Abstract
Purpose
To investigate if shotgun-sequencing method could be useful in detailed diagnosis of herpes simplex virus (HSV) infection and compare it with the conventional diagnostic method.
Observations
Using a sterile scraper, the infectious part of the ocular surface was scraped gently and placed on a glass slide for conventional diagnosis using PCR and histology and in RNA stabilizing reagent for shotgun sequencing respectively. Concentration of the DNA was determined using a sensitive fluorescence dye-based Qubit dsDNA HS Assay Kit. Shotgun-sequencing libraries were generated using the NEBNext DNA ultra II protocol. The samples were sequenced on the Illumina NextSeq 500 in high output mode with 2X150 bp paired-end sequencing. Taxonomic and functional profiles were generated.
Conventional diagnostic method suspected herpetic keratitis. The results indicated presence of an amplified product of 92 bp positive HSV-DNA. Conventional diagnostic method detected the presence of Herpes Simplex Virus DNA (type 1). Shotgun sequencing confirmed the diagnosis of HSV along with the taxonomical profiling of the virus. These results were achieved using 1.9 ng/μL of DNA concentration (114 ng in 60 μL) of the total sample volume.
Conclusions and importance
Shotgun sequencing is a hypothesis-free approach that identifies full taxonomic and functional profile of an organism. This technology is advantageous as it requires smaller sample size compared to conventional diagnostic methods.
Keywords: Sequencing, Shotgun, Diagnosis, Cornea, Infection, Virus
1. Introduction
Corneal infections can arise following a corneal transplant or due to other external causes. As cornea is avascular, it provides an immune privilege. Hence infections could possibly lead to a slower recovery.1 Often, an urgent treatment is required in cases of corneal infections to reduce any possible damage in the deeper layers of the tissue. Therefore, diagnosis of the primary causative agent becomes mandatory. Conventional methods for the detection of infectious microbes are limited to just defining the class of an organism. Shotgun sequencing allows a hypothesis-free approach to identify the full taxonomic and functional profile of an organism. As it amplifies the targeted sample, even low volumes of the samples could be sufficient to increase the diagnostic yield.
Herpes simplex virus (HSV) has been found to be a leading cause of infectious corneal blindness.2 Approximately 0.5 million people in the United States are currently infected with ocular HSV.3 Not only the prevalence, but also the costs associated with the diagnosis and treatment of this infection is significantly rising for both, unilateral and bilateral cases.4, 5, 6, 7 Primarily, this infection can be diagnosed by its clinical presentation, but uncommon cases of this infection can further obstruct accurate diagnosis and hence appropriate treatment.8 Other approaches like PCR, ELISA, immunofluorescent assays and viral cultures have been used currently for specific diagnosis, although with some limitations. Therefore, it is necessary to identify a diagnostic approach that should be more accurate in terms of sensitivity, specificity and reliability to prevent serious consequences following HSV infection.9 We recently published an article showing the advantages and disadvantages of next generation sequencing for the detection of microorganisms present in human donor corneal preservation medium, where we discussed the use of 16S and 18S methods in an eye bank setting.10 This article intends to investigate if shotgun-sequencing method could be useful clinically to carry out detailed diagnosis of a patient suffering from HSV infection and compare it with the conventional diagnostic method based on histology and PCR.
2. Materials and methods
As conventional diagnostic method is routinely performed clinically, no specific consent was obtained, but the patient was fully informed about the diagnostic procedures. The patient was diagnosed with keratoconus and treated with deep anterior lamellar keratoplasty in the left eye followed by right eye. The sutures were removed following which the patient had a visual acuity reduction and complained of superficial discomfort, which was treated with topical steroids. An increased intraocular pressure was observed with redness and pain. Oral herpes was detected and the ocular therapy was continued simultaneously with oral steroids. However, no improvement was observed and the patient was suspected for Herpetic Keratitis.
Routine eye check-up was performed using a slit lamp. A sterile scraper was used to scrape the infectious part of the ocular surface (not scraping too deep into the stroma) twice. The first scrape was gently placed on a glass slide for conventional diagnostic method using histology and PCR. Amplification of genomic fragments of HSV (specific region of type I and common region of type I and II) was carried out. Simultaneously, the second scrape was placed in RNA stabilizing reagent that was preserved at room temperature till shipped and analyzed using shotgun sequencing.
