Abstract
Background:
Emergence and prevalence of multi drug resistance strains such as Methicillin-Resistant Staphylococcus aureus (MRSA) call for new antibacterial option. Endolysins as a new option is suggested. The phage display technique is suggested for production of recombinant endolysins. The recombinant endolysins displayed nano phages specifically lysis bacteria, which penetrate to the depth of tissue and the effective dose is reduced.
Methods:
CHAPK gene was ligated in T7Select vector arms in T7Select10-3b cloning kit. To produce recombinant nano phages, ligation reaction was added directly to the packaging extract. Recombinant nano phages were amplified by Double Layer Agar assay (DLA). The recombinant nano phages were characterized using TEM. Size of recombinant nano phages was determined using DLS. The spot test was performed to confirm CHAPk -displayed on the surface of nano phages. The turbidimetry was used to investigate lytic activity of recombinant nano phages against MRSA ATCC No. 33591.
Results:
The results showed recombinant nano phages belonged to order Caudovirales and family Podoviridae with titer 2×107 PFU/ml. According to the results of DLS, size of recombinant nano phages was 71 nm. Formation inhibition zone confirmed the presence of CHAPk on the surface of nano phage phenotypically. The turbidimetry showed lytic activity recombinant nano phages against MRSA after 5 min.
Conclusion:
This study suggests that CHAPk -displayed nano phages can be effective in MRSA infections.
Keywords: Bacteriophages, Endolysin, Methicillin-resistant Staphylococcus aureus
Introduction
With developments in treatment of infectious diseases, Multi Drug Resistance (MDR) strains such as Methicillin-Resistant Staphylococcus aureus (MRSA) have emerged and expanded within the last 50 years 1–3. Prevalence of MDR strains and return of infections call for new antibacterial option. Bacteriophages and their endolysins as an interesting therapeutic option have attracted the attention of researchers 4–9.
Endolysins as hydrolytic enzymes are produced by bacteriophages which destroy the cell wall of bacteria at the final stage of lytic cycle. Lysines, specifically lysis bacteria, unlike antibiotics have the following features; resistance to them is not reported. They have no effect on normal microflora and no immune response is created against them 7,10–12. The truncated recombinant endolysins destroy gram-positive bacteria exogenously. In previous studies, cloning method was used to produce recombinant lysine such as LysK, phill, Twort, 187, P68, phiWMY, SAL-1, SAP-2, ClyS, and MV-L against Staphylococcus aureus (S. aureus) strains 8,11–15. In order to produce recombinant endolysins by nano-carrier technology, the phage display is suggested. This technique eliminates cloning method and high cost enzymatic reactions 16. By phage display, the peptide or protein gene is expressed so peptide or protein is displayed on the surface of nano-carrier. Passive and active targeting are caused by nano-carrier technology. In passive targeting, the recombinant endolysin penetrates to the depth of tissue. The effective dose is reduced, and there is no toxic effect on eukaryotic cells. Active targeting specifically lysis MDR strains have no effect on microflorabacteria 8,16–18.
One of the truncated recombinant lysine is cysteine- histidine-dependent amino hydrolase/peptidase, CHAPK, 18.6 kDa, the part of Lys K that specifically cleaves the peptide bonds between D-alanine and glycine in the pentaglycine cross bridges of S. aureus cell wall. The purpose of this study was to design and manufacture recombinant nano phage by phage display 8,15,19,20. With expression of CHAPK gene, the recombinant CHAPk -displayed on the surface of nano- carrier. The smart recombinant nano-phages specifically lysis MRSA with the least effective dose, have high penetration and no side effects. Also, they produce on a large scale with moderate cost 8,16.
To the best of our knowledge, production of recombinant nano-bacteriophages by phage display has not been performed. This study suggests that CHAPk-displayed nano phages can be effective in MRSA infections.
Materials and Methods
Preparation of MRSA strain
MRSA ATCC No. 33591 strain was confirmed using conventional methods such as microscopic examination of gram stain smear, tube coagulase, catalase, DNase, and mannitol fermentation tests were performed. To detect MRSA antibiotic resistance, the Kirby–Bauer technique was carried out using Methicillin and Oxacillin antibiotic discs 5,6,8.
Preparation of CHAPK gene
CHAPK gene was selected from the NCBI with sequence ID, KY974323.1, considering appropriate cutting positions by restriction enzymes, Hind III and EcoRI. This sequence with 501 bp was synthesized by Bioneer Company (South Korea). The CHAPK gene was cut in pBHA vector by double enzyme digestion. 10X Buffer Tango, 200 μg/ml inserted CHAPK gene in pBHA vector, 10 U/μl and EcoRI, 10 U/μl Hind III (Thermo scientific, U.S.A) and deionized water were combined and incubated at 37°C for 16 hr. To confirm double enzyme digestion reaction, electrophoresis was performed with TBE 1% and agarose 1%, 140 v for 45 min. Then, CHAPK gene was cut and extracted from gel by the EZ-10 Spin Column DNA Gel Extraction Kit (Bio Basic, Canada) 8,21–23.
