Abstract
Aims
To assess the effect of physical exercise (PE) on the histological and transcriptional characteristics of proteoglycan-induced arthritis (PGIA) in BALB/c mice.
Methods
Following PGIA, mice were subjected to treadmill PE for ten weeks. The tarsal joints were used for histological and genetic analysis through microarray technology. The genes differentially expressed by PE in the arthritic mice were obtained from the microarray experiments. Bioinformatic analysis in the DAVID, STRING, and Cytoscape bioinformatic resources allowed the association of these genes in biological processes and signalling pathways.
Results
Arthritic mice improved their physical fitness by 42.5% after PE intervention; it induced the differential expression of 2,554 genes. The bioinformatic analysis showed that the downregulated genes (n = 1,371) were significantly associated with cellular processes that mediate the inflammation, including Janus kinase-signal transducer and activator of transcription proteins (JAK-STAT), Notch, and cytokine receptor interaction signalling pathways. Moreover, the protein interaction network showed that the downregulated inflammatory mediators interleukin (IL) 4, IL5, IL2 receptor alpha (IL2rα), IL2 receptor beta (IL2rβ), chemokine ligand (CXCL) 9, and CXCL12 were interacting in several pathways associated with the pathogenesis of arthritis. The upregulated genes (n = 1,183) were associated with processes involved in the remodelling of the extracellular matrix and bone mineralization, as well as with the processes of aerobic metabolism. At the histological level, PE attenuated joint inflammatory infiltrate and cartilage erosion.
Conclusion
Physical exercise influences parameters intimately linked to inflammatory arthropathies. Research on the effect of PE on the pathogenesis process of arthritis is still necessary for animal and human models.
Cite this article: Bone Joint Res. 2020;9(1):36–48.
Keywords: Arthritis, Cytokine, Microarray
Article focus
What is the effect of physical exercise on the histological and transcriptional features of joints with arthritis?
What transcriptional modifications are induced by physical exercise in joints with arthritis?
What cellular processes are triggered by physical exercise in inflamed joints compared with non inflamed joints?
Key messages
Treadmill exercise downregulates genes associated with the inflammatory process and upregulates those associated with extracellular matrix remodelling and bone mineralization in the tarsal joints of proteoglycan-induced arthritic (PGIA) mice.
Treadmill exercise downregulates inflammation of the tarsal joints of proteoglycan-induced arthritic mice.
Treadmill exercise improves the physical fitness of PGIA mice, despite the presence of arthritis.
Strengths and limitations
The present investigation provides an overview of the insufficiently explored field which deals with the effect of physical exercise at the tissue level in an animal model of arthritis and its consequences in the process. The transcriptional modifications induced by physical exercise have not been previously described in models of inflammatory arthropathies, including the PGIA model.
The study is exploratory and preliminary since, although microarray analysis was internally validated, there is no external validation that includes another technique such as polymerase chain reaction.
The PGIA model developed mild to moderate histological arthritis scores, so the results should be circumscribed to these arthritis conditions.
Introduction
Rheumatic diseases (RDs) include many types of arthritides, which are defined by the inflammation of the synovial membrane and other joint structures. They are characterized by pain, swelling, stiffness, and progressive impairment of movement in the affected joint, including its destruction.1 The pathogenic mechanisms for several RDs have been partially defined. The description of key immune mediators has allowed the development of target-specific drug therapies that reduce the symptoms and prevent disease progression.2
The positive impact of physical exercise (PE) in the general population is evident. Some parallels in rheumatic patients have been shown3–5 and are considered an element in the treatment of inflammatory arthropathies.6–7 Physical exercise improves physical function, disease outcomes, and also cardiorespiratory function8,9 in RD patients. The European League Against Rheumatism (EULAR) has established recommendations for physical activity and exercise in people with inflammatory arthritis.10
However, it is not clear whether PE could also play a detrimental role by worsening joint destruction. We also ignore the intimate molecular mechanisms triggered by PE in the metabolic responses and inflammatory processes in the arthritis pathogenesis. Several conditions induced by PE such as hypoxia or mechanical stress are potentially inducers of inflammation.
Animal models of arthritis have allowed us to understand pathological processes in RD,11 and test different pharmacological treatments.12 The proteoglycan-induced arthritis (PGIA) model, which is started by the immunization of BALB/c mice with cartilage proteoglycan (PG), is a model with progressive polyarthritis, and is frequently accompanied by spondylitis.13,14 This model is suitable for analyzing the role of PE in the joint environment and to define its influence in the genetic expression.15,16
DNA microarray technology allows the assessment of genetic analyses on thousands of genes within a given sample and can provide a panoramic view of the intracellular signals influenced by PE in the pathogenic process of arthritis. Currently, the study of the gene-disease relationship is based on analyzing the behaviour of thousands of genes in a simultaneous form.17
The present study attempts to assess the effect of treadmill PE on inflammatory response and osteochondral metabolism in tarsal bones of PGIA mice through the analysis of their transcriptomes using a DNA microarray strategy.
Methods
Animals and study groups
The PGIA model was used in this study.18 Female BALB/c retired breeder mice aged eight to 11 months were randomly divided into four groups of eight mice each: healthy non-exercised mice; healthy exercised mice; PGIA non-exercised mice; and PGIA exercised mice. Mice were kept under controlled conditions of luminosity (12 hours light and 12 hours dark) and temperature (mean 23ºC, standard deviation (SD) 2ºC), and received food and water ad libitum.
Arthritis induction
The PGIA model was conducted as previously described18 with minor modifications. Mice from groups III and IV were intraperitoneally injected four times at 21-day intervals with 100 μl of an emulsion containing 100 μg of bovine PG extracted from the nasal septum and 1 mg of dimethyl dioctadecyl ammonium bromide (DDA) adjuvant. The analysis was performed at the end of the PE intervention (day 98 after the first injection) as described below.
Treadmill design
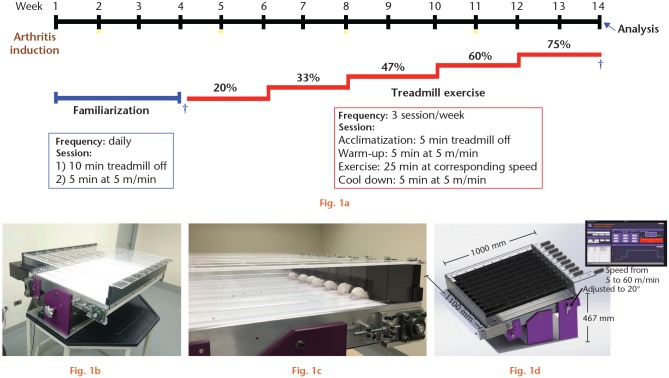
Physical exercise intervention was performed on a custom-designed and -built treadmill (Figure 1b) compliant with the American Physiological Society recommendations.19 The treadmill dimensions were 1000 mm length × 1100 mm width × 467 mm height, with nine lanes each measuring 115 mm in width, with separate access for individual mice handling. The belt speed could be set from 5 m/min to 60 m/min, and the belt slope could be adjusted manually to 20°. The treadmill was manufactured with a shock grid (0 mA to 2 mA) to provide an aversive stimulus to the animals; however, this stimulus was not used during our investigation. Additionally, the treadmill was complemented with a custom-designed software that allowed the automation of PE protocols. Specific times and speeds were programmed according to the routine.
