Highlights
-
•
A summary of the research progress in SeACoV (SADS-CoV) from 2017 to 2020.
-
•
Bat-derived SeACoV was most recently recognized prior to SARS-CoV-2 associated with COVID-19.
-
•
Focusing on the etiology, epidemiology, evolutionary perspective, potential for interspecies transmission, pathogenesis and diagnosis.
Keywords: Swine enteric alphacoronavirus (SeACoV), Swine acute diarrhea syndrome coronavirus (SADS-CoV), Etiology, Pathogenicity, Transmission, Diagnosis
Abstract
Discovered in 2017, swine enteric alphacoronavirus (SeACoV), also known as swine acute diarrhea syndrome coronavirus (SADS-CoV) or porcine enteric alphacoronavirus (PEAV), is the fifth porcine CoV identified in diarrheal piglets. The presumed name “SADS-CoV” may not be appropriate since current studies have not provided strong evidence for high pathogenicity of the virus. SeACoV was the most recently recognized CoV of potential bat origin prior to the novel human severe acute respiratory syndrome CoV 2 (SARS-CoV-2), associated with the pandemic CoV disease 2019 (COVID-19). Although SeACoV is recognized as a regional epizootic virus currently, it possesses the most extensive cell species tropism in vitro among known CoVs. This review summarizes the emergence of SeACoV and updates the research progress made from 2017 to early 2020, mainly focusing on the etiology, epidemiology, evolutionary perspective, potential for interspecies transmission, pathogenesis and diagnosis.
1. Introduction
Coronaviruses (CoVs; order Nidovirales, family Coronaviridae, subfamily Orthocoronavirinae) are single-stranded, positive-sense, enveloped RNA viruses that cause subclinical, mild or lethal respiratory and gastrointestinal diseases in humans and animals (de Groot et al., 2011). To date, there have been five different species of CoV identified which naturally infect pigs (Wang et al., 2019). These include three alphacoronaviruses: transmissible gastroenteritis virus (TGEV) together with its variant porcine respiratory virus (PRCV) (Pensaert et al., 1986), porcine epidemic diarrhea virus (PEDV) (Pensaert and de Bouck, 1978), and swine enteric alphacoronavirus (SeACoV) (Pan et al., 2017); one betacoronavirus: porcine hemagglutinating encephalomyelitis virus (PHEV) (Greig et al., 1962); and porcine deltacoronavirus (PDCoV) (Woo et al., 2012). TGEV, PEDV, PDCoV and SeACoV cause swine enteric coronavirus disease (SECD), characterized by acute diarrhea in neonatal piglets leading to moderate or high mortality and significant economic losses for the pork industry, in Asia since 2010 (Pan et al., 2012) and North America since 2013 (Huang et al., 2013; Wang et al., 2019).
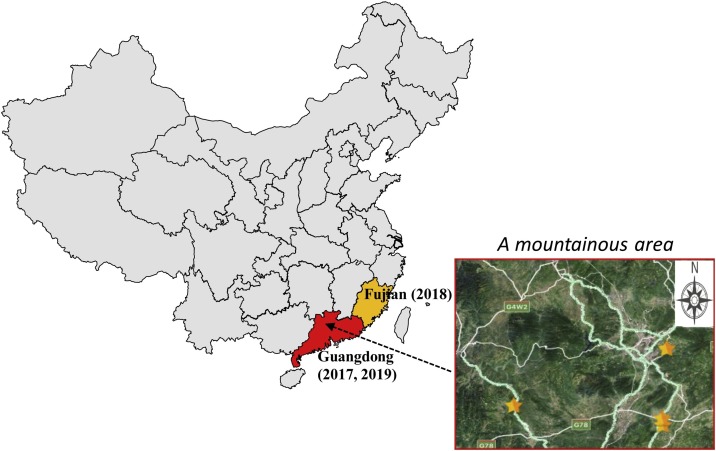
SeACoV was discovered in Guangdong province, China in 2017. Initially, outbreaks of severe diarrhea of suckling piglets occurred in four swine herds in a mountainous area of northern Guangdong (Fig. 1 ). Our laboratory was the first group to isolate and characterize this new virus in Vero cells, and determined its genome to be closely related to the species Rhinolophus bat coronavirus HKU2 in the subgenus Rhinacovirus (Pan et al., 2017). We also demonstrated that SeACoV was able to infect pigs orally (Pan et al., 2017). SeACoV is known by other names, such as porcine enteric alphacoronavirus (PEAV) (Gong et al., 2017), or swine acute diarrhea syndrome (SADS)-CoV (Zhou et al., 2018b). The latter nomenclature is attractive, likely due to name similarity to SARS-CoV (severe acute respiratory syndrome CoV of humans) which may make it easier to gain media attention. However, since the actual pathogenicity of this new virus is still controversial based on current evidence (see main text below), the presumed name “SADS” (also easily confused with “SARS”) may not be appropriate in the future in our opinion. Therefore, SeACoV is the name we will use to refer to this new virus in this review.
Fig. 1.
Map showing geographic location of farms with SeACoV outbreaks. SeACoV was first isolated from clinically sick animals in commercial pig herds of Guangdong province of China in 2017. There were reports of reemergence in Fujian in 2018 and again in Guangdong in 2019. The right panel showed the outbreak of newborn-piglet diarrhea first occurred in four commercial pig farms (marked by yellow stars) in northern Guangdong in 2017.
Very little is known about the molecular biology of SeACoV. This article will comprehensively review the current knowledge of SeACoV with regards to its origin, etiology, epidemiology, evolutionary perspective, potential for interspecies transmission, pathogenesis and diagnosis during the period from 2017 to early 2020.
2. Etiology
2.1. Virion and genome structure
Like the other CoVs, morphological observations by electron microscopy have revealed that SeACoV has the crown-like characteristics typical of the spike (S) protein distribution on the surface of the viral envelope (Pan et al., 2017; Yang et al., 2019a). SeACoV particles are round with a diameter of 100–120 nm (Fig. 2 A), and are covered with dimly visible trimers of S protein (Fig. 2A and B). Bat enteric alphacoronavirus HKU2 strains were first reported from Guangdong province and Hong Kong in 2004 and 2006 during the search for the source of SARS-CoV (Lau et al., 2007). The prototype SeACoV (CH/GD-01/2017 strain) shares 94.9 % nucleotide (nt) sequence identity with the four bat HKU2 strains (Pan et al., 2017). The SeACoV genome is 27,155 nt long with a 5ʹ cap and a 3ʹ polyadenylated tail, and contains a 5ʹ untranslated region (UTR) followed by nine open reading frames (ORFs) and a 3ʹ UTR (Fig. 2C). RNA synthesis in SeACoV is carried out by a replicase-transcriptase composed of 16 nonstructural proteins (Nsp1-16) encoded by ORF1a and ORF1b, which are located in the 5ʹ two-thirds of the genome (Pan et al., 2017; Zhou et al., 2018b). The 3ʹ third harbors ORFs that encode four structural proteins [S; envelope (E); membrane (M); nucleocapsid (N)], an accessory ORF3 between S and E, and two overlapping ORFs (NS7a and NS7b) following the N gene (Yang et al., 2019a; Zhou et al., 2018b) (Fig. 2B and C). These structural and accessory genes are expressed from six subgenomic mRNAs including a bicistronic mRNA containing the accessory NS7a and NS7b genes (Fig. 2D), which have been experimentally confirmed in SeACoV-infected cells (Yang et al., 2019a). The leader-body junction sequences of these subgenomic mRNAs are identical to the leader core sequence AACTAAA (Yang et al., 2019a).
