Abstract
Background:
Most of Gastric Cancer (GC) patients are diagnosed at an advanced stage with poor prognosis. Hypermethylations of several tumor suppressor genes in cell-free DNA of GC patients have been previously reported. In this study, an attempt was made to investigate the methylation status of P16, RASSF1A, RPRM, and RUNX3 and their potentials for early diagnosis of GC.
Methods:
Methylation status of the four tumor suppressor genes in 96 plasma samples from histopathologically confirmed gastric adenocarcinoma patients (Stage I–IV) and 88 healthy controls was determined using methylation-specific PCR method. Receiver operating characteristic curve analysis was performed and Area Under the Curve (AUC) was calculated. Two tailed p<0.05 were considered statistically significant.
Results:
Methylated P16, RASSF1A, RPRM, and RUNX3 were significantly higher in the GC patients (41.7, 33.3, 66.7, and 58.3%) compared to the controls (15.9, 0.0, 6.8, and 4.5%), respectively (p<0.001). Stratification of patients showed that RPRM (AUC: 0.70, Sensitivity: 0.47, Specificity: 0.93, and p<0.001) and RUNX3 (AUC: 0.77, Sensitivity: 0.59, Specificity: 0.95, and p<0.001) had the highest performances in detection of early-stage (I+II) GC. The combined methylation of RPRM and RUNX3 in detection of early-stage GC had a higher AUC of 0.88 (SE=0.042; 95% CI:0.793–0.957; p<0.001), higher sensitivity of 0.82 and reduced specificity of 0.89.
Conclusion:
Methylation analysis of RPRM and RUNX3 in circulating cell free-DNA of plasma could be suggested as a potential biomarker for detection of GC in early-stages.
Keywords: Biomarkers, Cell-free DNA, Gastric cancer, DNA methylation
Introduction
Gastric Cancer (GC) is the third leading cause of cancer-related mortality globally 1. Diagnosis of early-stage GC is still difficult, and in fact, most of them are diagnosed at an advanced stage with a poor prognosis 2. Currently, invasive endoscopy followed by pathological diagnosis is the gold standard for GC diagnosis 3. Single-lesion tumor-biopsy could not reflect the tumor heterogeneity, which could result in the treatment failure and drug resistance 4. In addition, the low sensitivity and specificity of available blood biomarkers are not satisfactory for early diagnosis of GC 2.
Epigenetic changes, including aberrant DNA methylation, are common in all types of cancers including gastrointestinal, and contribute to both initiation of cancer and progression 5. Deregulation of epigenetic modifications may actually even precede classical genetic changes in various oncogenes and tumor suppressor genes 6. Aberrant DNA methylation is not just a feature of advanced-stage, but also an early and driver event in GC 7, and could be non-invasively identified in cell-free DNA (cfDNA) of cancer patients 5.
Aberrant methylation of several tumor suppressor genes including RUNX3, P16, RASSF1A, ZIC1, RPRM, CDH1, and SOX17 as potential biomarkers for early detection of GC has been identified 8. However, to become a clinically approved test, a potential biomarker should be confirmed and validated in inter- and intra-laboratory studies using hundreds of specimens 9. The first step in finding a biomarker usually begins with studies of tumor tissues and non-tumor tissues 10. Previous studies showed that the methylation of P16 11,12, RASSF1A 13, RPRM 14, and RUNX3 11 was significantly higher in primary GC tissues compared to the corresponding normal gastric tissues.
P16, a cell cycle regulator, controls the G1 phase of cell cycle to S phase, and inhibits CDK4 and CDK6 15. RASSF1A, a putative tumor suppressor gene, plays an important role in regulation of cell cycle, apoptosis, and microtubule stability through the regulation of Ras signaling 16. RPRM in response to p53 expression arrests cell cycle at G2/M, and its expression is inversely associated with the cell proliferation and growth in GC 17. RUNX3 is a tumor suppressor gene considered as a downstream effector of the TGF-β signaling pathway 18.
Previous studies using the serum or plasma samples have shown that methylation of the RASSF1A 19, RUNX3 20, RPRM 21,22, and P16 23, as potential diagnostic biomarkers, could be suggested for early detection of GC. In this study, an attempt was made to investigate and validate the potential of these tumor suppressor genes as diagnostic biomarkers in the plasma of GC patients and normal controls.
Materials and Methods
Subjects
Altogether, 184 plasma samples from 88 normal healthy controls, and 96 histopathologically confirmed gastric adenocarcinoma patients with various Tumor-Node-Metastasis (TNM) stages (I–IV) were collected. The plasma samples were collected prior to surgery, chemotherapy, and/or radiotherapy. Participants were enrolled from the Department of Surgery, Cancer Institute, Tehran University of Medical Sciences, Tehran, Iran. The patients were followed-up until death or to the end of the study. The study was approved by the ethics committee of the Tehran University of Medical Sciences (Ethics code: IR.TUMS.VCR.REC.1395. 1078), and written informed consent was obtained from all patients.
