are displayed in a volcano plot with adjusted p value (y axis) versus protein expression Treg versus Tconv (x axis). 138 of 209 Foxp3-promoter-binding factors (fold-change log2>2, FDR<1%) were mapped to this dataset and highlighted in red. Key TF labeled.
(C) Analysis of TCF1 protein expression in Tcf7+/+ animals (top) and Tcf7−/− animals (below). Left, dot plots illustrating TCF1 expression in Treg cells (CD8−CD4+Foxp3+), Tconv cells (CD8−CD4+Foxp3-), and CD8 T cells (CD4−CD8+). Right, statistical analysis across replicates (n = 4, one-way ANOVA, error bars = standard deviation, ∗∗∗p < 0.001).
(D) Experiment overview: 2,000,000 Jurkat cells were electroporated with a Renilla normalization vector, Foxp3-Luciferase reporter vector, and a eukaryotic expression vector containing the transgene of interest; 24 h after electroporation, cells were stimulated with PMA/Ionomycin, and 20 h after stimulation, cells were lysed and Renilla as well as Luciferase enzyme activities were measured on a luminometer with automated injection of substrates.
(E) Jurkat cells were electroporated with a Renilla normalization vector, Foxp3-Luciferase reporter vector, and a GFP or TCF1 eukaryotic expression vector. Statistical testing with unpaired t test (n = 3–16, ∗p < 0.05). Data are derived from literature or two or more independent experiments.
TCF1 Overexpression Impairs Foxp3 Induction
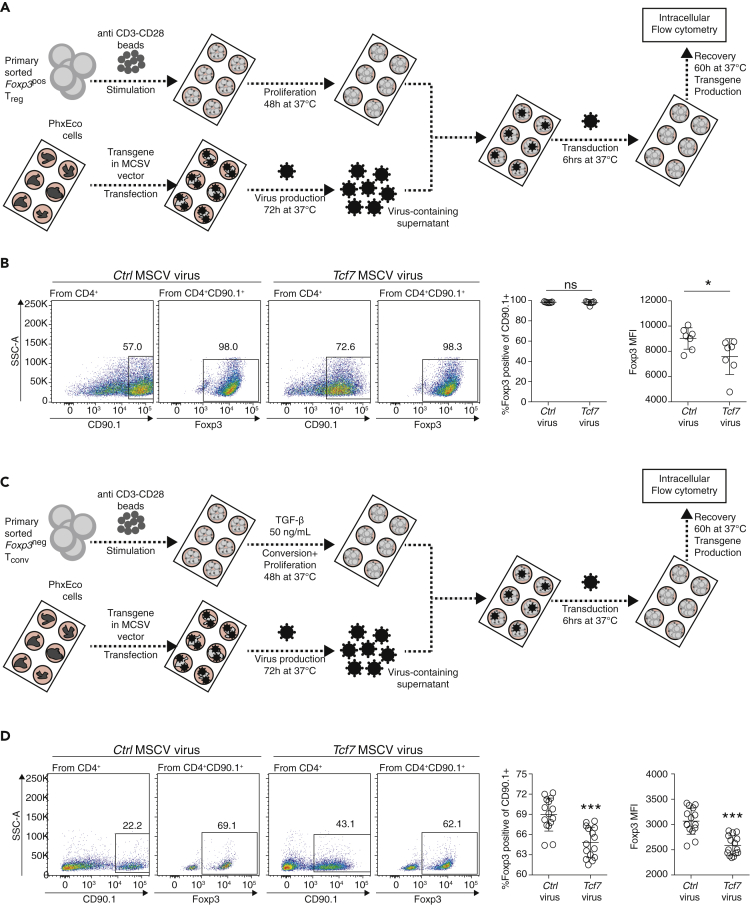
To validate the impact of TCF1 on Foxp3 gene expression in mature Treg cells, we transduced primary Treg cells with a Tcf7-MSCV retrovirus, with CD90.1 as a reporter for viral transduction and transgene expression (Figure 4A). The percentage of Foxp3 expression remained high in both Tcf7- and control-virus-transduced Treg cells and was not affected by TCF1 protein production. In contrast to this, the Foxp3 median fluorescence intensity (MFI) was significantly reduced in TCF1-overexpressing Treg cells, indicating that TCF1 can influence Foxp3 gene expression in mature Treg cells (Figure 4B). In line with this finding, we wanted to test the influence of TCF1 on the de novo induction of Foxp3 expression. To do so, we over-expressed TCF1 or a control protein in Tconv cells under TGF-β differentiation conditions, followed by measurement of intracellular Foxp3 levels (Figure 4C). Indeed, upon over-expressing TCF1, Foxp3 levels were reduced in percent and MFI, indicating that TCF1 also restrains Foxp3 induction (Figure 4D). Our data indicate that TCF1 impairs Foxp3 induction and maintenance in vitro.
Figure 4.
Effect of TCF1 Overexpression on Thymus-Derived thymic Treg or In Vitro Converted iTreg Cells
(A) Experiment overview: Primary murine CD4+CD25+Foxp3(GFP)+ Treg cells were purified by fluorescence-activated cell sorting (FACS), stimulated with anti-CD3/28 beads plus interleukin-2, and expanded for 48 h at 37°C. Then, virus-containing supernatant derived from PhxEco producer cells was harvested and T cells were virally transduced for 7 h at 37°C. T cells were allowed to recover and express the transgene for 60 h at 37°C, following intracellular flow cytometry to determine Foxp3 protein expression levels.
(B) Representative pseudocolor plots for ctrl MSCV virus and Tcf7-MSCV virus-treated Treg cells. Virus-transduced cells are CD90.1+, whereas non-transduced cells are CD90.1-. From CD90.1+ gate, Foxp3 expression can be determined. Numbers indicate positive cells in the gate in %. Axis labels indicate fluorescence intensity. MFI based on Foxp3 gate. Statistical quantification to the right (n = 7, unpaired t test, ∗p < 0.05 and ns p > 0.05).