Ligation of inserts in T7Select vector
The CHAPK gene was ligated in T7Select vector in T7Select10-3b Cloning Kit (Novagen, USA). The molar ratios 3:1, 500 ng/μl T7Select vector arms were added to 184 μg/ml CHAPK gene. T4 DNA ligase, 10x T4 DNA ligase buffer (Takara, Japan), and deionized water were added and incubated at 16°C for 3 hr 8,21–23.
In vitro packaging
The T7Select packaging extract in T7Select10-3b Cloning Kit (Novagen, USA) was thawed on ice. The ligation reaction was added directly per 25 μl packaging extract. Then, it was gently mixed by stirring with a pipette tip. The reaction was incubated at room temperature (22°C) for 2 hr. The reaction was stopped by adding 70 μl sterile Luria-Bertani medium. Then, 20 μl chloroform was added and mixed gently and the packaging reaction was stored at 4°C 8,21–23.
Amplification of the packaged phages
In order to determine the titer of recombinant nano phages and their amplification, Double Layer Agar (DLA) assay was done. The BL21 as host strain in T7Select10-3b Cloning Kit (Novagen, USA) was inoculated in M9 Luria-Bertani medium and shook at 37°C until the OD600 reached 1. The serial dilutions of CHAPk -displayed nano phages were prepared in 7 tubes using sterile Luria-Bertani medium. Tubes 8 and 9 were negative and positive controls which consist of M9 Luria-Bertani medium and BL21, respectively. 100 μl of diluted CHAPk -displayed nano phages were added to 250 μl host cell. Then, top agarose was added to each tube and poured on plate. The plates were allowed to sit undisturbed for several minutes until the top agar was hardened. They were incubated at room temperature overnight. After that, the plaques were counted and the CHAPk -displayed nano phages titers were calculated 5,6,8.
Confirmation of recombinant nano phages
PCR reaction was performed to approve ligation of CHAPk in T7Select vector. The plaques were removed with Pasteur pipettes and were transferred to sterile microtube. Then microtube was incubated for 10 min at 80°C. PCR reaction was performed. The sample consisted of 10 μl master mix, 1 μl up primer, 1 μl down primer, 4 μl deionize water, and 4 μl CHAPk in vector. The positive control consisted of 10 μl master mix, 1 μl up primer, 1 μl down primer, 4 μl deionize water, and 4 μl CHAPK gene in pBHA vector. The negative control consisted of 10 μl master mix, 1 μl up primer, 1 μl down primer, and 8 μl deionize water. PCR products were electrophoresed with TBE 1% and agarose 1%, 140 v for 45 min 8.
Characterization of recombinant nano phages using TEM
The recombinant nano phages were concentrated by centrifugation at 25,000×g for 60 min followed by two washes in 0.1 M neutral ammonium acetate. The recombinant nano phages were deposited on carbon-coated copper grids and stained by 2% uranyl acetate (pH=4–4.5). After staining, the recombinant nano phages were observed on a Zeiss EM 900 electron microscope at 150 Kv 5,6,8.
Determination of recombinant nano phages size using DLS
To determine the size of recombinant nano phages, DLS was done (ZEN3600, MALVERN). 1 ml of the recombinant nano phages was poured into quotes and the average size was determined 8.
Purification of recombinant nano phages
1 ml of BL21 as the host strain in T7Select10-3b Cloning Kit (Novagen, U.S.A) in 200 ml sterile M9 Luria-Bertani was shacked at 250 rpm 37°C until the OD600 reached 0.6–0.8. The culture was infected by adding 0.02 ml high titer T7 phage lysate and shaking continued at 37°C until 1–1.5 hr. Lysed culture was cooled at room temperature. Recombinant DNase I (Takara, Japan) was added and incubated for 30 min at room temperature. 1 M NaCl was added and the flask was left on ice for an hour. Cell debris was removed by centrifugation at 11000×g for 10 min at 4°C and super-natant was collected. Solid polyethylene glycol (PEG, Sigma Ltd., UK) was added to final concentration of 10% (w/v) and slowly dissolved at room temperature; then, the flask was incubated at 4°C for at least 1 hr. The precipitated bacteriophage particles were recovered by centrifugation at 11000×g for 10 min at 4°C and the pellet was re-suspended in SM buffer. An equal volume of chloroform was added and culture was centrifuged at 300×g for 15 min at 4°C. The upper aqueous phase was removed and CHAPk -displayed nano phages were pelleted by centrifugation at 11000 × g for 2 hr at 4°C. The CHAPk -displayed nano phages pellets were re-suspended in 1–2 ml of SM buffer at 4°C overnight 8,24.