DNA extraction from plasma samples
From each participant, 5 ml of peripheral blood was collected in 200 uL of 0.5 M EDTA. To separate the plasma, the blood samples were immediately centrifuged at 3000×g for 10 min at 4°C. The plasma was collected and transferred to new tubes and stored at −80°C. Circulating cfDNAs was extracted from 2 ml of plasma samples by the QIAamp Circulating Nucleic Acid Kit (Qiagen, Germany) according to the manufacture’s protocol. The concentrations of cfDNAs were measured using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). For all samples, before proceeding to sodium bisulfite conversion step, the accuracy of cfDNAs extractions were assessed by amplifying TBP (TATA-binding protein) gene using the forward 5′-CACAGACTCTCACA ACTGCAC-3′ and reverse primer 5′-ACAATCCCAG AACTCTCCGTAG-3′. The 115 bp PCR products of the TBP housekeeping gene were amplified in all cf- DNAs extracted in the plasma of GC patients as well as control samples.
Sodium bisulfite conversion
For each sample, 40 uL of cfDNA was modified with sodium bisulfite using the EpiTect Bisulfite Kit (Qiagen, Germany) according to the manufacture’s protocol of sodium bisulfite conversion of unmethylated cytosines in DNA from low-concentration solutions. Purified modified DNA was eluted in 30 μL of DDW and sorted at −80°C until use.
Methylation specific PCR
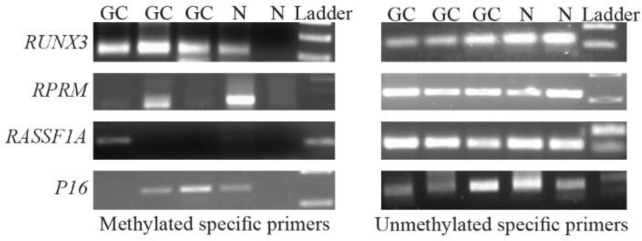
The methylation status of the P16, RASSF1A, RPRM, and RUNX3 promoters in cfDNA samples was detected by conventional methylation specific PCR (MSP) by specific primer pairs for both the methylated and unmethylated status (Table 1). Each MSP reaction was performed in a total volume of 12 uL. Briefly, 2 uL of sodium bisulfite converted DNA was added into a reaction mixture containing 6 uL of 2x master mix (Ampliqon, Denmark), and 1 uL of the corresponding forward and reverse primers (10 uM); finally ddw was added to a final volume of 12 uL. Amplification conditions for both methylated and unmethylated reactions were as follows: an initial denaturation of 95°C for 5 min, followed by 45 cycles of 95°C for 35 s, annealing temperature of 57–67°C for 35 s, and extension of 72°C for 35 s, and finally an extension of 72°C for 10 min. The MSP products were electrophoresed on 2% aga-rose gels, stained with ethidium bromide, and visualized under UV light (Figure 1).
Table 1.
Methylated and unmethylated specific primer pairs used in methylation specific PCR
| Gene | Forward primer 5′→3′ | Reverse primer 5′→3′ | Tm F/R | Product length (bp) |
|---|---|---|---|---|
| P16 | ||||
| Met | ATTAGAGGGTGGGGCGGATCGC | ACCCCGAACCGCGACCGTAA | 67/67 | 147 |
| Unmet | ATTAGAGGGTGGGGTGGATTGT | CAACCCCAAACCACAACCATAA | 61/60 | 149 |
| RASSF1A | ||||
| Met | GTTGGTATTCGTTGGGCGC | AACTACCGTATAAAATTACACGCG | 59/58 | 102 |
| Unmet | GGAGTTGGTATTTGTTGGGTGT | ACCAACTACCATATAAAATTACACACA | 58/57 | 108 |
| RPRM | ||||
| Met | TGCGAGTGAGCGTTTAGTTC | CTAATTACCTAAAACCGAATTCATCG | 58/58 | 120 |
| Unmet | AGTTTGTGAGTGAGTGTTTAGTTT | ATCTAATTACCTAAAACCAAATTCATCA | 57/57 | 126 |
| RUNX3 | ||||
| Met | ATAATAGCGGTCGTTAGGGCGTCG | GCTTCTACTTTCCCGCTTCTCGCG | 65/66 | 115 |
| Unmet | ATAATAGTGGTTGTTAGGGTGTTG | ACTTCTACTTTCCCACTTCTCACA | 57/59 | 115 |
The underlined nucleotides indicated the CpG sites.
Figure 1.
Agarose gel electrophoresis of methylation specific PCR products for RUNX3, RPRM, RASSF1A, and P16 in the plasma samples of gastric cancer (GC) patients and the controls (N). Met: Methylated-specific primers; UnMet: Unmethylated-specific primers. RUNX3 Met (115 bp); RUNX3 Unmet (115 bp); RPRM Met (120 bp); RPRM Unmet (126 bp); RASSF1A Met (102 bp); RASSF1A Unmet (108 bp); P16 Met (147 bp); P16 Unmet (149 bp).
