Abstract
The search for new therapies for the treatment of Arterial hypertension is a major concern in the scientific community. Here, we employ a computational biochemistry protocol to evaluate the performance of six compounds (Lig783, Lig1022, Lig1392, Lig2177, Lig3444 and Lig6199) to act as antihypertensive agents. This protocol consists of Docking experiments, efficiency calculations of ligands, molecular dynamics simulations, free energy, pharmacological and toxicological properties predictions (ADME-Tox) of the six ligands against Thermolysin. Our results show that the docked structures had an adequate orientation in the pocket of the Thermolysin enzymes, reproducing the X-ray crystal structure of Inhibitor-Thermolysin complexes in an acceptable way. The most promising candidates to act as antihypertensive agents among the series are Lig2177 and Lig3444. These compounds form the most stable ligand-Thermolysin complexes according to their binding free energy values obtained in the docking experiments as well as MM-GBSA decomposition analysis calculations. They present the lowest values of Ki, indicating that these ligands bind strongly to Thermolysin. Lig2177 was oriented in the pocket of Thermolysin in such a way that both OH of the dihydroxyl-amino groups to establish hydrogen bond interactions with Glu146 and Glu166. In the same way, Lig3444 interacts with Asp150, Glu143 and Tyr157. Additionally, Lig2177 and Lig3444 fulfill all the requirements established by Lipinski Veber and Pfizer 3/75 rules, indicating that these compounds could be safe compounds to be used as antihypertensive agents. We are confident that our computational biochemistry protocol can be used to evaluate and predict the behavior of a broad range of compounds designed in silicoagainst a protein target.
Keywords: Ligand efficiency, ADME-Tox, Antihypertensive, Molecular dynamics, MM-GBSA
1. Introduction
Blood pressure is the force exerted by circulating blood against the walls of vessels (arteries) when pumped by the heart. The higher the tension, the more effort the heart has to take to pump. Arterial hypertension, also called arterial high blood pressure, is a disorder in which blood vessels have persistently elevated blood pressure, resulting in their damage (Bhat et al., 2017).
Arterial hypertension constitutes one of the principal causes of the increase of cardiovascular diseases around the world. According to the World Health Organization, 9.4 millions deaths to year are due to complications with arterial hypertension (AH). Particularly in Chile; according to the latest health survey, 26.9% of the population had this condition and one in three deaths per year is caused of blood pressure increase (Petermann et al., 2017). Several authors proposed that around 20% of the adult world population, over 20 years old, are hypertensive; and in those older than 60 years, it is even greater: around 50%, currently reaching global epidemic magnitudes (3450 million people with this disease) (Gamboa, 2006, Petermann et al., 2017). In addition, approximately 90% of patients with severe arterial hypertension have an unknown etiology and consequently, it is diagnosed as primary or essential arterial hypertension (Calhoun and Sharma, 2010, Martins et al., 2011). The remaining patients (10%) are diagnosed as secondary arterial hypertension, characterized by an autonomous aldosterone production, caused by renal, neurological and/or endocrine diseases (Calhoun and Sharma, 2010, Martins et al., 2011, Petermann et al., 2017).
The principal target of arterial hypertension is the renin-angiotensin-aldosterone system (RAAS) (Riet et al., 2015). This is part of the most important hormonal mechanisms in the regulation of blood pressure, fluid volume and sodium-potassium balance in humans (Muñoz-Durango et al., 2016), an alteration in any of the molecules that make up this system (RAAS) could contribute to the development of arterial hypertension (Riet et al., 2015). RAAS system is based on renin generation, which is synthesized in the kidneys as their inactive form and released into the bloodstream as a response to low levels of physiological sodium (hypotension) (Muñoz-Durango et al., 2016). Then, renin is activated proteolytically to its active form, which catalyzes the formation of angiotensin I (Ang-I [1–10]), which is the substrate of the angiotensin-converting enzyme (ACE) to form angiotensin II (Ang-II [1–8]) (Choe et al., 2019, Putnam et al., 2012), while the neutral endopeptidase (NEP) takes as substrate the Ang-I to form angiotensin-[1–7] (Ang[1,7]) which, together with Ang-II, causes vasoconstriction in cardiac and vascular tissues, and therefore increase total peripheral resistance, causing an increment in blood pressure (Nehme et al., 2019, Ren et al., 2019, Rodrigues Prestes et al., 2017). Thus, Angiotensin-converting enzyme (ACE) and neutral endopeptidase (NEP) are very essential proteins in the control of blood pressure (Hubers and Brown, 2016, Patten et al., 2016, Stanisz et al., 2016). In this regard, several authors have focused on designind RAAS inhibitors using Thermolysin as a target model (Bohacek et al., 1996, Cañizares-Carmenate et al., 2019, Choe et al., 2019, DePriest et al., 1993, Spyroulias et al., 2004). Thermolysin (EC 3.4.24.27) is a metalloprotease, which contains in its active center and is classified within the family M4 metalloendopeptidases (Braunwald, 2015, Paulis et al., 2015). This protein has been used to construct the NEP and ACE models and design NEP/ACE inhibitors (Manzur et al., 2013) due to the structural and functional similarities between Thermolysin, NEP and ACE, so we can say that Thermolysin inhibitors may also inhibit ACE and NEP and be putative antihypertensives (Khan et al., 2009). Previously (Cañizares-Carmenate et al., 2019), we design and screen more than 200 compounds against Thermolysin, to find the most suitable compounds that can act as possible antihypertensive drugs using QSARIN methods. Here, we evaluate the 6 most promising compounds from our previous study (Lig-783, Lig-1022, Lig-1392, Lig-2177, Lig-3444, and Lig-6199)(Fig) to apply a computational biochemistry protocol consisting of docking experiments, efficiency calculations of ligands, molecular dynamics simulations, free energy, pharmacological and toxicological properties predictions (ADME-Tox) against Thermolysin, with the aim to evaluate if these compounds designed in silico could be good candidates as NEP/ACE inhibitors to be employed in an antihypertensive dual therapy.
2. Computational details
2.1. Data set
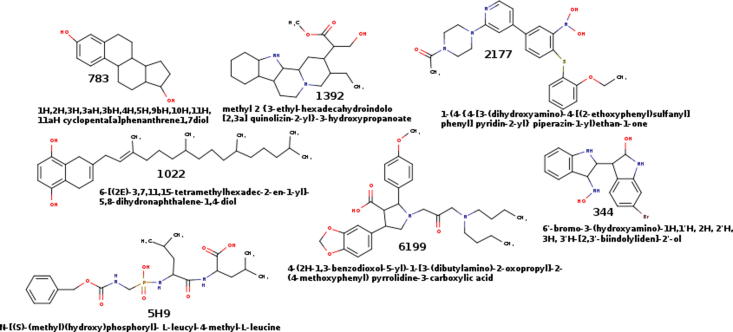
Here, we use six compounds selected as promising anti-hypertensive agents from a previous study (Fig. 1) (Cañizares-Carmenate et al., 2019). These molecules were designed in silico and evaluated using QSARINS and docking methodologies. Their chemical structures are shown in Fig. 1 and their molecular conformations were fully optimized to their ground state using Density Functional Theory (DFT) (Parr and Yang, 1984, Yang and Parr, 1985), employing the Perdew–Burke–Ernzerhof’s hybrid functional (PBE0) (Perdew et al., 1996, Vargas-Sánchez et al., 2015) in conjunction with the Pople’s 6-311++g(d,p) basis set for all atoms, as implemented in ORCA software version 4.1.2 (Neese, 2012, Neese, 2018). The ground state of each compound was verified by counting their imaginary frequencies. Molecules were sketched using Avogadro package version 1.2.0 (Hanwell et al., 2012). The optimized geometries were used for docking experiments, in order to examine the interactions established by these compounds in the Thermolysin pocket.
Fig. 1.