Confirmation of expressed CHAPk on the surface of nano phages
Phenotypically, to confirm CHAPk -displayed nano phages, spot test was performed. An overnight culture of the MRSA (100 μl) was mixed in soft agar. Then, the contents were poured in petri dishes containing bottom agar. Then, 10 μl of the recombinant nano phage was poured on the solidified soft agar. The petri dishes were incubated at 37°C overnight. The following day, formation of inhibition zone was checked 5,6,8.
Investigation of lytic activity of CHAPk -displayed nano phages
In order to investigate the lytic activity of CHAPk -displayed nano phages, the turbidimetry was used. The MRSA ATCC No. 33591 in Luria-Bertani medium was inoculated and shook at 37°C until the OD600 nm reached 0.1 (1.5×108 CFU/ml). Then serial dilutions of the MRSA bacteria were prepared in 6 tubes using sterile Luria-Bertani medium. The tube 0 consisted of MRSA bacteria and tube 7 consisted of Luria-Bertani medium which were positive control and negative control, respectively. Then, 100 μl from diluted 6 tubes was transferred to sterile tubes separately. 100 μl CHAPk -displayed nano phages (2×109 PFU/ml) were added to the 6 tubes. After 5 and 10 min, the concentration of MRSA ATCC No.33591 was detected by spectrophotometer at OD600 nm. Similar to the aforementioned method, natural bacteriophages (2×109 PFU/ml) were used as positive control group. Natural bacteriophages were isolated in our previous study from sewage in Sari Bouali Sina Hospital, Mazandaran province in north of Iran8.
Results
Preparation of CHAPK gene
The result of electrophoresis of inserted CHAPK genein pBHA vector after double digestion reaction by restriction enzymes Hind III and EcoRI showed that CHAPK gene was located in 500 bp (Figure 1).
Figure 1.
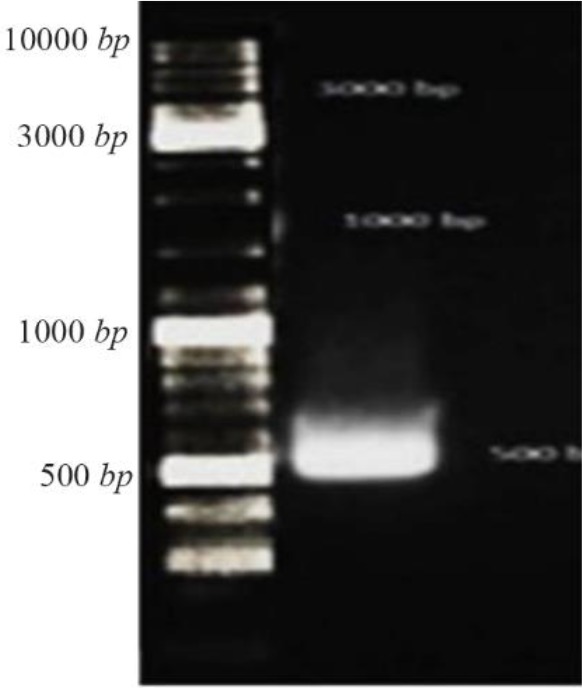
Gel electrophoresis CHAPK gene, CHAPK gene was located in 500 bp. Electrophoresis with TBE 1% and agarose 1%, 140 v for 40 min.
Amplification of the packaged phages
For amplification and detection of recombinant nano phages titer, DLA assay technique was used. The number of plaques was counted after 24 hr. Plaque-forming units per milliliter were calculated as follows: the number of plaques×10×inverse of the dilution factor. Titer of recombinant nano phages was 2×107 PFU/ml (Figure 2).
Figure 2.
A) Titer of recombinant nano phages in the first dilution from serial dilution, B) Titer of recombinant nano phages in the seventh dilution from serial dilution was calculated 2×107 PFU/ml.
Confirmation of recombinant nano phages
After ligation of CHAPK gene in T7Select vector, PCR reaction was performed. Then, PCR product was electrophoresed. The PCR product located at 600 bp (Figure 3).
Figure 3.