Fig. 1.
Homemade treadmill, experimental design and exercise routine. a) Experimental design of treadmill exercise intervention including arthritis induction, familiarization period, maximal exercise capacity tests (MECT), exercise, and analysis. b) Custom-designed and -built treadmill. c) Exercised mice on treadmill. d) Treadmill characteristics. *Arthritis induction. †MECTs.
Treadmill familiarization
For groups II and IV, a period of familiarization was included in order to minimize psychological stress on the animal. The mice were then subjected to treadmill running. The process lasted four weeks and was designed to promote visual/olfactory and sound/movement adaptation (Figure 1a). The animals were placed on the treadmill daily for a total of 15 minutes: ten minutes on the switched off treadmill and five minutes of a walk at the lowest speed. After the first week of familiarization, arthritis induction was started in PGIA mice (group IV) and familiarization continued for three more weeks.
Treadmill-running performance evaluation
On the last day of the familiarization period, treadmill-running performance was evaluated in mice from groups II and IV using an individual maximal exercise capacity test (MECT-1) (Figure 1a). Mice placed on the band ran for five minutes as a warm-up. After this time, the speed was increased by 3 m/min every two minutes until the mice stopped running. The maximal PE capacity (100%) was defined as the maximum speed reached by each animal. Each mouse individually underwent physical testing. The mean speed was estimated for each study group.
At the end of ten weeks of PE administration, a second treadmill-running performance was evaluated (MECT-2) (Figure 1a) to determine the physical fitness of the mice by comparing the maximum velocities reached pre- and post intervention.
Treadmill physical exercise intervention
After familiarization and MECT-1, the mice from groups II and IV that managed to adapt to the treadmill (seven mice per group), were exercised for ten weeks with a frequency of three sessions per week and 25 minutes for each PE session. To improve the physical fitness of the mice, the speed was gradually increased from 20% to 75% in increments of 13% (regarding the mean of maximum speeds reached per group in MECT-1) every six sessions. Each PE session included the following phases: acclimatization, when mice were placed for five minutes on the switched off treadmill; warm-up, when mice walked for five minutes at the lowest speed; PE, when mice ran for 25 minutes at the stage-specific speed; and cool down, when mice walked for five minutes at the lowest speed. The PE sessions were performed at the same time each day and mice were always placed in the same lane (Figure 1a).
Histological analysis
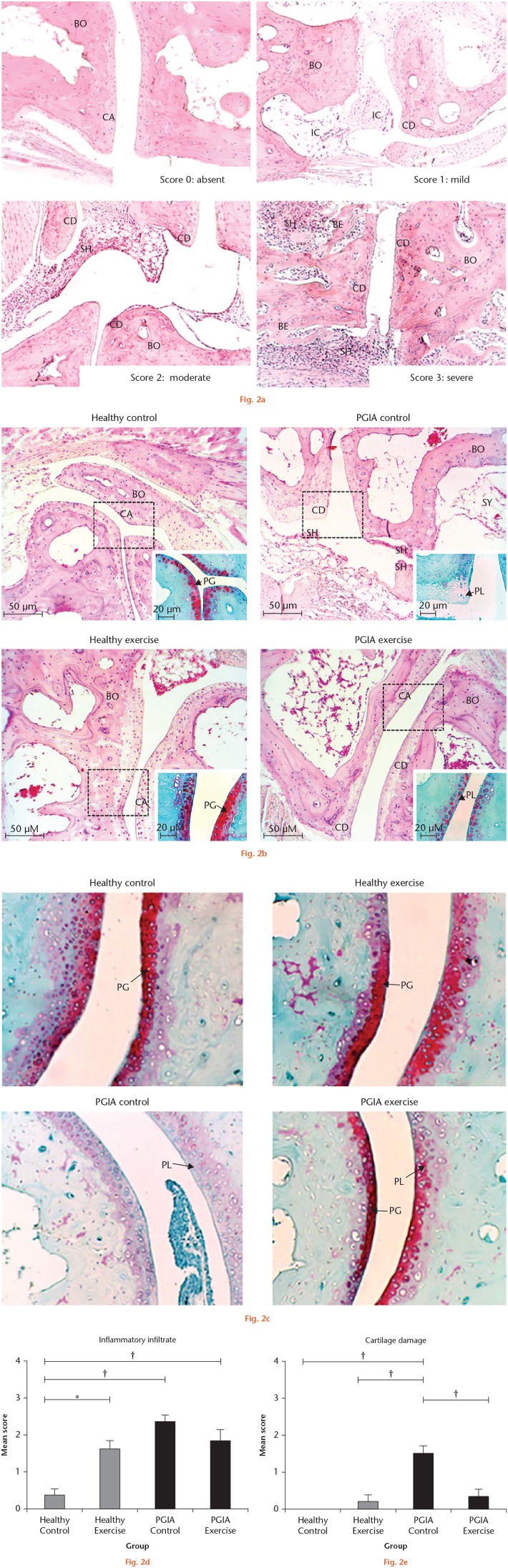
The mice were euthanized with isoflurane (Laboratorios PISA, S.A. de C.V, Mexico). The hind paws were dissected and fixed in 10% phosphate-buffered formalin for 48 hours. The samples were demineralized using 5% nitric acid for 24 hours, dehydrated in graded ethanol, and embedded in paraffin. Sections of 5 μm in thickness were obtained and placed on adhesive-coated glass slides.20 A histological assessment was carried out using haematoxylin and eosin (H&E) staining, in addition to Safranin-O/ Fast green staining, to unveil the cartilage PG. The images were acquired using a digital camera coupled to the optical microscope. The influence of PE on tarsal joint structures was semi-quantitatively evaluated in three slices of each sample, using the semi-quantitative scale: 0, absent; 1, mild; 2, moderate; and 3, severe (Figure 2a) to describe inflammatory infiltrate and cartilage erosion in the tarsal joints. The mean score was calculated for each group.
Fig. 2.
Effects of exercise on tarsal bone histological parameters in proteoglycan-induced arthritis (PGIA) and healthy mice. a) Representative images of the inflammatory infiltrate and cartilage damage scores in the tarsal joints of PGIA mice. The 0 (normal) score was established in the control group (healthy control), where the bone, cartilage, and synovium did not show alterations. The arthritis scores 1 (mild), 2 (moderate) and 3 (severe) were based on the inflammatory changes (presence of inflammatory cells and synovial hyperplasia) and structural remodelling (cartilage damage and bone erosion). The images were acquired with a 10× amplification. b) Representative images of histological findings in the tarsal joints of the four study groups at the end of the exercise intervention using haematoxylin and eosin (H&E) and Safranin-O/Fast green staining to unveil the cartilage proteoglycan. The proteoglycan loss (PL) allowed the assessment of cartilage damage. The rectangle delimits the section of the tissue stained with Safranin-O/Fast green which is shown in the lower right corner of each image. c) Representative images of cartilage damage in the four study groups using the Safranin-O/Fast green staining to unveil the proteoglycan content and its loss. d) and e) Joint involvement was scored by the semi-quantitative scale to describe inflammatory infiltrate and cartilage damage in the tarsal joints (8 mice per group). One-way analysis of variance (ANOVA) with post hoc Tukey’s test was used to compare histological measurements between groups. *p < 0.050. †p < 0.010. BE, bone erosion; BO, bone; CA, cartilage; CD, cartilage damage; IC, inflammatory cells; PG, proteoglycan; SH, synovial hyperplasia; SY, synovium.