2D chemical structure of ligand under study. The Ligand 5H9 is our ligand reference and were obtained from Protein Data Bank X-ray crystallography structure (PDB id: 5DPF).
2.2. Docking experiments
All docking experiments were done using Autodock Vina software (Koebel et al., 2016, Trott and Olson, 2010). X-ray Thermolysin crystallography structure was obtained from Protein Data Bank (Berman et al., 2000)(PDB Id:5 5DPF), resolved at 1.47 Å (Krimmer and Klebe, 2015). We used Thermolysin as a model for Angiotensin-converting enzyme (ACE) and neutral endopeptidase (NEP) since they are metallopeptidases that contain in its active center, exhibiting similar structures and catalytic behavior (Bohacek et al., 1996, Cañizares-Carmenate et al., 2019, Choe et al., 2019, DePriest et al., 1993, Spyroulias et al., 2004). Thermolysin was prepared by the addition of hydrogen atoms at physiological pH and the elimination of water molecules around the protein. The six ligands described in Fig. 1 were prepared to take into account the rotatable bonds as well as the protein using Autodock Tools (Morris et al., 2009). The size of the grid box was located in the mass center of (5H9) in the crystal structure of Thermolysin whose coordinates were x = 11,844 y=-40,329 and z = 6,420. In the docking experiments, the was kept into the Thermolysin pocket and the coordination geometry of this metal with His142, Glu166 and His231 were respected.
The grid spacing was 0.375 , the mode number was 10 and the energy rank was set up to 3 kcal/mol. The best docking poses were selected following two criteria: the relative total energy score and the positional root-mean-square deviation (RMSD) (Gohlke et al., 2000). The most energetically favorable conformations and lowest RMSD of each ligand-thermolysin complex were selected for ligand efficiency calculations and molecular dynamics simulations. The 5H9 ligand was re-docked using the same docking protocol of the other ligands, in order to validate the docking experiments. To get an idea of the docking experiments reproducibility, we calculate the RMSD of the ligands designed in silicotaking 5H9 compound as a reference from the crystallographic structure of the Protein Data Bank. These calculations were made using the LigRMSD server 1.0 program (Velázquez-Libera et al., 2020).
2.3. Ligand efficiency approach
Ligand efficiency calculations were performed by means of two parameters: and ligand efficiency (LE). The parameter corresponds to the dissociation constant between a ligand and the protein, their value indicates the bond strength between the ligand and the protein (Abad-Zapatero, 2013, Abad-Zapatero et al., 2010). Small values indicate a strong binding of the ligand to the protein. calculations were done using the following equations:
| (1) |
| (2) |
where corresponds to binding energy (kcal/mol) obtained from docking experiments, R is the gas constant whose value is and T is the temperature in Kelvin, in our case, its value was 298.15 K.
The ligand efficiency (LE) allows us to compare molecules according to their average binding energy (Reynolds et al., 2008, Abad-Zapatero, 2013). Thus, is determined as the ratio of binding energy per non-hydrogen atom, as follows (Abad-Zapatero, 2013, Abad-Zapatero et al., 2010, Cavalluzzi et al., 2017):
| (3) |
where is obtained from Eq. 2 and HAC corresponds to the number of non-hydrogen atoms (heavy atom counter) in a ligand.
2.4. Molecular dynamics simulations
The best poses for each ligand-thermolysin complex obtained in docking simulations were used as input for subsequent molecular dynamics simulations. The molecular geometry of these ligand-thermolysin complexes were placed into a water box of centered on the mass center of each ligand, using the TIP3P flexible water model (Boonstra et al., 2016, Lu et al., 2014). Parameters and topologies of the ligands were obtained by means of the SwissParam web server (Zoete et al., 2011). Thermolysin and ligands were described by using CHARMM36 and CGenFF force field respectively (MacKerell et al., 2004, Foloppe and MacKerell, 2000, Lee et al., 2016, Soteras Gutiérrez et al., 2016, Vanommeslaeghe et al., 2015).
To reduce any close contact of the complexes, we carry out 20000 steps for the energy minimization procedure using the conjugated gradient methodology. All simulations were done at 298.15 K, applying the weak coupling algorithm (Brian and Brooks, 1999) and Van der Waals cutoff was fixed to 12 Å. We apply a constraint to the backbone of ligand-thermolysin complexes using the NPT ensemble and the long ranges electrostatic forces were considered using the Particle Mesh Ewald (PME) approach (Onufriev et al., 2004). The velocity Verlet algorithm was used to solve the equations of motion with a time step of 1.0 fs. All the systems were subject to 2.0 ns of equilibration and 100 ns of production using the NAMD 2.10 software package (Tanner et al., 2012, Phillips et al., 2005).
2.5. Molecular Mechanics-Generalized Born Surface Area method (MM-GBSA)
MM-GBSA method use molecular dynamics simulation for obtaining the difference between the energy of the bound complex (ligand-thermolysin) with respect to the energy of the unbound protein (Thermolysin) and the ligand designed in silico (Adasme-Carreno et al., 2014, Cañizares-Carmenate et al., 2019, Gaillard et al., 2016, Massova and Kollman, 2000, Mena-Ulecia et al., 2015). From the 100 ns of molecular dynamics simulation we obtained 1000 snapshots to calculate the binding free energy of each ligand-thermolysin complex () through MM-GBSA calculations, as follows:
| (4) |
The above energies for the ligand-protein complex (), the unbound ligand () and unbound protein () were determined using the CHARMM36 force field with the generalized Born implicit solvent model, and the binding free energy of each ligand-thermolysin complex was calculated according to Eq. 4.
This binding free energy can be decomposed into three different terms, accounting for physical-chemical meaningful descriptors, as shown in the following equation:
| (5) |
where correspond to the interaction energy between the ligand and Thermolysin in the gas phase. Is given by the contributions of the electrostatic () and Van del Waals () terms, as well as the change of internal energy of thermolysin upon ligand complexation () according to Eq. 6. It must be noted that comprises the bond, angle and dihedral angles energy disturbances obtained from the MD trajectories.
| (6) |
On the other hand, the solvation energy () includes polar and non-polar contributions to the free energy and can be calculated using the following equation:
| (7) |
where corresponds to the polar solvation free energy, which was estimated using the Generalized Born solvent model that includes the electrostatic solute-solvent interactions according to:
| (8) |
where is the distance between the charge and . and are the Born radius and is a function which can be calculated as:
| (9) |
Another component of corresponds to non-polar free energy ()(Eq. 7). This energy was estimated based on the accessibility of the solvent to the surface area (SA) according to Eq. 10, where and depend of a parametrization radius used to calculate the surface. The surface area was estimated through a spherical probe of 1.4 Å that rolls on the Thermolysin surface.
| (10) |
2.6. ADME-Tox properties
The main goal to calculate ADME–Tox properties is to obtain a preliminary prediction of their potential pharmacological capacities of a compound to become a drug. The six ligands used in this study were submitted to the calculation of their absorption, distribution, metabolism, excretion and toxicological properties (ADME-Tox). Also, the physicochemical properties such as molecular weight (MW), octanol/water partition coefficient (LogP), hydrogen bond acceptor (HBA), hydrogen bond donor (HBD), topological polar surface area (TPSA) and rotatable bond count (RB) were calculated using SwissADME webserver (Daina et al., 2017). Ligand toxicological properties were analyzed taking into account the Lipinski, Veber and Pfizer toxicity empirical rules (Table 1).
Table 1.
Empirical rules for predicting oral availability and toxicity of a compound.
| Properties | Oral Availability |
Toxicity | |
|---|---|---|---|
| Lipinski Rules | Veber Rules | Pfizer 3/75 Rules | |
| MW | 500 | – | – |
| LogP | 5 | – | 3 |
| HBA | 10 | – | – |
| HBD | 5 | – | – |
| TPSA | – | 140 | 75 |
| RB | – | 10 | – |
MW: Molecular weight; LogP: Octanol/water partition coefficient; HBA: Hydrogen bond acceptor; HBD: Hydrogen bond donor; TPSA: Topological polar surface area; RB: Rotatable bond count.