(C) Experiment overview: Primary sorted Foxp3(GFP)-negative Tconv cells were FACS purified, stimulated with anti-CD3/28 beads, and differentiated with TGF-β for 48 h at 37°C. Then, virus-containing supernatant derived from PhxEco producer cells was harvested, and T cells were virally transduced for 7 h at 37°C. Afterward, T cells were allowed to recover and express the transgene for 60 h at 37°C, following intracellular flow cytometry to determine Foxp3 protein expression levels.
(D) Flow cytometry pseudocolor plots for Tconv cells treated with a control-MSCV virus or Tcf7-MSCV virus. From CD90.1+ gate, Foxp3 expression can be determined. To the right, percentage of Foxp3 expression and Foxp3 MFI are evaluated across experiments (unpaired t test, n = 15, ∗∗∗p < 0.001). Data are derived from two to three independent experiments with individual mice.
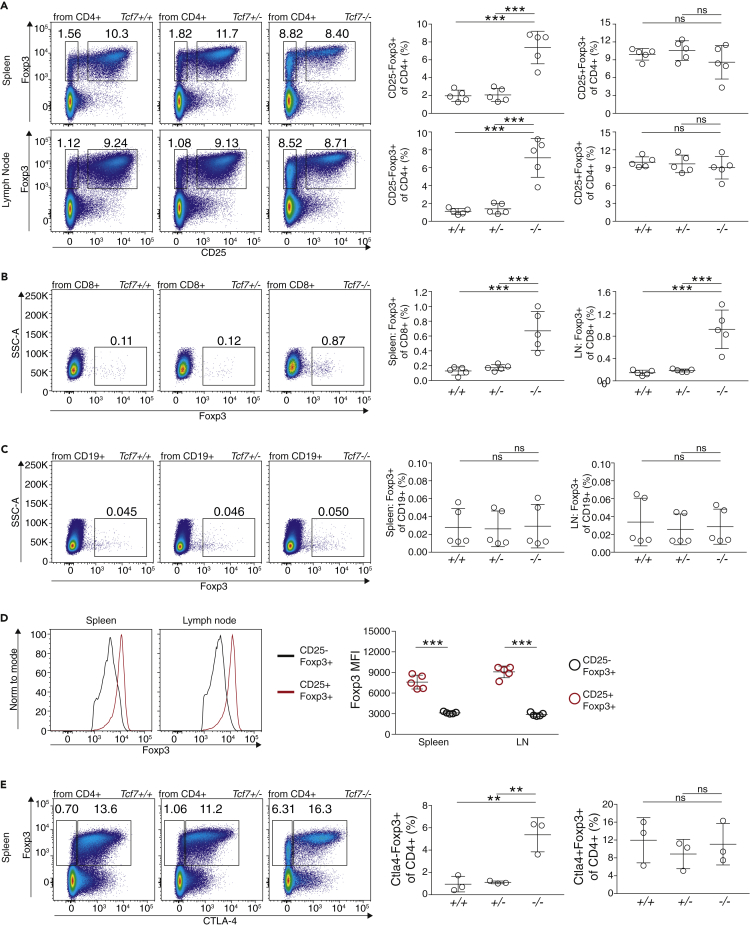
TCF1-Deficient Animals Have More CD25negFoxp3int T cells in the Periphery
To validate the relevance of TCF1 in vivo, we analyzed Tcf7−/− animals (Verbeek et al., 1995). First, we measured the frequency of CD4+CD25+Foxp3+ Treg cells in lymph node and spleen of Tcf7−/−, Tcf7+/−, and Tcf7+/+ (wild-type) animals and detected no obvious differences (Figure 5A). In contrast to this, we observed about 5-fold elevated numbers of CD25-negative Foxp3-intermediate (CD4+CD25-Foxp3int) T cells (1.56% in Tcf7+/+ versus 8.82% in Tcf7−/−). No dosage effect was observed, because heterozygous deletion of Tcf7 had no effect. Next, we were interested in whether only CD4+ T cells were affected by the deletion of TCF1. To answer this, we measured Foxp3 expression in CD8+ T cells and CD19+ B cells isolated from secondary lymphoid tissues of Tcf7−/−, Tcf7+/−, and Tcf7+/+ animals (Figures 5B and 5C). In line with our observations with CD4+ T cells, we could detect elevated numbers of Foxp3+ cells in the CD8+ population, although with a lower magnitude (Figure 5B). Interestingly, no effect was observed when analyzing Foxp3 expression in CD19+ B cells (Figure 5C).
Figure 5.
CD25-Negative Foxp3-Intermediate T cells in TCF1-Deficient Mice
(A) Analysis of spleen and lymph node in Tcf7−/−, Tcf7−/+, and Tcf7+/+ mice. CD4 T cells were identified (CD19−CD8−CD4+), and Foxp3 versus CD25 was plotted. Percentage of CD25-Foxp3+ and CD25+Foxp3+ of CD4+ T cells was calculated (n = 5, one-way ANOVA with Tukey's post-test, ∗∗∗p < 0.001).
(B) Analysis of spleen and lymph node in Tcf7−/−, Tcf7−/+, and Tcf7+/+ mice. CD8 T cells were identified (CD19−CD8+CD4-), and Foxp3 versus SSC-A was plotted. Percentage of Foxp3+ of CD8+ T cells was calculated (n = 5, one-way ANOVA with Tukey's post-test, ∗∗∗p < 0.001).
(C) Analysis of spleen and lymph node in Tcf7−/−, Tcf7−/+, and Tcf7+/+ mice. CD19 B cells were identified (CD19+CD8−CD4-) and Foxp3 versus SSC-A was plotted. Percentage of Foxp3+ of CD19+ B cells was calculated (n = 5, one-way ANOVA with Tukey's post-test, ns p > 0.05).