Statistical analysis
Statistical analysis of the data was performed using the SPSS version 16.0 (SPSS Inc, Chicago, IL). The methylation status and the other qualitative variables were expressed as frequencies and percentages. Continuous variables were compared by Student’s t test, while categorical data were checked by Chi-square or Fisher’s exact tests where appropriate. Receiver Operating Characteristic (ROC) curve and the Area Under the Curve (AUC) were used to assess the performance-of the biomarkers and the higher AUCs were considered as better diagnostic performance. A logistic regression model was performed to evaluate the diagnostic performance of the combination of the biomarkers. Survival rates were calculated with the Kaplan-Meier method and the statistical difference between survival curves was determined with log-rank test. Two tailed p-value <0.05 was considered statistically significant.
Results
Descriptive analysis of the subjects
The mean age of GC patients and healthy controls were 59.5±12.3 and 56.1±11.3, respectively. The female to male gender ratio in the control and GC groups were 26/62 and 34/62, respectively. No significant difference was found in the distribution of age (p=0.052) and gender (p=0.434) among the GC patients and the normal healthy controls. In the GC group, the ratio of males (n=62) to females (n=34) was nearly twice. Classification of the GC patients according to the TNM classification showed that 35.4% (34/96) and 64.6% (62/96) of the tumors were early (I+II) and advanced-stages (III+IV), respectively.
Methylation rates and performances of the candidate genes
The P16, RASSF1A, RPRM, and RUNX3 promoters were found to be methylated in 40 (41.7%), 32 (33.3%), 64 (66.7%), and 56 (58.3%) of the 96 GC samples, respectively. Alternatively, the P16, RASSF1A, RPRM, and RUNX3 promoters were found to be methylated in 14 (15.9%), 0 (0.0%), 6 (6.8%) and 4 (4.5%) of the 88 control samples, respectively (Table 2). Unmethylated-specific primers for the P16, RASSF1A, RPRM, and RUNX3 were amplified in all subjects. There was no significant association between methylation of four candidate genes and gender. In addition, stratification of subjects by ages (≤60 and >60) showed that there were no significant association between methylation of RASSF1A, RPRM, and RUNX3 and ages. However, methylated P16 was significantly higher in the subjects over 60 years old compared to the subjects under 60 years old (p=0.006).
Table 2.
Methylation frequencies of the P16, RASSF1A, RPRM, and RUNX3 in the subjects and their performances in detection of gastric cancer with various stages
| Gene | TNM stage | Gastric cancer (n=96) | Controls (n=88) | p-value | AUC | S | Sp | PPV | NPV | Accuracy |
|---|---|---|---|---|---|---|---|---|---|---|
| Met (%) | Met (%) | |||||||||
| P16 | ||||||||||
| I+II | 12/34 (35.3%) | 14/88 (15.9%) | 0.026 | 0.60 | 0.35 | 0.84 | 0.46 | 0.77 | 0.71 | |
| III+IV | 28/62 (45.2%) | p<0.001 | 0.65 | 0.45 | 0.84 | 0.67 | 0.69 | 0.68 | ||
| I–IV | 40/96 (41.7%) | p<0.001 | 0.63 | 0.42 | 0.84 | 0.74 | 0.57 | 0.62 | ||
| RASSF1A | ||||||||||
| I+II | 8/34 (23.5%) | 0/88 | p<0.001 * | 0.62 | 0.24 | 1.0 | 1.0 | 0.77 | 0.79 | |
| III+IV | 24/62 (38.7%) | p<0.001 | 0.69 | 0.39 | 1.0 | 1.0 | 0.70 | 0.75 | ||
| I–IV | 32/96 (33.3%) | p<0.001 | 0.67 | 0.33 | 1.0 | 1.0 | 0.58 | 0.65 | ||
| RPRM | ||||||||||
| I+II | 16/34 (47.1%) | 6/88 (6.8%) | p<0.001 | 0.70 | 0.47 | 0.93 | 0.73 | 0.82 | 0.80 | |
| III+IV | 48/62 (77.4%) | p<0.001 | 0.85 | 0.77 | 0.93 | 0.89 | 0.85 | 0.87 | ||
| I–IV | 64/96 (66.7%) | p<0.001 | 0.80 | 0.67 | 0.93 | 0.91 | 0.72 | 0.79 | ||
| RUNX3 | ||||||||||
| I+II | 20/34 (58.8%) | 4/88 (4.5%) | p<0.001 | 0.77 | 0.59 | 0.95 | 0.83 | 0.86 | 0.85 | |
| III+IV | 36/62 (58.1%) | p<0.001 | 0.76 | 0.58 | 0.95 | 0.90 | 0.76 | 0.80 | ||
| I–IV | 56/96 (58.3%) | p<0.001 | 0.77 | 0.58 | 0.95 | 0.93 | 0.68 | 0.76 | ||
TNM: Tumor-Node-Metastasis; AUC: Area under the curve; S: Sensitivity; Sp: Specificity; PPV: Positive predictive value; NPV: Negative predictive value.
Fisher’s exact tests.
