Abstract
Background
The high-temperature requirement factor A1 (HTRA1) gene located at 10q26 locus has been associated with age-related macular degenerative (AMD), with the significantly related polymorphism being (rs11200638, −625G/A), however, above association is not consistent. We investigated a comprehensive analysis to evaluate the correlations between rs11200638 polymorphism and AMD susceptibility thoroughly addressing this issue.
Methods
An identification was covered from the PubMed and Wanfang databases until 27th Jan, 2020. Odds ratios (OR) with 95% confidence intervals (CI) were applied to evaluate the associations. After a thorough and meticulous search, 35 different articles (33 case-control studies with HWE, 22 case-control studies about wet/dry AMD) were retrieved.
Results
Individuals carrying A-allele or AA genotype may have an increased risk to be AMD disease. For example, there has a significantly increased relationship between rs11200638 polymorphism and AMD both for Asians (OR: 2.51, 95%CI: 2.22–2.83 for allelic contrast) and Caucasians [OR (95%CI) = 2.63(2.29–3.02) for allelic contrast]. Moreover, a similar trend in the source of control was detected. To classify the type of AMD, increased association was also observed in both wet (OR: 3.40, 95%CI: 2.90–3.99 for dominant model) and dry (OR: 2.08, 95%CI: 1.24–3.48 for dominant model) AMD. Finally, based on the different genotyping methods, increased relationships were identified by sequencing, TaqMan, PCR-RFLP and RT-PCR.
Conclusions
Our meta-analysis demonstrated that HTRA1 rs11200638 polymorphism may be related to the AMD development, especially about individuals carrying A-allele or AA genotype, who may be as identified targets to detect and intervene in advance. Further studies using Larger sample size studies, including information about gene-environment interactions will be necessary to carry out.
Keywords: High-temperature requirement factor A1, Age-related macular degeneration, Polymorphism, Meta-analysis, Risk
Background
In both developed and developing countries, age-related macular degeneration (AMD) is the main cause of vision loss in the elderly people [1, 2]. By 2050, about 17.8 million people will be affected by AMD [3]. AMD’s visual loss is due to dead/non-functional photoreceptor cells and potential retinal pigment epithelium cells [4]. In clinical practice, early dryness that can develop into geographic atrophy (atrophic, non-exudative) and wet (exudative) AMD characterized by choroidal neovascularization (CNV) are two forms about AMD [5, 6].
Age, race, family history, smoking and sun exposure are common risk factors [7, 8]. Another main factor in the etiology of AMD is genetic susceptibility [9]. A genome-wide association study (GWAS) in 2005 confirmed the association between AMD risk and genetic variations, suggesting that AMD is a polygenic disease [10], and in the following 15 years triggered many studies involving AMD genetic association [11–13]. So far, polymorphism about age-related maculopathy susceptibility 2 (AMRS2) rs10490924, complement factor H (CFH), complement 2 (C2)/complement factor H (CFB), complement component C3 and apolipoprotein E (APOE) haplotypes have been demonstrated as associated factors with susceptibility to AMD [14–18].
As we all known, VEGF is contributed to the progression of wet AMD, because angiogenesis and the formation of vascular permeability can lead to fluid leakage in blood vessels, and eventually lead to loss of vision [19]. Anti-VEGF drugs (eg: ranibizumab and bevacizumab) have been widely used in clinics [20, 21]. In addition, they have been shown to be effective in slowing the development of CNV, however, individual differences and shorter treatment time have been observed [22]. It is assumed that genetic factors may be involved in this period of heterogeneous response, such as variants of complement factor F (CHF), VEGFA, ARMS2 and high-temperature requirement factor A1 (HTRA1) [23–26].
HTRA1 regulates the transforming growth factor-β (TGF-β), insulin-like growth factor, and its binding protein, which is considered as regulators for cell proliferation, angiogenesis and extracellular matrix deposition. Furthermore, the inhibition of TGF-β may result in the overexpression of HTRA1 gene in wet AMD [27] (https://www.ncbi.nlm.nih.gov/gene/5654).
One of common polymorphisms in HTRA1 gene is rs11200638 (wide allele G to mutation allele A) [28]. The A-allele can influence the overexpression of HTRA1 protein, which may affect the integrity of Bruch’s membrane and lead to promote the progression of CNV stage [29].
In view of the above, we are aware of the critial role of HTRA1 gene and its common rs11200638 polymorphism, and we conducted a comprehensive summary using meta-analysis methods, including 28 different publications (33 case-control studies) [26, 30–57].
Methods
Search strategy and criteria
Relative studies from PubMed and Wanfang databases before 27th Jan, 2020 were searched. The keywords were “age-related macular degeneration,” “AMD,” “polymorphism or variant,” and “HTRA1 or high-temperature requirement factor A1.” Included criteria were according with as follows: (1) studies were focused on the correlation between AMD and rs11200638 polymorphism; (2) studies were all case-control and retrospective studies, and (3) the (AA, AG, and GG) genotypes both in case and control groups must be listed in Tables. Excluded criteria were consistent with as follows: (1) just case samples were shown in studies; (2) the numbers for genotypes did not shown in Tables, and (3) some duplications studies [58]. After the above conditions of the layer-by-layer screening exclusion, finally, 35 different articles were identified.
Data extraction
Two authors (Ying Liu and Dong Wei) independently screened all papers that according using above criteria. The basic information collected from each study contains the first author, year of publication, country source for corresponding authors, race type, Hardy-Weinberg equilibrium (HWE) for control group, detecting methods and AMD disease types (dry and wet AMD) [58]..
Statistical analysis
Odds ratios (OR) with 95% confidence intervals (CI) were applied to assess associations for rs11200638 polymorphism and AMD [58, 59]. The statistical significance of the OR was evaluated by the Z-test [60]. The heterogeneity among studies was calculated using the Q-test. Mantel-Haenszel (fixed-effects model) was selected, when P-value for heterogeneity is more than 0.1, otherwise, the DerSimonian-Laird (random-effects model) was applied [61, 62]. Five genetic models were adopt: AA vs. AG + GG, A vs. G, AA + AG vs. GG, AA vs. GG and AG vs. GG. The stability of results was assessed by sensitivity analysis. The HWE of control group was assessed by the Pearson’s χ2 test [63]. The publication bias was appraised by both Begg’s test and Egger’s test [64]. All statistical calculation were performed through Stata software (version 10.0, StataCorp LP, College Station, TX, USA) [59]. The power for sample size was calculated by Power and Sample Size Calculation Program [65].
Gene-gene interaction involved HTRA1 from a network
The network of gene-gene interactions for HTRA1 gene was shown by String online server to more complete understanding of the role of HTRA1 in AMD [66].
Results
Study searching and their basic information
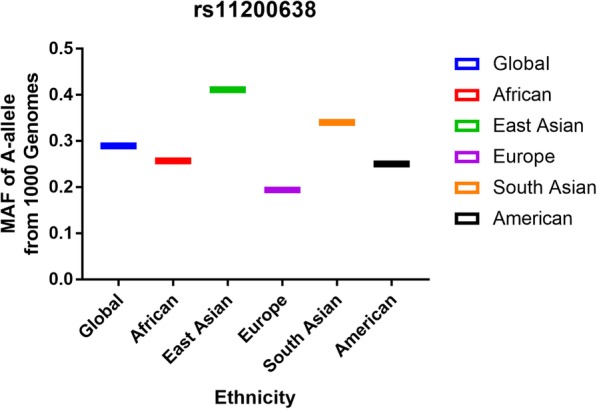
A number of 262 papers were garnered by a document from the PubMed (222 titles) and Wanfang (40 titles) databases. One hundred seventy-eight articles were deleted after reading over the abstract sections (Fig. 1). Forty-one articles were excluded due to duplication (7), meta-analysis or systematic analysis (26), clinical trial (10), randomized controlled trial (6). Finally, 35 different articles [26, 30–57, 67–71] were selected, including 38 studies about HTRA1 gene rs11200638 polymorphism and AMD risk (Table 1) and 27 case-control studies about HTRA1 gene rs11200638 polymorphism and wet or dry AMD risk (Table 2). Five case-control studies [67–71] were not consistent with HWE in control groups. To make our analysis to more strict, we deleted above five studies, so there were about 33 case-control studies (8101 cases and 7215 controls) for the whole AMD [26, 30–57], and 22 case-control studies for wet or dry (3938 cases and 4427 controls) studies [26, 30, 31, 33, 34, 40, 41, 43, 44, 46–48, 51, 53–56]. The A-allele frequency from case group was observed as higher in control individuals (54.2% vs. 36.5%) (Fig. 2, Supplementary Table 1). Nineteen studies were Asian samples, and 14 from Caucasian population; source of control in 22 studies were hospital-based (HB), and 11 were from population-based (PB); 17 case-control studies were about wet AMD disease, and 5 were about dry disease. Additional, the Minor Allele Frequency (MAF) about five main worldwide populations in the 1000 Genomes Browser was shown in Fig. 3: Global (0.290); East Asian (EAS = 0.411); European (EUR = 0.194); African (AFR = 0.257); American (AMR = 0.250); and South Asian (SAS = 0.340) (Supplementary Table 1).
Fig. 1.