The concentration of the DNA was determined using a sensitive fluorescence dye-based Qubit dsDNA HS Assay Kit. For the metagenomic analysis, shotgun-sequencing libraries were generated using the NEBNext DNA ultra II protocol. The samples were quality controlled and successfully sequenced on the Illumina NextSeq 500 next generation sequencing system in high output mode with 2X150 bp paired-end sequencing. Conversion to FastQ files and de-multiplexing of the reads according to their respective index was performed using the bcl2fastq software tool. The resulting reads were quality controlled, trimmed and mapped against the humane reference genome. The un-mapped reads were taxonomically classified using the Kraken software. Metagenomes were assembled from un-mapped reads using CLC genomics workbench and its microbial genomics module. Functional profiles were generated for these metagenomes separately for best Blast hit annotation, protein families (Pfam) domain annotation and GO term annotation respectively.
Meanwhile, the patient was treated with systemic antiviral tablet (acyclovir) and topical drops, betamethasone 0.2% plus chloramphenicol 0.5% (betabioptal) and sulfamethizole 5% plus tetracycline 1% (pensulvit) being given for post herpetic re-epithelialization. At 3 months, a slight improvement in the reduction of the infection was observed.
3. Results
Conventional diagnostic method suspected herpetic keratitis. Fibro-necrotic material incorporating ‘activated’ stromal cell elements were observed. The epithelium showed nuclear hyperchromatism of regenerative source, some of them multinucleated with nuclear ‘clarification’ aspects, probably secondary to viral cytopathic effect. The results indicated presence of an amplified product of 92 bp, positive HSV-DNA. This method detected the presence of Herpes Simplex Virus DNA (type 1). Histology and PCR analysis indicated that the causative agent was HSV Type 1. With a total of 32,029,553 cumulative sequenced reads in the sample, shotgun sequencing confirmed the diagnosis of HSV along with the taxonomical profiling of this virus as shown in Table 1. These results were achieved using 1.9 ng/μL of DNA concentration (114 ng in 60 μL) of the total sample volume. This indicates that especially for cases of corneal infections, where the amount of the acquired sample is a major limitation, techniques like shotgun sequencing that could analyze a complete database of the given microbiome with relatively small amount of the sample volume, could be highly advantageous.
Table 1.
Taxonomical profiling of HSV from the patient suffering from Herpetic Keratitis including the number of reads per sample.
| Taxonomy | Number of reads |
|---|---|
| Viruses|x__dsDNA_viruses, _no_RNA_stage|o__Herpesvirales|f__Herpesviridae|x__Alphaherpesvirinae|g__Simplexvirus | 124,439 |
| Viruses|x__dsDNA_viruses, _no_RNA_stage|o__Herpesvirales|f__Herpesviridae|x__Alphaherpesvirinae|g__Simplexvirus|s__Human_alphaherpesvirus_1 | 124,091 |
| Viruses|x__dsDNA_viruses, _no_RNA_stage|o__Herpesvirales|f__Herpesviridae|x__Alphaherpesvirinae|g__Simplexvirus|s__Human_alphaherpesvirus_2 | 36 |
4. Discussion
Target specific diagnosis of an infection is limited as it still relies on conventional microbial culture techniques for the identification of a pathogen. Shotgun approach has a potential to take over as a valid alternative to conventional diagnostic method, as it demonstrates the nature of a given infection. It further allows full taxonomical profiling of the infection, which is a huge limitation with conventional methodologies. Although conventional diagnostic method can be used for quick identification of the causative agent, a detailed taxonomy and function could be advantageous for target validation purposes and specific treatments. An empiric treatment is usually based on the susceptibility test of the isolated organism, which remains one of the technical challenges with the conventional methods, as it cannot target the identification of specific pathogens with complete details. Shotgun sequencing in such cases could therefore be useful to obtain detailed taxonomical and functional identification-based diagnosis.11