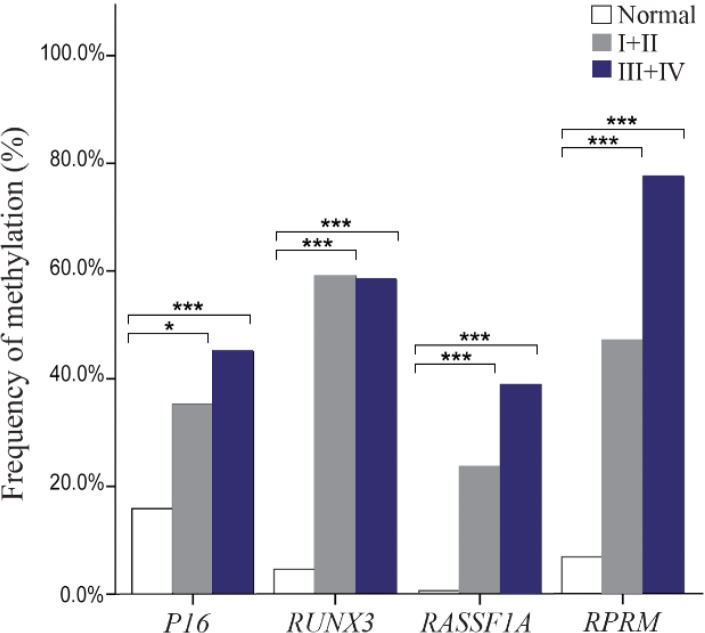
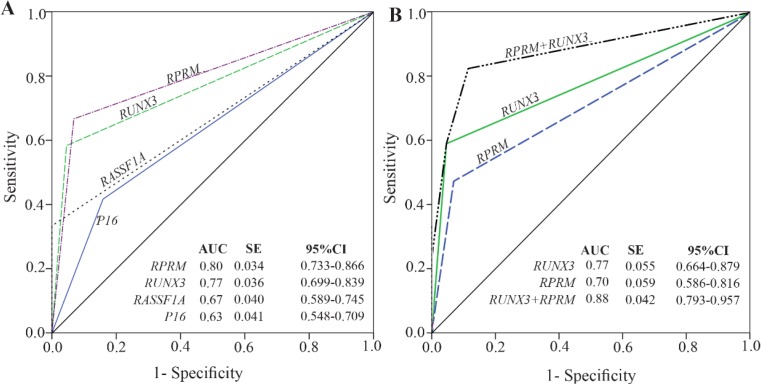
Methylation rates increased in the progression of gastric carcinogenesis from the controls to the early and advanced-stages GC samples for the P16, RASSF1A, and RPRM genes (Figure 2). Concurrent methylation in two or more genes was found in 77.1% (74/96) of plasma GC samples and 0.0% of normal plasma. On the other hand, 4.2% (4/96) of GC samples and 75% (66/88) of controls were methylation free for the P16, RASSF1A, RPRM, and RUNX3. Analysis of the biomarkers performances in patients (TM stages I–IV) showed that the AUC of RPRM, RUNX3, RASSF1A, and P16 were 0.80 (SE=0.034; 95% CI: 0.733–0.866), 0.77 (SE=0.036; 95% CI: 0.699–0.839), 0.67 (SE= 0.040; 95% CI: 0.589–0.745), and 0.63 (SE=0.041; 95% CI: 0.548–0.709), respectively (Figure 3).
Figure 2.
Frequency of methylated DNA in the plasma of controls and gastric cancer patients with early (I+II) and advanced-stage (III+IV). * and *** indicated p<0.05 and p<0.001, respectively.
Figure 3.
Performance of the candidate biomarkers. A) Receiver operating characteristic (ROC) for methylation status of the P16, RASSF1A, RPRM, and RUNX3 in detection of gastric cancer (I–IV). B) ROC analysis of the combined RPRM and RUNX3 methylation status in detection of early-stage gastric cancer (I+II). AUC: Area under curve; SE: Standard error; 95% CI: 95% Confidence interval.
To explore the potentials of methylation analysis of these genes in early detection of GC, the performances of RPRM, RUNX3, RASSF1A, and P16 in early-stage GC patients (I+II) were analyzed in comparison to the controls. The results showed that RUNX3 (p<0.001) with an AUC of 0.77 (SE=0.055; 95% CI: 0.664–0.879), sensitivity of 0.59, and specificity of 0.95, and RPRM (p<0.001) with an AUC of 0.70 (SE=0.059; 95% CI: 0.586–0.816), sensitivity of 0.47, and specificity of 0.93 could discriminate the early-stages GC patients from the normal controls with the highest performances (Table 2). Although methylation frequency of P16 (p=0.026) and RASSF1A (p<0.001) was significantly different between the early-stage GC patients and the healthy people, however, they were excluded from further analysis due to their low sensitivities of 0.35 and 0.24, respectively (Table 2).