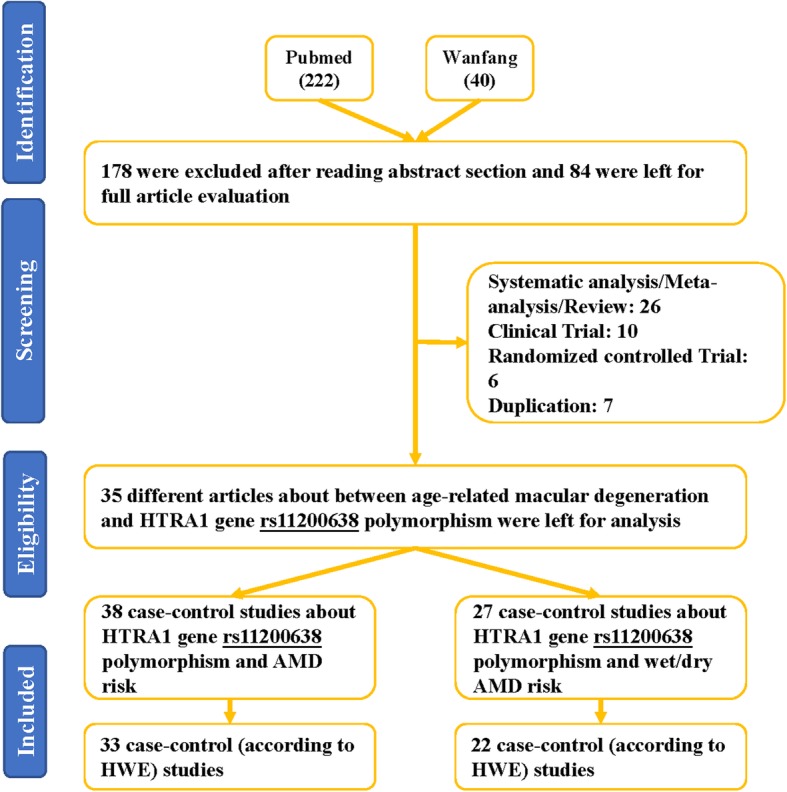
Flowchart for the search strategy was used to identify associated studies for rs11200638 polymorphism and AMD risk
Table 1.
Characteristics of included studies in HTRA1 rs11200638 polymorphism and AMD risk
| Author | Year | Country | Ethnicity | type | Case | Control | SOC | Cases | Controls | HWE | Genotype | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | AG | GG | AA | AG | GG | ||||||||||
| Tian | 2012 | China | Asian | AMD | 532 | 468 | HB | 255 | 193 | 84 | 104 | 224 | 140 | 0.423 | Typer 4.0 software |
| Ruamviboonsuk | 2017 | Thailand | Asian | wet | 377 | 1073 | PB | 125 | 164 | 88 | 146 | 490 | 437 | 0.643 | InfiniumOmniExpressExome-8 v1.3 platform |
| Chu | 2008 | China | Asian | wet | 144 | 126 | HB | 76 | 52 | 16 | 31 | 69 | 26 | 0.276 | PCR-RFLP |
| Losonczy | 2011 | Hungary | Caucasian | AMD | 103 | 95 | HB | 23 | 50 | 30 | 3 | 49 | 43 | 0.133 | PCR-RFLP |
| Tuo | 2008 | USA | Caucasian | AMD | 142 | 132 | PB | 33 | 60 | 49 | 7 | 51 | 74 | 0.638 | PCR-RFLP |
| Tuo | 2008 | USA | Caucasian | AMD | 330 | 191 | PB | 63 | 164 | 103 | 12 | 73 | 106 | 0.904 | PCR-RFLP |
| Tuo | 2008 | USA | Caucasian | AMD | 272 | 555 | PB | 15 | 122 | 135 | 20 | 186 | 349 | 0.431 | PCR-RFLP |
| Tuo | 2008 | USA | Caucasian | AMD | 46 | 22 | PB | 2 | 15 | 29 | 1 | 6 | 15 | 0.696 | PCR-RFLP |
| Chan | 2007 | USA | Caucasian | AMD | 52 | 13 | HB | 14 | 27 | 11 | 0 | 8 | 5 | 0.109 | RT-PCR |
| Kanda | 2007 | USA | Caucasian | AMD | 457 | 280 | HB | 102 | 183 | 172 | 11 | 90 | 179 | 0.94 | RT-PCR |
| Cheng | 2013 | China | Asian | wet | 93 | 93 | HB | 52 | 30 | 11 | 21 | 42 | 30 | 0.395 | Sequencing |
| Liang | 2012 | China | Asian | wet | 161 | 150 | HB | 61 | 83 | 17 | 30 | 72 | 48 | 0.751 | Sequencing |
| Jiang | 2008 | China | Asian | wet | 159 | 140 | HB | 99 | 47 | 13 | 31 | 67 | 42 | 0.662 | Sequencing |
| Lee | 2010 | Korea | Asian | wet | 137 | 187 | HB | 57 | 59 | 21 | 35 | 100 | 52 | 0.283 | Sequencing |
| Lin | 2008 | China-Taiwan | Asian | AMD | 95 | 90 | HB | 53 | 33 | 9 | 19 | 47 | 24 | 0.651 | Sequencing |
| Kaur | 2008 | India | Asian | AMD | 229 | 184 | HB | 90 | 89 | 50 | 21 | 85 | 78 | 0.765 | Sequencing |
| Xu | 2007 | China | Asian | wet | 121 | 132 | HB | 56 | 52 | 13 | 24 | 64 | 44 | 0.931 | Sequencing |
| Tam | 2008 | China-Hong Kong | Asian | wet | 163 | 183 | HB | 94 | 51 | 18 | 38 | 90 | 55 | 0.916 | Sequencing |
| Mori | 2007 | Japan | Asian | AMD | 123 | 133 | HB | 45 | 52 | 26 | 22 | 57 | 54 | 0.298 | Sequencing |
| Askari | 2015 | Iran | Asian | AMD | 120 | 120 | HB | 58 | 42 | 20 | 12 | 66 | 42 | 0.057 | Sequencing |
| Lana | 2018 | Brazil | Caucasian | AMD | 204 | 166 | HB | 73 | 89 | 42 | 22 | 77 | 67 | 0.987 | Sequencing |
| Kaur | 2013 | India | Asian | AMD | 198 | 145 | PB | 84 | 70 | 44 | 17 | 67 | 61 | 0.829 | Sequencing |
| Ng | 2016 | Hong Kong | Asian | wet | 194 | 183 | PB | 109 | 63 | 22 | 38 | 90 | 55 | 0.916 | Sequencing |
| Kaur | 2013 | India | Caucasian | AMD | 616 | 426 | PB | 130 | 292 | 194 | 17 | 138 | 271 | 0.913 | Sequencing |
| Chen | 2013 | China | Asian | AMD | 158 | 157 | HB | 28 | 74 | 56 | 21 | 77 | 59 | 0.599 | TaqMan |
| Kondo | 2007 | Japan | Asian | AMD | 73 | 94 | HB | 29 | 39 | 5 | 16 | 40 | 38 | 0.334 | TaqMan |
| Leveziel | 2007 | France | Caucasian | wet | 118 | 116 | HB | 32 | 57 | 29 | 5 | 41 | 70 | 0.743 | TaqMan |
| Weger | 2007 | Austria | Caucasian | wet | 242 | 157 | PB | 67 | 108 | 67 | 8 | 50 | 99 | 0.609 | TaqMan |
| Lu | 2007 | China | Asian | wet | 90 | 106 | HB | 53 | 34 | 3 | 15 | 63 | 28 | < 0.05 | PCR-RFLP |
| Cruz-González | 2013 | Spain | Caucasian | AMD | 121 | 91 | HB | 29 | 60 | 32 | 61 | 21 | 9 | < 0.05 | PCR-RFLP |
| Mohamad | 2019 | Malaysia | Asian | wet | 145 | 145 | HB | 79 | 47 | 19 | 48 | 82 | 15 | < 0.05 | PCR-RFLP |
| Yang | 2010 | China | Asian | wet | 109 | 150 | HB | 31 | 45 | 33 | 30 | 50 | 70 | < 0.05 | PCR-RFLP |
| Chen | 2008 | USA | Caucasian | AMD | 776 | 294 | HB | 131 | 400 | 245 | 10 | 128 | 156 | < 0.05 | Sequencing |
| Zeng | 2011 | China | Caucasian | AMD | 1335 | 509 | PB | 244 | 641 | 450 | 21 | 181 | 307 | 0.374 | SNaPshot |
| Li | 2015 | China | Asian | AMD | 146 | 145 | HB | 73 | 54 | 19 | 44 | 74 | 27 | 0.674 | MassARRAY MALDI-TOF |
| Yang | 2018 | China | Asian | AMD | 201 | 201 | HB | 103 | 74 | 24 | 42 | 100 | 59 | 0.975 | MassARRAY MALDI-TOF |
| Matuskova | 2020 | Czech Republic | Caucasian | wet | 307 | 191 | HB | 69 | 148 | 90 | 9 | 66 | 116 | 0.921 | SNaPshot Multiplex-System |
| Fritsche | 2008 | Germany | Caucasian | AMD | 760 | 549 | PB | 152 | 344 | 264 | 22 | 174 | 353 | 0.923 | Multiplex PCR |
HB hospital-based; PB population-based; SOC source of control; PCR-RFLP polymerase chain reaction followed by restriction fragment length polymorphism; RT-PCR real-time PCR; MALDI-TOF polymerase chain reaction–matrix-assisted laser desorption/ionization time-of-flight; HWE Hardy–Weinberg equilibrium of control group
Table 2.