(D) Expression of Foxp3 in CD25-Foxp3+ and CD25+Foxp3+ T cells from spleen and lymph node. Representative histograms to the left, and statistical quantification to the right (n = 5, one-way ANOVA with Sidak's post-test, ∗∗∗p < 0.001).
(E) Percentage of CD25-Ctla4+ and CD25+Ctla4+ in spleens of Tcf7−/−, Tcf7−/+, and Tcf7+/+ mice. Representative dot plots to the left, and statistical quantification to the right (n = 3, one-way ANOVA with Tukey's post-test, ∗∗p < 0.01 and ns p > 0.05). Data are derived from two or more independent experiments with individual mice.
Are CD25-Foxp3int T cells actually Tconv cells that express Foxp3 due to the absence of TCF1, a Foxp3-repressive factor? To investigate this, we first measured Foxp3 protein levels via flow cytometry and detected reduced Foxp3 MFI of CD25-Foxp3int Treg cells versus CD25+Foxp3+ “true” Treg cells (Figure 5D). Next, we stained CTLA-4, a critical factor for Treg identity and function (Figure 5E). Indeed, we also identified CTLA-4-Foxp3+ T cells, which are normally almost absent in wild-type animals (0.5% of CD4+), but increased in TCF1-deficient animals to about 4% of CD4+ T cells. The occurrence of this specific CD25−CTLA4-Foxp3int population could be the consequence of a less restricted Foxp3 induction potential in TCF1-deficient T cells.
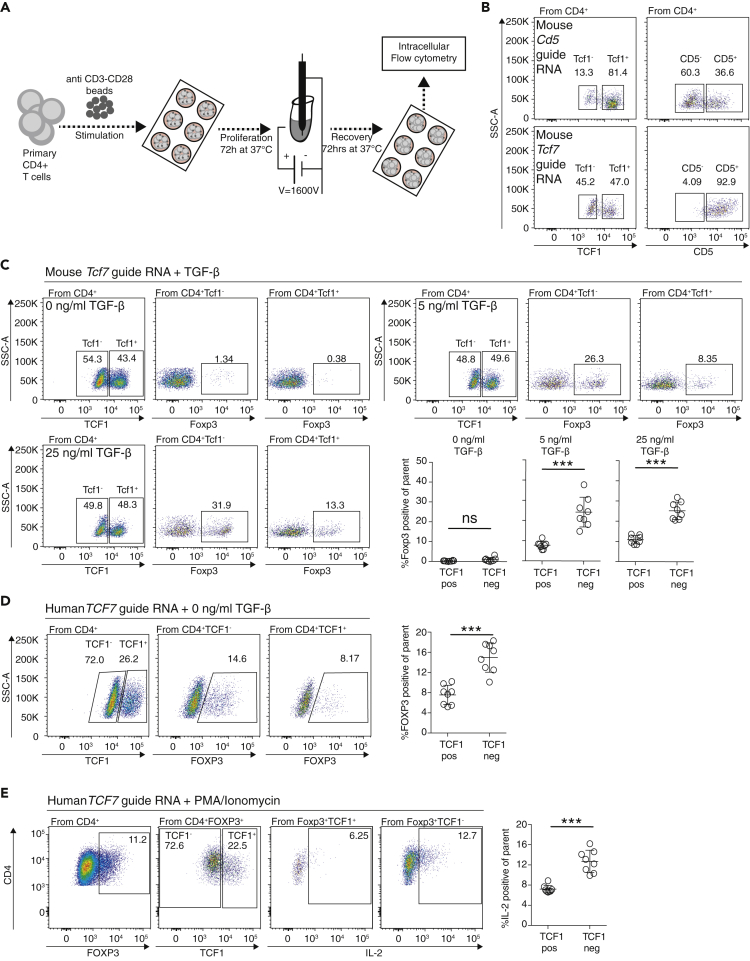
CRISPR-Cas9-Mediated Deletion of TCF1 Induces Foxp3 in Primary Tconv Cells
In Tcf7−/− animals, higher fractions of Foxp3-positive T cells were identified. However, these experiments did not tell us whether this was a consequence of an activation event in Tconv cells or a thymus-based selection process. To address the possibility that TCF1 restricts “unwanted” Foxp3 expression in Tconv cells, we deleted TCF1 in activated Tconv cells with CRISPR-Cas9 technology (Figure 6A). We calibrated the system by knocking out CD5 with a Cd5 guide RNA (crRNA) and achieved a loss of CD5 protein expression in about 60% TCR-activated primary CD4+ T cells (Figure 6B). Using Tcf7 crRNA, we observed TCF1 protein loss in about 50% primary CD4+ T cells (Figure 6B). Thus, individual wells contained equal ratios of TCF1-sufficient and TCF1-deficient CD4+ T cells, allowing us to directly compare the effect of TCF1 loss on Foxp3 expression under the same conditions. Using different concentrations of TGF-β to induce Foxp3 expression, we identified significantly increased Foxp3 protein expression in TCF1-deleted Tconv cells when compared with TCF1-sufficient Tconv cells (Figure 6C).
Figure 6.
CRISPR-Cas9 Knockout of TCF1 Induces Elevated Foxp3 Levels
(A) Experiment overview: Primary murine CD4+ T cells are stimulated with anti CD3-CD28 beads for 3 days in vitro. Then, cells are electroporated with a Cas9 protein, tracrRNA, and a Tcf7 guide RNA with the Neon electroporation system. Afterward, cells recovered for 3 days at 37°C. Different concentrations of TGF-β were used during proliferation and recovery phase.
(B) CRISPR-Cas9-based deletion of CD5 (left) and TCF1 (right) in CD4+ T cells.
(C) Results of CRISPR-Cas9-based deletion of TCF1 in murine CD4+ T cells. Pseudocolor dot plots illustrate TCF1-positive and TCF1-negative T cell populations after CRISPR-based TCF1 knockout and recovery for 72 h. Foxp3 expression for TCF1-negative and TCF1-positive populations shown to the right. A statistical verification across replicates is shown below (unpaired t test, n = 8, ∗∗∗p < 0.001 and ns p > 0.05).