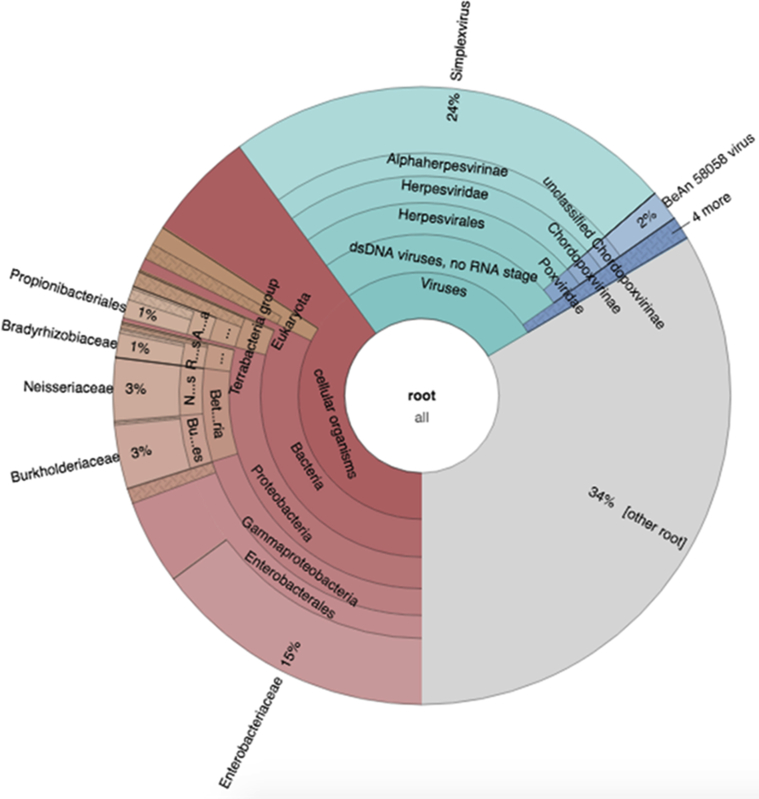
Traditional microbial cultures require huge volume of samples for analysis that are difficult to obtain in most of the clinical samples.12 Shotgun sequencing is advantageous in determining the total microbiome from as little as 1ng volume of the sample. Many studies have shown the clinical potential of this technique which includes deep characterization of microbiome13 for both, acute and chronic stages,14 human host response analyses and its applications in oncology. Although the number of reads could be useful in determining the likelihood of the presence of an organism, a control group is necessary to correlate with the ocular surface microbiome of a healthy subject. However, as the microbiome of each individual is different, this becomes fairly impossible unless a significant amount of samples are collected and analyzed. This was also observed from the Krona chart where the bacterial species can also be observed, possibly from the environment and the ocular surface (Fig. 1).
Fig. 1.
Krona chart showing the distribution of organisms found in the sample with a total number of 525,326 assigned reads.
These technologies heavily rely on the analysis of the downstream bioinformatics data. It becomes difficult for a clinician to analyze huge datasets on a routine patient-to-patient basis. The clinicians, however, could benefit from the decoded data from these datasets further increasing the specificity of diagnosis. Advanced bioinformatics software could allow processing only the required data with the resistance profile, which could potentially transform NGS into a widespread practically and clinically feasible technique. As most of these techniques are published as retrospective case reports and series, its true clinical potential therefore would be established after a prospective clinical trial.15,16 However, it's sensitivity and enrichment or depletion methods, laboratory workflow, reference standards for downstream analysis, bioinformatics challenges, regulatory issues, the associated costs, determining the presence of actual live organisms, turnover time could be considered as current limitations when such techniques are deemed for routine clinical practice.16 The turn-around time and costs are predicted to be significantly reduced with advances in this field. Conventional diagnostic method and metagenomics could hence be considered simultaneously to acquire data and create large databases that could be used in the future as a reference to correlate and make this technique highly specific both, in terms of diagnostics and treatments.
5. Conclusion
Shotgun sequencing allows a hypothesis-free approach to identify complete taxonomical and functional profile of an organism. This method confirmed the diagnosis of HSV along with the conventional diagnostic method. The results were achieved using 1.9 ng/μL of DNA concentration (114 ng in 60 μL) of the total sample volume thus showing advantages over conventional diagnostic method.
Patient consent
Written consent to publish this case has not been obtained. This report does not contain any personal identifying information. As conventional diagnostic method is routinely performed clinically, no specific consent was obtained, but the patient was fully informed about the diagnostic procedure.
Funding
The authors thank the European Society of Cataract & Refractive Surgeons for the META-COR grant (PI: Davide Borroni), funded through the ESCRS clinical research trial funding programme 2017-2018.