Fig. 2.
Schematic representations of SeACoV genome organization and virion structure. (A) Electron micrograph of a purified SeACoV virus particle, clearly showing the typical viral surface projections of S protein. The scale bar represents 100 nm. (B) Schematic diagram of SeACoV virion structure with color-coded protein components. (C) The structure of SeACoV genomic RNA (27,155 bp) is shown at the top with 5ʹ and 3ʹ UTRs. ORF1a and ORF1b are co-translationally or post-translationally processed to encode a replicase-transcriptase complex composed of 16 nonstructural proteins (Nsp1-16). Four structural proteins [spike (S); envelope (E); membrane (M); and nucleocapsid (N)] are encoded along with three accessory proteins (ORF3, NS7a and NS7b). (D) Genomic and subgenomic mRNAs containing the leader-body junction sites (LS) are shown with colors corresponding to the genome structure.
2.2. Viral proteins
Among the four viral structural proteins, the S glycoprotein of SeACoV is the major protein involved in the viral entry via cellular receptor binding, and it induces host immune responses through the induction of neutralizing antibodies. The SeACoV S protein shares 86.4 % amino acid (aa) identity with the bat HKU2/GD430 strain, supporting a common ancestry (Pan et al., 2017). Interestingly, SeACoV and HKU2 possess unique S genes that are closely related to the betacoronaviruses (Lau et al., 2007; Pan et al., 2017). Most recently, Yu et al reported cryo-EM structures for HKU2 and SeACoV S protein trimers, which are available in the open access preprint repository (Yu et al., 2020). The trimeric structures of HKU2 and SeACoV S proteins (resolutions of 2.38 Å and 2.83 Å, respectively) are very similar, with differences mainly found in the N- and C-terminal domains of the S1 subunit, which is responsible for cell attachment and receptor binding. Both the HKU2 and SeACoV S protein carboxy-terminal domain (CTD) structures have a single-layer core consisting of a twisted, five-stranded antiparallel β sheet similar to the betacoronaviruses (Yu et al., 2020). This suggests recombination between alpha- and betacoronavirus S glycoproteins, and reflects the evolutionary origin of the HKU2/SeACoV S protein at a structural level. Although the S2 subunit (which mediates membrane fusion) is also conserved, the conformation of the region after the fusion peptide of the HKU2-CoV and SeACoV S2 subunits are different from other CoVs. These results provide structural evidence of the close evolutionary relationship between HKU2-related CoV and betacoronavirus S proteins, and provide new insights into CoV evolution and cross-species transmission (Yu et al., 2020).
M and E proteins are major components of the viral envelope and are required for the viral assembly process (de Haan et al., 2000). Apart from that, the N protein is the most highly conserved structural protein among CoVs, and plays an important role in the packaging of virus particles via binding to the viral genomic RNA (Nelson et al., 2000). Specific polyclonal antibodies (pAbs) are usually generated based on viral structural proteins, and immunofluorescence assay using pAbs against N, M and S1 proteins have been used to detect SeACoV infection in cultured cells. Anti-N and anti-M pAbs have also been used for specific detection of SeACoV infection with expected molecular size (42- and 25-KDa, respectively) by western blot (Yang et al., 2019a) or immunohistochemistry (Yang et al., 2019b; Zhou et al., 2018b). In addition, rabbit antiserum against SeACoV Nsp3 acidic domain has been validated for use in the time course analysis of viral protein expression using confocal images, which detected the Nsp3 expression as early as 4 h post-infection in SeACoV-infected cells (Yang et al., 2019a). Accessory proteins play an important role in the immune modulation and viral pathogenesis of CoVs (Liu et al., 2014). Three accessory proteins (ORF3, NS7a and NS7b) have been identified within the SeACoV genome based on analysis of leader-body junctions in specific subgenomic mRNAs, though their functions remain to be confirmed experimentally (Yang et al., 2019a). Similar to the other CoVs like PEDV and TGEV (Ji et al., 2018; Ortego et al., 2003), ORF3 is dispensable for SeACoV infection in vitro since replacement with green fluorescent protein (GFP) in this position does not affect recovery of the recombinant virus (unpublished data).
2.3. Cell and species tropism
SeACoV was first isolated from the feces of piglets with severe watery diarrhea and high rates of mortality (Pan et al., 2017). Among the small intestine, the jejunum is the major target of SeACoV infection in vivo. Vero cells are routinely used for the isolation of novel CoVs; they were used to isolate SeACoV initially (Pan et al., 2017). Virus-positive samples were inoculated and blind-passaged on Vero cell lines plus exogenous trypsin, with cytopathic effect characterized by syncytia formation (Pan et al., 2017). A single-cycle of replication by SeACoV in Vero cells takes approximately 4–6 h (Yang et al., 2019a).
Furthermore, cell lines originating from various tissues of humans and several animal species including bats, mice, rats, gerbils, hamsters, pigs, chickens and nonhuman primates, were tested for viral susceptibility and species tropism. Twenty-one of the 24 tested cell lines showed significant susceptibility to SeACoV infection, defined by efficient viral antigen expression and increased viral RNA titers (Yang et al., 2019b). Notably, SeACoV infects both primary and passaged cell lines of rodent origin, suggesting that rodents may be susceptible to SeACoV infection. In most cases, exogenous trypsin enhanced the propagation of SeACoV in susceptible cell lines. HeLa cells showed the highest susceptibility based on viral load detected in culture supernatants by quantitative RT-PCR (Yang et al., 2019b). This study revealed a remarkably broad spectrum of SeACoV tropism in vitro, and suggested exogenous trypsin treatment and endogenous protease on the cell membrane may activate the fusion of SeACoV S protein for efficient cell entry and release (Yang et al., 2019b).