(D) Results of CRISPR-Cas9-based deletion of TCF1 in human CD4+ T cells. Left, pseudocolor dot plots illustrating TCF1-positive and TCF1-negative T cell populations after CRISPR-based TCF1 knockout (TCF7 AA guide RNA). Foxp3 expression for TCF1-negative and TCF1-positive populations is shown. A statistical verification across replicates is shown to the right (unpaired t test, n = 8, ∗∗∗p < 0.001).
(E) Human FOXP3-negative Tconv cells were treated with TCF7 AA guide RNA, followed by stimulation with PMA/Ionomycin and treatment with transport inhibitors. After 4 h, T cells were stained for intracellular expression of interleukin-2. A statistical verification across replicates is shown to the right (unpaired t test, n = 8, ∗∗∗p > 0.001). Data are derived from two independent experiments with individual samples.
To validate our findings in the human system, we used the CRISPR-Cas9 knockout approach to delete TCF1 in human CD4+ T cells. It has been described that a fraction of human CD4 Tconv cells spontaneously induces FOXP3 protein expression after TCR activation (Gavin et al., 2006, Walker et al., 2003). To test if TCF1 is relevant for this spontaneous FOXP3 expression, we stimulated human Tconv cells with anti-CD3 and anti-CD28 and observed the cells following CRISPR-Cas9 knockout of TCF1. Even without the addition of TGF-β, we detected significantly more FOXP3-expressing CD4+ T cells in TCF1-deleted when compared with TCF1-sufficient Tconv cells (Figure 6D). The FOXP3-positive fraction increased from about 8% of CD4-sufficient Tconv cells to about 16% in TCF1-deleted Tconv cells. To investigate the inflammatory phenotype of TCF1-deleted FOXP3-positive T cells, we stimulated CRISPR-Cas9-treated Tconv cells with a T cell stimulation cocktail containing PMA/IM and blocked cytokine secretion with a transport inhibitor. Interestingly, TCF1-deleted FOXP3-positive T cells produced more interleukin-2 compared with control cells, indicating that TCF1-deleted FOXP3-positive T cells might indeed produce FOXP3 as a bystander effect of TCR stimulation, but do not exert the regulatory phenotype associated with a classical Treg cell. Therefore, in summary, our data indicate that TCF1 protects Tconv cells from activation-induced FOXP3 expression.
Discussion
Several TFs can bind to the Foxp3 promoter. They modulate the downstream functions by direct binding to the promoter and initiation of transcription, via the recruitment of co-activators, by the selective displacement of repressive TFs, or by epigenetic modulation of the promoter (Delacher et al., 2014). It is believed that many factors co-operate in a context-dependent manner in a multiprotein network occupying the Foxp3 promoter (Rudra et al., 2012).
In this study, we used an unbiased approach (Mittler et al., 2009) to determine binding partners to specific regions of the Foxp3 gene promoter. As a source of protein, we used an EL4 T cell line. This cell line was shown to express Foxp3 mRNA only upon TCR triggering with CD3-CD28 stimulation and TGF-β supplementation to the medium (Tone et al., 2008). Because we neither stimulated the TCR nor added TGF-β during expansion of EL4 cells, we probably identified a multiprotein complex protecting this T cell line from the “side effects” of Foxp3 gene activity. Indeed, many of the candidate proteins were repressive factors actively down-regulating basic Foxp3 promoter activity in vitro. This is in line with three other recent reports identifying cyclin-dependent kinases 8 and 19, the protein Yin-Yang 1, or the long noncoding RNA Flicr as repressors of Foxp3 protein expression (Akamatsu et al., 2019, Hwang et al., 2016, Zemmour et al., 2017).
Having established that our identified binding partners were Foxp3 promoter suppressive in nature, we used a funnel approach to identify targets for further evaluation: we compared lists of factors that are down-regulated in Foxp3-expressing murine and human Treg cells with our Foxp3-promoter-binding TFs. One interesting protein identified in the cross-comparison with both human and murine datasets was TCF1, a factor well known for its importance during thymic T cell development (Barra et al., 2015a, Barra et al., 2015b, Verbeek et al., 1995, Weber et al., 2011). It has also been described that TCF1 protein associates with Foxp3 and, via the Wnt signaling pathway, impairs suppressive function of Treg cells (van Loosdregt et al., 2013). Testing TCF1 in a Luciferase screen, our data showed that TCF1 has Foxp3-promoter-suppressive capacity, and viral overexpression of TCF1 led to decreased Foxp3 expression in vitro. In a TCF1-deficient mouse strain, we detected an increase in a Foxp3-intermediate, but CD25-negative T cell population (CD25-Foxp3int). It is possible that, by deleting TCF1, we lowered the threshold for Foxp3 induction and thereby generated a population of CD25-Foxp3int T cells, otherwise almost absent in lymphoid tissues. As TCF1 has a strong influence on thymic differentiation of both T cells and Treg cells (Barra et al., 2015a, Barra et al., 2015b, Verbeek et al., 1995, Weber et al., 2011), we wanted a more formal proof that TCF1 is involved in inhibiting FOXP3 expression in Tconv cells. Therefore, we deleted TCF1 using CRISPR-Cas9 knockout technology in primary mouse Tconv cells, which resulted in more Foxp3-expressing cells in the TCF1-deleted CD4+ T cell fraction.
Unlike in mice, in the human system, a certain percentage of T cells express FOXP3 upon activation in vitro (Gavin et al., 2006, Walker et al., 2003, Wang et al., 2007). To study whether activation-induced FOXP3 expression is TCF1 dependent, we used CRISPR-Cas9 technology to knockout TCF1 in human CD4 Tconv cells. We saw that spontaneous FOXP3 induction in activated human CD4+ T cells was significantly higher in TCF1-deficient T cells. This finding indicates an important function of TCF1 to suppress FOXP3 in activated non-Treg CD4+ T cells. TCF1 expression also favors memory formation in T cells (Nish et al., 2017, Utzschneider et al., 2016, Zhou et al., 2010). Whether TCF1 expression protects memory T cells from unintentional expression of FOXP3 has to be further evaluated.