DNA microarray and bioinformatic analysis
RNA was obtained from tarsal bones and joints, including ligaments. Tissues were immediately disrupted in liquid nitrogen using a mortar and pestle. Total RNA was purified using the RNeasy Lipid Tissue Mini Kit (Qiagen, Hilden, Germany) extraction kit, following the manufacturer's protocol, which was pooled with equal amounts of RNA from each mouse’s study group.
The microarray was carried out in the Instituto de Fisiologia Celular, Universidad Autónoma de Mexico (UNAM), Mexico. Briefly, the reverse transcription polymerase chain reaction (RT-PCR) was performed, and the resulting complementary DNA (cDNA) from group IV was labelled with Cy5 (Thermo Fisher, Waltham, Massachusetts, USA) while the cDNA from group III was labelled with Cy3 (Thermo Fisher). Hybridization was performed using a M22K_01 (UNAM, Mexico City, Mexico) chip containing 22,000 genes from the mouse genome. The scan and acquisition of the signal were developed using the ScanArray 4000 (Packard BioChips Technologies, Billerica, Massachusetts, USA). The analysis of the microarray scanning was done using GenArise Microarray Analysis Tool software (UNAM), and the lists of differentially expressed genes (DEGs) (Z-score ≥ 1.5 SD) were obtained. The DEG in greater magnitude (Z-score ≥ 3 SD) was searched for its biological processes and molecular functions using the gene database of the National Center for Biotechnology Information (Bethesda, Maryland, USA) according to Mouse Genome Informatics’ (MGI) gene ontology.
The bioinformatic analysis included the use of the platform DAVID Bioinformatics Resources 6.8 (Laboratory of Human Retrovirology and Immunoinformatics, Frederick, Maryland, USA) in order to analyze the associations of deregulated genes in biological processes and the Kyoto Encyclopedia of Genes and Genomes (KEGG) signalling pathways.21 In addition, the STRING 11.0 database22 was used to obtain the analysis and integration of direct and indirect protein-protein interactions (IPP) based on the functional associations.23 The DEGs identified in the microarray were loaded and the interactions were selected with minimal confidence (interaction score > 0.4). The obtained IPP network was analyzed to obtain primary clusters of subnetworks using the Cytoscape24 software v. 3.7.0 with the Molecular Complex Detection (MCODE) complement.25,26
Statistical analysis
The statistical analysis was carried out in SPSS Statistics v. 22.0 (IBM, Armonk, New York, USA). Measures of central tendency were estimated for each variable. A paired t-test was used to evaluate the physical performance of the mice by comparing the speed reached by the mice in MECT 1 and 2.
One-way analysis of variance (ANOVA) with post hoc Tukey’s range test was used to evaluate the effect of PE on histological parameters (inflammatory infiltrate and cartilage erosion). Statistical significance was set at p ≤ 0.05. GraphPad software (La Jolla, California, USA) was used to generate the graphic in the histological analysis.
Results
Treadmill-running performance evaluation
Exercised healthy and PGIA mice significantly improved their physical fitness. Healthy mice had a mean initial MECT of 22.1 m/min (SD 5.5) and finished with 31.5 m/min (SD 5.6) after ten weeks of exercise (41.3% increase; p = 0.047, paried t-test.), while the PGIA mice had an initial MECT of 19.6 m/min (SD 4.5) and finished with 27.7 m/min (SD 1.9) (42.5% increase; p = 0.018, paired t-test ). These data prove the effectiveness of the PE programme to improve aerobic capacity in mice.
The behaviour of the mice in both the healthy group and the arthritic group was monitored during the PE intervention (in all its phases). At the end of the intervention, the MECT and body mass of the mice were evaluated. In the familiarization stage, it was evident how the animals adapted to the treadmill. No clear differences were observed in the behaviour of the animals in the two groups, even after the arthritis induction in the PGIA mice. Likewise, in the quantitative measures, there were no statistically significant differences between the two groups at the beginning and at the end of the PE intervention in the physical test or in their body mass.
Histological analysis
The effect of PE was histologically evaluated in the hind paws of the mice using H&E and Safranin-O/Fast green staining (Figures 2b and 2c). The inflammatory joint infiltrate and cartilage erosion scores were obtained from each mouse and the mean scores were calculated for each group. Physical exercise had a differential effect on histological markers depending on the study group (Figures 2d and 2e). In the PGIA mice, PE decreased the inflammatory infiltrate and cartilage erosion. However, only the decrease in cartilage damage was statistically significant. Additionally, an increase in the amount of PG in PGIA mice due to exercise was observed by Safranin-O/Fast green staining (Figures 2b and 2c). In healthy mice, PE significantly increased the inflammatory infiltrate and there was no detectable influence of PE in the cartilage damage or in the PG content (Figures 2d and 2e).
Microarray and Bioinformatic analysis
Physical exercise induced the differential expression of 2,554 genes; 1,183 were upregulated and 1,371 were downregulated (Z-score ≥ 1.5 SD) in PGIA mice. Microarray data were deposited in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) database with the accession number GSE103686.
Using the NCBI gene database and MGI gene ontology, the DEGs in Z-Score ≥ 3 SD were classified according to their previously associated biological process and/or molecular function (Supplementary Figure a). These DEGs were mainly clustering in the biological processes of cell cycle, cell signalling, and cell metabolism. Interestingly, 11 DEGs had a previously associated immune function. Moreover, genes of relevance in joint structures such as type XI and type VI collagen showed an expression level higher than 4 SD and 3 SD, respectively.
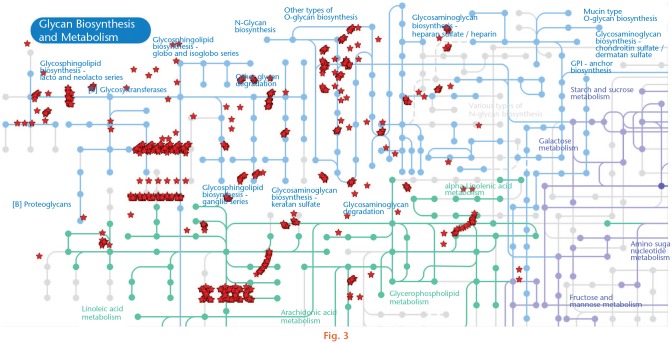
According to bioinformatics analysis in the DAVID platform, the DEGs induced by PE (Z-score ≥ 1.5 SD) had a significant association in 116 biological processes: 41 upregulated and 75 downregulated (Supplementary Table i), and in 26 KEGG signalling pathways: 11 upregulated and 15 downregulated (Table I). In this analysis, the upregulation of the metabolic pathway in which 61 genes related to the glycan biosynthesis were associated was remarkable (Figure 3).
Table I.