3. Results and discussion
A set of six designed in silico compounds were selected from a previous study, due to their possible use as anti-hypertensive agents. These compounds, presented in Fig. 1, titled as Lig783, Lig1022, Lig1392, Lig2177, Lig3444, and Lig6199 were subjected to a thorough analysis using a computational biochemistry protocol consisting of: (i) docking experiments; (ii) ligand efficiency calculations; (iii) molecular dynamics simulations; (iv) MM-GBSA energy decomposition analysis and (v) pharmacological and toxicological properties predictions. All these analyses are employed to evaluate the possibility that the present series of designed in silico compounds behave as good NEP/ACE inhibitors, with the aim to be employed in a dual antihypertensive therapy.
3.1. Docking
Molecular The Molecular docking Docking procedure is a computational tool that determines the structure and position of minimum energy of a protein-ligand complex. It is commonly used due to their its usefulness for the design of drugs (Cañizares-Carmenate et al., 2019, Mena-Ulecia et al., 2015, Mena-Ulecia et al., 2018). In this article, the docking protocol was used to determine the ligand-binding mode into the Thermolysin pocket.
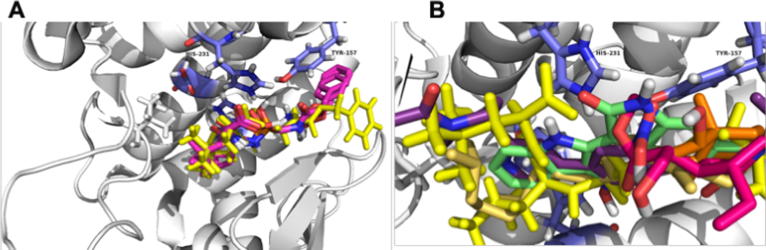
In Fig. 2 is shown that the docked structures fitted in an acceptable way with available inhibitor X-ray crystal structures; all the inhibitors were adequately oriented in Thermolysin active center. All the ligands present binding energies () between −7.0 and −8.5 kcal/mol (Table 2) with less negative energies than 5H9, which means that the Termolysin-5H9 complex is more stable than inhibitors designed in silico.
Fig. 2.
Alignment of all docked ligands in complex with Thermolysin. A represents the re-docked result of 5H9, in yellow the 5H9 obtained from Protein Data Bank X-ray crystallography structure (PDB id: 5DPF); in magenta the 5H9 re-docked poses. B represents the best docking poses of other Thermolysin inhibitors.
Table 2.
Calculated binding free energies (kcal/mol) and RMSD (Å) of the firsts ranked Autodock Vina poses of thermolysin complexes.
| Ligand | Docking Poses |
|||||||
|---|---|---|---|---|---|---|---|---|
| Dock-1 |
Dock-2 |
Dock-3 |
Dock-4 |
|||||
| Binding | RMSD | Binding | RMSD | Binding | RMSD | Binding | RMSD | |
| a | −9.2 | 1.67 | −9.2 | 1.83 | −9.2 | 1.04 | −9.1 | 1.13 |
| Lig-783 | −7.9 | 0.97 | −7.9 | 0.82 | −7.9 | 0.97 | −7.6 | 0.85 |
| Lig-1022 | −7.1 | 0.32 | −7.1 | 0.61 | −7.0 | 1.00 | −6.9 | 0.93 |
| Lig-1392 | −7.2 | 0.36 | −7.1 | 0.28 | −7.0 | 0.82 | −7.0 | 0.77 |
| Lig-2177 | −8.5 | 2.43 | −8.5 | 2.26 | −8.3 | 1.39 | −8.0 | 1.41 |
| Lig-3444 | −8.4 | 3.22 | −8.3 | 4.51 | −8.3 | 4.64 | −8.1 | 5.09 |
| Lig-6199 | −7.0 | 3.16 | −7.0 | 3.17 | −6.8 | 3.05 | −6.5 | 3.09 |
The N-[(S)-([(benzyloxy) carbonyl]amino methyl)(hydroxy) phosphoryl]-L-leucyl-4-methyl-L-leucine (5H9) was re-docked with the same docking procedure of the ligand studied.
In addition, most of the ligands present RMSD lower than 2 Å with respect to the X-ray crystal structure of the thermolysin-inhibitor complex obtained from the Protein Data Bank. This reference value identifies either correct or incorrect resolution of the docking (Gohlke et al., 2000). These two and parameters ( and RMSD) suggesting that ligands that fulfill both criteria can be considered as good Thermolysin inhibitors (Velázquez-Libera et al., 2020).
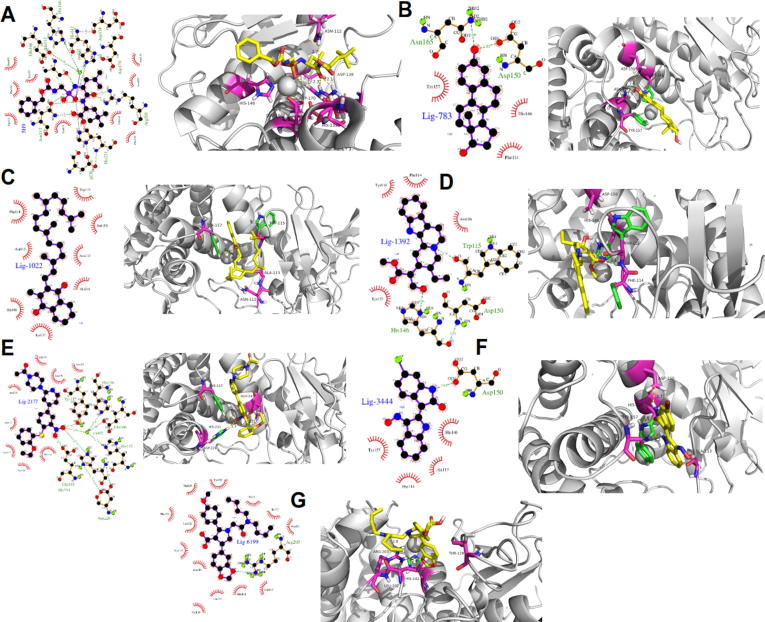
A detailed analysis of our docking results reveals that Lig2177 presents the most negative value. This is due to the presence of stabilizing interactions of this ligand with several amino acids into the Thermolysin pocket. As shown in Fig. 1, the phenyl-dihydroxyl amino group of the Lig2177 present hydrogen bonds (H-bond) interactions with Glu143 (2,87 Å), Asp226 (3,14 Å) and Tyr157 (3,04 Å). Moreover, the negative charge density of the O1 and O2 oxygen atoms of the hydroxyl amino group present attractive electrostatic interaction with the metallic center in the Thermolysin pocket which enhances the stability of the Lig2177-thermolysin complex. Additionally, Lig2177 presents the lowest RMSD value along with the series, which is congruent with the value for the Lig2177-thermolysin complex.
A similar performance is observed for Lig3444, which has the second most negative binding energy and the second-lowest RMSD, suggesting great stability for the Lig3444-thermolysin complex. This stability is given by the presence of H-bond interactions between the pyrrolic ring and Asp150 (2,95 Å) as well as the hydrophobic interaction between indol-hydroxylamine group, which is located in a hydrophobic zone of the Thermolysin pocket constituted by Ala113, Phe114, His146, and Tyr157, as can be observed from Fig. 3. The Lig783 occupies the third position considering both, and RMSD. Unlike the two compounds examined above, this ligand establishes weak H-bond interactions between their phenolic OH and Asp150 (2.81 Å) and Asp165 (3.09 Å), see Fig. 1. The foremost feature of Lig783 is that it can form hydrophobic interactions with Thermolysin. The fused hydrocarbon ring skeleton is oriented in a hydrophobic pocket formed by Phe114, His146 and Tyr157, which confer the relative stability to Lig783-thermolysin complex (Fig. 3).