CoVs are known to utilize different primary cellular receptors to gain entry into host cells, including angiotensin converting enzyme 2 (ACE2) for SARS-CoV and SARS-CoV-2, dipeptidyl peptidase 4 (DPP4) for Middle East respiratory syndrome CoV (MERS-CoV), aminopeptidase N (APN) for TGEV and PDCoV and mouse carcinoembryonic antigen-related cell adhesion molecule 1a (mCEACAM1a) for mouse hepatitis virus (MHV) (Li et al., 2003; Raj et al., 2013; Wang et al., 2018a; Williams et al., 1991; Zhou et al., 2020). However, it has been demonstrated that SeACoV does not utilize any of the known CoV protein receptors for cellular entry (Yang et al., 2019b; Zhou et al., 2018b). Though the receptor for SeACoV has not yet been identified, the results have revealed the most extensive cell tropism in vitro among known CoVs, implying that the functional receptor(s) for SeACoV is likely to be a very common molecule (Yang et al., 2019b). Receptor discovery is a critical direction for future research.
2.4. Reverse genetics
Recovery of infectious virus from plasmids or RNA transcripts encoding cloned cDNAs corresponding to the viral genome is one of the most important experimental platforms for CoV research (Almazan et al., 2014). Recently, our lab developed a DNA-launched reverse genetics system for SeACoV (the infectious clone “pSEA”) (Yang et al., 2019a). The full-length consensus cDNA sequence of SeACoV (27,155 nt) was assembled and inserted into a modified bacterial artificial chromosome (BAC) backbone plasmid that contains a cytomegalovirus promoter, a hepatitis delta virus ribozyme sequence and a bovine growth hormone polyadenylation sequence and terminator. In addition, two silent mutations (A24222T and G24223C) in ORF3 were introduced in pSEA as a genetic marker to differentiate the parental virus. The recombinant SeACoV with the genetic marker can be recovered by direct transfection of cells with pSEA and a helper plasmid expressing the N gene, with similar growth kinetics to the parental virus (Yang et al., 2019a). This plasmid-based system was applied to determine whether a SeACoV non-susceptible cell line, MDCK (canine kidney cells), could confer SeACoV replication competency by transfection of pSEA (Yang et al., 2019b). By introducing desired mutations into viral structural and nonstructural genes, the SeACoV reverse genetics system will facilitate functional analysis of replication and packaging signals, and can be exploited to engineer recombinant SeACoV for design of attenuated vaccines. Most recently, a reporter virus expressing GFP by replacement of ORF3 was generated to track SeACoV in cell culture based on this system in our lab (unpublished data), which will also allow for high-throughout screening of antivirals in the future.
3. Molecular epidemiology
3.1. The emergence of SeACoV in Guangdong, 2017
Beginning from February 2017, a remarkable outbreak of diarrhea in newborn piglets occurred at four pig farms located in Guangdong province of southern China (Fig. 1). The clinical signs of affected pigs were similar to other swine enteric CoVs, characterized by acute vomiting and watery diarrhea. The original diarrheal samples analyzed by RT-PCR were negative for PEDV, TGEV, PDCoV and PHEV, and the pigs had been vaccinated against PEDV. Subsequently a bat-HKU2 origin SeACoV was detected by a pan-CoV RT-PCR assay, and was responsible for a large-scale outbreak that killed 24,693 piglets in commercial farms (Gong et al., 2017; Pan et al., 2017; Zhou et al., 2018b). The four affected farms were distributed around the foot of a mountain (Fig. 1), and there were reports of bat populations nearby.
During early outbreaks, the SeACoV genome sequences identified from three independent groups were 99.8–100 % identical to each other (Fig. 3 ). These SeACoV strains from the same source include “PEAV” GDS04 and GDS04-P12 (GenBank accession nos. MF167434 and MH697599) (Gong et al., 2017; Xu et al., 2019), “SeACoV” CH/GD-01/2017 (MF370205) (Pan et al., 2017), and “SADS-CoV” CN/GDWT/2017 (MG557844), Farm-A to Farm-D (MF094681-MF094684) (Zhou et al., 2018b). Except for an extra 7 nt (ACATGGG) at the 5′-terminus of the genome, no insertions/deletions were found among the five SADS-CoV strains in comparison with SeACoV and its cell adapted strain P10 (MK977618) which is 27,155 nt in genomic length (Fig. 2C). Since SeACoV-p10 could be rescued from the infectious cDNA clone pSEA (Yang et al., 2019a), we suggest that MK977618 may represent a reference strain for sequence comparison. MK977618 is 6 nt longer than the prototype bat CoV HKU2/GD430 strain (27,149 nt), including a 3-nt (TTG) insertion at nt 4,555-4,557 in the nsp3 region, a 6-nt (GGCCTC) insertion at nt 20,508-20,513 in the S gene, and a 3-nt (GTT) deletion at nt 24,780- 24,781 in the M gene.
Fig. 3.
Phylogenetic analysis of the species Rhinolophus bat coronavirus HKU2 and other representative coronaviruses based on nucleotide sequences of the complete genomes. The tree was constructed by the neighbor-joining method using the MEGA X 10.1 software. Bootstrap values are indicated for each node from 1000 resamplings. The names of the viruses and strains as well as their GenBank accession numbers are depicted. The black solid circle indicates the reference sequence SeACoV p10 corresponding to the infectious clone capable of recovering the virus.
Subsequently, a retrospective investigation of SeACoV at 45 pig farms from Guangdong province based on 236 diarrhea samples showed that SeACoV had emerged in Guangdong at least as early as August 2016 (Zhou et al., 2018a). This study also determined two additional genomic sequences CN/GDGL/2017 and CN/GDLS/2017 (MG605090 and MG605091) that shared nearly 100 % nt identity with the sequence CN/GDWT/2017 (MG557844) based on our analysis (Fig. 3).
Since 2010, PEDV variant strains associated with large-scale outbreaks of diarrhea in neonatal piglets have emerged in Mainland China (Huang et al., 2013; Pan et al., 2012). The successive discovery of PDCoV in 2015 and SeACoV in 2017 has taught us that the cause of piglet diarrhea in China may not be as simple as PEDV or TGEV. At present, there remains a lack of nationwide epidemiological investigation of newly emerging porcine enteric CoVs beyond PEDV and TGEV, and thus comprehensive, systematic research is urgently needed.
3.2. Current situation
From May 2017 to January 2019, there were no new SeACoV cases arising in pig herds in Guangdong beyond the first outbreaks. In 2018, a new SeACoV strain (CH/FJWT/2018) was identified in pig stool and small intestine samples from seven pig farms in Fujian outside the Guangdong province (Fig. 1) (Li et al., 2018a). The complete genome (GenBank accession no. MH615810) was 27,168 nt in length, and there were a total of 13 nt insertions compared with the reference strain SeACoV-p10 (99.5 % nt identity) (Fig. 3), including an extra seven nt (ACATGGG) at the 5′-end, a 3-nt (GGC) insertion at nt 22,473-22,474 in the S gene, and a 3-nt (GTA) insertion at nt 24,780- 24,781 in the M gene.