KEGG pathways dysregulated by physical exercise in mice with proteoglycan-induced arthritis.
| KEGG pathway (number of genes) | Genes (p-value*) |
|---|---|
| Upregulated | |
| Spliceosome (12) | Ddx39b, Thoc3, Cdc5l, Hspa1a, Hnrnpa1, Prpf18, Prpf6, Snrnp27, Snrpe, Sf3a2, Sf3a3, Smndc1 (p = 0.010)† |
| Nonalcoholic fatty liver disease (13) | Ndufa1, Ndufa12, Ndufa7, Ndufab1, Ndufc1, Ndufs3, Ndufs4, Cox8b, Cox5a, Eif2s1, Jun, Ppara, Uqcrq (p = 0.130)† |
| Oxidative phosphorylation (12) | Atp5j, Atp5l, Ndufa1, Ndufa12, Ndufa7, Ndufab1, Ndufc1, Ndufs3, Ndufs4, Cox8b, Cox5a, Uqcrq (p = 0.014)† |
| Huntington's disease (15) | Atp5j, Ndufa1, Ndufa12, Ndufa7, Ndufab1, Ndufc1, Ndufs3, Ndufs4, Cltb, Cox8b, Cox5a, Ift57, Pparg, Polr2i, Uqcrq (p = 0.015)† |
| Glutathione metabolism (7) | Anpep, Ggct, Ggt1, Gsto2, Gsta3, Gpx5, Mgst3 (p = 0.015)† |
| Parkinson's disease (12) | Atp5j, Ndufa1, Ndufa12, Ndufa7, Ndufab1, Ndufc1, Ndufs3, Ndufs4, Cox8b, Cox5a, Uqcrq, Uba1y (p = 0.022)† |
| Metabolic pathways (61) | Papss2, Atp5j, Atp5l, Coasy, Ndufa1, Ndufa12, Ndufa7, Ndufab1, Ndufc1, Ndufs3, Ndufs4, B3gnt2, Acaa1a, Acaa2, Acsm3, Ada, Anpep, Aldh1a3, Aldh1a7, Acy1, Alox12b, Arg1, Aanat, Bckdha, Cmpk1, Cyp2c68, Cyp24a1, Cox8b, Cox5a, Coq7, Dgke, Enpp1, Extl3, Galc, Ggt1, Gne, Gad2, Gpat4, Inpp1, Idh3g, Kmo, Lpcat1, Mecr, Ebp, Pigc, Pmvk, Pola2, Pold1, Pold2, Pole4, Polr2i, Prodh, Sds, Sphk1, Tyms, Tkt, Tpi1, Uqcrq, Upp1, Uro (p = 0.025)† |
| Pyrimidine metabolism (9) | Cmpk1, Entpd8, Pola2, Pold1, Pold2, Pole4, Polr2i, Tyms, Upp1 (p = 0.036)† |
| Renin-angiotensin system (5) | Atp6ap2, Anpep, Agtr2, Ctsg, Lnpep (p = 0.039)† |
| DNA replication (5) | Pola2, Pold1, Pold2, Pole4, Ssbp1 (p = 0.039)† |
| Mismatch repair (4) | Msh2, Pold1, Pold2, Ssbp1 (p = 0.046)† |
| Downregulated | |
| Haematopoietic cell lineage (14) | Cd2, Cd44, Cd59a, Gp1bb, Il11ra1, Il11, Il2rα, Il3, Il4ra, Il4, Il5, Il6ra, Kitl, Tfrc (p < 0.001)† |
| JAK-STAT signalling pathway (16) | Crebbp, Ctf1, Il11ra1, Il11, Il2rα, Il2rβ, Il3, Il4ra, Il4, Il5, Il6ra, Lif, Stam2, Socs4, Socs5, Akt (p = 0.004)† |
| Notch signalling pathway (8) | Crebbp, Aph1c, Dll3, Dtx2, Dvl2, Dvl3, Numbl, Rbpj (p = 0.008)† |
| GABAergic synapse (11) | Cacna1s, Gabra2, Gabrb1, Gabrb3, Gabrg2, Gabrg3, Gnb5, Gng12, Gng2, Gngt2, Slc32a1 (p = 0.009)† |
| Retrograde endocannabinoid signalling (12) | Cacna1s, Gabra2, Gabrb1, Gabrb3, Gabrg2, Gabrg3, Gnb5, Gng12, Gng2, Gngt2, Slc17a7, Slc32a1. (p = 0.010)† |
| Nicotine addiction (7) | Gabra2, Gabrb1, Gabrb3, Gabrg2, Gabrg3, Slc17a7, Slc32a1 (p = 0.012)† |
| Calcium signalling pathway (17) | Adra1b, Cacna1s, Camk2a, Camk2d, Camk4, Ednra, Ednrb, Erbb4, Gnal, Lhcgr, Plcd3, Plcd4, Phka1, Ptafr, P2rx4, Ryr1, Vdac1 (p = 0.013)† |
| Cytokine-cytokine receptor interaction (21) | Cd40lg, Acvr2b, Bmpr1a, Ctf1, Ccl27a, Ccr6, Cxcl12, Cxcl9, Edar, Il11ra1, Il11, Il2rα, Il2rβ, Il3, Il4ra, Il4, Il5, Il6ra, Kitl, Lif, Tnfrsf17 (p = 0.014)† |
| Signalling pathways regulating pluripotency of stem cells (14) | Bmi1, Acvr2b, Bmpr1a, Dvl2, Dvl3, Esrrb, Esx1, Fgfr2, Fzd7, Hand1, Lif, Pcgf5, Akt3 (p = 0.015)† |
| Asthma (5) | Cd40lg, H2-Aa, Il3, Il4, Il5 (p = 0.026)† |
| Lysosome (12) | Abca2, Atp6v0a2, Gnptab, Ap1g2, Ap3b1, Ap3b2, Ap3d1, Ap4m1, Galns, Lamp2, Manba, Ppt1 (p = 0.032)† |
| Bacterial invasion of epithelial cells (9) | Cd2ap, Rac1, Arhgef26, Was, Wasl, Cdh1, Ilk, Sept11, Sept3 (p = 0.033)† |
| Morphine addiction (10) | Gabra2, Gabrb1, Gabrb3, Gabrg2, Gabrg3, Gnb5, Gng12, Gng2, Gngt2, Slc32a1 (p = 0.034)† |
| Wnt signalling pathway (13) | Crebbp, Rac1, Wif1, Camk2a, Camk2d, Dvl2, Dvl3, Fosl1, Fzd7, Nkd1, Nkd2, Sfrp1, Tbl1xr1 (p = 0.038)† |
| Intestinal immune network for immunoglobin A production (6) | Cd40lg, Cxcl12, H2-Aa, Il4, Il5, Tnfrsf17 (p = 0.049)† |
Fisher’s exact test.
Significant association (p < 0.05) of differentially expressed genes (Z-score ≥ 1.5) by physical exercise in mice with proteoglycan-induced arthritis was obtained using the DAVID Bioinformatics Resources 6.8 platform (Laboratory of Human Retrovirology and Immunoinformatics, Frederick, Maryland, USA).
GABA, γ-aminobutyric acid; JAK-STAT, Janus kinase-signal transducer and activator of transcription proteins; KEGG, Kyoto Encyclopedia of Genes and Genomes; PGIA, proteoglycan-induced arthritis.
Fig. 3.
Upregulation of the metabolic pathway glycan biosynthesis and metabolism by physical exercise. The image shows the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway in which 61 upregulated genes were associated using the DAVID Bioinformatics Resources 6.8 platform (Laboratory of Human Retrovirology and Immunoinformatics, Frederick, Maryland, USA).