Fig. 3.
Graphical representation of the ligands binding modes designed in silico into the Thermolysin pocket: (A) 5H9 reference ligand, (B) Lig-783, (C) Lig-1022, (D) Lig-1392, (E) Lig-2177, (F) Lig-3444 and (G) Lig-6199.
The remaining ligands present very similar values around 7.0 kcal/mol. Their relative stability is explained as follows: Lig1392 form weak H-bond interactions with Trp115 (3.26 Å) and His146 (3.06 Å); Lig1022 is exclusively stabilized in the Thermolysin pocket via hydrophobic interactions of the hydrocarbon chain and the hydrophobic pocket formed by Asn112, Ala113, Phe114, Trp115 and Asn116 in Thermolysin; Lig6199 is oriented in a hydrophobic area formed by Asn111, Asn112, Leu202, and Leu133, where stablish a unique H-bond with Arg203 (3.18 Å), see Fig. 3. Owing to the low number of H-bonds formed by Lig1392, Lig1022, and Lig6199 in the Thermolysin pocket, these ligands present the highest values for RMSD, as can be observed in Table 2.
3.2. Ligand efficiency analysis
We use two parameters, and LE (Ligand Efficiency), to compare the affinity of the ligands studied here and Thermolysin, corresponds to the dissociation constant of a ligand-thermolysin complex, its values are a measure of the protein-ligand interaction strength. Thus, very small values indicate that the compound binds strongly to the protein. LE represents averaged binding energy per non-hydrogen atoms, resulting in standardized values that allow us to compare molecules with different sizes (Table 3).
Table 3.
Ligand efficiency calculation of the firsts ranked Autodock Vina poses of thermolysin complexes.
| Ligand | Docking Experiments |
Ligand Efficiency Calculations |
||
|---|---|---|---|---|
| (Rank) | RMSD Å | LE (kcal/mol) | ||
| Lig-783 | −7.9(1) | 0.97 | 0.395 | |
| Lig-1022 | −7.1(1) | 0.32 | 0.215 | |
| Lig-1392 | −7.2(1) | 0.36 | 0.300 | |
| Lig-2177 | −8.5(2) | 2.26 | 0.253 | |
| Lig-3444 | −8.4(1) | 3.22 | 0.382 | |
| Lig-6199 | −7.0(1) | 3.16 | 0.189 | |
The ligands under study that exhibit the lowest values are Lig3444 and Lig2177, which means that these ligand-thermolysin complexes are the most stables in the series. These results are consistent with those obtained in the docking experiments in which these ligand-thermolysin complexes were the most stable according to their values.
The proposed tolerable values of LE for drug candidates are HAC. According to this reference value, compounds Lig783, Lig1392, and Lig3444 are excellent prospects to be used as Thermolysin inhibitors. It must be thought that studies on medications that are already on sale, present a LE value of 0.39 to 0.52 for oral medications, indicating that the results obtained for the ligands mentioned above are close to drugs already established in the market (Cavalluzzi et al., 2017, Hopkins et al., 2004, Hopkins et al., 2014).
3.3. Molecular dynamics simulation
-
•
RMSD
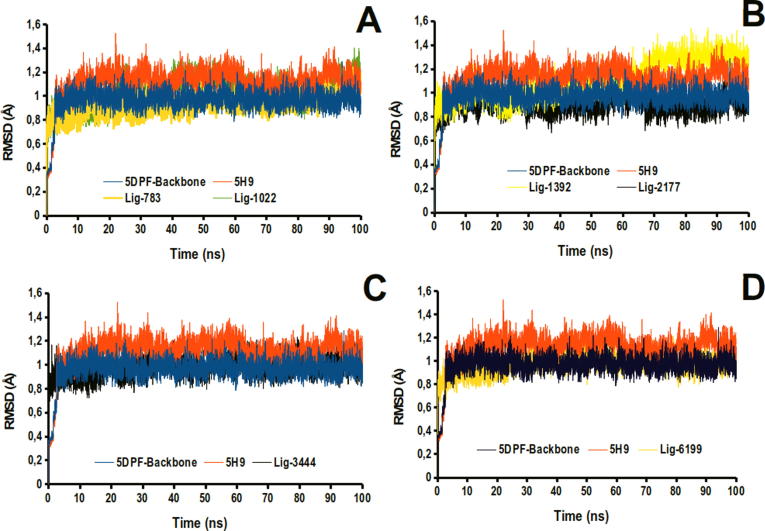
Molecular dynamics (MD) simulation is a very used tool to obtain trajectories that contain all structural information about the stability and relevance of molecular interactions on the ligand-protein complexes and their evolution through time. As a criterion of stability of the complexes studied in the simulation time, we analyze the RMSD parameter during the 100 ns of molecular dynamics, after the first 2.0 ns of equilibration. As presented in Fig. 4, all the complexes studied had RMSD values lower than 1.6 Å with very similar values with respect to our reference ligand 5H9, indicating that the ligands of this study form time-stable complexes with Thermolysin.
Fig. 4.
Plots of RMSD values against simulation time corresponding to molecular dynamics of the ligand-thermolysin complexes. A: Represent the comparative study between 5H9 and Lig-783 and 1022. B: Represent the comparative study between 5H9 and Lig-1392 and 2177. C: Represent the comparative study between 5H9 and Lig-3444 and D: Represent the comparative study between 5H9 and Lig-6199.
The Lig2177-thermolysin complex is the most stable among the series since it presents the lowest RMSD values (). This result is consistent with those obtained through docking experiments; therefore, we can expect that this molecule could be a good candidate as an antihypertensive agent. On the contrary, the highest RMSD values were obtained for Lig1392-thermolysin () and Lig1022-thermolysin () complexes, which indicates that these ligands were the least stable of all those studied. These results also agree with previous results from docking experiments.
-
•
H-Bond
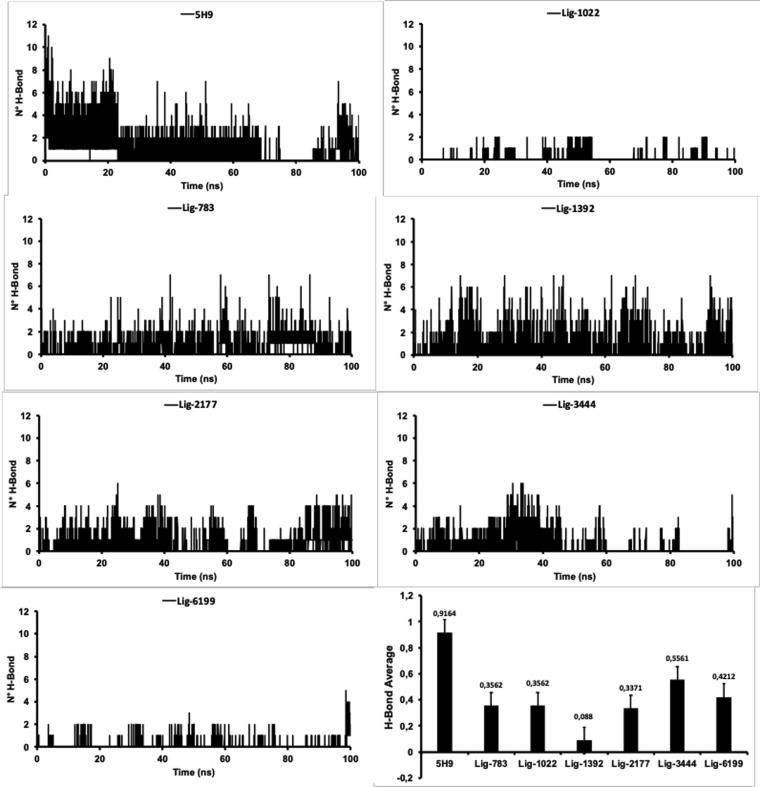
According to the results obtained above with respect to the RMSD parameter, it is necessary to obtain an explanation about the stability of these complexes at the molecular level and their evolution through time. For this, we quantify the number of hydrogen bonds presented by the ligands designed in the Thermolysin pocket and their behavior during the 100 ns of simulation, after the first 2.0 ns of equilibration.