Characteristics of included studies in HTRA1 rs11200638 polymorphism and wet/dry AMD risk, respectively
| Author | Year | Country | Ethnicity | type | Case | Control | SOC | Cases | Controls | HWE | Genotype | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | AG | GG | AA | AG | GG | ||||||||||
| Lin | 2008 | China-Taiwan | Asian | dry | 52 | 90 | HB | 28 | 19 | 5 | 19 | 47 | 24 | 0.651 | Sequencing |
| Chan | 2007 | USA | Caucasian | dry | 18 | 13 | HB | 4 | 10 | 4 | 0 | 8 | 5 | 0.109 | RT-PCR |
| Mori | 2007 | Japan | Asian | dry | 19 | 116 | HB | 4 | 7 | 8 | 5 | 41 | 70 | 0.743 | Sequencing |
| Askari | 2015 | Iran | Asian | dry | 32 | 120 | HB | 11 | 12 | 9 | 12 | 66 | 42 | 0.576 | Sequencing |
| Ruamviboonsuk | 2017 | Thailand | Asian | wet | 377 | 1073 | PB | 125 | 164 | 88 | 146 | 490 | 437 | 0.643 | InfiniumOmniExpressExome-8 v1.3 platform |
| Cheng | 2013 | China | Asian | wet | 93 | 93 | HB | 52 | 30 | 11 | 21 | 42 | 30 | 0.395 | Sequencing |
| Ng | 2016 | Hong Kong | Asian | wet | 194 | 183 | PB | 109 | 63 | 22 | 38 | 90 | 55 | 0.915 | Sequencing |
| Liang | 2012 | China | Asian | wet | 161 | 150 | HB | 61 | 83 | 17 | 30 | 72 | 48 | 0.75 | Sequencing |
| Chu | 2008 | China | Asian | wet | 144 | 126 | HB | 76 | 52 | 16 | 31 | 69 | 26 | 0.276 | PCR-RFLP |
| Jiang | 2008 | China | Asian | wet | 159 | 140 | HB | 99 | 47 | 13 | 31 | 67 | 42 | 0.662 | Sequencing |
| Lee | 2010 | Korea | Asian | wet | 137 | 187 | HB | 57 | 59 | 21 | 35 | 100 | 52 | 0.283 | Sequencing |
| Lin | 2008 | China-Taiwan | Asian | wet | 43 | 90 | HB | 25 | 14 | 4 | 19 | 47 | 24 | 0.651 | Sequencing |
| Xu | 2007 | China | Asian | wet | 121 | 132 | HB | 56 | 52 | 13 | 24 | 64 | 44 | 0.931 | Sequencing |
| Tam | 2008 | China-Hong Kong | Asian | wet | 163 | 183 | HB | 94 | 51 | 18 | 38 | 90 | 55 | 0.916 | Sequencing |
| Chan | 2007 | USA | Caucasian | wet | 31 | 13 | HB | 8 | 16 | 7 | 0 | 8 | 5 | 0.109 | RT-PCR |
| Leveziel | 2007 | France | Caucasian | wet | 118 | 116 | HB | 32 | 57 | 29 | 5 | 41 | 70 | 0.743 | TaqMan |
| Mori | 2007 | Japan | Asian | wet | 104 | 116 | HB | 41 | 45 | 18 | 5 | 41 | 70 | 0.743 | Sequencing |
| Askari | 2015 | Iran | Asian | wet | 88 | 120 | HB | 47 | 30 | 11 | 12 | 66 | 42 | 0.576 | Sequencing |
| Weger | 2007 | Austria | Caucasian | wet | 242 | 157 | PB | 67 | 108 | 67 | 8 | 50 | 99 | 0.609 | TaqMan |
| Lu | 2007 | China | Asian | wet | 90 | 106 | HB | 53 | 34 | 3 | 15 | 63 | 28 | < 0.05 | PCR-RFLP |
| Mohamad | 2019 | Malaysia | Asian | wet | 145 | 145 | HB | 79 | 47 | 19 | 48 | 82 | 15 | < 0.05 | PCR-RFLP |
| Yang | 2010 | China | Asian | wet | 109 | 150 | HB | 31 | 45 | 33 | 30 | 50 | 70 | < 0.05 | PCR-RFLP |
| Chen | 2008 | USA | Caucasian | wet | 470 | 294 | HB | 76 | 245 | 149 | 10 | 128 | 156 | < 0.05 | Sequencing |
| Chen | 2008 | USA | Caucasian | dry | 306 | 294 | HB | 55 | 155 | 96 | 10 | 128 | 156 | < 0.05 | Sequencing |
| Zeng | 2011 | China | Caucasian | dry | 341 | 509 | PB | 60 | 166 | 115 | 21 | 181 | 307 | 0.374 | SNaPshot |
| Zeng | 2011 | China | Caucasian | wet | 994 | 509 | PB | 184 | 475 | 335 | 21 | 181 | 307 | 0.374 | SNaPshot |
| Matuskova | 2020 | Czech Republic | Caucasian | wet | 307 | 191 | HB | 69 | 148 | 90 | 9 | 66 | 116 | 0.921 | SNaPshot Multiplex-System |
Fig. 2.
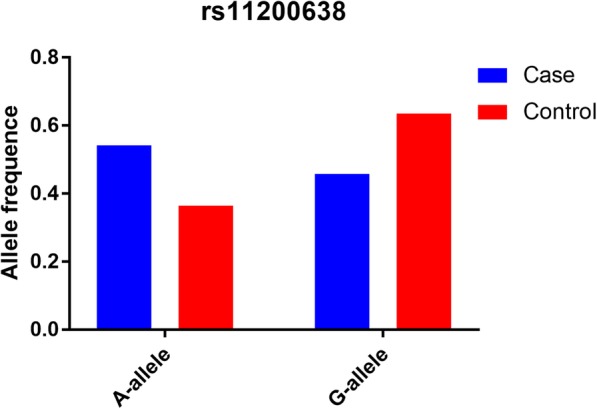
The MAF of minor-allele (mutant-allele) for HTRA1 gene rs11200638 polymorphism from the 1000 Genomes online database and present analysis. EAS: East Asian; EUR: European; AFR: African; AMR: American; SAS: South Asian
Fig. 3.
Frequency about A-allele for the rs11200638 polymorphism among cases or controls by ethnicity
Quantitative synthesis
Total analysis
Increasing relationships were found for rs11200638 and AMD risk in all models (eg: AA vs. GG: OR = 5.45, 95CI% = 4.26–6.98, P < 0.001) (Table 3). In order to make this study more convincing and reliable, we detected five studies, which were not according with HWE, finally, we tested the 33 case-control studies. Also significantly increasing correlations were observed in whole models [for example: allelic contrast: OR (95%CI): 2.56(2.34–2.80), P < 0.001; AA+AG vs. GG: OR (95%CI): 2.80 (2.49–3.15), P < 0.001] (Fig. 4) (Table 3).
Table 3.