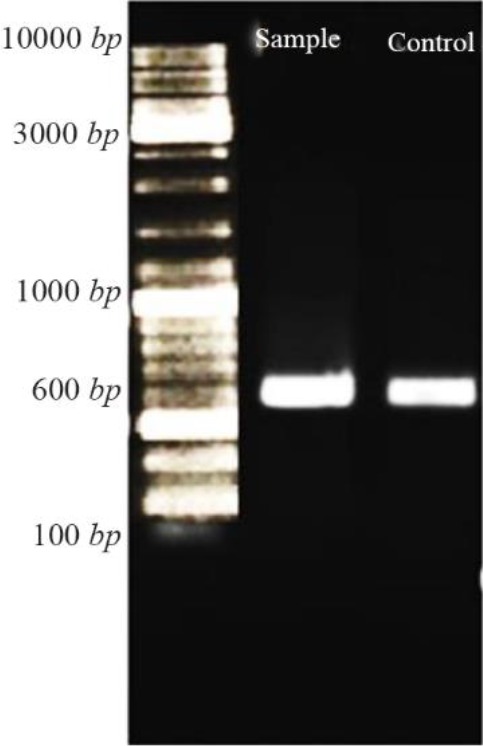
Confirmation of ligation CHAPk in T7Select vector using PCR reaction. After electrophoresis, the PCR product and positive control were located at 600 bp.
Characterization of recombinant nano phages using TEM
Electron microscopy was performed by negative staining of 2% uranyl acetate (pH=4–4.5) at 150 Kv voltage. The results showed one recombinant nano phage group with an icosahedral head (50 nm) and a short non-contractile tail (28 nm). The recombinant nano phage group belonged to order Caudovirales and family Podoviridae (Figure 4).
Figure 4.
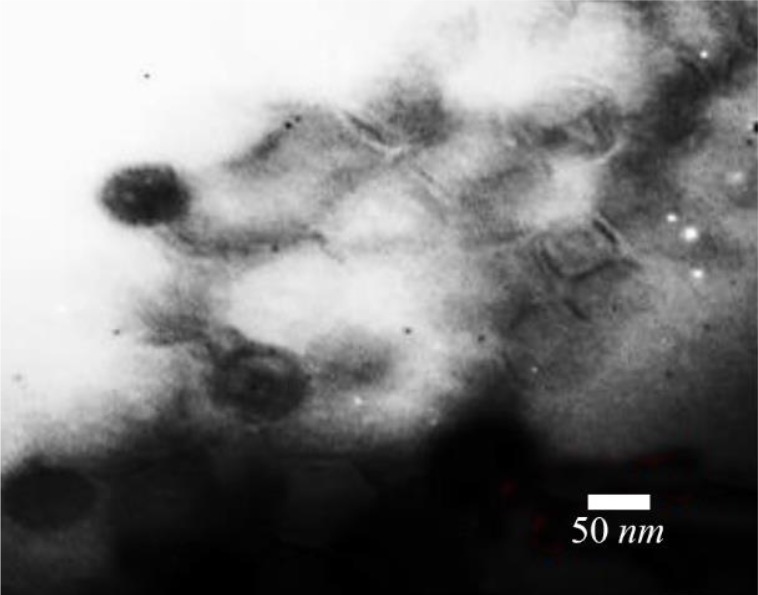
Electron micrographs showed recombinant nano phage belonged to Podoviridae family. Negatively stained with 2% uranyl acetate (pH=4–4.5). Voltage 150 Kv, the scale bar represents 50 nm.
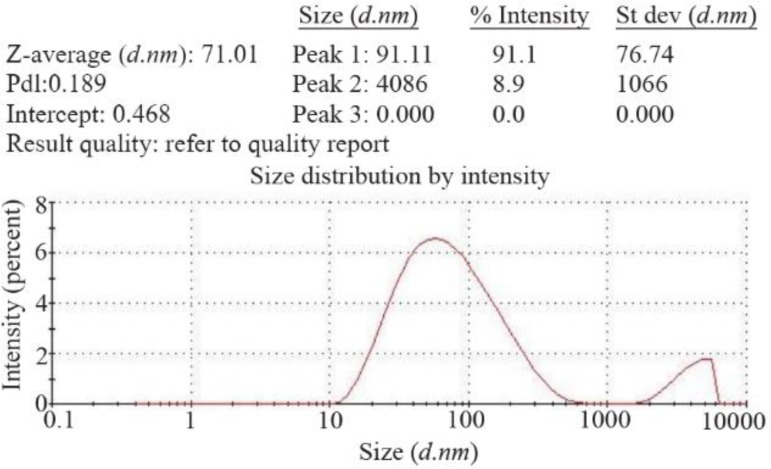
Determination of recombinant nano phages size using DLS
To determine the size of recombinant nano phage, DLS technique was used by ZEN3600, MALVERN. This instrument with back scattering detector (Laser wavelength) was used for measuring the size of recombinant nano phages (diameter) in batch mode at 25°C in a quartz cuvette. According to the results, the size of recombinant nano phages was 71 nm (Figure 5).