Since the sensitivity of a single gene methylation was still unsatisfying, combined detection of several genes might be a solution. A logistic regression model was performed to evaluate the diagnostic performance of the combination of the RPRM, and RUNX3 biomarkers in discriminating between early-stage GC (I+ II) and normal controls (Figure 3). Our results showed that combination of RPRM and RUNX3 increased the AUC to 0.88 (SE=0.042; 95% CI: 0.793–0.957), and sensitivity to 0.82; however, the specificity decreased to 0.89 (p<0.001).
Correlations between methylation status and survival
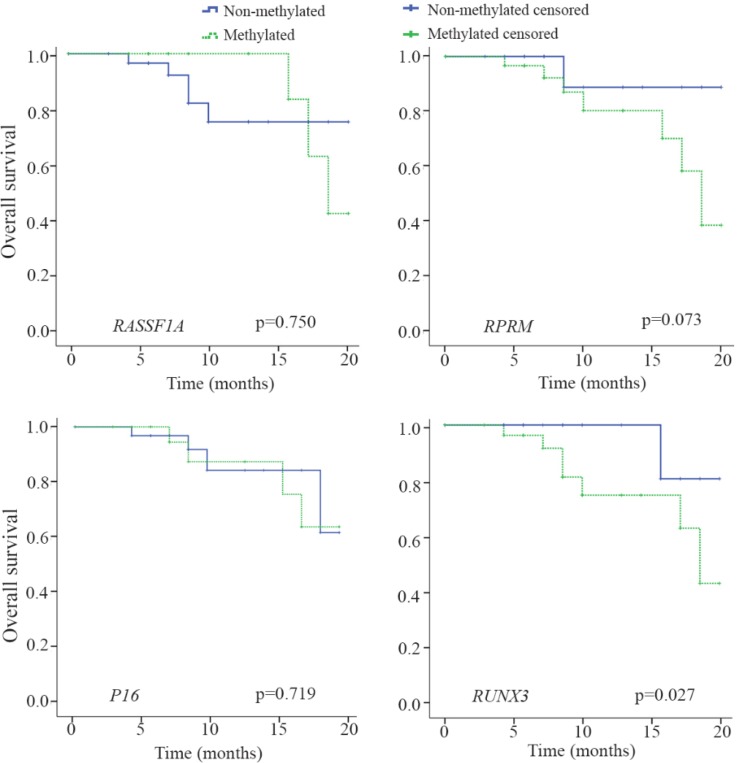
After a median follow up period of 20 months, 16.7% (16/96) of patients died because of the disease progression. The P16, RASSF1A, RPRM, and RUNX3 methylation were detected in 50% (8/16), 37.5% (6/16), 87.5% (14/16), and 87.5% (14/16) of these patients, respectively. Among the entire cohort, the mean survival time was 18.1 months (SE=0.422; 95% CI: 17.2–18.9).
Patients’ survival as depicted in figure 4 was significantly associated with methylation status of RUNX3 (p=0.027). On the other hand, patients’ survival was not significantly associated with methylation status of P16 (p=0.719), RASSAF1A (p=0.750), RPRM (p= 0.073). Mean survival time (months) of patients with a methylated P16, RASSF1A, RPRM, and RUNX3 were 18.0±0.61 (95% CI: 16.8–19.2), 18.9±0.34 (95% CI: 18.2–19.6), 17.7±0.53 (95% CI: 16.6–18.7), and 17.4± 0.59 (95% CI: 16.2–18.5), respectively. In addition, mean survival time (months) of patients with an unmethylated P16, RASSF1A, RPRM, and RUNX3 were 18.2±0.58 (95% CI: 17.1–19.3), 17.9±0.58 (95% CI: 16.8–19.1), 19.1±0.59 (95% CI: 17.9–20.3), and 19.4± 0.38 (95% CI: 18.7–20.1), respectively.
Figure 4.
Kaplan-Meier estimate of overall survival for the gastric cancer patients with or without methylation of P16, RASSF1A, RPRM, and RUNX3. Log-rank statistics are shown as p-values.
Discussion
In the present study, an attempt was made to investigate the methylation status of four potential candidate genes in circulating cfDNA of plasma in GC patients and healthy controls, and their possible correlations with tumor stage, gender, age, and survival were examined. Our results showed that P16, RASSF1A, RPRM, and RUNX3 promoters were methylated in 41.7, 33.3, 66.7, and 58.3% of the GC patients (I–IV), and in 15.9, 0.0, 6.8 and 4.5% of the healthy controls, respectively (p<0.001). No significant correlation was found between the methylation status of four candidate genes and gender. Analysis of methylation status with the ages (≤60 versus >60) showed that methylated P16 was significantly higher in the subjects over 60 years old compared to the subjects under 60 years old (p=0.006). Analysis of overall survival with methylation status of the four candidate biomarkers showed that patients’ survival was significantly associated with methylation status of RUNX3 (p=0.027).