As can be observed in Fig. 5, the number of H-bond interactions remained low during 100 ns of molecular dynamics simulation. It should be noted that on average, none of the systems analyzed in this manuscript exceeded the amount of one H-bond. The highest amount of H-bonds was presented by the Lig3444-thermolysin complex with an average of . From these results, we can conclude that hydrogen-bonds interactions were not stable over time nor are they responsible for the stability of the systems studied across the MD simulation time.
Fig. 5.
The number of H-bond between Thermolysin and the ligand studied during 100 ns of simulation time.
To verify this initial approach, an analysis of H-bond occupancy (%) was performed during the simulation time. The term occupancy refers to the time at which the interaction was maintained at a distance of 3 Å (cutoff criteria) during the 100 ns of molecular dynamics. As a criterion of stability of the H-bond, a 50% higher occupancy was taken.
In none of the complexes studied, the hydrogen bond occupancy exceeded 25%, indicating that h-bond interactions were not stable over time. The highest occupancy was obtained with Lig783 through heir interaction, which was maintained at a distance of less than 3 Å for 23.26% of the simulation time. The second highest value was obtained with Lig2177 through their interaction with occupancy of 14.16%. These results demonstrate that hydrogen bond interactions were not stable over time and therefore, this type of interaction is not the determinant for the stabilization of the ligand-thermolysin complexes presented here.
-
•
Radius of Gyration ()
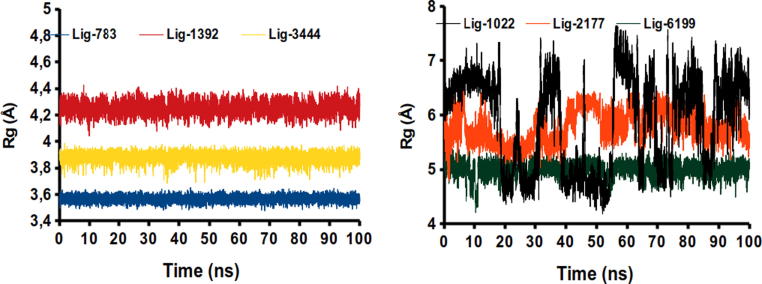
The quantity and occupancy of the H-bond interactions could not explain the stability of ligand-thermolysin complexes. Thus, it is necessary to analyze the radius of gyration parameter () to understand the level of compaction that Thermolysin structure presents when the ligands are included inside their active pocket. The is defined as the mean quadratic distance of the mass of a collection of atoms from a common center of mass. As can be observed in Fig. 6, all systems studied exceeded 3.5 Å of . It must be noted that these values are superior to other systems previously studied.
Fig. 6.
The number of H-bond and average between Thermolysin and the ligand studied during 100 ns of simulation time.
Among the analyzed complexes, the lowest value of Rg along the simulation time was obtained for the Lig783-thermolysin complex, as depicted in Fig. 4. This behavior was expected to occur, since this compound was the one that presented the highest H-bond occupancy, in addition to being the third most stable complex in the docking experiments. Indicating that Lig783 has the lowest fluctuations over time being, therefore, the most compact of all the series. The pattern established by Lig783 across time is observed for Lig1392 and Lig3444, with 3.9 and 4.1 Å of respectively, being concordant with H-bond interactions established by this ligand in the pocket of Thermolysin.
The other systems exceeded the value of 4 Å in the . The higher values were obtained for Lig1022 and Lig2177-thermolysin complexes indicating less stability of the interaction into the pocket of Thermolysin. This can be explained as Lig1022 is mainly stabilized via hydrophobic interactions. On the other hand, Lig2177 establish several H-bond interactions with different amino acids and electrostatic interactions with , which can be responsible for the high mobility inside the thermolysin pocket. These results are consistent with those obtained in the quantification and occupancy of H-bonds, indicating that h-bond interactions are key in the stabilization of ligand-protein complexes.
3.4. MM-GBSA
The results obtained above have not been able to give a completely satisfactory explanation about the stability of the ligand-thermolysin complexes in the simulation time, Thus, it is necessary to get a deeper understanding at the molecular level to determine the factors that contribute to the stabilization or destabilization of the ligand-thermolysin complexes. For this purpose, we have performed a free energy decomposition analysis using the MM-GBSA method (Hou et al., 2011, Hou et al., 2011, Saíz-Urra et al., 2013, Vergara-Jaque et al., 2013) whose results are presented in Table 4.
Table 4.
Predicted binding free energies (kcal/mol) and individual energy terms calculated from molecular dynamics simulation through the MM-GBSA protocol for Thermolysin complexes.
| Ligand | Calculated Free Energy Decomposition (kcal/mol |
||||
|---|---|---|---|---|---|
| Lig-783 | |||||
| Lig-1022 | |||||
| Lig-1392 | |||||
| Lig-2177 | |||||
| Lig-3444 | |||||
| Lig-6199 | |||||
According to MM-GBSA methodology, first, we calculate the binding free energy () from the snapshots of the molecular dynamics trajectories. This binding free energy is then averaged and decomposed into Van der Waals (), electrostatic () and solvation () contributions for all ligand-thermolysin complexes.
As can be observed in Table 4, according to calculated values, the most stable ligand-thermolysin complexes are those that contain Lig3444 and Lig2177. This result is in line with those obtained through docking experiments and confirms that they are the most stable complexes among the series. It must be noted that all other ligand-thermolysin complexes exhibit values >10 kcal/mol, denoting their high stability.
When considering the gas phase interaction energy () that corresponds to the sum of Van der Waals () and electrostatic terms (), Lig2177 is the one that exhibits the major stabilizing contribution to , which is followed by Lig783, Lig1392 and Lig1022 with alues around . Ligand-thermolysin complexes formed with Lig 3444 and Lig 6199 exhibit the lowest values. It must be noted that do not exist a clear trend when analyzing and terms, due to the different topologies of the studied ligands, where different functional groups establish different types of interactions in the thermolysin pocket. However the gas phase-electrostatic interactions do not favor the ligand complexation, maybe to a poor desolvation penalty of charged and polar groups, especially those forming hydrogen-bonds that can form stabilizing interactions with water molecules in a solvated media. This explanation can be reinforced, when analyzing the solvation energy term (), where Lig3444 exhibit the largest stabilizing contribution to , followed by Lig6199. The rest of the Ligands exhibit similar values around , with the exception of Lig1022 where is −4.52 kcal/mol.
3.5. ADME-Tox properties
Computational models based on pharmacokinetic properties (absorption, distribution, metabolism, and elimination), as well as toxicological parameters (ADME-Tox) are essential for drug design (Kauthale et al., 2018, Kumar et al., 2017, Tsaioun et al., 2016, Speck-Planche and Cordeiro, 2017, Teotia et al., 2018). In order to assess whether ligands can be selected as potential antihypertensive agents; we have calculated some structural properties as, molecular weight (MW in g/mol), octanol/water partition coefficient (LogP), hydrogen bond acceptor number (HBA), hydrogen bond donor number (HBD), topological polar surface area (TPSA in ) and rotatable bond count (RB), see Table 5.
Table 5.
ADME molecular descriptors of compounds designed to inhibit Termolysin.
| Ligand | MW (g/mol) | LogP | HBA | HBD | TPSA | RB |
|---|---|---|---|---|---|---|
| Lig-783 | 272.38 | 3.40 | 2 | 2 | 40,46 | 0 |
| Lig-1022 | 450.70 | 8.31 | 2 | 0 | 34.14 | 14 |
| Lig-1392 | 375.47 | 2,60 | 3 | 3 | 74.35 | 5 |
| Lig-2177 | 480.58 | 3.17 | 5 | 2 | 114.67 | 0 |
| Lig-3444 | 356.17 | 2.62 | 3 | 3 | 73.72 | 0 |
| Lig-6199 | 510.62 | 3.49 | 7 | 1 | 88.54 | 13 |
MW: Molecular weight; LogP: Octanol/water partition coefficient; HBA: Hydrogen bond acceptor; HBD: Hydrogen bond donor; TPSA: Topological polar surface area; RB: Rotatable bond count.