The biological processes resulting from the analysis in DAVID that had some association with the arthritis pathogenesis are shown in Table II. The downregulation of the immune response process was remarkable, and various interleukins, cytokines and chemokines were associated. The upregulation of the bone mineralization process could be observed where the genes of fibroblast growth factor (Fgf)-23 and osteomodulin (Omd) were included. Bone homeostasis and arthritis pathways were dysregulated. These included the canonical and non canonical Wnt pathway, as well as the bone morphogenetic proteins (BMP) and the mitogen-activated protein kinase (MAPK) pathways.
Table II.
Differentially expressed genes by physical exercise associated with biological process of interest in arthritis pathogenesis.
| Process (number of genes) | Upregulated genes | Downregulated genes |
|---|---|---|
| Negative regulation of cell proliferation (24) | Wt1, Brd7, Ctnnb1, Cdc73, Cer1, Fntb, Fth1, Fgfr3, Hspa1a, Ing1, Insm1, Jarid2, Jun, Lbx1, Myocd, Pth1r, Pparg, Ppp1r15a, Rara, Slfn3, Tob2, Tob1, Trim35, Twist2 | N/A |
| Positive regulation of cell proliferation (43) | N/A | Cnot7, Cd81, E2f3, Gli1, Mlxipl, Rasip1, Sox11, Tiam1, Adcyap1, Akr1c18, Calr, Cep131, Cxcl12, Cul4a, Ddr2, Ednra, Ednrb, Erbb4, Fer, Fgf15, Fgfr2, Folr2, Hmga2, Insr, Itgav, Ilk, Il11ra1, Il11, Il3, Il4, Il5, Il6ra, Kitl, Lif, Nudt16, Prox1, Romo1, Rbpj, Rarb, Rarg, Runx2, Sfrp1, Tet1 |
| Cell differentiation (46) | Abcb5, Dazap1, Lhx3, Rorc, Racgap1, Thoc5, Zpr1, Anpep, Asz1, Ctnnb1, Catsper1, Cdx2, Cdx4, Cdc5l, Chrdl1, Cand1, Cyfip1, Dab1, Dlx2, Fgf23, Fgfr3, Gmcl1, Gadd45g, Gap43, Insm1, Jarid2, Lbx1, Myod1, Myo7b, Nedd4l, Neurod6, Odf3, Sema4d, Sema5a, Sema6a, Srrm4, Slc22a16, Spata24, Spata5, Sra1, Sdc2, Tll1, Tlk2, Tnp2, Tnfsf11, Twist2 | N/A |
| Osteoblast differentiation (10) | N/A | Gli1, Pcp4, Rbmx, Fignl1, Lgr4, Rdh14, Runx2, Sfrp1, Trp53inp2, Vcan |
| Bone mineralization (4) | Fgf23, Ifitm5, Matn1, Omd | N/A |
| Positive regulation of Wnt signalling pathway (5) | Atp6ap2, Smarca4, Cdc73, Dvl1, Mbd2 | N/A |
| Wnt signalling pathway (17) | N/A | Bcl7b, Cd44, Cyld, Rtf1, Wif1, Amotl1, Dvl2, Dvl3, Fzd7, Hbp1, Invs, Kremen1, Lgr4, Nkd1, Nkd2, Peg12, Sfrp1 |
| Non-canonical Wnt signalling pathway (4) | N/A | Rac1, Dvl2, Dvl3, Fzd7 |
| Negative regulation of canonical Wnt signalling pathway (10) | N/A | Cyld, Gli1, Wif1, Cdh1, Egr1, Invs, Nkd1, Nkd2, Pfdn5, Sfrp1 |
| Negative regulation of BMP signalling pathway (6) | Cer1, Chrdl1, Fstl3, Gdf3, Sostdc1, Tob1 | N/A |
| Positive regulation of MAPK cascade (10) | N/A | Gpr37l1, Traf7, Adra1b, Fgfr2, Insr, Itgav, Ilk, Il11, Il6ra, Lif |
| Immune response (22) | N/A | Clec4e, Cd40lg, Was, Bmpr1a, Xcl1, Ccl27a, Ccr6, Cxcl12, Cxcl9, Hfe, H2-Q5, H2-Aa, Iltifb, Il2rα, Il3, Il4, Il5, Lif, Nfil3, Psg17, Prg3, Tlr6 |
Significant association (p < 0.05) of differentially expressed genes (Z-score ≥ 1.5) by physical exercise in PGIA-mice were obtained using the DAVID Bioinformatics Resources 6.8 platform.
BMP, bone morphogenic proteins; MPAK, mitogen-activated protein kinase; N/A, no associations.
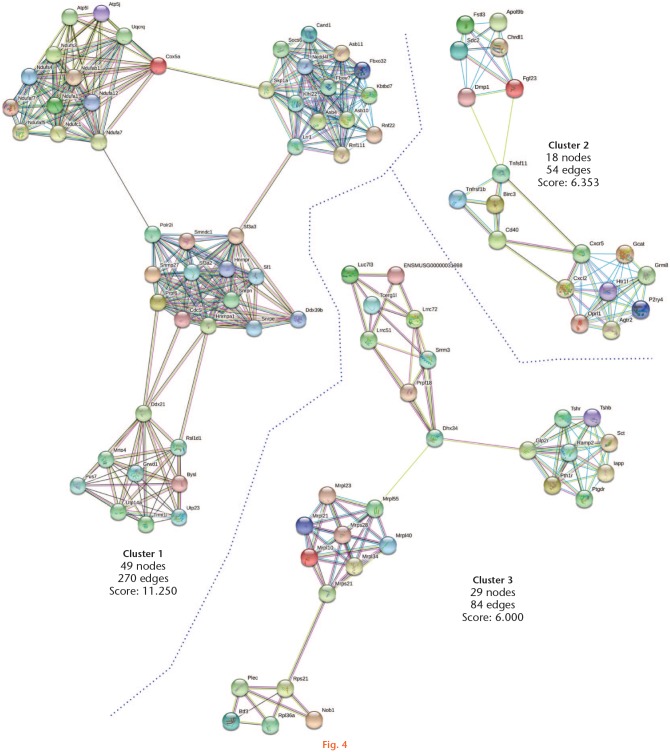
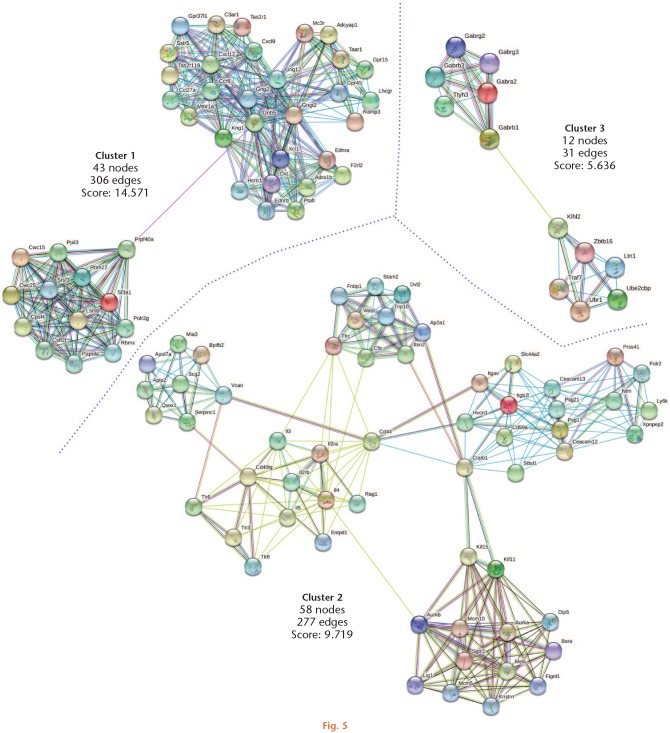
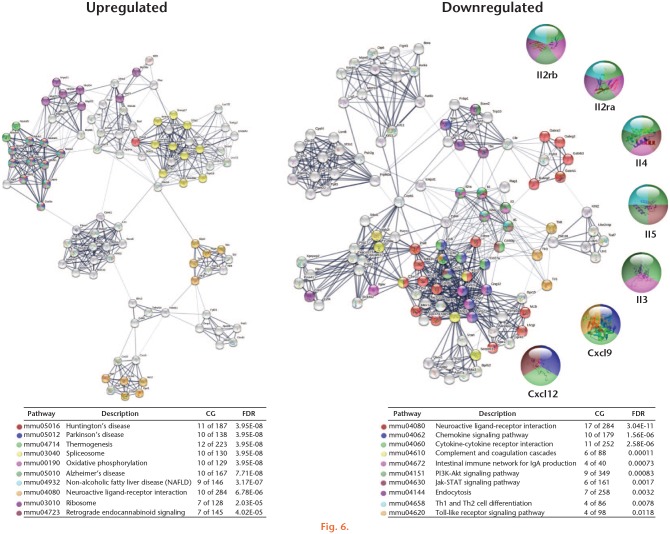
We analyzed the primary clusters of protein subnetworks for upregulated and downregulated genes using STRING and Cytoscape platforms. Figures 4 and 5 show the networks of the first three clusters obtained with the MCODE complement for both the upregulated and downregulated genes, respectively. The genes from the first three clusters were loaded in STRING to identify the associated KEGG signalling pathways. Those relevant to arthritis were selected and marked with different colours (Figure 6). The pathways considered as relevant included cytokine-cytokine receptor interaction, the Janus kinase-signal transducer and activator of transcription proteins (JAK-STAT) signalling pathway, and the chemokine signalling pathway. The analysis of the protein networks plausibly linked to arthritis pathogenesis eventually unveiled candidate proteins which showed multiple functional connections as crossroads in these processes and turned out to be the inflammatory mediators. These proteins included interleukin (IL) 3, IL4, IL5, IL2 receptor alpha (IL2rα), IL2 receptor beta (IL2rβ), chemokine ligand (CXCL) 9, and CXCL12. In every case, they were downregulated by the effect of PE.
Fig. 4.
Protein-protein interactions of upregulated genes by physical exercise. Upregulated genes (Z-score ≥ 1.5 SD) were analyzed on the STRING and Cytoscape platforms. The primary clusters of subnetworks were obtained using the Molecular Complex Detection (MCODE) complement (cut-off = 0.3). Line colour indicates the type of interaction evidence; coloured nodes indicate query proteins and first shell of interactors; white nodes indicate second shell of interactors.
Fig. 5.