The methylation rates in our study were similar to the results of previous studies. The results of previous studies on serum/plasma, which have evaluated the diagnostic potentials of the candidate genes methylation, are summarized in table 3. According to the previous studies by serum and plasma samples, the mean methylation rates of P16 23–25, RASSF1A 19,26,27, RPRM 21,22,28,29, and RUNX3 20,30,31 were 52.8, 61.9, 84.5, and 47.5% in the GC patients and 0.8, 1.8, 6.2, and 0.0% in the control groups, respectively. Evaluation of their findings revealed that variations in the results were lower in the controls compared to the GC groups. The methylation rates of previous studies in the GC groups for the P16 23,24, RASSF1A 26,27, RPRM 21,28 and RUNX3 30,31 ranged from 26.9–79.7, 34.0–83.2, 62.0–95.3, and 29.0–70.8%, respectively. These discrepancies could be explained by several factors. First, various methods with different sensitivity including MSP, qMSP, and bisufite sequencing have been used for analysis of methylation status of the relevant gene in each study. Second, methylation specific primers have been designed in different CpG sites for each gene. Analysis of eight CpG sites in RUNX3 promoter by quantitative pyrosequencing has shown that methylation frequencies of only six specific sites were different between GC and normal gastric tissues 32.
Table 3.
Methylation rates of the P16, RASSAF1A, RPRM, and RUNX3 in diagnosis of gastric cancer using serum or plasma samples
| Gene | Source | Gastric cancer | Control | Country | Method | Reference |
|---|---|---|---|---|---|---|
| P16 | ||||||
| Serum | 51.9% | 0.0% | Hong Kong | MSP | 25 | |
| Serum | 26.9% | 0.0% | Iran | MSP | 24 | |
| Serum | 79.7% | 2.5% | China | MSP | 23 | |
| Plasma | 41.7% | 15.9% | Iran | MSP | This Study | |
| RASSF1A | ||||||
| Serum | 34.0% | 0.0% | China | MSP | 27 | |
| Serum | 68.5% | 0.0% | Greece | MSP | 19 | |
| Plasma | 83.2% | 5.5% | Thailand | MSP | 26 | |
| Plasma | 33.3% | 0.0% | Iran | MSP | This Study | |
| RPRM | ||||||
| Plasma | 95.3% | 9.7% | Chile | MSP | 28 | |
| Plasma | 62.0%% | 0.0% | China | MSP | 21 | |
| Serum | 94.3% | 7.1% | China | MS-MCA | 22 | |
| Plasma | 86.3% | 7.9% | China | BS | 29 | |
| Plasma | 66.7% | 6.8% | Iran | MSP | This Study | |
| RUNX3 | ||||||
| Serum | 29.0% | 0.0% | Japan | qMSP | 31 | |
| Serum | 70.8% | 0.0% | China | qMSP | 30 | |
| Plasma | 42.7% | 0.0% | China | MSP | 20 | |
| Plasma | 58.3% | 4.5% | Iran | MSP | This Study | |
MS-MCA: Methylation sensitive melt curve analysis; MSP: Methylation specific PCR; q-MSP: quantitative MSP; BS: Bisufite sequencing.
Our results showed that the methylation rates increased in the progression of gastric carcinogenesis from the control to the early and advanced-stage GC for the P16, RASSF1A, and RPRM genes. Increasing the methylation rate could be explained by the fact that more cfDNA gets into systemic circulation with enhancement of the disease. In addition, concurrent methylations in two or more genes were found in 77.1% of plasma GC samples and 0.0% of normal plasma. On the other hand, 4.2% of GC patients and 75.0% of controls were methylation free for all four genes.
Also, the performances of candidate biomarkers in early-stage GC patients (I+II) were analyzed. The results showed that RUNX3 with an AUC of 0.77, sensitivity of 0.59, and specificity of 0.95, and RPRM with an AUC of 0.70, sensitivity of 0.47, and specificity of 0.93 could discriminate the early-stage GC from normal controls with the highest performances (p<0.001). Although P16 (p=0.026) and RASSF1A (p<0.001) could also discriminate the early-stage patients from the controls, their sensitivities were very low as 0.35 and 0.24, respectively. For that reason, they were excluded from further analysis.
The sensitivity and specificity of RUNX3 hypermethylation by qMSP, in serum samples of GC patients and normal controls in a study by Lu et al were 70.8 and 99.8%, respectively 30. In another study by Lin et al, the sensitivity and specificity of RUNX3 hypermethylation were 42.7 and 79.2%, respectively 20. Our results were similar to the study by Lin et al which used MSP as the method for methylation detection 20. The higher performance of RUNX3 methylation in the study by Lu et al might be explained by the fact that qMSP is more sensitive than MSP 30.
The use of only a single gene to discriminate cancer patients from the healthy people has several drawbacks. First, the maximum sensitivity of a test by only a single gene could be as high as the rate of methylation for that gene. Second, non-cancerous tissues could be occasionally methylated at the same gene locus as cancerous tissue 33. Furthermore, methylation of a single gene locus can occur in different cancers. For example, in addition to GC, aberrant methylation of P16 and RASSF1A in serum of breast cancer patients 34, hypermethylation of RASSF1A in hepatocellular carcinoma tissues 35, and hypermethylation of RUNX3 in serum of colorectal cancer patients have also been reported 36. For that reason, a panel of hypermethylated genes, instead of a single gene, could be more effective to guarantee that the biomarker is specific to a cancer.