Figure 5.
Size of recombinant nano phages detected by DLS method (ZEN3600, MALVERN). The size of recombinant nano phages was 71 nm.
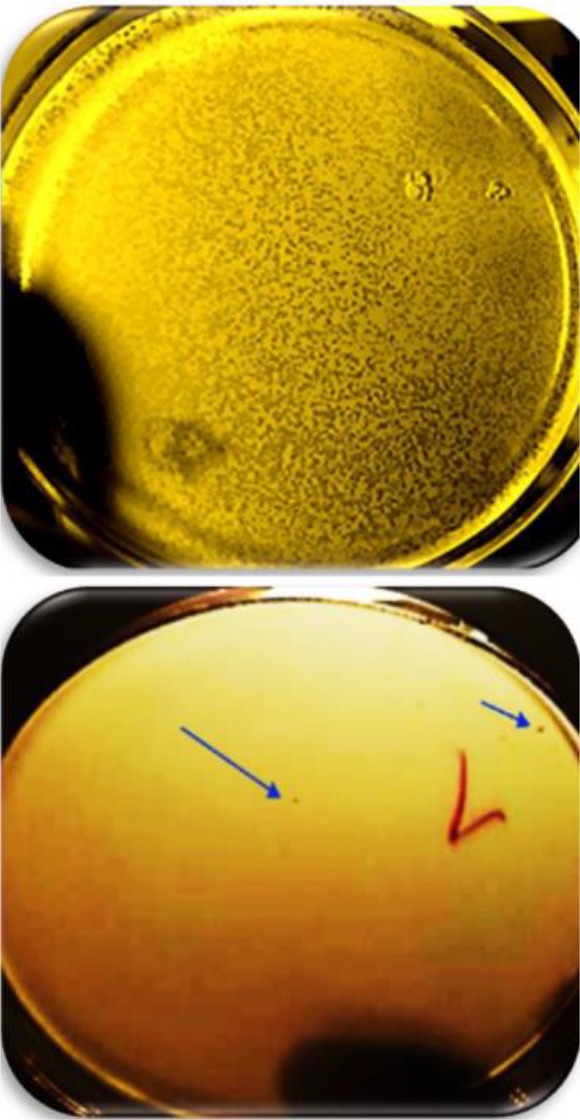
Confirmation of expressed CHAPk on the surface of nano phages
The spot test was used to confirm CHAPk displayed on the surface of nano phages. The inhibition zone was observed. This demonstrated that the CHAPk displayed on the surface of nano phages. The recombinant nano phages had lytic activity against MRSA ATCC No. 33591 (Figure 6).
Figure 6.
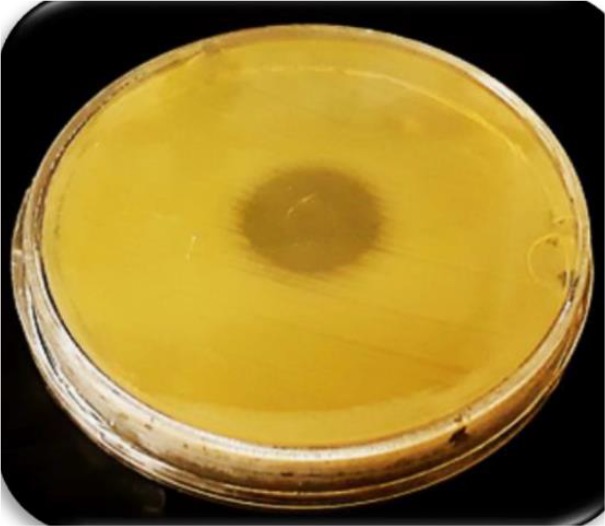
The inhibition zone in the spot test confirmed CHAPk displayed on the surface of nano phages. The recombinant nano phages had lytic activity against MRSA ATCC No. 33591.