Protein-protein interactions of downregulated genes by physical exercise. Downregulated genes (Z-score ≥ 1.5 SD) were analyzed on the STRING and Cytoscape platforms. The primary clusters of subnetworks were obtained using the Molecular Complex Detection (MCODE) complement (cut-off = 0.3). Line colour indicates the type of interaction evidence; coloured nodes indicate query proteins and first shell of interactors; white nodes indicate second shell of interactors.
Fig. 6.
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways associated with protein-protein interactions of genes dysregulated by physical exercise. The set of genes present in the first three clusters for upregulated and downregulated genes were analyzed in the STRING database that showed the associated KEGG signalling pathways. The signalling pathways of relevance in arthritis were selected and marked with different colours. Line thickness indicates the strength of data support. CG, count in gene set; FDR, false discovery rate.
Discussion
The influence that PE can exert on patients with arthritis remains a potential and relevant research topic. Exercise is heterogeneous in its multiple modalities and so are its effects on different physiological processes. The effect of exercise could differ in the same patient if exerted at times of different disease activity, as well as in patients with preserved joints at early stages, as opposed to those with chronic changes. Using microarray methodology, our study explores and provides descriptive data of the effect of PE on the differential gene expression and histological changes in the tarsal bones and joints of PGIA mice. Our data show that low- to moderate-intensity exercise can decrease joint inflammation while promoting anabolism on cartilage and improving the physical fitness of PGIA mice.
Studies assessing the effects of PE on joint structures in animal models of inflammatory arthropathies are scarce and focus mostly on osteoarthritis (OA).27–29 The present study provides an overview of the PE-induced transcriptional modifications in a model of inflammatory arthropathy as it is in the PGIA model. Our findings may be relevant to human patients as they demonstrate a potential positive effect of PE in reducing the joint inflammation and define molecular pathways which explain this effect.
The immune system can be modulated by PE, including both the innate and adaptive immune responses.30 However, its influence can be defined by several parameters, including its type, intensity, and volume.31,32 Our bioinformatic analysis showed that the treadmill PE downregulated immune response genes, among them chemokines and cytokines. Several downregulated genes, including IL3, IL4, IL2rα and IL2rβ, were associated with the JAK-STAT, chemokine and cytokine-cytokine receptor interaction signalling pathways. All of them have previously been linked directly to the pathogenesis of several forms of human arthritis.33,34
The anti-inflammatory effect of PE has been previously recognized in humans and animal models.35,36 Findings similar to ours have been reported in the monoiodoacetate-induced arthritis (MIA) model in rats, where treadmill PE suppresses genes associated with the inflammation process in the affected knees, especially in the early stages of arthritis.37
The results in the microarray were confirmed by the comparison of the histological findings among the groups. Exercised PGIA mice had lower joint inflammation scores. Similar findings have been reported in the adjuvant-induced arthritis (AIA) model in rats,38,40 where PE reduced the synovial leukocyte count39 and decreased oedema and cell migration.40
Mechanical loading, including PE, induces bone formation particularly in the load-bearing areas.41 In our study, the treadmill PE dysregulated biological processes which are relevant to bone homeostasis including Wnt, BMP, and MAPK signalling.42–45 Wnt signalling has been implicated in several arthropathies, including rheumatoid arthritis,46 osteoarthritis,47 and ankylosing spondylitis.48,49 The link between inflammation and mechanical demand by PE on this signalling has not been previously described. Our study shows that PE in an inflammatory scenario can upregulate and downregulate genes of the Wnt pathway at the level of ligands, transducers, and transcripts, unveiling the complexity of its potential effects.
Physical exercise also modulates the physiological turnover of the cartilage components, including PGs.50,51 In our study, PE upregulated genes in the signalling pathway for ECM biosynthesis, including glycerophospholipids, glycoesphingolipids, PGs, glycosaminoglycans, and linoleic acid derivatives. Moreover, the histological analysis showed an increase in the number of PGs, measured by Safranin-O staining, in response to PE. Similar results have been demonstrated in the MIA model in rats.37 These findings provide evidence of the relevance of PE to preserve and stimulate cartilage integrity.