Results of the meta-analysis on HTRA1 rs11200638 polymorphism and AMD risk in total and types of subgroups
| Variables | N | Case/ | A-allele vs. G-allele | AG vs. GG | AA+AG vs. GG | AA vs. GG | AA vs. AG + GG |
|---|---|---|---|---|---|---|---|
| Control | OR(95%CI) PhP | OR(95%CI) PhP | OR(95%CI) PhP | OR(95%CI) PhP | OR(95%CI) PhP | ||
| Total | 38 | 8582/7452 | 2.39(2.12–2.69)0.000 0.000 | 1.91(1.69–2.16)0.000 0.000 | 2.63(2.30–3.01)0.000 0.000 | 5.45(4.26–6.98)0.000 0.000 | 3.75(3.04–4.63)0.000 0.000 |
| HWE | 33 | 8101/7215 | 2.56(2.34–2.80)0.000 0.000 | 1.98(1.76–2.23)0.001 0.000 | 2.80(2.49–3.15)0.000 0.000 | 6.13(5.09–7.38)0.000 0.000 | 4.10(3.55–4.73)0.000 0.000 |
| Ethnicity | |||||||
| Asian | 19 | 3424/4004 | 2.51(2.22–2.83)0.000 0.000 | 1.67(1.47–1.88)0.167 0.000 | 2.68(2.25–3.19)0.009 0.000 | 5.36(4.31–6.66)0.003 0.000 | 3.70(3.16–4.35)0.009 0.000 |
| Caucasian | 14 | 4677/3211 | 2.63(2.29–3.02)0.000 0.000 | 2.37(2.15–2.61)0.140 0.000 | 2.94(2.51–3.45)0.004 0.000 | 7.52(5.62–10.07)0.014 0.000 | 5.00 (3.91–6.40)0.070 0.000 |
| SOC | |||||||
| HB | 22 | 3589/3273 | 2.56(2.28–2.88)0.000 0.000 | 1.86(1.58–2.19)0.025 0.000 | 2.81(2.38–3.31)0.005 0.000 | 5.99(4.74–7.59)0.001 0.000 | 3.93(3.31–4.68)0.005 0.000 |
| PB | 11 | 4512/33942 | 2.55(2.18–2.99)0.000 0.000 | 2.16(1.84–2.53)0.021 0.000 | 2.80(2.35–3.35)0.001 0.000 | 6.37(4.64–8.76)0.001 0.000 | 4.43(3.41–5.75)0.009 0.000 |
| AMD type | |||||||
| Wet | 17 | 3476/3579 | 3.03(2.59–3.55)0.000 0.000 | 2.11(1.81–2.46)0.138 0.000 | 3.40(2.90–3.99)0.073 0.000 | 7.65(5.73–10.21)0.009 0.000 | 4.65(3.72–5.82)0.008 0.000 |
| Dry | 5 | 462/848 | 2.36(1.71–3.24)0.750 0.000 | 1.33(0.76–2.32)0.705 0.316 | 2.08(1.24–3.48)0.618 0.005 | 6.01(3.05–11.87)0.889 0.000 | 4.77(2.79–8.16)0.964 0.000 |
| Genotyping | |||||||
| Others | 5 | 3004/2599 | 2.55(2.22–2.94)0.027 0.000 | 2.14(1.67–2.73)0.006 0.000 | 2.85(2.38–3.43)0.055 0.000 | 6.25(4.26–9.15)0.005 0.000 | 4.18 (3.14–5.57)0.033 0.000 |
| Sequencing | 14 | 2613/2332 | 2.84(2.61–3.09)0.237 0.000 | 2.08(1.81–2.41)0.252 0.000 | 3.19(2.79–3.65)0.710 0.000 | 7.00(5.84–8.39)0.677 0.000 | 4.51(3.90–5.21)0.169 0.000 |
| TaqMan | 4 | 591/524 | 2.66(1.43–4.94)0.000 0.002 | 2.79(1.31–5.91)0.000 0.008 | 3.61(1.52–8.60)0.000 0.004 | 7.52(2.05–27.68)0.000 0.002 | 3.86(1.66–8.98)0.002 0.002 |
| PCR-RFLP | 6 | 1037/1121 | 1.98(1.72–2.26)0.105 0.000 | 1.76(1.45–2.14)0.611 0.000 | 2.08(1.73–2.50)0.411 0.000 | 4.30(2.51–7.35)0.073 0.000 | 3.33(2.09–5.31)0.095 0.000 |
| RT-PCR | 2 | 509/293 | 2.91(2.30–3.69)0.755 0.000 | 2.08(1.51–2.86)0.643 0.000 | 2.90(2.15–3.92)0.734 0.000 | 9.83(5.18–18.65)0.817 0.000 | 7.19(3.84–13.45)0.806 0.000 |
| MassARRAY MALDI-TOF | 2 | 347/346 | 2.18(1.39–3.49)0.043 0.001 | 1.46(0.95–2.24)0.213 0.088 | 2.22(1.13–4.38)0.099 0.021 | 3.84(1.53–9.63)0.044 0.004 | 3.05(1.78–5.23)0.097 0.000 |
Ph: value of Q-test for heterogeneity test; P: Z-test for the statistical significance of the OR
Fig. 4.
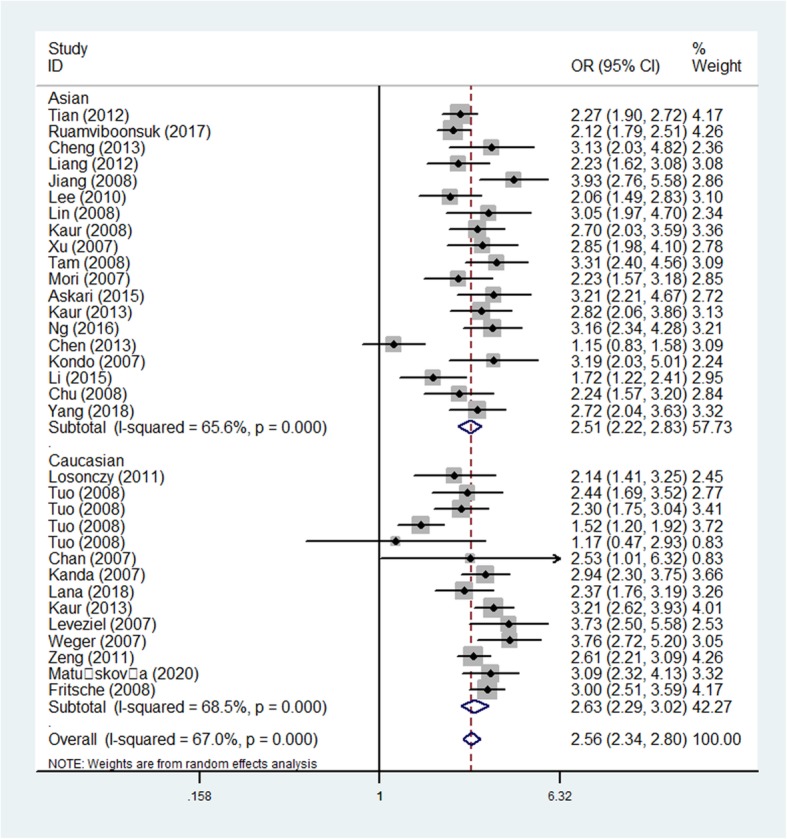
Forest plot of AMD risk associated with HTRA1 gene rs11200638 polymorphism (A-allele vs. G-allele) by ethnicity subgroup
Subgroup analysis
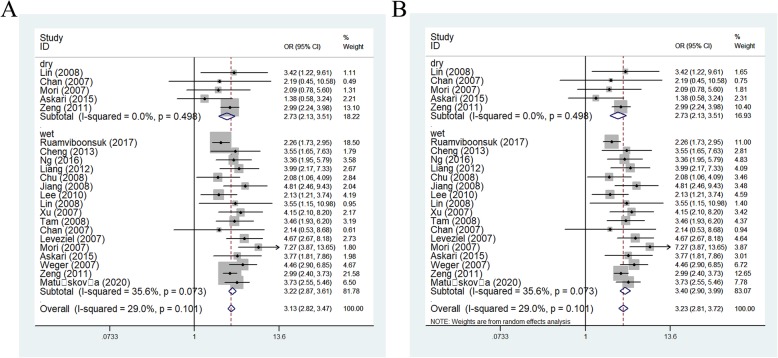
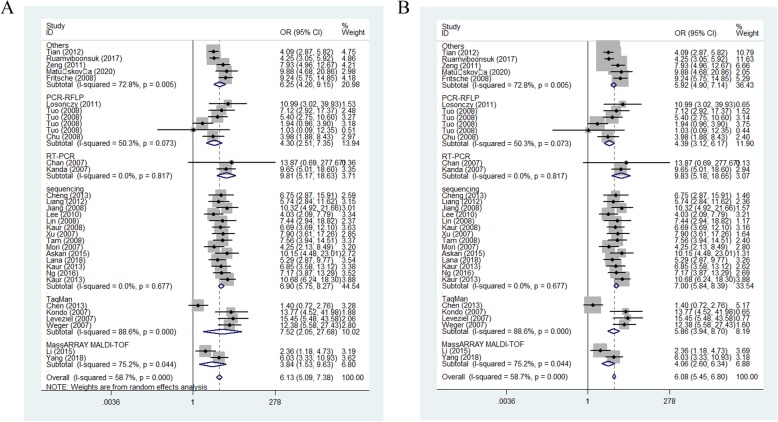
Coming up, we all know that the frequency of A-allele in different races was not the same, so we tried to analysis the relationships by ethnicity subgroups in further, which indicated an incremental statistically association between this polymorphism and both in Asians [OR(95% CI) = 2.51(2.22–2.83), P(heterogeneity) < 0.001, P < 0.001 in allelic contrast, Fig. 4; AA vs. AG + GG: OR (95% CI) = 3.70(3.16–4.35), P(heterogeneity) = 0.009, P < 0.001] and Caucasian populations [OR (95% CI) = 2.94(2.51–3.45), P(heterogeneity) < 0.001 in dominant model; OR = 2.37, 95% CI = 2.15–2.61, P(heterogeneity) < 0.001 in heterozygote comparison; allelic comparison, OR = 2.63, 95% CI = 2.29–3.02, Pheterogeneity < 0.001, P < 0.001, Fig. 4) (Table 3). In addition, regular analysis by source of control, also similar results were detected in both PB and HB studies [AG vs. GG: OR (95% CI) = 1.86(1.58–2.19), Pheterogeneity = 0.025, P < 0.001 in HB; AG vs. GG: OR (95% CI) = 2.16(1.84–2.53), P(heterogeneity) = 0.021, P < 0.001 in PB] (Table 3) (Fig. 5). AMD have different types and stages, the different of clinical presentation for dry and wet AMD is completely different, so we firmly believed that the correlations existed should be evaluated separately, significant positive associations were found both for dry (eg. AA+AG vs. GG: OR (95% CI) = 2.73(2.13–3.51), P(heterogeneity) = 0.498, P < 0.001, Fig. 6a) and wet AMD (for example in AA+AG vs. GG model: OR = 3.40, 95% CI = 2.90–3.99, Pheterogeneity = 0.073, P < 0.001, Fig. 6b). Finally, we tried to in each method, whether associations may exist in our analysis, we found some positive results in some methods (such as in AA vs. GG model: OR = 7.52, 95% CI = 2.05–27.68, Pheterogeneity < 0.001, P = 0.002 about TaqMan; OR = 4.30, 95% CI = 2.51–7.35, Pheterogeneity = 0.073 about PCR-RFLP, OR = 3.84, 95% CI = 1.53–9.63, Pheterogeneity = 0.044 about MassARRAY MALDI-TOF, Fig. 7a; OR = 7.00, 95% CI = 5.84–8.39, Pheterogeneity = 0.677 about sequencing, OR = 9.83, 95% CI = 5.18–18.65, Pheterogeneity = 0.817 about sequencing RT-PCR, Fig. 7b) (Table 3).