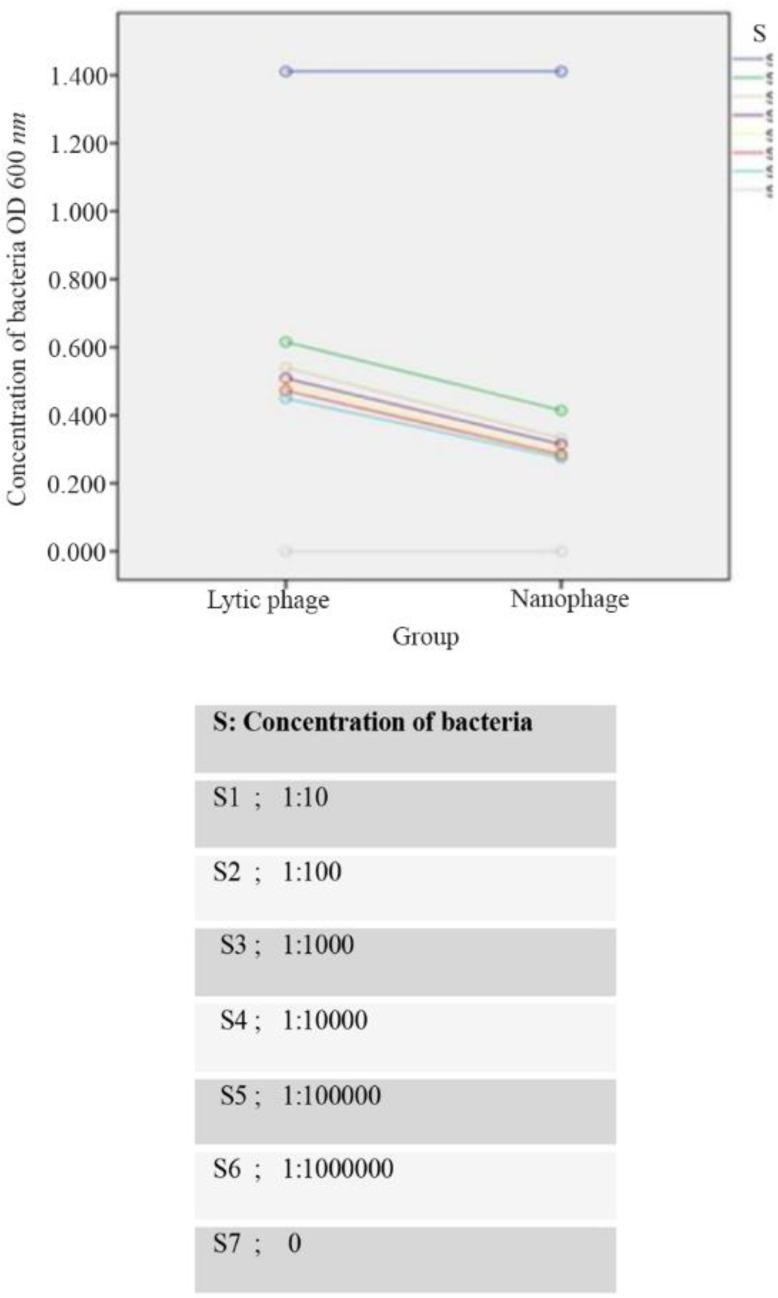
Investigation of lytic activity of CHAPk -displayed nano phages
The turbidimetry was used to investigate the lytic activity of CHAPk -displayed nano phages against MRSA ATCC No. 33591. The results showed CHAPk -displayed nano phages (2×107 PFU/ml) had lytic activity against MRSA after 5 min of incubation (Figure 7).
Figure 7.
Comparison of bacterial concentration in two groups, natural phage and recombinant nano phage.
Discussion
Currently, recombinant endolysins are suggested as a novel class of antimicrobial 7,10,11. Recombinant lysine exogenously destroys gram-positive bacteria, has no effect on normal microflora, and so far no forms of resistance to them has been identified 8,13–15.
Nano carrier technology is a safe and efficient approach for encapsulation of recombinant lysine. It creates recombinant nano phages with active and passive targeting. Manipulated recombinant nano phages with these features specifically lysis bacterial cells with the least effective dose, have high penetrating power, and no side effects 8,16,18.
In order to generate recombinant nano phages, the phage display technique is suggested 8,17,18. In this method, nano phages act as nano-carriers in which peptides or proteinsare displayed on their surface caused by expression of peptides or protein gene in a low-cost production system such as Escherichia coli (E. coli). Phage display is an inexpensive technology since phages are self-replicating, thus, the recombinant peptides or proteins are produced in a large scale. In phage display technique, recombinant peptide or protein is purified by the sensitive and specific bioppaning procedure, so, there would be no problem in pharma- ceutical manipulations 8,17,18,24.
In this study, CHAPk -displayed nano phages against MRSA strains were produced by phage display technique. The CHAPK gene was selected from reference sequence database in the NCBI. CHAPk-displayed nano phages were fused to the C-terminal of gp10B capsomers using expression vector containing CHAPK gene by T7 bacteriophage. CHAPk -displayed nano phages belonged to the Podoviridae family (71 nm, titer of 2×109 PFU/ml).