The characteristics of the training programme influence the magnitude of the skeletal muscle and cardiovascular and integrative adaptations to PE.52 The PE programme designed for our study allowed an increase of about 40% of the physical fitness of both healthy and PGIA mice. At the transcriptome level, the electron transport chain signalling pathway and the oxidative phosphorylation were found to be upregulated by PE, evidencing an increase of aerobic metabolism. Although we cannot contrast these findings with studies in animal models of inflammatory arthropathies, studies in humans with different types of arthritis have shown that despite the pathological process, PE improves respiratory capacity.53,54
In conclusion, the present study explored the genetic and histological parameters of arthropathies and PE that have not yet been thoroughly researched in animal models. Our results describe an exploratory and preliminary scenario in which PE influences parameters intimately linked to inflammatory arthropathies, including immune and inflammatory response, remodelling of ECM, bone metabolism, and physical fitness, among others. Further research on the effect of PE on the pathogenesis process of arthritis is still necessary for animal models and humans. Clarifying the mechanisms by which PE modulates an established inflammatory process could contribute to the design of PE programmes with greater effectiveness for patients, an aspect that to date has not been established.
Acknowledgments
The authors thank Lorena Chávez, José Luis Santillán, Simón Guzmán, and Jorge Ramírez for the microarray analysis. Likewise, the authors thank Rogelio González and Adrián Chávez for their support in the design and construction of the treadmill and software used in this research.
Footnotes
Author contributions: S. A. González-Chávez: Performed the experiment, Carried out the histological, microarray and bioinformatic analysis, Wrote the manuscript.
C. Pacheco-Tena: Carried out the research, Obtained the funding, Carried out the histological analysis, Analyzed the data, Wrote the manuscript.
C. M. Quiñonez-Flores: Performed the experiment, Carried out the microarray analysis, Revised the manuscript.
G. P. Espino-Solis: Analyzed the data, Revised the manuscript.
J. I. Burrola-De Anda: Performed the experiment.
P. M. Muñoz-Morales: Performed the experiment.
Ethical review statement: The entire protocol, including manipulation of the animals, complied with the institutional ethics committee and Institutional Animal Care and Use Committee (IACUC), ID number: FM-FM-B-276/14.
Follow us @BoneJointRes
Supplementary Material
Figure showing the classification of genes by previously associated biological process and/or molecular function and table showing bioinformatics analysis in the DAVID platform.
Funding statement
No benefits in any form have been received or will be received from a commercial party related directly or indirectly to the subject of this article.
References
- 1. Kiltz U, van der Heijde D. Health-related quality of life in patients with rheumatoid arthritis and in patients with ankylosing spondylitis. Clin Exp Rheumatol. 2009;27(4)(Suppl 55):S108-S111. [PubMed] [Google Scholar]
- 2. Cantini F, Niccoli L, Nannini C, et al. Second-line biologic therapy optimization in rheumatoid arthritis, psoriatic arthritis, and ankylosing spondylitis. Semin Arthritis Rheum. 2017;47(2):183-192. [DOI] [PubMed] [Google Scholar]
- 3. Scarvell J, Elkins MR. Aerobic exercise is beneficial for people with rheumatoid arthritis. Br J Sports Med. 2011;45(12):1008-1009. [DOI] [PubMed] [Google Scholar]
- 4. Barker AL, Talevski J, Morello RT, Brand CA, Rahmann AE, Urquhart DM. Effectiveness of aquatic exercise for musculoskeletal conditions: a meta-analysis. Arch Phys Med Rehabil. 2014;95(9):1776-1786. [DOI] [PubMed] [Google Scholar]
- 5. Thomas JL. Helpful or harmful? Potential effects of exercise on select inflammatory conditions. Phys Sportsmed. 2013;41(4):93-100. [DOI] [PubMed] [Google Scholar]
- 6. Agca R, Heslinga SC, Rollefstad S, et al. EULAR recommendations for cardiovascular disease risk management in patients with rheumatoid arthritis and other forms of inflammatory joint disorders: 2015/2016 update. Ann Rheum Dis. 2017;76(1):17-28. [DOI] [PubMed] [Google Scholar]
- 7. Sieper J, Rudwaleit M, Baraliakos X, et al. The Assessment of SpondyloArthritis international Society (ASAS) handbook: a guide to assess spondyloarthritis. Ann Rheum Dis. 2009;68(Suppl 2):ii1-ii44. [DOI] [PubMed] [Google Scholar]
- 8. O’Dwyer T, O’Shea F, Wilson F. Exercise therapy for spondyloarthritis: a systematic review. Rheumatol Int. 2014;34(7):887-902. [DOI] [PubMed] [Google Scholar]
- 9. Baillet A, Zeboulon N, Gossec L, et al. Efficacy of cardiorespiratory aerobic exercise in rheumatoid arthritis: meta-analysis of randomized controlled trials. Arthritis Care Res (Hoboken). 2010;62(7):984-992. [DOI] [PubMed] [Google Scholar]
- 10. Millner JR, Barron JS, Beinke KM, et al. Exercise for ankylosing spondylitis: an evidence-based consensus statement. Semin Arthritis Rheum. 2016;45(4):411-427. [DOI] [PubMed] [Google Scholar]
- 11. Braem K, Lories RJ. Insights into the pathophysiology of ankylosing spondylitis: contributions from animal models. Joint Bone Spine. 2012;79(3):243-248. [DOI] [PubMed] [Google Scholar]
- 12. Alves CH, Farrell E, Vis M, Colin EM, Lubberts E. Animal Models of Bone Loss in Inflammatory Arthritis: from Cytokines in the Bench to Novel Treatments for Bone Loss in the Bedside-a Comprehensive Review. Clin Rev Allergy Immunol. 2016;51(1):27-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Glant TT, Mikecz K. Proteoglycan aggrecan-induced arthritis: a murine autoimmune model of rheumatoid arthritis. Methods Mol Med. 2004;102:313-338. [DOI] [PubMed] [Google Scholar]
- 14. Glant TT, Mikecz K, Arzoumanian A, Poole AR. Proteoglycan-induced arthritis in BALB/c mice. Clinical features and histopathology. Arthritis Rheum. 1987;30(2):201-212. [DOI] [PubMed] [Google Scholar]
- 15. Kurkó J, Besenyei T, Laki J, Glant TT, Mikecz K, Szekanecz Z. Genetics of rheumatoid arthritis - a comprehensive review. Clin Rev Allergy Immunol. 2013;45(2):170-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tseng H-W, Pitt ME, Glant TT, et al. Inflammation-driven bone formation in a mouse model of ankylosing spondylitis: sequential not parallel processes. Arthritis Res Ther. 2016;18:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cigudosa JC. The microarray revolution in biomedical research: types of platforms, uses and perspectives in oncology. An Sist Sanit Navar. 2004;27(1):11-20. (Article in Spanish) [DOI] [PubMed] [Google Scholar]
- 18. Ishikawa LLW, Colavite PM, da Rosa LC, et al. Commercial bovine proteoglycan is highly arthritogenic and can be used as an alternative antigen source for PGIA model. Biomed Res Int. 2014;2014:148594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kregel KC, Allen DL, Booth FW, et al. Resource Book for the Design of Animal Exercise Protocols. American Physiological Society; 2006. [Google Scholar]