A recent study looked at the effects of TCF1 deletion on Foxp3 expression during thymic development. The authors could show that TCF1-deficient mice harbor an increased number of Foxp3-expressing double-negative cells in the thymus (Barra et al., 2015b). In line with our data, this publication reports that TCF1 is required to prevent premature expression of Foxp3 in the thymus. Another set of publications investigated the effect of a Treg-specific knockout of TCF1 and Lef1 (Xing et al., 2019, Yang et al., 2019). Both studies showed that, whereas single-gene knockouts of Lef1 or TCF1 in Treg cells had no catastrophic systemic effect, deleting both genes led to autoimmune disease by impairing the immunosuppressive function of Treg cells. One of these studies closely examined the effect of a Treg cell-specific TCF1 deletion on Treg cell homeostasis and reported no perturbation (Xing et al., 2019). In contrast to this, we saw changes in the CD4 T cell compartment with TCF1 global knockout animals. Our animals, which also lack TCF1 in Tconv cells, showed the appearance of a Foxp3-intermediate, but CD25-negative T cell population (CD25-Foxp3int).
Whether our observation that TCF1 inhibits activation-induced FOXP3 expression in human and mouse conventional CD4+ T cells is also transferable to other T cell subsets, specifically CD8+ T cells, has to be further evaluated. Our mouse data showed an increased fraction of CD8+ T cells expressing Foxp3 in TCF1-deficient versus TCF1-sufficient animals, although the overall magnitude was lower compared with the CD4+ T cell compartment. Indeed, another study investigating TCF1−/−Lef1−/− double knockout animals also reported an increased frequency of CD8+Foxp3+ T cells in the thymus (Xing et al., 2016). The authors reported intrinsic histone deacetylase activity of the TCF1 protein. Therefore, TCF1 could change the epigenetic accessibility of the Foxp3 gene locus to prevent unwanted Foxp3 expression.
In summary, our study shows that TCF1 might be a key factor to protect Tconv cells from inadvertent Foxp3 expression. This allows unperturbed effector function of T cells while preventing the regulatory phenotype associated with Foxp3 expression.
Limitations of the Study
In this study, we present data about potential binding partners to the Foxp3 promoter region, identified by mass spectrometry. Data were generated with a cell line (EL4), whereas follow-up experiments were conducted with primary human or murine T cells. In this article, we investigated TCF1 and the effects of overexpression or deletion on T cell biology, which could be extended by the analysis of other proteins identified with our screen.
Resource Availability
Lead Contact
Lead contact for this publication is Prof. Dr. Markus Feuerer.
Materials Availibility
Materials are available upon request by Prof. Dr. Markus Feuerer.
Data and Code Availability
Mass spectrometry data have been deposited to the ProteomeXchange Consortium under the PRIDE identifier PXD018764.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
The Foxp3 promoter Luciferase vector was a kind gift of Prof. Dr. Akihiko Yoshimura (Keio University, Japan). TCF1-deficient animals were a gift from Prof. Dr. Hans Clevers (Hubrecht Institute, Utrecht, The Netherlands). We thank the DKFZ core facilities Preclinical Research, Flow Cytometry, and Genomics & Proteomics, and the RCI flow cytometry core facility for technical support. We thank Sabine Schmitt (DKFZ), Marina Wuttke, Brigitte Ruhland, K.S., and Veronika Hofmann (all RCI) for technical support. We thank Dr. Simone Thomas for help with human samples. This work was supported by grants from the European Research Council (ERC-CoG, #648145 REGiREG) to M.F., and by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation)—Projektnummer 324392634—TRR 221 to M.F., M.D. was supported by the German-Israeli Helmholtz Research School in Cancer Biology.
Author Contributions
M.D., M.M.B., J.K., J.A., and M.F. designed experiments; M.D., M.M.B., Y.H., K.E., M.-R.R., D.M.R., U.T., A.-C.H., A.K., K.L.B., M.G.., L.W., and K.S. performed the experiments; M.D., K.E., C.D.I., J.A., J.K., and M.F. analyzed data; M.D. and M.F. wrote the manuscript.
Declaration of Interests
The authors declare no competing financial interests.
Published: May 22, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2020.101127.