SeACoV reemerged in pig herds in Guangdong starting in February 2019 (Zhou et al., 2019a), with an outbreak of diarrhea that caused the death of about 2000 pigs at another pig farm near the original farms with the first outbreaks. This strain (CN/GDLX/2019, GenBank accession no. MK651076) shared approximately 100 % nt identity with the “Farm-D” strain, and had an S gene that shares high nt identity (99.2–99.9 %) with previously reported SeACoV strains from Guangdong in 2017, but lower identity (97.5 %) with CH/FJWT/2018 from Fujian in 2018 (Fig. 3).
SeACoV has not been reported in other provinces in China or in any other regions in the world. All the identified sequences in pigs to date belong to a sublineage “SeACoV/PEAV/SADS-CoV” which shares >99.5 % nt identities, and is closely related to the bat “HKU2-like CoV” sublineage, together forming the species Rhinolophus bat coronavirus HKU2 in the genus Alphacoronavirus (Fig. 3). Interestingly, some globally distributed rodent CoVs are phylogenetically related to this species (Fig. 3) (Tsoleridis et al., 2019). Taken together, it is not clear whether the sporadic emergences of SeACoV in Guangdong and Fujian from 2016 to 2019 resulted from multiple instances of bat HKU2-like CoVs spillover. Thus, it is necessary to track the evolution of the species Rhinolophus bat coronavirus HKU2 in order to better understand its epidemiology and prevent further outbreaks and spread in China. Further discussion is in Section 5: Transmission.
4. Pathogenesis
The molecular mechanism of how SeACoV antagonizes the host innate immune response has been preliminarily studied. SeACoV interfered with the activity of IPS-1 and RIG-I to inhibit the phosphorylation and nuclear translocation of IRF3 to block IFN-β production (Zhou et al., 2019b). In addition, SeACoV can induce apoptosis via caspase-dependent FasL and mitochondria-mediated apoptotic pathways (Zhang et al., 2020). These studies provide insights into the comprehensive understanding of the interactions between SeACoV and susceptible hosts. A mouse infection model has been established for SeACoV with active viral replication in splenic dendritic cells, which suggests that SeACoV may be susceptible to rodents (Yang et al., 2019b). In this study, mice exhibited only subclinical infection, and seem to be more susceptible by the intraperitoneal route, with modest replication in intestinal tissues and titers declining around 3 days post-infection (Yang et al., 2019b).
The pathogenicity of SeACoV in pigs remains controversial. Initially, it was believed that SeACoV causes enteric disease with a mortality rate over 35 % in piglets less than 10 days old (Zhou et al., 2018b). Its clinical symptoms were similar to PEDV and TGEV, including severe diarrhea, vomiting, and dehydration (Pan et al., 2017; Zhou et al., 2018b). Although the viruses isolated from different groups were able to infect piglets by the oral route, they each caused mild or severe diarrhea symptom (Pan et al., 2017; Xu et al., 2019; Zhou et al., 2018b). In a challenge experiment on 4-day-old piglets using a cell culture strain GDS04-P12, clinical signs developed gradually and the virus was shed in the feces throughout the experimental period (Xu et al., 2019). All piglets (n = 10) challenged with the virus died from 5 to 12 days post-infection (dpi). Examination of the affected pigs revealed that the small intestine had accumulated a large amount of yellow liquid in the colon and the cecum, the intestine was inflated, and the intestinal wall was thin and transparent. SeACoV antigen was detected in the cytoplasm of the villous enterocytes of the challenged piglets by immunohistochemical analysis. Histopathology revealed that the intestinal villi of the duodenum, jejunum, and other places had varying degrees of atrophy, and the intestinal epithelium also showed necrosis (Xu et al., 2019). These results indicated that GDS04-P12 is highly pathogenic to the newborn piglets. A pathogenesis study using the cell culture strain CN/GDWT/2017 from the group that designated the virus “SADS-CoV” also showed high mortality (3/6) in the inoculated 3-day-old piglets (Zhou et al., 2018b). However, immunohistochemical analysis displayed only a few jejunal epithelial cells that were stained with viral antigen in challenged piglets, which may not be in line with the 50 % mortality (Zhou et al., 2018b). We have conducted a couple of experimental infection of neonatal piglets with a moderate or high dose of purified SeACoV (isolate CH/GD-01/2017), and they resulted only in mild-moderate diarrheal signs or subclinical infection compared to the first outbreaks in pig herds [(Pan et al., 2017) and unpublished data], implying a lower pathogenicity of SeACoV.
The discrepancies in the results from different challenged studies are very confusing since these SeACoV strains were obtained from the same source in the first outbreaks. One explanation may be attributed to the sequence differences in the SeACoV cell adapted strains used for challenge. By comparative analysis, we found that the sequences of CH/GD-01/2017 and CN/GDWT/2017 are almost identical at the genome level (99.9 %), with no insertions and deletions, whereas GDS04-P12 shares 99.7 % or 99.8 % nt identity with CH/GD-01/2017 or CN/GDWT/2017, repectively. GDS04-P12 has 14 nt in gaps, including a 1-nt deletion (A) at nt 179, a 10-nt insertion (GACTAGAGCC) at nt 12,483-12,494, a 1-nt insertion (A) at nt 27,058-27,059, a 1-nt insertion (T) at nt 27,067-27,068, and a 1-nt insertion (A) in nt 27,074-27,075. The other factors, such as the age of the piglets (all were 3- or 4-day-old), the source of the piglets (conventional status), and the geographical environment where these experiments were performed (all were done in Guangdong), may not contribute to the observed differences.
Therefore, future studies based on our recently developed SeACoV reverse genetics system are warranted to investigate this important question. We hope to reconstruct the wild-type strain from the original sample by using an infectious clone, and conduct a comparative challenge experiment to further clarify the true pathogenicity of SeACoV.
5. Transmission
Like other porcine enteric viruses, SeACoV is transmitted through the fecal-oral route. This has been demonstrated by challenge experiments in pigs using the cell-cultured SeACoV strains (Pan et al., 2017; Xu et al., 2019; Zhou et al., 2018b). Diarrhea or vomitus from infected piglets is likely the major transmission route of SeACoV. Other swine CoVs may be able to utilize different routes of infection. For example, PEDV invasion through the nasal cavity (respiratory tract) can also cause intestinal mucosal disease (Li et al., 2018b), and PRCV is a pathogenic respiratory mutant of TGEV (Wesley and Lager, 2003). Whether aerosolized SeACoV is also infectious should be investigated in the future.
