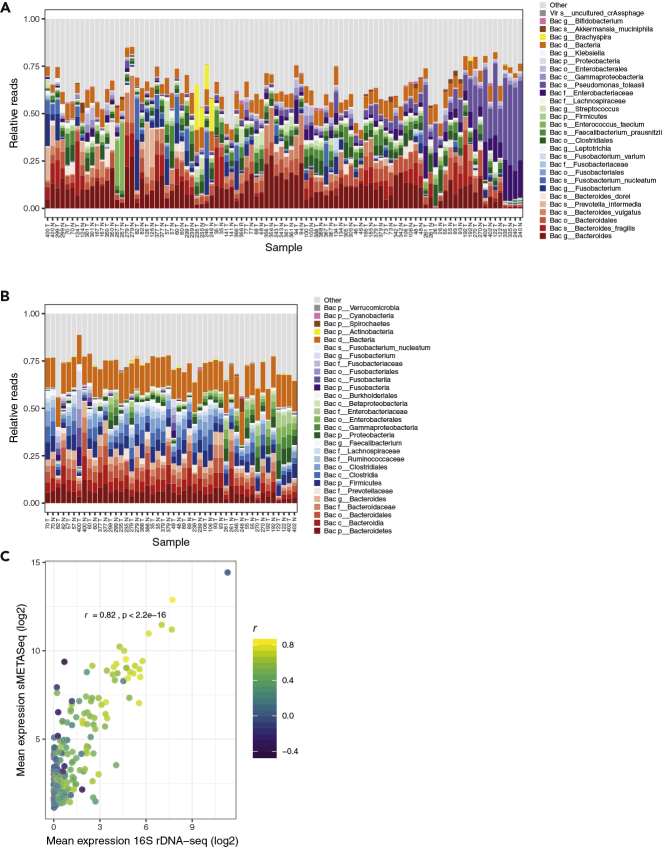
Figure 3.
OTUs in Colon Tissue as Identified by sMETASeq and 16S rDNA-seq
(A) Detected OTUs by sMETASeq in 96 colon tissue samples. The samples are ordered from left to right with respect to the number of bacteria reads identified. OTUs within the same taxonomic lineage are indicated with different saturation of the same color. The taxonomic level for each OTU is indicated as “d,” domain; “p,” phylum; “c,” class; “o,” order; “g,” genus; “s,” species, and bacteria are indicated as “Bac” and virus as “Vir.” Reads aligned to other OTUs than those listed are indicated as “other.” Samples are indicated with patient number and “T” and “N” for tumor and normal, respectively.
(B) Detected OTUs by 16S rDNA-seq in 48 colon tissue samples. See (A) for a description of the plot.
(C) Correlation plot showing the mean expression of OTUs across samples as identified by sMETASeq (y axis) and 16S rDNA-seq (x axis). The color of the points indicates the correlation coefficient (Pearson's) and the axis shows the expression. p < 2.2e-16.