Supplemental Information
References
- Akamatsu M., Mikami N., Ohkura N., Kawakami R., Kitagawa Y., Sugimoto A., Hirota K., Nakamura N., Ujihara S., Kurosaki T. Conversion of antigen-specific effector/memory T cells into Foxp3-expressing Treg cells by inhibition of CDK8/19. Sci. Immunol. 2019;4:eaaw2707. doi: 10.1126/sciimmunol.aaw2707. [DOI] [PubMed] [Google Scholar]
- Andersen K.G., Nissen J.K., Betz A.G. Comparative genomics reveals key gain-of-function events in Foxp3 during regulatory T cell evolution. Front. Immunol. 2012;3:113. doi: 10.3389/fimmu.2012.00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barra M.M., Richards D.M., Hansson J., Hofer A.C., Delacher M., Hettinger J., Krijgsveld J., Feuerer M. Transcription factor 7 limits regulatory T cell generation in the thymus. J. Immunol. 2015;195:3058–3070. doi: 10.4049/jimmunol.1500821. [DOI] [PubMed] [Google Scholar]
- Barra M.M., Richards D.M., Hofer A.C., Delacher M., Feuerer M. Premature expression of Foxp3 in double-negative thymocytes. PLoS One. 2015;10:e0127038. doi: 10.1371/journal.pone.0127038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett C.L., Christie J., Ramsdell F., Brunkow M.E., Ferguson P.J., Whitesell L., Kelly T.E., Saulsbury F.T., Chance P.F., Ochs H.D. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat. Genet. 2001;27:20–21. doi: 10.1038/83713. [DOI] [PubMed] [Google Scholar]
- Brunkow M.E., Jeffery E.W., Hjerrild K.A., Paeper B., Clark L.B., Yasayko S.A., Wilkinson J.E., Galas D., Ziegler S.F., Ramsdell F. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat. Genet. 2001;27:68–73. doi: 10.1038/83784. [DOI] [PubMed] [Google Scholar]
- Bruno L., Mazzarella L., Hoogenkamp M., Hertweck A., Cobb B.S., Sauer S., Hadjur S., Leleu M., Naoe Y., Telfer J.C. Runx proteins regulate Foxp3 expression. J. Exp. Med. 2009;206:2329–2337. doi: 10.1084/jem.20090226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burchill M.A., Yang J., Vogtenhuber C., Blazar B.R., Farrar M.A. IL-2 receptor beta-dependent STAT5 activation is required for the development of Foxp3+ regulatory T cells. J. Immunol. 2007;178:280–290. doi: 10.4049/jimmunol.178.1.280. [DOI] [PubMed] [Google Scholar]
- Cuadrado E., van den Biggelaar M., de Kivit S., Chen Y.Y., Slot M., Doubal I., Meijer A., van Lier R.A.W., Borst J., Amsen D. Proteomic analyses of human regulatory T cells reveal adaptations in signaling pathways that protect cellular identity. Immunity. 2018;48:1046–1059.e6. doi: 10.1016/j.immuni.2018.04.008. [DOI] [PubMed] [Google Scholar]
- Delacher M., Imbusch C.D., Hotz-Wagenblatt A., Mallm J.P., Bauer K., Simon M., Riegel D., Rendeiro A.F., Bittner S., Sanderink L. Precursors for nonlymphoid-tissue Treg cells reside in secondary lymphoid organs and are programmed by the transcription factor BATF. Immunity. 2020;52:295–312.e11. doi: 10.1016/j.immuni.2019.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delacher M., Imbusch C.D., Weichenhan D., Breiling A., Hotz-Wagenblatt A., Trager U., Hofer A.C., Kagebein D., Wang Q., Frauhammer F. Genome-wide DNA-methylation landscape defines specialization of regulatory T cells in tissues. Nat. Immunol. 2017;18:1160–1172. doi: 10.1038/ni.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delacher M., Schmidl C., Herzig Y., Breloer M., Hartmann W., Brunk F., Kagebein D., Trager U., Hofer A.C., Bittner S. Rbpj expression in regulatory T cells is critical for restraining TH2 responses. Nat. Commun. 2019;10:1621. doi: 10.1038/s41467-019-09276-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delacher M., Schreiber L., Richards D.M., Farah C., Feuerer M., Huehn J. Transcriptional control of regulatory T cells. Curr. Top. Microbiol. Immunol. 2014;381:83–124. doi: 10.1007/82_2014_373. [DOI] [PubMed] [Google Scholar]
- Floess S., Freyer J., Siewert C., Baron U., Olek S., Polansky J., Schlawe K., Chang H.D., Bopp T., Schmitt E. Epigenetic control of the foxp3 locus in regulatory T cells. PLoS Biol. 2007;5:e38. doi: 10.1371/journal.pbio.0050038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontenot J.D., Gavin M.A., Rudensky A.Y. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat. Immunol. 2003;4:330–336. doi: 10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- Gavin M.A., Torgerson T.R., Houston E., DeRoos P., Ho W.Y., Stray-Pedersen A., Ocheltree E.L., Greenberg P.D., Ochs H.D., Rudensky A.Y. Single-cell analysis of normal and FOXP3-mutant human T cells: FOXP3 expression without regulatory T cell development. Proc. Natl. Acad. Sci. U S A. 2006;103:6659–6664. doi: 10.1073/pnas.0509484103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada Y., Harada Y., Elly C., Ying G., Paik J.H., DePinho R.A., Liu Y.C. Transcription factors Foxo3a and Foxo1 couple the E3 ligase Cbl-b to the induction of Foxp3 expression in induced regulatory T cells. J. Exp. Med. 2010;207:1381–1391. doi: 10.1084/jem.20100004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang S.S., Jang S.W., Kim M.K., Kim L.K., Kim B.S., Kim H.S., Kim K., Lee W., Flavell R.A., Lee G.R. YY1 inhibits differentiation and function of regulatory T cells by blocking Foxp3 expression and activity. Nat. Commun. 2016;7:10789. doi: 10.1038/ncomms10789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janson P.C., Winerdal M.E., Marits P., Thorn M., Ohlsson R., Winqvist O. FOXP3 promoter demethylation reveals the committed Treg population in humans. PLoS One. 2008;3:e1612. doi: 10.1371/journal.pone.0001612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josefowicz S.Z., Niec R.E., Kim H.Y., Treuting P., Chinen T., Zheng Y., Umetsu D.T., Rudensky A.Y. Extrathymically generated regulatory T cells control mucosal TH2 inflammation. Nature. 