Considering that the zoonotic SARS-CoV and MERS-CoV originated from bats and jumped between intermediate hosts (civets and camels, respectively), SeACoV may pose a similar risk to human health through transmission from pigs or other potential intermediate hosts (Yang et al., 2019b). The zoonotic potential of SeACoV was assessed in vitro, showing that SeACoV was able to grow efficiently in nine human and monkey cell lines. As a conserved receptor across different animal species for SeACoV cell entry has been proposed, SeACoV may pose a similar risk to human health through interspecies transmission from pigs in the future (Fig. 4 ).
Fig. 4.
Potential routes of interspecies transmission between HKU2-origin SeACoV and different hosts. The ability of SeACoV to infect various rodent cell lines and replicate in a murine infection model suggests rodents as a potentially susceptible host. The solid arrow indicates the known SeACoV transmission event from bat to pig that occurred in 2017. The dashed arrows with question marks indicate hypothetical or potential transmissions from bat to rodent, pig to rodent or vice versa, and pig to human. See Section 5 for details of the proposed model.
The direct progenitor of SeACoV is not the prototype HKU2-CoV identified in Rhinolophus affinis in 2004 and 2006, due to a low sequence identity (86 %) of the S gene (Pan et al., 2017; Zhou et al., 2018b). Rather, a couple of HKU2-like CoV sequences, designated SADSr-CoV (Fig. 3), were determined in several species of horseshoe bats such as Rhinolophus affinis, Rhinolophus sinicus, and Rhinolophus rex from seven different locations in Guangdong province between 2013 and 2016 (Zhou et al., 2018b). The S proteins of two SADSr-CoVs identified from Rhinolophus affinis were more closely related to that of SeACoV (∼98 % aa sequence identity), indicating that horseshoe bats are reservoirs of HKU2-like CoVs. The phylogenetic, haplotype network and recombination analyses suggested that SeACoV had probably been introduced from these HKU2-like CoVs independently, and that some strains might have undergone further intragenotypic recombination (Zhou et al., 2018b). However, it is still not clear how the virus was transmitted from bats to pigs. In fact, it is hardly possible that pigs in captivity could contact flying bats directly in the four pig farms with disease outbreaks.
A possible scenario for such transmission may include bats infected with HKU2-like CoVs preying on insects near pig facilities, dropping contaminated feces that were later introduced into the pens somehow, by pig feed or some kind of animal (Fig. 4). According to our onsite observation in 2017, rodents were frequently visible in these pig farms. Notably, bat HKU2-like CoVs are clustered with rat CoVs in the genus Alphacoronavirus (Fig. 3). As we found that SeACoV infects different cell lines originating from rodents, and mice may be susceptible to SeACoV experimental infection (Yang et al., 2019b), we hypothesize that in such field conditions, rodents (especially wild rats) in the farms may eat pig feed contaminated by bat feces, becoming carriers of SeACoV (Fig. 4). Alternatively, if pigs became infected and shed SeACoV-positive feces, the virus could begin circulating in pig facilities. Contamination of pig feed, pig feces and water supplies by rodents could accumulate and develop into outbreaks of diarrhea in neonatal piglets (Fig. 4). Future studies on identifying HKU2-like CoV positive samples in rodents near pig farms are warranted to test this hypothesis.
6. Diagnostic methods
With increasing throughput and decreasing costs, next-generation sequencing (NGS) technology is increasingly being used for diagnosis of pathogens in clinical samples. In fact, except for initial isolation (Pan et al., 2017), complete genome sequences of SeACoV strains have been determined by assembling and mapping sequencing reads with NGS data initially by two independent groups (Gong et al., 2017; Zhou et al., 2018b). Current SeACoV diagnostic assays can be divided into two categories: molecular and serological methods. Molecular methods detect viral nucleic acid, while serological assays detect the antibodies induced in host immune responses to infection. Since the initial outbreak of SeACoV in 2017, multiple detection methods have been established to monitor the epidemic. A summary of current SeACoV diagnostic assays and their properties are presented in Table 1, Table 2 .
Table 1.
Summary of virological methods for SeACoV RNA detection.
| Method | Target Gene | Primer or Probe | Sequences (5'–3') | Limit of Detection | Principle | Reference |
|---|---|---|---|---|---|---|
| RT-PCR assay | N gene | SEAF | ATGGATAAACCTGAATGGAAGCG | Not defined | Identified SeACoV from fecal samples. | Pan et al. (2017) |
| SEAR | CACCATCTCAACCTCTTCCTCAG | |||||
| M gene | Forward primer | GGTCCCTGTGACCGAAGTTTTAG | Not defined | Retrospective detection of SeACoV in Guangdong Province. | Zhou et al. (2018a) | |
| Reverse primer | GCGTTCTGCGATAAAGCTTAAAACTATTA | |||||
| LAMP-RT-PCR assay | N gene | Outer primers: SADS-F3 | CAGCCTTCTAACTGGCACTT | 10 RNA copies/μL | A simple and rapid detected method for the result can be directly observed. | Wang et al. (2018b) |
| Outer primers: SADS-B3 | ACAGTCAGGTCTGGTGGTAA | |||||
| Inner primers: SADS -FIP | CGTCAACAGCGACCCAATGCATCCTCACGCAGATGCTCC | |||||
| Inner primers: SADS -BIP | AACTAGCCCCACAGGTCTTGGTAACCCAAACTGAGGTGTAGC | |||||
| Loop primers: SADS -LB | TCGCAATCGTAACAAAGAACCT | |||||
| Loop primers: SADS -LF | CACCCTGAATCCGTTTCCTG | |||||
| TaqMan-based real-time RT-PCR assay | N gene | qSADS-N-F | CTGACTGTTGTTGAGGTTAC | 30 DNA copies/μL | More sensitive than the conventional PCR. | Zhou et al. (2018a) |
| qSADS-N-R | TCTGCCAAAGCTTGTTTAAC | |||||
| Probe | FAM-TCACAGTCTCGTTCTCGCAATCA-TAMRA | |||||
| N gene | Forward primer | CTAAAACTAGCCCCACAGGTC | 100 RNA copies/μL | To detect viral RNA in feces and various tissues of the mouse model experiment. | Yang et al. (2019b) | |
| Reverse primer | TGATTGCGAGAACGAGACTG | |||||
| Probe | FAM-GAAACCCAAACTGAGGTGTAGCAGG-TAMRA | |||||
| N gene | Forward primer | GCACTTTTATTACCTTGGTA | Not defined | To detect viral RNA in feces and various tissues of the piglets challenge experiment. | Xu et al. (2019) | |
| Reverse primer | GTAGCAGGTTCTTTGTTAC | |||||
| Probe | FAM-TCCTCACGCAGATGCTCCTT-TAMRA | |||||
| TaqMan-based Multiplex real-time RT-PCR assay | N gene | Forward primer | TCTCGGCTTACTCTAAACCC | 110 RNA copies/μL | A multiplex real-time RT-qPCR assay for detection of swine enteric coronaviruses. | Huang et al. (2019) |
| Reverse primer | CATCCACCATCTCAACCTC | |||||
| Probe | TexasRed-AAGACCTAAATGCTGATGCCCCA-BHQ2 | |||||
| SYBR green-based real-time RT-PCR assay | N gene | SADS-N-F | CTAAAACTAGCCCCACAGGTC | Not defined | To detect SeACoV in acutely sick piglets and sows with diarrhea. | Zhou et al. (2018b) |
| SADS-N-R | TGATTGCGAGAACGAGACTG | |||||
| RdRp | SADS-RdRp-F | GTTGATTGTAAGGCTTGGCG | Not defined | To detect bat anal swabs samples and determined SADSr-CoV. | Zhou et al. (2018b) | |
| SADS-RdRp-R | AACCACACTTCCACTCAGC | |||||
| M gene | Forward primer | ATGTGGCTCCTATGGCCCTT | 10 RNA copies/μL | More sensitive than probe-based RT-qPCR. | Ma et al. (2018) | |
| Reverse primer | ACGTTCGCGTTCTGCGATAA | |||||
| RdRp | qSADS-CoV-ORF1 F | AGTGAAAGACCACAGCAAACAG | Not defined | To detect multiple-shRNA-mediated inhibition of viral RNA replication. | Li et al. (2019) | |
| qSADS-CoV-ORF1 R | TAACATACGCCCAGCAACATAG | |||||
| S gene | qSADS-CoV-S F | TTGTGAGAGTGATGAATTGGGT | Not defined | |||
| qSADS-CoV-S R | GATTTTCTGGTTTGTAAAGGTT |
Table 2.