These results were contrasted against Lipinski (Lipinski et al., 2001), Veber (Veber et al., 2002) and Pfizer toxicity empirical rules (Hughes et al., 2008). If any of the compounds only comply with two of the Lipinski rules, we take that compound as a precaution (Lipinski et al., 2001), if it complies with only one rule then this molecule is not a good drug candidate. According to the Veber rules, if a compound does not meet any of the two parameters then it is not a good drug candidate (Veber et al., 2002). The Pfizer toxicity 3/75 rules (Hughes et al., 2008) were also taken into account, if any of our ligands does not comply with both parameters, then it is not a good drug candidate.
The analysis performed on ADME-Tox properties presented in Table 5 shown that Lig2177, Lig3444, and Lig1392 fulfill all the requirements established by Lipinski Veber and Pfizer 3/75 rules, indicating that these compounds could be safe compounds to be used as antihypertensive agents. It should be pointed out that Lig2177 and Lig3444 were found the most promising compounds from docking experiments, molecular dynamics simulation and MM-GBSA decomposition analysis. Compound Lig-783 presents a violation of Pfizer 3/75 rules, due to their octanol/water partition coefficient is greater than the admissible limit, making this substance liposoluble, which suggests a tendency to be more toxic and less selective to their target. The same behavior was obtained for Lig1022 and Lig6199, which present violations to Lipinski and Verber rules.
4. Conclusions
High blood pressure is one of the main causes of cardiovascular disease around the world. Currently, more than 9 million people die every year from this cause, which has motivated many researchers to design safer drugs to treat this condition. Our research group has designedin silicoseveral compounds as possible antihypertensive agents using the QSARINS method taking Thermolysin as the target for these drug candidates. We have selected the six most promising candidates from this study. In this article, we apply a computational biochemistry protocol to determine by means of docking experiments, molecular dynamics, MM-GBSA and ADMe-Tox properties whether these compounds are suitable to be employed in a dual antihypertensive therapy.
From the results of this computational biochemistry protocol, we conclude that these ligands designed in silico present an adequate orientation in the pocket of Thermolysin. Lig2177 and Lig3444 establish the most favorable interactions with amino acids Asp150, Glu143 and Tyr157 in addition to interacting with in the active center of Thermolysin. Although these interactions did not remain constant over time, it must be pointed out that MM-GBSA calculations indicate that Van der Waals and solvation interactions were the most stabilizing terms for binding energy across time. Lig2177 and Lig3444 in conjunction with Lig-1392 were the only molecules that fulfill all the ADME-Tox requirements. The information here presented suggests that Lig2177 and Lig3444 are the most promising candidates to be used as antihypertensive agents.
Funding
This work was supported by FONDECYT Iniciación grant N° 11180650.
References
- Abad-Zapatero C. Ligand efficiency indices for drug discovery. Ligand Efficiency Indices Drug Discov. 2013;10(7):469–488. doi: 10.1517/17460441.2.4.469. [DOI] [PubMed] [Google Scholar]
- Abad-Zapatero C., Perišić O., Wass J., Bento A.P., Overington J., Al-Lazikani B., Johnson M.E. Ligand efficiency indices for an effective mapping of chemico-biological space: the concept of an atlas-like representation. Drug Discov. Today. 2010;15(19–20):804–811. doi: 10.1016/j.drudis.2010.08.004. [DOI] [PubMed] [Google Scholar]
- Adasme-Carreno F., Munoz-Gutierrez C., Caballero J., Alzate-Morales J.H. Performance of the MM/GBSA scoring using a binding site hydrogen bond network-based frame selection: the protein kinase case. Phys. Chem. Chem. Phys. 2014;16(27):14047–14058. doi: 10.1039/c4cp01378f. [DOI] [PubMed] [Google Scholar]
- Berman H., Westbrook Z., Feng G., Gilliland T., Bhat H., Weissig I., Shindyalov P.E., Bourne P. The Protein Data Bank. Nucl. Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat Z.F., Kumar S., Bhat H.F. Antihypertensive peptides of animal origin: a review. Crit. Rev. Food Sci. Nutr. 2017;57(3):566–578. doi: 10.1080/10408398.2014.898241. [DOI] [PubMed] [Google Scholar]
- Bohacek R., De Lombaert S., McMartin C., Priestle J., Grütter M. Three-dimensional models of ACE and NEP inhibitors and their use in the design of potent dual ACE/NEP inhibitors. J. Am. Chem. Soc. 1996;118(35):8231–8249. [Google Scholar]
- Boonstra S., Onck P.R., van der Giessen E. CHARMM TIP3P water model suppresses peptide folding by solvating the unfolded state. J. Phys. Chem. B. 2016;120(15):3692–3698. doi: 10.1021/acs.jpcb.6b01316. [DOI] [PubMed] [Google Scholar]
- Braunwald E. The path to an angiotensin receptor antagonist-neprilysin inhibitor in the treatment of heart failure. J. Am. Coll. Cardiol. 2015;65(10):1029–1041. doi: 10.1016/j.jacc.2015.01.033. [DOI] [PubMed] [Google Scholar]
- Brian, N.D., Brooks, Charles L., 1999. Development of a generalized born model parametrization for proteins and nucleic acids. J. Chem. Phys. B 103(8), 3765–3773.
- Calhoun D.A., Sharma K. The role of aldosteronism in causing obesity-related cardiovascular risk. Cardiol. Clin. 2010;28(3):517–527. doi: 10.1016/j.ccl.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cañizares-Carmenate Y., Mena-Ulecia K., Perera-Sardiña Y., Torrens F., Castillo-Garit J.A. An approach to identify new antihypertensive agents using Thermolysin as model: in silico study based on QSARINS and docking. Arab. J. Chem. 2019;12(8):4861–4877. [Google Scholar]
- Cavalluzzi M.M., Mangiatordi G.F., Nicolotti O., Lentini G. Ligand efficiency metrics in drug discovery: the pros and cons from a practical perspective. Expert Opin. Drug Discov. 2017;12(11):1087–1104. doi: 10.1080/17460441.2017.1365056. [DOI] [PubMed] [Google Scholar]
- Choe J., Seol K.-H., Kim H.-J., Hwang J.-T., Lee M., Jo C. Isolation and identification of angiotensin I-converting enzyme inhibitory peptides derived from thermolysin-injected beef M. longissimus. Asian-Austral. J. Anim. Sci. 2019;32(3):430–436. doi: 10.5713/ajas.18.0455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daina A., Michielin O., Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017;7(1):42717. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner David E., Phillips James C., Schulten K., Tanner D.E., Phillips J.C., Schulten K., Tanner David E., Phillips James C., Schulten K. GPU/CPU algorithm for generalized born/solvent-accessible surface area implicit solvent calculations. J. Chem. Theory Comput. 2012;8(7):2521–2530. doi: 10.1021/ct3003089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePriest S.A., Mayer D., Naylor C.B., Marshall G.R. 3D-QSAR of angiotensin-converting enzyme and thermolysin inhibitors: a comparison of CoMFA models based on deduced and experimentally determined active site geometries. J. Am. Chem. Soc. 1993;115(13):5372–5384. [Google Scholar]
- Foloppe N., MacKerell A.D., Jr. All-atom empirical force field for nucleic acids: I. Parameter optimization based on small molecule and condensed phase macromolecular target data. J. Comput. Chem. 2000;21(2):86–104. [Google Scholar]
- Gaillard T., Panel N., Simonson T. Protein side chain conformation predictions with an MMGBSA energy function. Proteins: Struct., Funct., Bioinf. 2016;84(6):803–819. doi: 10.1002/prot.25030. [DOI] [PubMed] [Google Scholar]
- Gamboa, R., 2006. Fisiopatología de la hipertensión arterial esencial. In: Simposio: Hipertensión Arterial, vol. 23.