2012;482:395–399. doi: 10.1038/nature10772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.P., Leonard W.J. CREB/ATF-dependent T cell receptor-induced FoxP3 gene expression: a role for DNA methylation. J. Exp. Med. 2007;204:1543–1551. doi: 10.1084/jem.20070109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa Y., Ohkura N., Kidani Y., Vandenbon A., Hirota K., Kawakami R., Yasuda K., Motooka D., Nakamura S., Kondo M. Guidance of regulatory T cell development by Satb1-dependent super-enhancer establishment. Nat. Immunol. 2017;18:173–183. doi: 10.1038/ni.3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klunker S., Chong M.M., Mantel P.Y., Palomares O., Bassin C., Ziegler M., Ruckert B., Meiler F., Akdis M., Littman D.R., Akdis C.A. Transcription factors RUNX1 and RUNX3 in the induction and suppressive function of Foxp3+ inducible regulatory T cells. J. Exp. Med. 2009;206:2701–2715. doi: 10.1084/jem.20090596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantel P.Y., Ouaked N., Ruckert B., Karagiannidis C., Welz R., Blaser K., Schmidt-Weber C.B. Molecular mechanisms underlying FOXP3 induction in human T cells. J. Immunol. 2006;176:3593–3602. doi: 10.4049/jimmunol.176.6.3593. [DOI] [PubMed] [Google Scholar]
- Mayor C., Brudno M., Schwartz J.R., Poliakov A., Rubin E.M., Frazer K.A., Pachter L.S., Dubchak I. Vista : visualizing global DNA sequence alignments of arbitrary length. Bioinformatics. 2000;16:1046–1047. doi: 10.1093/bioinformatics/16.11.1046. [DOI] [PubMed] [Google Scholar]
- Mittler G., Butter F., Mann M. A SILAC-based DNA protein interaction screen that identifies candidate binding proteins to functional DNA elements. Genome Res. 2009;19:284–293. doi: 10.1101/gr.081711.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouly E., Chemin K., Nguyen H.V., Chopin M., Mesnard L., Leite-de-Moraes M., Burlen-defranoux O., Bandeira A., Bories J.C. The Ets-1 transcription factor controls the development and function of natural regulatory T cells. J. Exp. Med. 2010;207:2113–2125. doi: 10.1084/jem.20092153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nish S.A., Zens K.D., Kratchmarov R., Lin W.W., Adams W.C., Chen Y.H., Yen B., Rothman N.J., Bhandoola A., Xue H.H. CD4+ T cell effector commitment coupled to self-renewal by asymmetric cell divisions. J. Exp. Med. 2017;214:39–47. doi: 10.1084/jem.20161046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh-Hora M., Komatsu N., Pishyareh M., Feske S., Hori S., Taniguchi M., Rao A., Takayanagi H. Agonist-selected T cell development requires strong T cell receptor signaling and store-operated calcium entry. Immunity. 2013;38:881–895. doi: 10.1016/j.immuni.2013.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkura N., Hamaguchi M., Morikawa H., Sugimura K., Tanaka A., Ito Y., Osaki M., Tanaka Y., Yamashita R., Nakano N. T cell receptor stimulation-induced epigenetic changes and Foxp3 expression are independent and complementary events required for Treg cell development. Immunity. 2012;37:785–799. doi: 10.1016/j.immuni.2012.09.010. [DOI] [PubMed] [Google Scholar]
- Ouyang W., Beckett O., Ma Q., Paik J.H., DePinho R.A., Li M.O. Foxo proteins cooperatively control the differentiation of Foxp3+ regulatory T cells. Nat. Immunol. 2010;11:618–627. doi: 10.1038/ni.1884. [DOI] [PubMed] [Google Scholar]
- Ruan Q., Kameswaran V., Tone Y., Li L., Liou H.C., Greene M.I., Tone M., Chen Y.H. Development of Foxp3(+) regulatory t cells is driven by the c-Rel enhanceosome. Immunity. 2009;31:932–940. doi: 10.1016/j.immuni.2009.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudensky A.Y. Regulatory T cells and Foxp3. Immunol. Rev. 2011;241:260–268. doi: 10.1111/j.1600-065X.2011.01018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudra D., deRoos P., Chaudhry A., Niec R.E., Arvey A., Samstein R.M., Leslie C., Shaffer S.A., Goodlett D.R., Rudensky A.Y. Transcription factor Foxp3 and its protein partners form a complex regulatory network. Nat. Immunol. 2012;13:1010–1019. doi: 10.1038/ni.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadlon T.J., Wilkinson B.G., Pederson S., Brown C.Y., Bresatz S., Gargett T., Melville E.L., Peng K., D'Andrea R.J., Glonek G.G. Genome-wide identification of human FOXP3 target genes in natural regulatory T cells. J. Immunol. 2010;185:1071–1081. doi: 10.4049/jimmunol.1000082. [DOI] [PubMed] [Google Scholar]
- Schlenner S.M., Weigmann B., Ruan Q., Chen Y., von Boehmer H. Smad3 binding to the foxp3 enhancer is dispensable for the development of regulatory T cells with the exception of the gut. J. Exp. Med. 2012;209:1529–1535. doi: 10.1084/jem.20112646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidl C., Delacher M., Huehn J., Feuerer M. Epigenetic mechanisms regulating T-cell responses. J. Allergy Clin. Immunol. 2018;142:728–743. doi: 10.1016/j.jaci.2018.07.014. [DOI] [PubMed] [Google Scholar]
- Schuster M., Glauben R., Plaza-Sirvent C., Schreiber L., Annemann M., Floess S., Kuhl A.A., Clayton L.K., Sparwasser T., Schulze-Osthoff K. IkappaB(NS) protein mediates regulatory T cell development via induction of the Foxp3 transcription factor. Immunity. 2012;37:998–1008. doi: 10.1016/j.immuni.2012.08.023. [DOI] [PubMed] [Google Scholar]