- 20. González-Chávez SA, Pacheco-Tena C, Macías-Vázquez CE, Luévano-Flores E. Assessment of different decalcifying protocols on Osteopontin and Osteocalcin immunostaining in whole bone specimens of arthritis rat model by confocal immunofluorescence. Int J Clin Exp Pathol. 2013;6(10):1972-1983. [PMC free article] [PubMed] [Google Scholar]
- 21. Huang DW, Sherman BT, Tan Q, et al. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35(Web Server issue):W169-W175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jensen LJ, Kuhn M, Stark M, et al. STRING 8—a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37(Database issue):D412-D416. doi: 10.1093/nar/gkn760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Szklarczyk D, Morris JH, Cook H, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45(D1):D362-D368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498-2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Isserlin R, Merico D, Voisin V, Bader GD. Enrichment Map – a Cytoscape app to visualize and explore OMICs pathway enrichment results. F1000Res. 2014;3:141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bader GD, Hogue CWV. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 2003;4:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Iijima H, Aoyama T, Ito A, et al. Exercise intervention increases expression of bone morphogenetic proteins and prevents the progression of cartilage-subchondral bone lesions in a post-traumatic rat knee model. Osteoarthritis Cartilage. 2016;24(6):1092-1102. [DOI] [PubMed] [Google Scholar]
- 28. Blazek AD, Nam J, Gupta R, et al. Exercise-driven metabolic pathways in healthy cartilage. Osteoarthritis Cartilage. 2016;24(7):1210-1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu S-S, Zhou P, Zhang Y. Abnormal expression of key genes and proteins in the canonical Wnt/β-catenin pathway of articular cartilage in a rat model of exercise-induced osteoarthritis. Mol Med Rep. 2016;13(3):1999-2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Walsh NP, Gleeson M, Shephard RJ, et al. Position statement. Part one: immune function and exercise. Exerc Immunol Rev. 2011;17:6-63. [PubMed] [Google Scholar]
- 31. Malm C. Exercise immunology: a skeletal muscle perspective. Exerc Immunol Rev. 2002;8:116-167. [PubMed] [Google Scholar]
- 32. Malm C. Exercise immunology: the current state of man and mouse. Sports Med. 2004;34(9):555-566. [DOI] [PubMed] [Google Scholar]
- 33. Villarino AV, Kanno Y, Ferdinand JR, O’Shea JJ. Mechanisms of Jak/STAT signaling in immunity and disease. J Immunol. 2015;194(1):21-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shang Y, Smith S, Hu X. Role of Notch signaling in regulating innate immunity and inflammation in health and disease. Protein Cell. 2016;7(3):159-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gleeson M, Bishop NC, Stensel DJ, Lindley MR, Mastana SS, Nimmo MA. The anti-inflammatory effects of exercise: mechanisms and implications for the prevention and treatment of disease. Nat Rev Immunol. 2011;11(9):607-615. [DOI] [PubMed] [Google Scholar]
- 36. Petersen AMW, Pedersen BK. The anti-inflammatory effect of exercise. J Appl Physiol (1985). 2005;98(4):1154-1162. [DOI] [PubMed] [Google Scholar]
- 37. Nam J, Perera P, Liu J, et al. Transcriptome-wide gene regulation by gentle treadmill walking during the progression of monoiodoacetate-induced arthritis. Arthritis Rheum. 2011;63(6):1613-1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gomes RP, Bressan E, Silva TM, Gevaerd Mda S, Tonussi CR, Domenech SC. Standardization of an experimental model suitable for studies on the effect of exercise on arthritis. Einstein (Sao Paulo). 2013;11(1):76-82. (Article in English, Portuguese) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gomes RP, Bressan E, da Silva TM, Domenech SC, Tonussi CR. Evidências de que um protocolo de atividade física pode reduzir a contagem de leucócitos sinoviais de ratos artríticos. Rev Bras Med Esporte. 2013;19(1):70-73. [Google Scholar]
- 40. Gomes RP, Bressan E, Silva TM, Gevaerd Mda S, Tonussi CR, Domenech SC. Effects of one minute and ten minutes of walking activity in rats with arthritis induced by complete Freund’s adjuvant on pain and edema symptoms. Rev Bras Reumatol. 2014;54(2):83-89. (Article in English, Portuguese) [PubMed] [Google Scholar]
- 41. Heinemeier KM, Olesen JL, Haddad F, et al. Expression of collagen and related growth factors in rat tendon and skeletal muscle in response to specific contraction types. J Physiol. 2007;582(Pt 3):1303-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kjaer M, Jørgensen NR, Heinemeier K, Magnusson SP. Exercise and Regulation of Bone and Collagen Tissue Biology. In: Teplow DB, Ed. Progress in Molecular Biology and Translational Science. Vol. 168 Elsevier; 2015:259-291. [DOI] [PubMed] [Google Scholar]
- 43. Galli C, Passeri G, Macaluso GM. Osteocytes and WNT: the mechanical control of bone formation. J Dent Res. 2010;89(4):331-343. [DOI] [PubMed] [Google Scholar]
- 44. Yu J, Virshup DM. Updating the Wnt pathways. Biosci Rep. 2014;34(5):e00142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kopf J, Paarmann P, Hiepen C, Horbelt D, Knaus P. BMP growth factor signaling in a biomechanical context. Biofactors. 2014;40(2):171-187. [DOI] [PubMed] [Google Scholar]
- 46. Miao CG, Yang YY, He X, et al. Wnt signaling pathway in rheumatoid arthritis, with special emphasis on the different roles in synovial inflammation and bone remodeling. Cell Signal. 2013;25(10):2069-2078. [DOI] [PubMed] [Google Scholar]
- 47. Sassi N, Laadhar L, Allouche M, et al. WNT signaling and chondrocytes: from cell fate determination to osteoarthritis physiopathology. J Recept Signal Transduct Res. 2014;34(2):73-80. [DOI] [PubMed] [Google Scholar]
- 48. Xie W, Zhou L, Li S, Hui T, Chen D. Wnt/β-catenin signaling plays a key role in the development of spondyloarthritis. Ann N Y Acad Sci. 2016;1364:25-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. González-Chávez SA, Quiñonez-Flores CM, Pacheco-Tena C. Molecular mechanisms of bone formation in spondyloarthritis. Joint Bone Spine. 2016;83(4):394-400. [DOI] [PubMed] [Google Scholar]
- 50. Kjaer M. Role of extracellular matrix in adaptation of tendon and skeletal muscle to mechanical loading. Physiol Rev. 2004;84(2):649-698. [DOI] [PubMed] [Google Scholar]
- 51. Hae Yoon J, Brooks R, Hwan Kim Y, Terada M, Halper J. Proteoglycans in chicken gastrocnemius tendons change with exercise. Arch Biochem Biophys. 2003;412(2):279-286. [DOI] [PubMed] [Google Scholar]
- 52. MacInnis MJ, Gibala MJ. Physiological adaptations to interval training and the role of exercise intensity. J Physiol. 2017;595(9):2915-2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hsieh LF, Wei JCC, Lee HY, Chuang CC, Jiang JS, Chang KC. Aerobic capacity and its correlates in patients with ankylosing spondylitis. Int J Rheum Dis. 2016;19(5):490-499. [DOI] [PubMed] [Google Scholar]
- 54. Jennings F, Oliveira HA, de Souza MC, Cruz V, da G, Natour J. Effects of Aerobic Training in Patients with Ankylosing Spondylitis. J Rheumatol. 2015;42(12):2347-2353. [DOI] [PubMed] [Google Scholar]