Fig. 5.
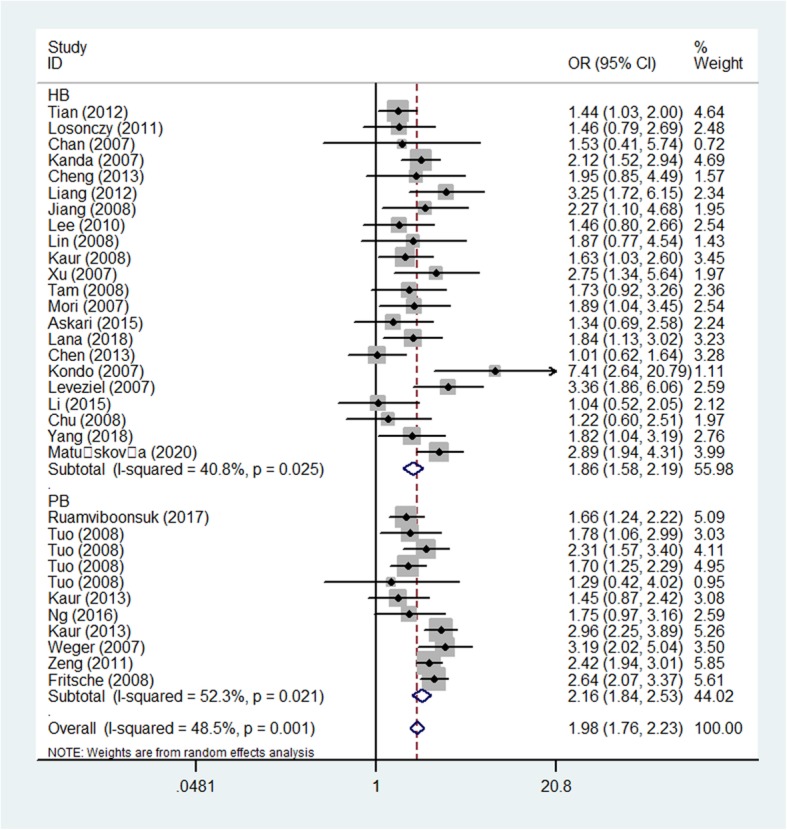
Forest plot of AMD risk associated with HTRA1 gene rs11200638 polymorphism (AG vs. GG) by source of control subgroup
Fig. 6.
Forest plot of AMD risk associated with HTRA1 gene rs11200638 polymorphism (AA+AG vs. GG) by AMD type subgroup. a: wet AMD; b: dry AMD
Fig. 7.
Forest plot of AMD risk associated with HTRA1 gene rs11200638 polymorphism (AA vs. GG) by genotyping methods subgroup. a: random model; b: fixed model
Bias diagnosis for publication and sensitivity analysis
Begg’s funnel plot and Egger’s test were applied to assess publication bias. At beginning, the funnel plots seemed asymmetrical in allele comparison for rs11200638 by Begg’s test, suggesting no publication bias was existed. Then, Egger’s test was applied to assess the funnel plot symmetry. As a result, no publication bias was observed [eg. allelic contrast, Egger’s test (t = 0.89, P = 0.38); Begg’s test (z = 0.85, P = 0.396), Supplementary Figure 1A,B] (Table 4).
Table 4.
Publication bias tests (Begg’s funnel plot and Egger’s test for publication bias test) for HTRA1 rs11200638 polymorphism
| Egger’s test | Begg’s test | ||||||
|---|---|---|---|---|---|---|---|
| Genetic type | Coefficient | Standard error | t | P value | 95%CI of intercept | z | P value |
| A-allele vs. G-allele | 0.211 | 0.924 | 0.23 | 0.820 | (−1.673–2.096) | 0.42 | 0.676 |
| AG vs. GG | −0.031 | 0.500 | −0.06 | 0.951 | (−1.051–0.989) | 0.29 | 0.768 |
| AA+AG vs. GG | −0.045 | 0.532 | −0.08 | 0.933 | (−1.130–1.040) | 0.26 | 0.792 |
| AA vs. GG | 0.297 | 0.382 | 0.78 | 0.441 | (−0.481–1.076) | 0.36 | 0.722 |
| AA vs. AG + GG | 0.365 | 0.438 | 0.83 | 0.412 | (−0.529–1.258) | 0.60 | 0.546 |
To assess the power and stability of whole study and each study, the sensitive analysis was adopt to carry out, as a result, no significant showing were found (Supplementary Figure 2).
Gene-gene network from string online site
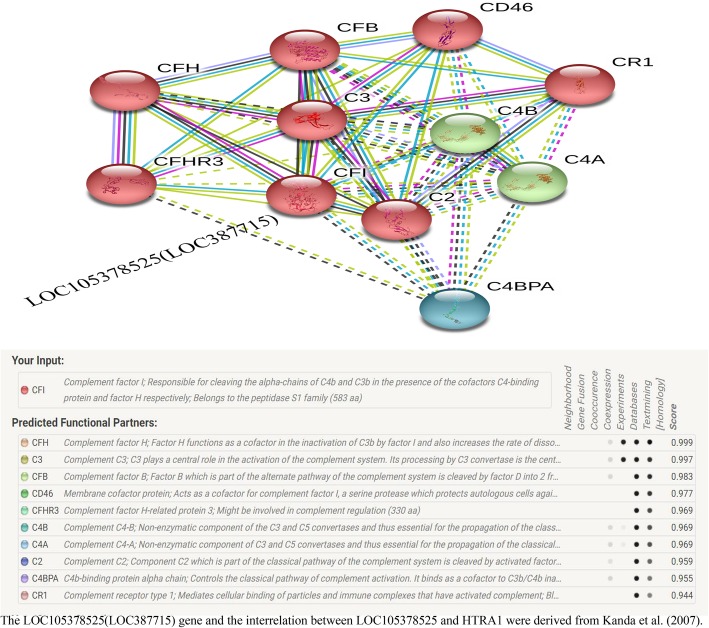
The HTRA1 gene may interacts with numerous genes from String online server (Fig. 8).
Fig. 8.
Interactions network about human HTRA1 gene and other related genes from String server. ACAN: aggrecan core protein; ARMS2: age-related maculopathy susceptibility; CLPP: ATP-dependent Clp protease proteolytic subunit; CTRC: chymotrypsin-C; YME1L1: ATP-dependent zine metalloprotease YME1L1; CFH: complement factor H; HSPD1: 60 kDa heat shock protein; RPL34: 60S ribosomal protein L34; CLPX: ATP-dependent Clp protease ATP-binding subunit clpX-like; PLEKHG4: Puratrophin-1
Discussion
Due to the severe consequences of vision loss caused by AMD, especially advanced AMD (atrophic/dry or neovascular/wet), it is necessary to study its etiology and mechanism, and then develop early diagnostic methods and effective treatments. Today, VEGF inhibitors have been widely regarded as effective drugs in clinical application for CNV (wet AMD) [3, 72, 73]. Therefore, identifying some novel detection markers and target drugs for some different types of AMD is the focus of current and future research. In the introduction, we clarified that genetic factors may help us to search for AMD in potential high-risk groups, which can be prevented and treated in advance.
In our analysis, we selected the HTRA1 gene that can regulate certain growth factors. The rs11200638 polymorphism in HTRA1 is the most common single nucleotide polymorphism (SNP) and has been received attention. However, Kanda et al. [35] demonstrated that there was no HTRA1 gene involved in AMD related SNPs, and its rs11200638 polymorphism did not appear to have an effect on the transcripts. Instead, they found a putative mitochondrial protein (LOC105378525) that may be expressed in the retina in the negative strand, which may be a candidate gene. In fact, they showed that rs11200638 and rs10490924 are in a strong linkage disequilibrium, which is a predicted non-synonymous A69S change in a protein named LOC105378525(LOC387715)/ARMS2. According to their research, rs10490924 was a strong candidate SNP associated with AMD risk, not rs11200638. In addition, Bonyadi et al. [74] conducted a meta-analysis of rs10490924 and found that the combined cigarette smoking and rs10490924 polymorphism may have significant association with AMD risk. We believed rs10490924 was a valuable SNP for AMD, nevertheless, conclusions based on a single study may not be negated by the potential functions for HTRA1 and its SNPs, which need more evidences and support from published and future researches.
Mori et al. before all others reported the correlations for rs11200638 polymorphism and AMD [47]. After that, researchers imitated similar works involving different ethnicities and different types about AMD. Nevertheless, each conclusion was indecipherable, even same population, though two published meta-analysis. As we all know, meta-analysis offers a method combining all related studies to acquire a powerful genetic effects for disease susceptibility [75].