- Gohlke H., Hendlich M., Klebe G. Knowledge-based scoring function to predict protein-ligand interactions. J. Mol. Biol. 2000;295(2):337–356. doi: 10.1006/jmbi.1999.3371. [DOI] [PubMed] [Google Scholar]
- Hanwell M.D., Curtis D.E., Lonie D.C., Vandermeersch T., Zurek E., Hutchison G.R. Avogadro: an advanced semantic chemical editor, visualization, and analysis platform. J. Cheminformat. 2012;4(1):17. doi: 10.1186/1758-2946-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins A.L., Groom C.R., Alex A. Ligand efficiency: a useful metric for lead selection. Drug Discov. Today. 2004;9(10):430–431. doi: 10.1016/S1359-6446(04)03069-7. [DOI] [PubMed] [Google Scholar]
- Hopkins A.L., Keserü G.M., Leeson P.D., Rees D.C., Reynolds C.H. The role of ligand efficiency metrics in drug discovery. Nat. Rev. Drug Discov. 2014;13(2):105–121. doi: 10.1038/nrd4163. [DOI] [PubMed] [Google Scholar]
- Hou T., Wang J., Li Y., Wang W. Assessing the performance of the MM/PBSA and MM/GBSA methods. 1. The accuracy of binding free energy calculations based on molecular dynamics simulations. J. Chem. Inform. Model. 2011;51(1):69–82. doi: 10.1021/ci100275a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou T., Wang J., Li Y., Wang W. molecular mechanics/Poisson Boltzmann surface area and molecular mechanics/generalized Born surface area methods. II. The accuracy of ranking poses generated. J. Comput. 2011;32(5):866–877. doi: 10.1002/jcc.21666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubers S.A., Brown N.J. Combined angiotensin receptor antagonism and neprilysin inhibition. Circulation. 2016;133(11) doi: 10.1161/CIRCULATIONAHA.115.018622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes J.D., Blagg J., Price D.A., Bailey S., DeCrescenzo G.A., Devraj R.V., Ellsworth E., Fobian Y.M., Gibbs M.E., Gilles R.W., Greene N., Huang E., Krieger-Burke T., Loesel J., Wager T., Whiteley L., Zhang Y. Physiochemical drug properties associated with in vivo toxicological outcomes. Bioorgan. Med. Chem. Lett. 2008;18(17):4872–4875. doi: 10.1016/j.bmcl.2008.07.071. [DOI] [PubMed] [Google Scholar]
- Kauthale S., Tekale S., Damale M., Sangshetti J., Pawar R. Synthesis, biological evaluation, molecular docking, and ADMET studies of some isoxazole-based amides. Med. Chem. Res. 2018;27(2):429–441. [Google Scholar]
- Khan M.T.H., Fuskevåg O.-M., Sylte I. Discovery of potent thermolysin inhibitors using structure based virtual screening and binding assays. J. Med. Chem. 2009;52(1):48–61. doi: 10.1021/jm8008019. [DOI] [PubMed] [Google Scholar]
- Koebel M.R., Schmadeke G., Posner R.G., Sirimulla S. AutoDock VinaXB: implementation of XBSF, new empirical halogen bond scoring function, into AutoDock Vina. J. Cheminformat. 2016;8(1):27. doi: 10.1186/s13321-016-0139-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krimmer S., Klebe G. Thermodynamics of protein-ligand interactions as a reference for computational analysis: how to assess accuracy, reliability and relevance of experimental data. J. Comput. Aided Mol. Des. 2015;29:867–883. doi: 10.1007/s10822-015-9867-y. [DOI] [PubMed] [Google Scholar]
- Kumar N., Goel N., Chand Yadav T., Pruthi V. Quantum chemical, ADMET and molecular docking studies of ferulic acid amide derivatives with a novel anticancer drug target. Med. Chem. Res. 2017;26(8):1822–1834. [Google Scholar]
- Lee J., Cheng X., Swails J.M., Yeom M.S., Eastman P.K., Lemkul J.A., Wei S., Buckner J., Jeong J.C., Qi Y., Jo S., Pande V.S., Case D.A., Brooks C.L., MacKerell A.D., Klauda J.B., Im W. CHARMM-GUI Input Generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM Simulations Using the CHARMM36 Additive Force Field. J. Chem. Theory Comput. 2016;12(1):405–413. doi: 10.1021/acs.jctc.5b00935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski C.A., Lombardo F., Dominy B.W., Feeney P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001;46(1–3):3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- Lu J., Qiu Y., Baron R., Molinero V. Coarse-graining of TIP4P/2005, TIP4P-Ew, SPC/E, and TIP3P to monatomic anisotropic water models using relative entropy minimization. J. Chem. Theory Comput. 2014;10(9):4104–4120. doi: 10.1021/ct500487h. [DOI] [PubMed] [Google Scholar]
- MacKerell, A.D., Feig, M., Brooks, C.L., 2004. Improved treatment of the protein backbone in empirical force fields. J. Am. Chem. Soc. 11. MacKer(3), 698–699. [DOI] [PubMed]
- Manzur F., Villarreal T., Moneriz C. Inhibición dual de la neprilisina y el receptor de angiotensina II: nueva estrategia prometedora en el tratamiento de la enfermedad cardiovascular. Revista Colombiana de Cardiología. 2013;20(6):386–393. [Google Scholar]
- Martins L.C., Figueiredo V.N., Quinaglia T., Boer-Martins L., Yugar-Toledo J.C., Martin J.F.V., Demacq C., Pimenta E., Calhoun D.A., Moreno H. Characteristics of resistant hypertension: ageing, body mass index, hyperaldosteronism, cardiac hypertrophy and vascular stiffness. J. Hum. Hypertens. 2011;25(9):532–538. doi: 10.1038/jhh.2010.95. [DOI] [PubMed] [Google Scholar]
- Massova I., Kollman P. Combined molecular mechanical and continuum solvent approach (MM-PBSA/GBSA) to predict ligand binding. Perspect. Drug Discov. Des. 2000;18:113–135. [Google Scholar]
- Mena-Ulecia K., Gonzalez-Norambuena F., Vergara-Jaque A., Poblete H., Tiznado W., Caballero J. Study of the affinity between the protein kinase PKA and homoarginine-containing peptides derived from kemptide: Free energy perturbation (FEP) calculations. J. Comput. Chem. 2018;39(16):986–992. doi: 10.1002/jcc.25176. [DOI] [PubMed] [Google Scholar]
- Mena-Ulecia K., Tiznado W., Caballero J. Study of the differential activity of thrombin inhibitors using docking, QSAR, molecular dynamics, and MM-GBSA. Plos One. 2015;10(11):e0142774. doi: 10.1371/journal.pone.0142774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30(16):2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Durango N., Fuentes C.A., Castillo A.E., González-Gómez L.M., Vecchiola A., Fardella C.E., Kalergis A.M. Role of the renin-angiotensin-aldosterone system beyond blood pressure regulation: molecular and cellular mechanisms involved in end-organ damage during arterial hypertension. Int. J. Mol. Sci. 2016;17(7):1–17. doi: 10.3390/ijms17070797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neese F. The ORCA program system. Wiley Interdiscipl. Rev.: Comput. Mol. Sci. 2012;2(1):73–78. [Google Scholar]
- Neese F. Software update: the ORCA program system, version 4.0. Wiley Interdisciplinary Reviews: Computational Molecular. Science. 2018;8(1):e1327. [Google Scholar]
- Nehme A., Zouein F.A., Zayeri Z.D., Zibara K. An update on the tissue renin angiotensin system and its role in physiology and pathology. J. Cardiovasc. Develop. Dis. 2019;6(2):14. doi: 10.3390/jcdd6020014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onufriev A., Bashford D., Case D.A. Exploring protein native states and large-scale conformational changes with a modified generalized born model. Proteins: Struct., Funct., Bioinf. 2004;55(2):383–394. doi: 10.1002/prot.20033. [DOI] [PubMed] [Google Scholar]