Authorship
All authors attest that they meet the current ICMJE criteria for Authorship.
Acknowledgements
None.
Declaration of competing interest
The following authors have no financial disclosures: MP, VR, AF, PL, FB, DB, SBK, DP, SA, SF.
References
- 1.Varley G.A., Meisler D.M. Complications of penetrating keratoplasty: graft infections. Refract Corneal Surg. 1991;7(1):62–66. [PubMed] [Google Scholar]
- 2.Darougar S., Wishart M.S., Viswalingam N.D. Epidemiological and clinical features of primary herpes simplex virus ocular infection. Br J Ophthalmol. 1985;69(1):2–6. doi: 10.1136/bjo.69.1.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Farooq A.V., Shukla D. Herpes simplex epithelial and stromal keratitis: an epidemiologic update. Surv Ophthalmol. 2012;57(5):448–462. doi: 10.1016/j.survophthal.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lairson D.R., Begley C.E., Reynolds T.F., Wilhelmus K.R. Prevention of herpes simplex virus eye disease: a cost-effectiveness analysis. Arch Ophthalmol. 2003;121(1):108–112. doi: 10.1001/archopht.121.1.108. [DOI] [PubMed] [Google Scholar]
- 5.Liesegang T.J. Herpes simplex virus epidemiology and ocular importance. Cornea. 2001;20(1):1–13. doi: 10.1097/00003226-200101000-00001. [DOI] [PubMed] [Google Scholar]
- 6.Tsatsos M., MacGregor C., Athanasiadis I., Moschos M.M., Hossain P., Anderson D. Herpes simplex virus keratitis: an update of the pathogenesis and current treatment with oral and topical antiviral agents. Clin Exp Ophthalmol. 2016;44(9):824–837. doi: 10.1111/ceo.12785. [DOI] [PubMed] [Google Scholar]
- 7.Souza P.M., Holland E.J., Huang A.J. Bilateral herpetic keratoconjunctivitis. Ophthalmology. 2003;110(3):493–496. doi: 10.1016/S0161-6420(02)01772-4. [DOI] [PubMed] [Google Scholar]
- 8.McGilligan V.E., Moore J.E., Tallouzi M. A comparison of the clinical and molecular diagnosis of herpes simplex keratitis. Open J Ophthalmol. 2014;4(3):65–74. [Google Scholar]
- 9.Azher T.N., Yin X.T., Tajfirouz D., Huang A.J.W., Stuart P.M. Herpes simplex keratitis: challenges in diagnosis and clinical management. Clin Ophthalmol. 2017;11:185–191. doi: 10.2147/OPTH.S80475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parekh M., Borroni D., Romano V. Next-generation sequencing for the detection of microorganisms present in human donor corneal preservation medium. BMJ Open Ophthalmol. 2019;4(1) doi: 10.1136/bmjophth-2018-000246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eguchi H., Hotta F., Kuwahara T. Diagnostic approach to ocular infections using various techniques from conventional culture to next-generation sequencing analysis. Cornea. 2017;36(1):S46–S52. doi: 10.1097/ICO.0000000000001338. [DOI] [PubMed] [Google Scholar]
- 12.Hariya T., Maruyama K., Sugita S. Multiplex polymerase chain reaction for pathogen detection in donor/recipient corneal transplant tissue and donor storage solution. Sci Rep. 2017;7(1):5973. doi: 10.1038/s41598-017-06344-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jovel J., Patterson J., Wang W. Characterization of the gut microbiome using 16S or shotgun metagenomics. Front Microbiol. 2016;7:459. doi: 10.3389/fmicb.2016.00459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Young V. The role of the microbiome in human health and disease: an introduction for clinicians. BMJ. 2017;356:j831. doi: 10.1136/bmj.j831. [DOI] [PubMed] [Google Scholar]
- 15.Simner P.J., Miller S., Carroll K.C. Understanding the promises and hurdles of metagenomic next-generation sequencing as a diagnostic tool for infectious diseases. Clin Infect Dis. 2018;66(5):778–788. doi: 10.1093/cid/cix881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chiu C.Y., Miller S.A. Clinical metagenomics. Nat Rev Genet. 2019;20(6):341–343. doi: 10.1038/s41576-019-0113-7. [DOI] [PMC free article] [PubMed] [Google Scholar]