- Sekiya T., Kashiwagi I., Inoue N., Morita R., Hori S., Waldmann H., Rudensky A.Y., Ichinose H., Metzger D., Chambon P., Yoshimura A. The nuclear orphan receptor Nr4a2 induces Foxp3 and regulates differentiation of CD4+ T cells. Nat. Commun. 2011;2:269. doi: 10.1038/ncomms1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiya T., Kashiwagi I., Yoshida R., Fukaya T., Morita R., Kimura A., Ichinose H., Metzger D., Chambon P., Yoshimura A. Nr4a receptors are essential for thymic regulatory T cell development and immune homeostasis. Nat. Immunol. 2013;14:230–237. doi: 10.1038/ni.2520. [DOI] [PubMed] [Google Scholar]
- Tone Y., Furuuchi K., Kojima Y., Tykocinski M.L., Greene M.I., Tone M. Smad3 and NFAT cooperate to induce Foxp3 expression through its enhancer. Nat. Immunol. 2008;9:194–202. doi: 10.1038/ni1549. [DOI] [PubMed] [Google Scholar]
- Utzschneider D.T., Charmoy M., Chennupati V., Pousse L., Ferreira D.P., Calderon-Copete S., Danilo M., Alfei F., Hofmann M., Wieland D. T cell factor 1-expressing memory-like CD8(+) T cells sustain the immune response to chronic viral infections. Immunity. 2016;45:415–427. doi: 10.1016/j.immuni.2016.07.021. [DOI] [PubMed] [Google Scholar]
- van Loosdregt J., Fleskens V., Tiemessen M.M., Mokry M., van Boxtel R., Meerding J., Pals C.E., Kurek D., Baert M.R., Delemarre E.M. Canonical Wnt signaling negatively modulates regulatory T cell function. Immunity. 2013;39:298–310. doi: 10.1016/j.immuni.2013.07.019. [DOI] [PubMed] [Google Scholar]
- Verbeek S., Izon D., Hofhuis F., Robanus-Maandag E., te Riele H., van de Wetering M., Oosterwegel M., Wilson A., MacDonald H.R., Clevers H. An HMG-box-containing T-cell factor required for thymocyte differentiation. Nature. 1995;374:70–74. doi: 10.1038/374070a0. [DOI] [PubMed] [Google Scholar]
- Walker M.R., Kasprowicz D.J., Gersuk V.H., Benard A., Van Landeghen M., Buckner J.H., Ziegler S.F. Induction of FoxP3 and acquisition of T regulatory activity by stimulated human CD4+CD25- T cells. J. Clin. Invest. 2003;112:1437–1443. doi: 10.1172/JCI19441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Ioan-Facsinay A., van der Voort E.I., Huizinga T.W., Toes R.E. Transient expression of FOXP3 in human activated nonregulatory CD4+ T cells. Eur. J. Immunol. 2007;37:129–138. doi: 10.1002/eji.200636435. [DOI] [PubMed] [Google Scholar]
- Wang Y., Su M.A., Wan Y.Y. An essential role of the transcription factor GATA-3 for the function of regulatory T cells. Immunity. 2011;35:337–348. doi: 10.1016/j.immuni.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber B.N., Chi A.W., Chavez A., Yashiro-Ohtani Y., Yang Q., Shestova O., Bhandoola A. A critical role for TCF-1 in T-lineage specification and differentiation. Nature. 2011;476:63–68. doi: 10.1038/nature10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildin R.S., Ramsdell F., Peake J., Faravelli F., Casanova J.L., Buist N., Levy-Lahad E., Mazzella M., Goulet O., Perroni L. X-linked neonatal diabetes mellitus, enteropathy and endocrinopathy syndrome is the human equivalent of mouse scurfy. Nat. Genet. 2001;27:18–20. doi: 10.1038/83707. [DOI] [PubMed] [Google Scholar]
- Wohlfert E.A., Grainger J.R., Bouladoux N., Konkel J.E., Oldenhove G., Ribeiro C.H., Hall J.A., Yagi R., Naik S., Bhairavabhotla R. GATA3 controls Foxp3(+) regulatory T cell fate during inflammation in mice. J. Clin. Invest. 2011;121:4503–4515. doi: 10.1172/JCI57456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Stubbington M.J., Nissen J.K., Andersen K.G., Hebenstreit D., Teichmann S.A., Betz A.G. The regulatory T cell lineage factor Foxp3 regulates gene expression through several distinct mechanisms mostly independent of direct DNA binding. PLoS Genet. 2015;11:e1005251. doi: 10.1371/journal.pgen.1005251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing S., Gai K., Li X., Shao P., Zeng Z., Zhao X., Zhao X., Chen X., Paradee W.J., Meyerholz D.K. Tcf1 and Lef1 are required for the immunosuppressive function of regulatory T cells. J. Exp. Med. 2019;216:847–866. doi: 10.1084/jem.20182010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing S., Li F., Zeng Z., Zhao Y., Yu S., Shan Q., Li Y., Phillips F.C., Maina P.K., Qi H.H. Tcf1 and Lef1 transcription factors establish CD8(+) T cell identity through intrinsic HDAC activity. Nat. Immunol. 2016;17:695–703. doi: 10.1038/ni.3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B.H., Wang K., Wan S., Liang Y., Yuan X., Dong Y., Cho S., Xu W., Jepsen K., Feng G.S. TCF1 and LEF1 control Treg competitive survival and Tfr development to prevent autoimmune diseases. Cell Rep. 2019;27:3629–3645.e6. doi: 10.1016/j.celrep.2019.05.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z., Kanno Y., Kerenyi M., Stephens G., Durant L., Watford W.T., Laurence A., Robinson G.W., Shevach E.M., Moriggl R. Nonredundant roles for Stat5a/b in directly regulating Foxp3. Blood. 2007;109:4368–4375. doi: 10.1182/blood-2006-11-055756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemmour D., Pratama A., Loughhead S.M., Mathis D., Benoist C. Flicr, a long noncoding RNA, modulates Foxp3 expression and autoimmunity. Proc. Natl. Acad. Sci. U S A. 2017;114:E3472–E3480. doi: 10.1073/pnas.1700946114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Josefowicz S., Chaudhry A., Peng X.P., Forbush K., Rudensky A.Y. Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature. 2010;463:808–812. doi: 10.1038/nature08750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Yu S., Zhao D.M., Harty J.T., Badovinac V.P., Xue H.H. Differentiation and persistence of memory CD8(+) T cells depend on T cell factor 1. Immunity. 2010;33:229–240. doi: 10.1016/j.immuni.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Mass spectrometry data have been deposited to the ProteomeXchange Consortium under the PRIDE identifier PXD018764.