Some researchers produced truncated recombinant CHAPK against S. aureus strain Xen29 (3.7×103 CFU/ml) and S. aureus strain DPC5246 (1.29×106 CFU/ml) by cloning method with pQE60 expression vector and overexpression in E. coli XL1-Blue host cell. It was then purified by ion-exchange chromatography. 31.25 μg/ml CHAPK was incubated with 200 μl S. aureus DPC5246 (1.29×106 CFU/ml) in TSBg at 37°C for 24 hr, so, it completely prevented the formation of S. aureus biofilms. CHAPK can be used as a spray for elimination of S. aureus from the surface of mammalian skin in 30 min. Fenton et al reported that 10 μg/ml of CHAPK reduced 108 (CFU/ml) S. aureus by 4-log in 15 min at pH=9 19,20,25,26.
But, in the current study, CHAPk-displayed nano phages were generated by phage display technique with expression of CHAPK gene on the surface of T7 phages. Efficiency of phage display technique is more than that of the cloning method because phages are self-replicating, so, the highest amount of recombinant proteins is produced in minimum time. In our study, CHAPk -displayed nano phages (2×109 PFU/ml) were incubated at 37°C in Luria-Bertani broth. Recombinant nano phage showed lytic activity effect against MRSA ATCC No. 33591(1.5×108 CFU/ml). After 5 min, due to its nano dimensions and its greater permeability, lytic phage showed lytic effect against MRSA (1.5×108 CFU/ml) after 20 min.
In our study, according to pI=6.7 of CHAPK and gp10B capsomers (achieved from http://www.expasy.org), lytic activity of CHAPk -displayed nano phages was assayed at pH=6.7–7. In some studies, the specific activities of LysK and CHAPK were assayed. CHAPK enhances lytic activity by at least twofold. The CHAPk domain (165 amino acid) is more active against live S. aureus than LysK (495 amino acid).In turbidity reduction assays, 10 μg/ml of CHAPK reduced 108 CFU/ml S. aureus by 4-log in 15 min 20. But, in our study with turbidity assay, 2×109 PFU/ml of CHAPk -displayed nano phages reduced 1.5×108 CFU/ml of MRSA ATCC No.33591 by 5-log in 5 min.
Donovan et al found that the activity of CHAPK was threefold higher than that of the native enzyme K and amidase domain. Also, lytic activity CHAPK in addition to MRSA bacteria was reported on negative coagulase staphas such as S. warneri, S. simulans, and S. epidermiditis. The CHAPk domain alone can exhibit activity against live Staphylococcibut other S. aureus phage endolysins such as PlyTW and Ply187 lysis exhibit activity only in heat-killed S. aureus 14,27. In the present study, for confirmation of expressed CHAPk on the surface of nano phages, methods such as SDS-Page and ELISA cannot be used due to limited funding.
Conclusion
To the best of our knowledge, in this study, phage display technique was used in nano medicine to design and produce CHAPk -displayed nano phages against MRSA ATCC No. 33591 in vitro. Antibacterial effects of CHAPk -displayed nano phages are higher than truncated recombinant CHAPk against MRSA strain. CHAPk -displayed nano phages reduced the effective dose and lytic time.
Acknowledgement
This study was supported by the deputy of research and technology of Mazandaran University of Medical Sciences.
Footnotes
Conflict of Interest
All authors declared no conflict of interest in the present manuscript.
References
- 1.Basak S, Singh P, Rajurkar M. Multidrug resistant and extensively drug resistant bacteria: A study. J Pathog 2016;2016:4065603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harsha P, Yogeshkumar V, Vyas BRM, Sumitra C. Emergence of methicillin-resistant staphylococcus aureus (MRSA) as a public-health threat and future directions of antibiotic therapy for MRSA infections. Antiinfect Agents 2012;10(2):149–157. [Google Scholar]
- 3.Magiorakos AP, Srinivasan A, Carey R, Carmeli Y, Falagas M, Giske C, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 2012;18(3): 268–281. [DOI] [PubMed] [Google Scholar]
- 4.Nobrega FL, Costa AR, Kluskens LD, Azeredo J. Revisiting phage therapy: new applications for old resources. Trends Microbiol 2015;23(4):185–191. [DOI] [PubMed] [Google Scholar]
- 5.Rahimzadeh G, Gill P, Rezai MS. Characterization and lytic activity of methicillin-resistant Staphylococcus aureus (MRSA) phages isolated from NICU. Australas Med J 2016;9(6):169. [Google Scholar]
- 6.Rahimzadeh G, Gill P, Rezai MS. Characterization of methicillin-resistant Staphylococcus aureus (MRSA) phages from sewage at a tertiary pediatric hospital. Arch Pediatr Infect Dis 2016;5(1):e39615. [Google Scholar]
- 7.Rahimzadeh G, Gill P, Rezai MS. Endolysins of bacteriophages as an anti-methicillin resistant Staphylococcus aureus infection in children: a narrative review. J Pediatr Rev 2018;6(1):36–43. [Google Scholar]
- 8.Rezai MS, Rahimzadeh G, Gill P. Fabrication of phage nanobioparticles against burn wound methicillin-resistant Staphylococcus aureus (MRSA) infections. WO 2019/162972 A1.