Summary of serological methods for SeACoV antibody detection.
| Method | Target protein | Principle | Reference |
|---|---|---|---|
| IFA | Live virus | To examine SeACoV antibody from pig sera in SeACoV-infected cells | Pan et al. (2017) |
| ELISA | S1 domain | Antibody assay based on the S1 domain of the S protein using a luciferase immunoprecipitation system | Zhou et al. (2018b) |
| Virus particles | Purified virus particles as antigen to detect SeACoV antibodies in the serum from the infected mice | Yang et al. (2019b) | |
| Not defined | Serological investigation of recent SADS cases implied that SADS-CoV, rather than PEDV, may directly contribute to the diarrhea | Zhou et al. (2019a) |
6.1. PCR-based assays
PCR is one of the most commonly used rapid detection methods in clinical diagnosis. In order to identify the etiological agent associated with outbreaks of diarrhea in newborn piglets, a pair of conserved primers was used to confirm a 251bp fragment in ORF1b by conventional RT-PCR of a region 100 % identical to the corresponding part of four bat HKU2-CoV strains (Pan et al., 2017). After the complete genome of SeACoV was determined, multiple rapid PCR detection methods were subsequently developed (Table 1). Reverse transcription loop-mediated isothermal amplification (RT-LAMP) and real-time RT-PCR assays were established for the detection of SeACoV (Wang et al., 2018b; Zhang et al., 2019; Zhou et al., 2018a). Real-time RT-PCR is more powerful than RT-LAMP, with greater sensitivity and reproducibility for better quantification of pathogen. SYBR green-based and TaqMan-based methods have been developed for several SeACoV isolates (Ma et al., 2018). Since the S gene is more prone to mutation and recombination, PCR-based methods mainly target the relatively conserved N and M gene. In addition there is a multiplex real-time RT-qPCR assay for simultaneous detection of swine enteric PEDV, PDCoV (both targeting the M gene), TGEV and SeACoV (both targeting the N gene) (Huang et al., 2019).
6.2. Serological assays
The most commonly used serological assays include the indirect fluorescent assay (IFA) and enzyme linked immunosorbent assay (ELISA). The IFA detection method is mainly used in laboratory research on the mechanism of viral replication and infection. SeACoV is antigenically distinct from PEDV, TGEV and PDCoV, and antibodies against the N protein do not cross-react (Pan et al., 2017). In addition, our preliminary result showed that virus-specific sera from SeACoV, PEDV, TGEV and PDCoV-infected pigs do not cross-react in IFA tests (unpublished data). Since SeACoV was first isolated in pigs vaccinated with PEDV vaccines (Pan et al., 2017; Zhou et al., 2018b), this evidence suggests that the existing porcine CoV vaccines may not be effective against SeACoV infection.
While ELISA can be used to monitor changes of antibody levels in field herds under clinical situations, it can also be used retrospectively to detect viruses and IgA/IgG levels in serum to provide guidance for vaccine use. The ELISA detection methods for SeACoV are only available for use in the laboratory, for the epidemiological investigation of the initial virus outbreak and reemergence (Table 2). The ELISA uses a luciferase immunoprecipitation system based on the S1 protein, and has been used to investigate possible zoonotic transmission (Zhou et al., 2018b). In addition, there is a virion-based ELISA detection method designed for sample detection in mouse model experiments, which can also be used for pig serum samples (Yang et al., 2019b).
7. Conclusion
As early as 2004, the HKU2-related Rhinolophus bat CoV was identified (Lau et al., 2007), and the first outbreak of acute diarrhea caused by SeACoV in pigs appeared in 2017 (Pan et al., 2017). Although the virus reemerged in Fujian in 2018 and Guangdong in 2019, there have not been large-scale losses reported, which is positive news for its prevention and control. However, it is strange that SeACoV has not resulted in a large-scale epidemic in the pig industry in Guangdong in recent years, similar to the way SARS-CoV emerged in 2003 and then basically disappeared (Ksiazek et al., 2003) until the emergence of SARS-CoV-2 at the end of 2019 (Zhou et al., 2020). This outbreak of the novel SARS-CoV-2 and current pandemic also requires strict control to ensure the health of people around the world. Since all CoVs have the capability for recombination, no CoV epidemic can be taken lightly, even those in pigs (Opriessnig and Huang, 2020). To prevent future outbreaks, unified detection methods are needed to establish better surveillance of SeACoV, including virological and serological methods.
More research is needed to understand the infectious properties of SeACoV and HKU2-related CoVs, especially given their broad ability to infect cell lines from various animal origins (Yang et al., 2019b). The presumed name “SADS-CoV” may not be appropriate since the available pathogenicity studies have not provided strong evidence of high virulence. The reverse genetic system for SeACoV has been established successfully in our lab (Yang et al., 2019a), which will facilitate future studies exploring the molecular mechanisms of SeACoV replication and pathogenicity. With the structural analysis of SeACoV and HKU2 S glycoproteins, it is also important to identify the primary receptor and study the mechanism of receptor binding. It is clear now more than ever that CoV-induced diseases not only pose a huge threat to the pig industry but also to human health. If HKU2-related CoVs emerge again, causing unknown damage to the pig industry, such basic research on SeACoV will be necessary to prevent and control future epidemics.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (31872488 and 32041003), Zhejiang University special scientific research fund for COVID-19 prevention and control (2020XGZX084), and the Fundamental Research Funds for the Central Universities of China (2019FZA6014).