Two previous meta-analysis [76, 77] about rs11200638 polymorphism and AMD have been reported, however, each study has its limitations. For example, Tang et al. just included fourteen case-control studies, two studies [67, 69] were not consistent with HWE, and Tuo et al. actually reported four-source case-control studies, which shouldn’t be combined together [77]. Chen et al. also performed a same study in the same year including 14 case-control studies, similar limitations were existed [76]. After year of 2008, newly added studies have been published, and to perfect the above deficiencies, we carried out a comprehensive analysis to come to a more convincing conclusion about HTRA1 gene rs11200638 polymorphism and AMD susceptibility.
Our current research is the comprehensive analysis about the associations between HTRA1 gene rs11200638 polymorphism and AMD, involving 8101 AMD samples and 7215 controls. Increased associations were found in the whole group, in Asian and Caucasian subgroups, source of control subgroup, and dry/wet sub-types of AMD, different genotyping methods (Sequencing, TaqMan, PCR-RFLP, RT-PCR and MassARRAY MALDI-TOF), which means that A-allele or AA genotype is the risk factor for AMD, in other words, if individuals carry on this SNP from peripheral blood test, which may indicate that it is possible to increase the occurrence of AMD for them in present time or at some point in the future. Therefore, this polymorphism may be helpful in screening vulnerable populations for AMD in advance. In addition, the power of present study was 1.00, which suggested our conclusions were stable and convincing.
In addition, the online String website was used to make a forecast several potential and functional genes associated with HTRA1. As a result, ten genes were predicted. Among them, the highest score of association was ACAN (0.943), however, so far, no research has been reported between this gene and AMD and interaction between this gene and HTRA1. Future research should be payed attention to above information, which may be in favor of AMD early detection/prevention and intervention. In other partners, ARMS2 and CFH have been shown to associate with AMD. Both ARMS2 and HTRA1 genes have a linkage disequilibrium, which is located nearby form 10q26 chromosome. ARMS2 rs10490924 was related to response to ranibizumab treatment among wet AMD patients [70]. CFH gene T1277C polymorphism is strong associated with both wet and dry AMD and may be contribute to the inflammation in the pathogenesis of AMD [78]. As for the rest interaction genes (CLPP, CTRC, YME1L1, HSPD1, RPL34, CLPX and PLEKHG4) both had moderate score and no literature to support. It seems that above ten genes associated with HTRA1 came from text mining scores, which were derived from the co-occurrence of gene/protein names in related abstracts. In addition, it was important considered the occurrence of the LOC105378525 (LOC387715) and its polymorphism (A69S, rs10490924) as the main factor for AMD reported by Kanda et al. (2007) [35], which should be added in the network of HTRA1 related genes. In a word, we should deep explore these partners of HTRA1 gene, and gene-gene interactions in the development of AMD in the next step.
Some limitations should be declared. First of all, Mixed and African individuals should be paid more attention in future studies, which was vacant in present analysis. Second, analysis about gene-gene/gene-environment interactions should be added, because some specific environmental and lifestyle factors may influence associations between rs11200638 polymorphism and AMD (such as hypertension, familial history, age range, diabetes stage, smoking status). Third, vision is the most concerned-clinical indicator of AMD, future studies should include the value of the vision and analyze the relationships between rs11200638 polymorphism and the degree of visual impairment, which may help us to better detect disease progression.
Conclusion
In conclusion, our present analysis demonstrated HTRA1 rs11200638 polymorphism may play a risk factor for the susceptibility of AMD, larger and more comprehensive studies should be performed in the future.
Supplementary information
Additional file 1: Table S1. Allele Frequency from 1000 Genomes Browser and present study.
Additional file 2: Figure S1. A: Begg’s funnel plot for publication bias test (A-allele vs. G-allele). Each point represents a separate study for the indicated association. B: Egger’s publication bias plot (A-allele vs. G-allele).
Additional file 3: Figure S2. Sensitivity analysis between HTRA1 gene rs11200638 polymorphism and AMD risk (A-allele vs. G-allele).
Acknowledgements
Not applicable.
Abbreviations
- AMD
Age-related macular degeneration
- GWAS
Genome-wide association studies
- HTRA1
High-temperature requirement factor A1
- CNV
Choroidal neovascularization
- VEGF
Vascular endothelial growth factor
- AREDS
Age-Related Eye Disease Study
- SNP
Single nucleotide polymorphism
- HB
Hospital-based
- PB
Population-based
- SOC
Source of control
- PCR-RFLP
Polymerase chain reaction followed by restriction fragment length polymorphism
- MALDI-TOF
A chip-based matrix-assisted laser-desorption/ionization time-of-flight
Authors’ contributions
YL and HJ designed the study and drafted the manuscript; DW extracted, analyzed, interpreted the data, and collected the clinical data; DW and WL performed the targeted sequencing, analyzed and interpreted the data; DW and WL participated in the study coordination and revised the manuscript. All authors read and approved the final version of the manuscript.
Funding
This study was supported by Heilongjiang Health and Family Planning Commission Research Project (No. 2016–379). The funding body play no direct role in the design of the study, and collection, analysis, and interpretation of data, and in writing the manuscript.
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12881-020-01047-5.
References
- 1.Klein BE, Klein R. Forecasting age-related macular degeneration through 2050. Jama. 2009;301(20):2152–2153. doi: 10.1001/jama.2009.729. [DOI] [PubMed] [Google Scholar]
- 2.Klein R, Chou CF, Klein BE, Zhang X, Meuer SM, Saaddine JB. Prevalence of age-related macular degeneration in the US population. Arch Ophthal (Chicago, Ill : 1960) 2011;129(1):75–80. doi: 10.1001/archophthalmol.2010.318. [DOI] [PubMed] [Google Scholar]
- 3.Khanna S, Komati R, Eichenbaum DA, Hariprasad I, Ciulla TA, Hariprasad SM. Current and upcoming anti-VEGF therapies and dosing strategies for the treatment of neovascular AMD: a comparative review. BMJ open ophthalmology. 2019;4(1):e000398. doi: 10.1136/bmjophth-2019-000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blasiak J. Senescence in the pathogenesis of age-related macular degeneration. Cell Mol Life Sci. 2020;77(5):789. doi: 10.1007/s00018-019-03420-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rao P, Lum F, Wood K, Salman C, Burugapalli B, Hall R, Singh S, Parke DW, 2nd, Williams GA. Real-world vision in age-related macular degeneration patients treated with single anti-VEGF drug type for 1 year in the IRIS registry. Ophthalmology. 2018;125(4):522–528. doi: 10.1016/j.ophtha.2017.10.010. [DOI] [PubMed] [Google Scholar]
- 6.Cukras C, Fine SL. Classification and grading system for age-related macular degeneration. Int Ophthalmol Clin. 2007;47(1):51–63. doi: 10.1097/IIO.0b013e31802bd785. [DOI] [PubMed] [Google Scholar]
- 7.Chakravarthy U, Wong TY, Fletcher A, Piault E, Evans C, Zlateva G, Buggage R, Pleil A, Mitchell P. Clinical risk factors for age-related macular degeneration: a systematic review and meta-analysis. BMC Ophthalmol. 2010;10:31. doi: 10.1186/1471-2415-10-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jager RD, Mieler WF, Miller JW. Age-related macular degeneration. N Engl J Med. 2008;358(24):2606–2617. doi: 10.1056/NEJMra0801537. [DOI] [PubMed] [Google Scholar]
- 9.Shahid H, Khan JC, Cipriani V, Sepp T, Matharu BK, Bunce C, Harding SP, Clayton DG, Moore AT, Yates JR. Age-related macular degeneration: the importance of family history as a risk factor. Br J Ophthalmol. 2012;96(3):427–431. doi: 10.1136/bjophthalmol-2011-300193. [DOI] [PubMed] [Google Scholar]