- Parr R.G., Yang W. Density functional approach to the frontier-electron theory of chemical reactivity. J. Am. Chem. Soc. 1984;106(14):4049–4050. [Google Scholar]
- Patten G.S., Abeywardena M.Y., Bennett L.E. Inhibition of angiotensin converting enzyme, angiotensin II receptor blocking, and blood pressure lowering bioactivity across plant families. Crit. Rev. Food Sci. Nutr. 2016;56(2):181–214. doi: 10.1080/10408398.2011.651176. [DOI] [PubMed] [Google Scholar]
- Paulis L., Rajkovicova R., Simko F. New developments in the pharmacological treatment of hypertension: dead-end or a glimmer at the horizon? Curr. Hypertens. Rep. 2015;17(6) doi: 10.1007/s11906-015-0557-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdew J.P., Burke K., Ernzerhof M. Generalized gradient approximation made simple. Phys. Rev. Lett. 1996;77(18):3865–3868. doi: 10.1103/PhysRevLett.77.3865. [DOI] [PubMed] [Google Scholar]
- Petermann F., Durán E., Labraña A.M., Martínez M.A., Leiva A.M., Garrido A., Poblete F., Díaz X., Salas C., Celis C. Risk factors associated with hypertension. Analysis of the 2009–2010 Chilean health survey. Revista médica de Chile. 2017;145(8):996–1004. doi: 10.4067/s0034-98872017000800996. [DOI] [PubMed] [Google Scholar]
- Phillips J.C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R.D., Kalé L., Schulten K., Brauna R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R.D., Kale L., Phillips J.C., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26(16):1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam K., Shoemaker R., Yiannikouris F., Cassis L.A. The renin-angiotensin system: a target of and contributor to dyslipidemias, altered glucose homeostasis, and hypertension of the metabolic syndrome. Am. J. Physiol.-Heart Circul. Physiol. 2012;302(6):H1219–H1230. doi: 10.1152/ajpheart.00796.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren L., Lu X., Danser A.H.J. Revisiting the brain renin-angiotensin system—focus on novel therapies. Curr. Hypertens. Rep. 2019;21(4):28. doi: 10.1007/s11906-019-0937-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds C.H., Tounge B.A., Bembenek S.D. Ligand binding efficiency: trends, physical basis, and implications. J. Med. Chem. 2008;51(8):2432–2438. doi: 10.1021/jm701255b. [DOI] [PubMed] [Google Scholar]
- Rodrigues Prestes T.R., Rocha N.P., Miranda A.S., Teixeira A.L., Simoes-e Silva A.C. The anti-inflammatory potential of ACE2/angiotensin-(1–7)/Mas receptor axis: evidence from basic and clinical research. Curr. Drug Targets. 2017;18(11) doi: 10.2174/1389450117666160727142401. [DOI] [PubMed] [Google Scholar]
- Saíz-Urra, L., Pérez, M.A.C., Froeyen, M., Sa⧹’⧹iz-Urra, L., Pérez, M.A.C., Froeyen, M., Saíz-Urra, L., Pérez, M.A.C., Froeyen, M., 2013. Thermodynamic computational approach to capture molecular recognition in the binding of different inhibitors to the DNA gyrase B subunit from Escherichia coli. J. Mol. Model. [DOI] [PubMed]
- Soteras Gutiérrez I., Lin F.-Y., Vanommeslaeghe K., Lemkul J.A., Armacost K.A., Brooks C.L., MacKerell A.D. Parametrization of halogen bonds in the CHARMM general force field: Improved treatment of ligand–protein interactions. Bioorgan. Med. Chem. 2016;24(20):4812–4825. doi: 10.1016/j.bmc.2016.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speck-Planche A., Cordeiro M.N.D. De novo computational design of compounds virtually displaying potent antibacterial activity and desirable in vitro ADMET profiles. Med. Chem. Res. 2017;26(10):2345–2356. [Google Scholar]
- Spyroulias G., Galanis A., Pairas G., Manessi-Zoupa E., Cordopatis P. Structural features of angiotensin-i converting enzyme catalytic sites: conformational studies in solution, homology models and comparison with other zinc metallopeptidases. Curr. Top. Med. Chem. 2004;4(4):403–429. doi: 10.2174/1568026043451294. [DOI] [PubMed] [Google Scholar]
- Stanisz B., Regulska K., Regulski M. The angiotensin converting enzyme inhibitors – alternative clinical applications. Journal of Medical Science. 2016;83(1):57–61. [Google Scholar]
- Te Riet L., Van Esch J.H., Roks A.J., Van Den Meiracker A.H., Danser A.H. Hypertension: renin-angiotensin-aldosterone system alterations. Circ. Res. 2015;116(6):960–975. doi: 10.1161/CIRCRESAHA.116.303587. [DOI] [PubMed] [Google Scholar]
- Teotia P., Prakash Dw S., Dwivedi N. In silico molecular docking and ADME/Tox study on benzoxazole derivatives against inosine 5’-monophosphate dehydrogenase. Asian J. Biotechnol. 2018;10(1):1–10. [Google Scholar]
- Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsaioun K., Blaauboer B.J., Hartung T. Evidence-based absorption, distribution, metabolism, excretion (ADME) and its interplay with alternative toxicity methods. Altex. 2016;33(4):343–358. doi: 10.14573/altex.1610101. [DOI] [PubMed] [Google Scholar]
- Vanommeslaeghe K., Yang M., MacKerell A.D. Robustness in the fitting of molecular mechanics parameters. J. Comput. Chem. 2015;36(14):1083–1101. doi: 10.1002/jcc.23897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas-Sánchez R., Mendoza-Wilson A., Balandrán-Quintana R., Torrescano-Urrutia G., Sánchez-Escalante A. Study of the molecular structure and chemical reactivity of pinocembrin by DFT calculations. Comput. Theoret. Chem. 2015;1058:21–27. [Google Scholar]
- Veber D.F., Johnson S.R., Cheng H.-Y., Smith B.R., Ward K.W., Kopple K.D. Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 2002;45(12):2615–2623. doi: 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]
- Velázquez-Libera J.L., Durán-Verdugo F., Valdés-Jiménez A., Núñez-Vivanco G., Caballero J. LigRMSD: a web server for automatic structure matching and RMSD calculations among identical and similar compounds in protein-ligand docking. Bioinformatics. 2020 doi: 10.1093/bioinformatics/btaa018. [DOI] [PubMed] [Google Scholar]
- Vergara-Jaque A., Comer J., Monsalve L., González-Nilo F.D., Sandoval C. Computationally efficient methodology for atomic-level characterization of dendrimer-drug complexes: a comparison of amine- and acetyl-terminated PAMAM. J. Physi. Chem. B. 2013;117(22):6801–6813. doi: 10.1021/jp4000363. [DOI] [PubMed] [Google Scholar]
- Yang W., Parr R.G. Hardness, softness, and the fukui function in the electronic theory of metals and catalysis. Proc. Nat. Acad. Sci. 1985;82(20):6723–6726. doi: 10.1073/pnas.82.20.6723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoete V., Cuendet M.A., Grosdidier A., Michielin O., Vincent Z., A. C.M., Aurélien G., Olivier M., Zoete V., Cuendet M.A., Grosdidier A., Michielin O. SwissParam: A fast force field generation tool for small organic molecules. J. Comput. Chem. 2011;32(11)::2359–2368. doi: 10.1002/jcc.21816. [DOI] [PubMed] [Google Scholar]