Also, the combination of RPRM and RUNX3 in distinguishing the early-stage GC (I+II) from the controls was analyzed. The results showed a higher AUC of 0.88, with a higher sensitivity of 0.82 and reduced specificity of 0.89 (p<0.001). Although the specificity of combined detection of RPRM and RUNX3 methylation was lower than that of single gene assays, the sensitivity was increased.
Conclusion
In conclusion, combined detection of plasma RPRM, and RUNX3 methylation could be suggested as a potential strategy for early diagnosis of GC; however, further studies for validation of the panel are required.
Acknowledgement
The study was supported by a grant from the Hematology, Oncology, and Stem Cell Transplantation Research Centre, Shariati hospital, Tehran University of Medical Sciences, Tehran, Iran.
Footnotes
Conflict of Interest
The authors have no conflicts of interest to declare.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136(5):E359–386. [DOI] [PubMed] [Google Scholar]
- 2.Uchôa Guimarães CT, Ferreira Martins NN, Cristina da Silva Oliveira K, Almeida CM, Pinheiro TM, Gigek CO, et al. Liquid biopsy provides new insights into gastric cancer. Oncotarget 2018;9(19):15144–15156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levy I, Gralnek IM. Complications of diagnostic colonoscopy, upper endoscopy, and enteroscopy. Best Pract Res Clin Gastroenterol 2016;30(5):705–718. [DOI] [PubMed] [Google Scholar]
- 4.Gerlinger M, Rowan AJ, Horswell S, Math M, Larkin J, Endesfelder D, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 2012;366(10):883–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vedeld HM, Goel A, Lind GE. Epigenetic biomarkers in gastrointestinal cancers: The current state and clinical perspectives. Semin Cancer Biol 2018;51:36–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Toiyama Y, Okugawa Y, Goel A. DNA methylation and microRNA biomarkers for noninvasive detection of gastric and colorectal cancer. Biochem Biophys Res Commun 2014;455(1–2):43–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Padmanabhan N, Ushijima T, Tan P. How to stomach an epigenetic insult: the gastric cancer epigenome. Nat Rev Gastroenterol Hepatol 2017;14(8):467–478. [DOI] [PubMed] [Google Scholar]
- 8.Hu W, Zheng W, Liu Q, Chu H, Chen S, Kim JJ, et al. Diagnostic accuracy of DNA methylation in detection of gastric cancer: a meta-analysis. Oncotarget 2017;8(68): 113142–113152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Issaq HJ, Waybright TJ, Veenstra TD. Cancer biomarker discovery: Opportunities and pitfalls in analytical methods. Electrophoresis 2011;32(9):967–975. [DOI] [PubMed] [Google Scholar]
- 10.Pepe MS, Etzioni R, Feng Z, Potter JD, Thompson ML, Thornquist M, et al. Phases of biomarker development for early detection of cancer. J Natl Cancer Inst 2001; 93(14):1054–1061. [DOI] [PubMed] [Google Scholar]
- 11.Hu SL, Kong XY, Cheng ZD, Sun YB, Shen G, Xu WP, et al. Promoter methylation of p16, Runx3, DAPK and CHFR genes is frequent in gastric carcinoma. Tumori 2010;96(5):726–733. [DOI] [PubMed] [Google Scholar]
- 12.do Nascimento Borges B, Burbano RM, Harada ML. Analysis of the methylation patterns of the p16 INK4A, p15 INK4B, and APC genes in gastric adenocarcinoma patients from a Brazilian population. Tumour Biol 2013; 34(4):2127–2133. [DOI] [PubMed] [Google Scholar]
- 13.Ye M, Xia B, Guo Q, Zhou F, Zhang X. Association of diminished expression of RASSF1A with promoter methylation in primary gastric cancer from patients of central China. BMC Cancer 2007;7:120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ooki A, Yamashita K, Yamaguchi K, Mondal A, Nishimiya H, Watanabe M. DNA damage-inducible gene, reprimo functions as a tumor suppressor and is suppressed by promoter methylation in gastric cancer. Mol Cancer Res 2013;11(11):1362–1374. [DOI] [PubMed] [Google Scholar]
- 15.Sperka T, Wang J, Rudolph KL. DNA damage checkpoints in stem cells, ageing and cancer. Nat Rev Mol Cell Biol 2012;13(9):579–590. [DOI] [PubMed] [Google Scholar]
- 16.Qu Y, Dang S, Hou P. Gene methylation in gastric cancer. Clin Chim Acta 2013;424:53–65. [DOI] [PubMed] [Google Scholar]
- 17.Amigo JD, Opazo JC, Jorquera R, Wichmann IA, Garcia-Bloj BA, Alarcon MA, et al. The reprimo gene family: a novel gene lineage in gastric cancer with tumor suppressive properties. Int J Mol Sci 2018;19(7). E1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen F, Liu X, Bai J, Pei D, Zheng J. The emerging role of RUNX3 in cancer metastasis (Review). Oncol Rep 2016;35(3):1227–1236. [DOI] [PubMed] [Google Scholar]