References
- Almazan F., Sola I., Zuniga S., Marquez-Jurado S., Morales L., Becares M., Enjuanes L. Coronavirus reverse genetic systems: infectious clones and replicons. Virus Res. 2014;189:262–270. doi: 10.1016/j.virusres.2014.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot R.J., Baker S.C., Baric R., Enjuanes L., Gorbalenya A.E., Holmes K.V., Perlman S., Poon L., Rottier P.J.M., Talbot P.J., Woo P.C.Y., Ziebuhr J. Coronaviridae. In: King A.M.Q., Adams M.J., Carstens E.B., Lefkowitz E.J., editors. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press; London: 2011. pp. 806–828. [Google Scholar]
- de Haan C.A., Vennema H., Rottier P.J. Assembly of the coronavirus envelope: homotypic interactions between the M proteins. J. Virol. 2000;74(11):4967–4978. doi: 10.1128/jvi.74.11.4967-4978.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong L., Li J., Zhou Q., Xu Z., Chen L., Zhang Y., Xue C., Wen Z., Cao Y. A new Bat-HKU2-like coronavirus in swine, China, 2017. Emerg. Infect. Dis. 2017;23(9):1607–1609. doi: 10.3201/eid2309.170915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greig A.S., Mitchell D., Corner A.H., Bannister G.L., Meads E.B., Julian R.J. A hemagglutinating virus producing encephalomyelitis in baby pigs. Can. J. Comp. Med. Vet. Sci. 1962;26(3):49–56. [PMC free article] [PubMed] [Google Scholar]
- Huang X., Chen J., Yao G., Guo Q., Wang J., Liu G. A TaqMan-probe-based multiplex real-time RT-qPCR for simultaneous detection of porcine enteric coronaviruses. Appl. Microbiol. Biotechnol. 2019;103(12):4943–4952. doi: 10.1007/s00253-019-09835-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.W., Dickerman A.W., Pineyro P., Li L., Fang L., Kiehne R., Opriessnig T., Meng X.J. Origin, evolution, and genotyping of emergent porcine epidemic diarrhea virus strains in the United States. MBio. 2013;4(5):e00737–00713. doi: 10.1128/mBio.00737-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji C.M., Wang B., Zhou J., Huang Y.W. Aminopeptidase-N-independent entry of porcine epidemic diarrhea virus into Vero or porcine small intestine epithelial cells. Virology. 2018;517:16–23. doi: 10.1016/j.virol.2018.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A.E., Humphrey C.D., Shieh W.J., Guarner J., Paddock C.D., Rota P., Fields B., DeRisi J., Yang J.Y., Cox N., Hughes J.M., LeDuc J.W., Bellini W.J., Anderson L.J., Group S.W. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348(20):1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- Lau S.K., Woo P.C., Li K.S., Huang Y., Wang M., Lam C.S., Xu H., Guo R., Chan K.H., Zheng B.J., Yuen K.Y. Complete genome sequence of bat coronavirus HKU2 from Chinese horseshoe bats revealed a much smaller spike gene with a different evolutionary lineage from the rest of the genome. Virology. 2007;367(2):428–439. doi: 10.1016/j.virol.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K., Li H., Bi Z., Gu J., Gong W., Luo S., Zhang F., Song D., Ye Y., Tang Y. Complete genome sequence of a novel swine acute diarrhea syndrome coronavirus, CH/FJWT/2018, isolated in Fujian, China, in 2018. Microb. Resour. Announc. 2018;7(22) doi: 10.1128/MRA.01259-18. pii: e01259-01218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Wu Q., Huang L., Yuan C., Wang J., Yang Q. An alternative pathway of enteric PEDV dissemination from nasal cavity to intestinal mucosa in swine. Nat. Commun. 2018;9(1):3811. doi: 10.1038/s41467-018-06056-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K., Li H., Bi Z., Song D., Zhang F., Lei D., Luo S., Li Z., Gong W., Huang D., Ye Y., Tang Y. Significant inhibition of re-emerged and emerging swine enteric coronavirus in vitro using the multiple shRNA expression vector. Antiviral Res. 2019;166:11–18. doi: 10.1016/j.antiviral.2019.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D.X., Fung T.S., Chong K.K., Shukla A., Hilgenfeld R. Accessory proteins of SARS-CoV and other coronaviruses. Antiviral Res. 2014;109:97–109. doi: 10.1016/j.antiviral.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Zeng F., Cong F., Huang B., Huang R., Ma J., Guo P. Development of a SYBR green-based real-time RT-PCR assay for rapid detection of the emerging swine acute diarrhea syndrome coronavirus. J. Virol. Methods. 2018;265:66–70. doi: 10.1016/j.jviromet.2018.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson G.W., Stohlman S.A., Tahara S.M. High affinity interaction between nucleocapsid protein and leader/intergenic sequence of mouse hepatitis virus RNA. J. Gen. Virol. 2000;81(Pt. 1):181–188. doi: 10.1099/0022-1317-81-1-181. [DOI] [PubMed] [Google Scholar]
- Opriessnig T., Huang Y.W. Coronavirus disease 2019 (COVID-19) outbreak: could pigs be vectors for human infections? Xenotransplantation. 2020;27(2):e12591. doi: 10.1111/xen.12591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortego J., Sola I., Almazan F., Ceriani J.E., Riquelme C., Balasch M., Plana J., Enjuanes L. Transmissible gastroenteritis coronavirus gene 7 is not essential but influences in vivo virus replication and virulence. Virology. 2003;308(1):13–22. doi: 10.1016/S0042-6822(02)00096-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y., Tian X., Li W., Zhou Q., Wang D., Bi Y., Chen F., Song Y. Isolation and characterization of a variant porcine epidemic diarrhea virus in China. Virol. J. 2012;9:195. doi: 10.1186/1743-422X-9-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y., Tian X., Qin P., Wang B., Zhao P., Yang Y.L., Wang L., Wang D., Song Y., Zhang X., Huang Y.W. Discovery of a novel swine enteric alphacoronavirus (SeACoV) in southern China. Vet. Microbiol. 2017;211:15–21. doi: 10.1016/j.vetmic.2017.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pensaert M., Callebaut P., Vergote J. Isolation of a porcine respiratory, non-enteric coronavirus related to transmissible gastroenteritis. Vet. Q. 1986;8(3):257–261. doi: 10.1080/01652176.1986.9694050. [DOI] [PubMed] [Google Scholar]