- 10.Klein RJ, Zeiss C, Chew EY, Tsai JY, Sackler RS, Haynes C, Henning AK, SanGiovanni JP, Mane SM, Mayne ST, et al. Complement factor H polymorphism in age-related macular degeneration. Science (New York, NY) 2005;308(5720):385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arakawa S, Takahashi A, Ashikawa K, Hosono N, Aoi T, Yasuda M, Oshima Y, Yoshida S, Enaida H, Tsuchihashi T, et al. Genome-wide association study identifies two susceptibility loci for exudative age-related macular degeneration in the Japanese population. Nat Genet. 2011;43(10):1001–1004. doi: 10.1038/ng.938. [DOI] [PubMed] [Google Scholar]
- 12.Fritsche LG, Fariss RN, Stambolian D, Abecasis GR, Curcio CA, Swaroop A. Age-related macular degeneration: genetics and biology coming together. Annu Rev Genomics Hum Genet. 2014;15:151–171. doi: 10.1146/annurev-genom-090413-025610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neale BM, Fagerness J, Reynolds R, Sobrin L, Parker M, Raychaudhuri S, Tan PL, Oh EC, Merriam JE, Souied E, et al. Genome-wide association study of advanced age-related macular degeneration identifies a role of the hepatic lipase gene (LIPC) Proc Natl Acad Sci U S A. 2010;107(16):7395–7400. doi: 10.1073/pnas.0912019107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Black JR, Clark SJ. Age-related macular degeneration: genome-wide association studies to translation. Genet Med. 2016;18(4):283–289. doi: 10.1038/gim.2015.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duvvari MR, Paun CC, Buitendijk GH, Saksens NT, Volokhina EB, Ristau T, Schoenmaker-Koller FE, van de Ven JP, Groenewoud JM, van den Heuvel LP, et al. Analysis of rare variants in the C3 gene in patients with age-related macular degeneration. PLoS One. 2014;9(4):e94165. doi: 10.1371/journal.pone.0094165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fritsche LG, Chen W, Schu M, Yaspan BL, Yu Y, Thorleifsson G, Zack DJ, Arakawa S, Cipriani V, Ripke S, et al. Seven new loci associated with age-related macular degeneration. Nat Genet. 2013;45(4):433–439. doi: 10.1038/ng.2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu F, Liu S, Hao Q, Liu L, Zhang J, Chen X, Hu W, Huang P. Association between complement factor C2/C3/CFB/CFH polymorphisms and age-related macular degeneration: a meta-analysis. Genet Testing Mol Biomarkers. 2018;22(9):526–540. doi: 10.1089/gtmb.2018.0110. [DOI] [PubMed] [Google Scholar]
- 18.Xiying M, Wenbo W, Wangyi F, Qinghuai L. Association of Apolipoprotein E Polymorphisms with age-related macular degeneration subtypes: an updated systematic review and meta-analysis. Arch Med Res. 2017;48(4):370–377. doi: 10.1016/j.arcmed.2017.08.002. [DOI] [PubMed] [Google Scholar]
- 19.Lim LS, Mitchell P, Seddon JM, Holz FG, Wong TY. Age-related macular degeneration. Lancet (London, England) 2012;379(9827):1728–1738. doi: 10.1016/S0140-6736(12)60282-7. [DOI] [PubMed] [Google Scholar]
- 20.Moja L, Lucenteforte E, Kwag KH, Bertele V, Campomori A, Chakravarthy U, D'Amico R, Dickersin K, Kodjikian L, Lindsley K, et al. Systemic safety of bevacizumab versus ranibizumab for neovascular age-related macular degeneration. Cochrane Database Syst Rev. 2014;9:Cd011230. doi: 10.1002/14651858.CD011230.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Solomon SD, Lindsley K, Vedula SS, Krzystolik MG, Hawkins BS. Anti-vascular endothelial growth factor for neovascular age-related macular degeneration. Cochrane Database Syst Rev. 2014;8:Cd005139. doi: 10.1002/14651858.CD005139.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ba J, Peng RS, Xu D, Li YH, Shi H, Wang Q, Yu J. Intravitreal anti-VEGF injections for treating wet age-related macular degeneration: a systematic review and meta-analysis. Drug Design Dev Ther. 2015;9:5397–5405. doi: 10.2147/DDDT.S86269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kloeckener-Gruissem B, Barthelmes D, Labs S, Schindler C, Kurz-Levin M, Michels S, Fleischhauer J, Berger W, Sutter F, Menghini M. Genetic association with response to intravitreal ranibizumab in patients with neovascular AMD. Invest Ophthalmol Vis Sci. 2011;52(7):4694–4702. doi: 10.1167/iovs.10-6080. [DOI] [PubMed] [Google Scholar]
- 24.Park UC, Shin JY, McCarthy LC, Kim SJ, Park JH, Chung H, Yu HG. Pharmacogenetic associations with long-term response to anti-vascular endothelial growth factor treatment in neovascular AMD patients. Mol Vis. 2014;20:1680–1694. [PMC free article] [PubMed] [Google Scholar]
- 25.Ratnapriya R, Chew EY. Age-related macular degeneration-clinical review and genetics update. Clin Genet. 2013;84(2):160–166. doi: 10.1111/cge.12206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu Y, Guan N, Xu J, Yang X, Ma K, Zhou H, Zhang F, Snellingen T, Jiao Y, Liu X, et al. Association of CFH, LOC387715, and HTRA1 polymorphisms with exudative age-related macular degeneration in a northern Chinese population. Mol Vis. 2008;14:1373–1381. [PMC free article] [PubMed] [Google Scholar]
- 27.Tosi GM, Caldi E, Neri G, Nuti E, Marigliani D, Baiocchi S, Traversi C, Cevenini G, Tarantello A, Fusco F, et al. HTRA1 and TGF-beta1 concentrations in the aqueous humor of patients with Neovascular age-related macular degeneration. Invest Ophthalmol Vis Sci. 2017;58(1):162–167. doi: 10.1167/iovs.16-20922. [DOI] [PubMed] [Google Scholar]
- 28.Yang Z, Camp NJ, Sun H, Tong Z, Gibbs D, Cameron DJ, Chen H, Zhao Y, Pearson E, Li X, et al. A variant of the HTRA1 gene increases susceptibility to age-related macular degeneration. Science (New York, NY) 2006;314(5801):992–993. doi: 10.1126/science.1133811. [DOI] [PubMed] [Google Scholar]
- 29.Dewan A, Liu M, Hartman S, Zhang SS, Liu DT, Zhao C, Tam PO, Chan WM, Lam DS, Snyder M, et al. HTRA1 promoter polymorphism in wet age-related macular degeneration. Science (New York, NY) 2006;314(5801):989–992. doi: 10.1126/science.1133807. [DOI] [PubMed] [Google Scholar]
- 30.Askari M, Nikpoor AR, Gorjipour F, Mazidi M, Sanati MH, Aryan H, Irani A, Ghasemi Falavarjani K, Nazari H, Mousavizadeh K. Association of Htra1 gene polymorphisms with the risk of developing AMD in Iranian population. Rep Biochemistry Mol Biol. 2015;4(1):43–49. [PMC free article] [PubMed] [Google Scholar]
- 31.Chan CC, Shen D, Zhou M, Ross RJ, Ding X, Zhang K, Green WR, Tuo J. Human HtrA1 in the archived eyes with age-related macular degeneration. Trans Am Ophthalmol Soc. 2007;105:92–97. [PMC free article] [PubMed] [Google Scholar]
- 32.Chen JH, Yang Y, Zheng Y, Qiu M, Xie M, Lin W, Zhang M, Pang CP, Chen H. No association of age-related maculopathy susceptibility protein 2/HtrA serine peptidase 1 or complement factor H polymorphisms with early age-related maculopathy in a Chinese cohort. Mol Vis. 2013;19:944–954. [PMC free article] [PubMed] [Google Scholar]
- 33.Cheng Y, Huang L, Li X, Zhou P, Zeng W, Zhang C. Genetic and functional dissection of ARMS2 in age-related macular degeneration and polypoidal choroidal vasculopathy. PLoS One. 2013;8(1):e53665. doi: 10.1371/journal.pone.0053665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chu J, Zhou CC, Lu N, Zhang X, Dong FT. Genetic variants in three genes and smoking show strong associations with susceptibility to exudative age-related macular degeneration in a Chinese population. Chin Med J. 2008;121(24):2525–2533. [PubMed] [Google Scholar]
- 35.Kanda A, Chen W, Othman M, Branham KE, Brooks M, Khanna R, He S, Lyons R, Abecasis GR, Swaroop A. A variant of mitochondrial protein LOC387715/ARMS2, not HTRA1, is strongly associated with age-related macular degeneration. Proc Natl Acad Sci U S A. 2007;104(41):16227–16232. doi: 10.1073/pnas.0703933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaur I, Cantsilieris S, Katta S, Richardson AJ, Schache M, Pappuru RR, Narayanan R, Mathai A, Majji AB, Tindill N, et al. Association of the del443ins54 at the ARMS2 locus in Indian and Australian cohorts with age-related macular degeneration. Mol Vis. 2013;19:822–828. [PMC free article] [PubMed] [Google Scholar]
- 37.Kaur I, Katta S, Hussain A, Hussain N, Mathai A, Narayanan R, Hussain A, Reddy RK, Majji AB, Das T, et al. Variants in the 10q26 gene cluster (LOC387715 and HTRA1) exhibit enhanced risk of age-related macular degeneration along with CFH in Indian patients. Invest Ophthalmol Vis Sci. 2008;49(5):1771–1776. doi: 10.1167/iovs.07-0560. [DOI] [PubMed] [Google Scholar]