- 19.Balgkouranidou I, Matthaios D, Karayiannakis A, Bolanaki H, Michailidis P, Xenidis N, et al. Prognostic role of APC and RASSF1A promoter methylation status in Cell-free circulating DNA of operable gastric cancer patients. Mutat Res 2015;778:46–51. [DOI] [PubMed] [Google Scholar]
- 20.Lin Z, Luo M, Chen X, He X, Qian Y, Lai S, et al. Combined detection of plasma ZIC1, HOXD10 and RUNX3 methylation is a promising strategy for early detection of gastric cancer and precancerous lesions. J Cancer 2017;8 (6):1038–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu L, Yang X. Implication of reprimo and hMLH1 gene methylation in early diagnosis of gastric carcinoma. Int J Clin Exp Pathol 2015;8(11):14977–14982. [PMC free article] [PubMed] [Google Scholar]
- 22.Wang H, Zheng Y, Lai J, Luo Q, Ke H, Chen Q. Methylation-sensitive melt curve analysis of the reprimo Gene methylation in gastric cancer. PLoS One 2016;11(12): e0168635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu YC, Lv P, Han J, Yu JL, Zhu X, Hong LL, et al. Enhanced serum methylated p16 DNAs is associated with the progression of gastric cancer. Int J Clin Exp Pathol 2014;7(4):1553–1562. [PMC free article] [PubMed] [Google Scholar]
- 24.Abbaszadegan MR, Moaven O, Sima HR, Ghafarzadegan K, A’rabi A, Forghani MN, et al. p16 promoter hypermethylation: a useful serum marker for early detection of gastric cancer. World J Gastroenterol 2008;14(13): 2055–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee TL, Leung WK, Chan MW, Ng EK, Tong JH, Lo KW, et al. Detection of gene promoter hypermethylation in the tumor and serum of patients with gastric carcinoma. Clin Cancer Res 2002;8(6):1761–1766. [PubMed] [Google Scholar]
- 26.Pimson C, Ekalaksananan T, Pientong C, Promthet S, Putthanachote N, Suwanrungruang K, et al. Aberrant methylation of PCDH10 and RASSF1A genes in blood samples for non-invasive diagnosis and prognostic assessment of gastric cancer. Peer J 2016;4:e2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang YC, Yu ZH, Liu C, Xu LZ, Yu W, Lu J, et al. Detection of RASSF1A promoter hypermethylation in serum from gastric and colorectal adenocarcinoma patients. World J Gastroenterol 2008;14(19):3074–3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bernal C, Aguayo F, Villarroel C, Vargas M, Díaz I, Ossandon FJ, et al. Reprimo as a potential biomarker for early detection in gastric cancer. Clin Cancer Res 2008; 14(19):6264–6269. [DOI] [PubMed] [Google Scholar]
- 29.Lai J, Wang H, Luo Q, Huang S, Lin S, Zheng Y, et al. The relationship between DNA methylation and Reprimo gene expression in gastric cancer cells. Oncotarget 2017; 8(65):108610–108623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu XX, Yu JL, Ying LS, Han J, Wang S, Yu QM, et al. Stepwise cumulation of RUNX3 methylation mediated by Helicobacter pylori infection contributes to gastric carcinoma progression. Cancer 2012;118(22):5507–5517. [DOI] [PubMed] [Google Scholar]
- 31.Sakakura C, Hamada T, Miyagawa K, Nishio M, Miyashita A, Nagata H, et al. Quantitative analysis of tumor-derived methylated RUNX3 sequences in the serum of gastric cancer patients. Anticancer Res 2009;29 (7):2619–2625. [PubMed] [Google Scholar]
- 32.Wang N, Sui F, Ma J, Su X, Liu J, Yao D, et al. Site-specific hypermethylation of RUNX3 predicts poor prognosis in gastric cancer. Arch Med Res 2016;47(4):285–292. [DOI] [PubMed] [Google Scholar]
- 33.Phé V, Cussenot O, Rouprêt M. Methylated genes as potential biomarkers in prostate cancer. BJU Int 2010;105(10):1364–1370. [DOI] [PubMed] [Google Scholar]
- 34.Shan M, Yin H, Li J, Li X, Wang D, Su Y, et al. Detection of aberrant methylation of a six-gene panel in serum DNA for diagnosis of breast cancer. Oncotarget 2016;7 (14):18485–18494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu B, Di J, Wang Z, Han X, Li Z, Luo X, et al. Quantitative analysis of RASSF1A promoter methylation in hepatocellular carcinoma and its prognostic implications. Biochem Biophys Res Commun 2013;438(2):324–338. [DOI] [PubMed] [Google Scholar]
- 36.Nishio M, Sakakura C, Nagata T, Komiyama S, Miyashita A, Hamada T, et al. RUNX3 promoter methylation in colorectal cancer: its relationship with microsatellite instability and its suitability as a novel serum tumor marker. Anticancer Res 2010;30(7):2673–2682. [PubMed] [Google Scholar]