- Pensaert M.B., de Bouck P. A new coronavirus-like particle associated with diarrhea in swine. Arch. Virol. 1978;58(3):243–247. doi: 10.1007/BF01317606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj V.S., Mou H., Smits S.L., Dekkers D.H., Muller M.A., Dijkman R., Muth D., Demmers J.A., Zaki A., Fouchier R.A., Thiel V., Drosten C., Rottier P.J., Osterhaus A.D., Bosch B.J., Haagmans B.L. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495(7440):251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsoleridis T., Chappell J.G., Onianwa O., Marston D.A., Fooks A.R., Monchatre-Leroy E., Umhang G., Muller M.A., Drexler J.F., Drosten C., Tarlinton R.E., McClure C.P., Holmes E.C., Ball J.K. Shared common ancestry of rodent alphacoronaviruses sampled globally. Viruses. 2019;11(2) doi: 10.3390/v11020125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Liu Y., Ji C.M., Yang Y.L., Liang Q.Z., Zhao P., Xu L.D., Lei X.M., Luo W.T., Qin P., Zhou J., Huang Y.W. Porcine deltacoronavirus engages the transmissible gastroenteritis virus functional receptor porcine aminopeptidase N for infectious cellular entry. J. Virol. 2018;92(12):e00318–318. doi: 10.1128/JVI.00318-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Cong F., Zeng F., Lian Y., Liu X., Luo M., Guo P., Ma J. Development of a real time reverse transcription loop-mediated isothermal amplification method (RT-LAMP) for detection of a novel swine acute diarrhea syndrome coronavirus (SADS-CoV) J. Virol. Methods. 2018;260:45–48. doi: 10.1016/j.jviromet.2018.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Vlasova A.N., Kenney S.P., Saif L.J. Emerging and re-emerging coronaviruses in pigs. Curr. Opin. Virol. 2019;34:39–49. doi: 10.1016/j.coviro.2018.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley R.D., Lager K.M. Increased litter survival rates, reduced clinical illness and better lactogenic immunity against TGEV in gilts that were primed as neonates with porcine respiratory coronavirus (PRCV) Vet. Microbiol. 2003;95(3):175–186. doi: 10.1016/S0378-1135(03)00150-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams R.K., Jiang G.S., Holmes K.V. Receptor for mouse hepatitis virus is a member of the carcinoembryonic antigen family of glycoproteins. Proc. Natl. Acad. Sci. U. S. A. 1991;88(13):5533–5536. doi: 10.1073/pnas.88.13.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Lam C.S., Lau C.C., Tsang A.K., Lau J.H., Bai R., Teng J.L., Tsang C.C., Wang M., Zheng B.J., Chan K.H., Yuen K.Y. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012;86(7):3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Zhang Y., Gong L., Huang L., Lin Y., Qin J., Du Y., Zhou Q., Xue C., Cao Y. Isolation and characterization of A Highly Pathogenic Strain of Porcine enteric alphacoronavirus Causing Watery Diarrhea and High Mortality in Newborn Piglets. Transbound. Emerg. Dis. 2019;66(1):119–130. doi: 10.1111/tbed.12992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.L., Liang Q.Z., Xu S.Y., Mazing E., Xu G.H., Peng L., Qin P., Wang B., Huang Y.W. Characterization of a novel bat-HKU2-like swine enteric alphacoronavirus (SeACoV) infection in cultured cells and development of a SeACoV infectious clone. Virology. 2019;536:110–118. doi: 10.1016/j.virol.2019.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.L., Qin P., Wang B., Liu Y., Xu G.H., Peng L., Zhou J., Zhu S.J., Huang Y.W. Broad cross-species infection of cultured cells by bat HKU2-related swine acute diarrhea syndrome coronavirus and identification of its replication in murine dendritic cells in vivo highlight its potential for diverse interspecies transmission. J. Virol. 2019;93(24):e01448–1419. doi: 10.1128/JVI.01448-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Qiao S., Guo R., Wang X. Cryo-EM structures of HKU2 and SADS-CoV spike glycoproteins and insights into coronavirus evolution. bioRxiv. 2020 doi: 10.1038/s41467-020-16876-4. 2020.2002.2023.961912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Luo S., Gu J., Li Z., Li K., Yuan W., Ye Y., Li H., Ding Z., Song D., Tang Y. Prevalence and phylogenetic analysis of porcine diarrhea associated viruses in southern China from 2012 to 2018. BMC Vet. Res. 2019;15(1):470. doi: 10.1186/s12917-019-2212-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Han Y., Shi H., Chen J., Zhang X., Wang X., Zhou L., Liu J., Zhang J., Ji Z., Jing Z., Ma J., Shi D., Feng L. Swine acute diarrhea syndrome coronavirus-induced apoptosis is caspase- and cyclophilin D- dependent. Emerg. Microbes Infect. 2020;9(1):439–456. doi: 10.1080/22221751.2020.1722758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Li Q.N., Su J.N., Chen G.H., Wu Z.X., Luo Y., Wu R.T., Sun Y., Lan T., Ma J.Y. The re-emerging of SADS-CoV infection in pig herds in Southern China. Transbound. Emerg. Dis. 2019;66(5):2180–2183. doi: 10.1111/tbed.13270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Sun Y., Lan T., Wu R.T., Chen J.W., Wu Z.X., Xie Q.M., Zhang X.B., Ma J.Y. Retrospective detection and phylogenetic analysis of swine acute diarrhea syndrome coronavirus in pigs in southern China. Transbound. Emerg. Dis. 2018;66(2):687–695. doi: 10.1111/tbed.13008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Fan H., Lan T., Yang X.L., Shi W.F., Zhang W., Zhu Y., Zhang Y.W., Xie Q.M., Mani S., Zheng X.S., Li B., Li J.M., Guo H., Pei G.Q., An X.P., Chen J.W., Zhou L., Mai K.J., Wu Z.X., Li D., Anderson D.E., Zhang L.B., Li S.Y., Mi Z.Q., He T.T., Cong F., Guo P.J., Huang R., Luo Y., Liu X.L., Chen J., Huang Y., Sun Q., Zhang X.L., Wang Y.Y., Xing S.Z., Chen Y.S., Sun Y., Li J., Daszak P., Wang L.F., Shi Z.L., Tong Y.G., Ma J.Y. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature. 2018;556(7700):255–258. doi: 10.1038/s41586-018-0010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Sun Y., Yan X., Tang X., Li Q., Tan Y., Lan T., Ma J. Swine acute diarrhea syndrome coronavirus (SADS-CoV) antagonizes interferon-beta production via blocking IPS-1 and RIG-I. Virus Res. 2019;278 doi: 10.1016/j.virusres.2019.197843. [DOI] [PMC free article] [PubMed] [Google Scholar]