- 9.Rahimzadeh G, Saeedi M, Farshidi F, Rezai MS. Phage therapy in treatment of gram-negative bacterial infections: a systematic review. J Mazandaran Univ Med Sci 2018;28(165):203–212. [Google Scholar]
- 10.Borysowski J, Weber-Dąbrowska B, Górski A. Bacteriophage endolysins as a novel class of antibacterial agents. Exp Biol Med (Maywood) 2006;231(4):366–377. [DOI] [PubMed] [Google Scholar]
- 11.Fischetti VA. Bacteriophage lysins as effective antibacterials. Curr Opin Microbiol 2008;11(5):393–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fischetti VA. Bacteriophage endolysins: a novel anti-infective to control Gram-positive pathogens. Int J Med Microbiol 2010;300(6):357–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daniel A, Euler C, Collin M, Chahales P, Gorelick KJ, Fischetti VA. Synergism between a novel chimeric lysin and oxacillin protects against infection by methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 2010;54(4):1603–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donovan DM, Abaev IV. Staphylococcal Phage2638A endolysin amidase domain is lytic for Staphylococcus aureus. Google Patents; 2015.
- 15.Fenton M, Casey PG, Hill C, Gahan CG, McAuliffe O, O’Mahony J, et al. The truncated phage lysin CHAPk eliminates Staphylococcus aureus in the nares of mice. Bioeng Bugs 2010;1(6):404–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mozafari MR. Nanocarrier technologies: frontiers of nano therapy. 1st edition. Netherlands: Springer; 2006. 226 p. [Google Scholar]
- 17.Bair CL, Oppenheim A, Trostel A, Prag G, Adhya S. A phage display system designed to detect and study protein–protein interactions. Mol Microbiol 2008;67(4):719–728. [DOI] [PubMed] [Google Scholar]
- 18.Pande J, Szewczyk MM, Grover AK. Phage display: concept, innovations, applications and future. Biotechnol Adv 2010;28(6):849–858. [DOI] [PubMed] [Google Scholar]
- 19.Hathaway H, Ajuebor J, Stephens L, Coffey A, Potter U, Sutton JM, et al. Thermally triggered release of the bacteriophage endolysin CHAPK and the bacteriocin lysostaphin for the control of methicillin resistant Staphylococcus aureus (MRSA). J Control Release 2017;245:108–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horgan M, O’Flynn G, Garry J, Cooney J, Coffey A, Fitzgerald GF, et al. Phage lysin LysK can be truncated to its CHAP domain and retain lytic activity against live antibiotic-resistant staphylococci. Appl Environ Microbiol 2009;75(3):872–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burdon RH, van Knippenberg P. Laboratory techniques in biochemistry and molecular biology: practice and theory of enzyme immunoassays. Vol. 15 Amsterdam: Elsevier; 1985. 279 p. [Google Scholar]
- 22.Mamiatis T, Fritsch E, Sambrook J, Engel J. Molecular cloning-A laboratory manual. New York: Cold Spring Harbor Laboratory; 1982. 545 p. [Google Scholar]
- 23.Son M, Hayes SJ, Serwer P. Concatemerization and packaging of bacteriophage T7 DNA in vitro: determination of the concatemers’ length and appearance kinetics by use of rotating gel electrophoresis. Virology 1988;162 (1):38–46. [DOI] [PubMed] [Google Scholar]
- 24.Ghaemi A, Soleimanjahi H, Gill P, Hassan Z, Jahromi SRM, Roohvand F. Recombinant lambda-phage nanobioparticles for tumor therapy in mice models. Genet Vaccine Ther 2010;8:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fenton M, Keary R, McAuliffe O, Ross RP, O’Mahony J, Coffey A. Bacteriophage-derived peptidase eliminates and prevents Staphylococcal biofilms. Int J Microbiol 2013;2013:625341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nelson D, Loomis L, Fischetti VA. Prevention and elimination of upper respiratory colonization of mice by group A streptococci by using a bacteriophage lytic enzyme. Proc Natl Acad Sci USA 2001;98(7):4107–4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Donovan DM, Lardeo M, Foster-Frey J. Specific lysis of Staphylococcal pathogens by bacteriophage PHIL 1 endolysin. FEMS Microbiol Lett 2006;265(1):133–139. [DOI] [PubMed] [Google Scholar]