- 38.Kondo N, Honda S, Ishibashi K, Tsukahara Y, Negi A. LOC387715/HTRA1 variants in polypoidal choroidal vasculopathy and age-related macular degeneration in a Japanese population. Am J Ophthalmol. 2007;144(4):608–612. doi: 10.1016/j.ajo.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 39.Lana TP, da Silva Costa SM, Ananina G, Hirata FE, Rim PHH, Medina FM, de Vasconcellos JPC, de Melo MB. Association of HTRA1 rs11200638 with age-related macular degeneration (AMD) in Brazilian patients. Ophthalmic Genet. 2018;39(1):46–50. doi: 10.1080/13816810.2017.1354382. [DOI] [PubMed] [Google Scholar]
- 40.Lee SJ, Kim NR, Chin HS. LOC387715/HTRA1 polymorphisms, smoking and combined effects on exudative age-related macular degeneration in a Korean population. Clin Exp Ophthalmol. 2010;38(7):698–704. doi: 10.1111/j.1442-9071.2010.02316.x. [DOI] [PubMed] [Google Scholar]
- 41.Leveziel N, Souied EH, Richard F, Barbu V, Zourdani A, Morineau G, Zerbib J, Coscas G, Soubrane G, Benlian P. PLEKHA1-LOC387715-HTRA1 polymorphisms and exudative age-related macular degeneration in the French population. Mol Vis. 2007;13:2153–2159. [PubMed] [Google Scholar]
- 42.Li WJ, Sheng XL, Zhuang WJ, Li HP. Associations of SNPs in ARMS2/HTRA1 with age-related macular degeneration. Ningxia Medical University Thesis for Master's Degree. 2015. [Google Scholar]
- 43.Liang X, Cui L, Gu H, Zhou HY, Xu J, Liu NP. Interaction of susceptibility genes in patients with exudative age-related macular degeneration. Chin J Ophthalmol. 2012;48(3):241–245. [PubMed] [Google Scholar]
- 44.Lin JM, Wan L, Tsai YY, Lin HJ, Tsai Y, Lee CC, Tsai CH, Tsai FJ, Tseng SH. HTRA1 polymorphism in dry and wet age-related macular degeneration. Retina (Philadelphia, Pa) 2008;28(2):309–313. doi: 10.1097/IAE.0b013e31814cef3a. [DOI] [PubMed] [Google Scholar]
- 45.Losonczy G, Fekete A, Voko Z, Takacs L, Kaldi I, Ajzner E, Kasza M, Vajas A, Berta A, Balogh I. Analysis of complement factor H Y402H, LOC387715, HTRA1 polymorphisms and ApoE alleles with susceptibility to age-related macular degeneration in Hungarian patients. Acta Ophthalmol. 2011;89(3):255–262. doi: 10.1111/j.1755-3768.2009.01687.x. [DOI] [PubMed] [Google Scholar]
- 46.Matuskova V, Zeman T, Ewerlingova L, Hlinomazova Z, Soucek J, Vlkova E, Goswami N, Balcar VJ, Sery O. An association of neovascular age-related macular degeneration with polymorphisms of CFH, ARMS2, HTRA1 and C3 genes in Czech population. Acta Ophthalmol. 2020. [DOI] [PubMed]
- 47.Mori K, Horie-Inoue K, Kohda M, Kawasaki I, Gehlbach PL, Awata T, Yoneya S, Okazaki Y, Inoue S. Association of the HTRA1 gene variant with age-related macular degeneration in the Japanese population. J Hum Genet. 2007;52(7):636–641. doi: 10.1007/s10038-007-0162-1. [DOI] [PubMed] [Google Scholar]
- 48.Ng TK, Liang XY, Lai TY, Ma L, Tam PO, Wang JX, Chen LJ, Chen H, Pang CP. HTRA1 promoter variant differentiates polypoidal choroidal vasculopathy from exudative age-related macular degeneration. Sci Rep. 2016;6:28639. doi: 10.1038/srep28639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tian J, Qin X, Fang K, Chen Q, Hou J, Li J, Yu W, Chen D, Hu Y, Li X. Association of genetic polymorphisms with response to bevacizumab for neovascular age-related macular degeneration in the Chinese population. Pharmacogenomics. 2012;13(7):779–787. doi: 10.2217/pgs.12.53. [DOI] [PubMed] [Google Scholar]
- 50.Tuo J, Ross RJ, Reed GF, Yan Q, Wang JJ, Bojanowski CM, Chew EY, Feng X, Olsen TW, Ferris FL, et al. The HtrA1 promotor polymorphism, smoking, and age-related macular degeneration in multiple case-control samples. Ophthalmology. 2008;115(11):1891–1898. doi: 10.1016/j.ophtha.2008.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weger M, Renner W, Steinbrugger I, Kofer K, Wedrich A, Groselj-Strele A, El-Shabrawi Y, Schmut O, Haas A. Association of the HTRA1 -625G>a promoter gene polymorphism with exudative age-related macular degeneration in a central European population. Mol Vis. 2007;13:1274–1279. [PubMed] [Google Scholar]
- 52.Yang N, Xing J, Shao Y, Zhu Z, Ba YY, Wei W. Relationship between HTRA1 polymorphism and genetic susceptibility of wet age-related macular degeneration in Han population. Guoji Yanke Zazhi. 2018;18(5):815–818. [Google Scholar]
- 53.Zeng JX, Tang LS, Zhang K. Assessing susceptibility to age-related macular gegeneration with genetic makers and environment factors. Doctoral Dissertation of Zhongnan University. 2011. [Google Scholar]
- 54.Tam PO, Ng TK, Liu DT, Chan WM, Chiang SW, Chen LJ, DeWan A, Hoh J, Lam DS, Pang CP. HTRA1 variants in exudative age-related macular degeneration and interactions with smoking and CFH. Invest Ophthalmol Vis Sci. 2008;49(6):2357–2365. doi: 10.1167/iovs.07-1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jiang H, Qu Y, Dang G, Zhang X, Yin N, Zhang Y, Bi H, Pan X, Xu X, Zhou F, et al. Analyses of single nucleotide polymorphisms and haplotype linkage of LOC387715 and the HTRA1 gene in exudative age-related macular degeneration in a Chinese cohort. Retina (Philadelphia, Pa) 2009;29(7):974–979. doi: 10.1097/IAE.0b013e3181a3b90e. [DOI] [PubMed] [Google Scholar]
- 56.Ruamviboonsuk P, Tadarati M, Singhanetr P, Wattanapokayakit S, Kunhapan P, Wanitchanon T, Wichukchinda N, Mushiroda T, Akiyama M, Momozawa Y, et al. Genome-wide association study of neovascular age-related macular degeneration in the Thai population. J Hum Genet. 2017;62(11):957–962. doi: 10.1038/jhg.2017.72. [DOI] [PubMed] [Google Scholar]
- 57.Fritsche LG, Loenhardt T, Janssen A, Fisher SA, Rivera A, Keilhauer CN, Weber BH. Age-related macular degeneration is associated with an unstable ARMS2 (LOC387715) mRNA. Nat Genet. 2008;40(7):892–896. doi: 10.1038/ng.170. [DOI] [PubMed] [Google Scholar]
- 58.Pan HY, Mi YY, Xu K, Zhang Z, Wu H, Zhang W, Yuan W, Shi L, Zhang LF, Zhu LJ, et al. Association of C-reactive protein (CRP) rs1205 and rs2808630 variants and risk of cancer. J Cell Physiol. 2020. [DOI] [PubMed]
- 59.Zhang LF, Xu K, Tang BW, Zhang W, Yuan W, Yue C, Shi L, Mi YY, Zuo L, Zhu LJ. Association between SOD2 V16A variant and urological cancer risk. Aging. 2020;12(1):825–843. doi: 10.18632/aging.102658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21(11):1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 61.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 62.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22(4):719–748. [PubMed] [Google Scholar]
- 63.Napolioni V. The relevance of checking population allele frequencies and Hardy-Weinberg Equilibrium in genetic association studies: the case of SLC6A4 5-HTTLPR polymorphism in a Chinese Han Irritable Bowel Syndrome association study. Immunology Letters. 2014;162(1 Pt A):276–278. doi: 10.1016/j.imlet.2014.08.009. [DOI] [PubMed] [Google Scholar]
- 64.Hayashino Y, Noguchi Y, Fukui T. Systematic evaluation and comparison of statistical tests for publication bias. J Epidemiology. 2005;15(6):235–243. doi: 10.2188/jea.15.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yu Q, Zhu J, Yao Y, Sun C. Complement family member CFI polymorphisms and AMD susceptibility from a comprehensive analysis. Biosci Rep. 2020;40:4. doi: 10.1042/BSR20200406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shao HB, Ren K, Gao SL, Zou JG, Mi YY, Zhang LF, Zuo L, Okada A, Yasui T. Human methionine synthase A2756G polymorphism increases susceptibility to prostate cancer. Aging. 2018;10(7):1776–1788. doi: 10.18632/aging.101509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen H, Yang Z, Gibbs D, Yang X, Hau V, Zhao P, Ma X, Zeng J, Luo L, Pearson E, et al. Association of HTRA1 polymorphism and bilaterality in advanced age-related macular degeneration. Vis Res. 2008;48(5):690–694. doi: 10.1016/j.visres.2007.10.014. [DOI] [PubMed] [Google Scholar]
- 68.Cruz-Gonzalez F, Cieza-Borrella C, Lopez Valverde G, Lorenzo-Perez R, Hernandez-Galilea E, Gonzalez-Sarmiento R. CFH (rs1410996), HTRA1 (rs112000638) and ARMS2 (rs10490923) gene polymorphisms are associated with AMD risk in Spanish patients. Ophthalmic Genet. 2014;35(2):68–73. doi: 10.3109/13816810.2013.781193. [DOI] [PubMed] [Google Scholar]
- 69.Lu F, Hu J, Zhao P, Lin Y, Yang Y, Liu X, Fan Y, Chen B, Liao S, Du Q, et al. HTRA1 variant increases risk to neovascular age-related macular degeneration in Chinese population. Vis Res. 2007;47(24):3120–3123. doi: 10.1016/j.visres.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 70.Mohamad NA, Ramachandran V, Mohd Isa H, Chan YM, Ngah NF, Ching SM, Hoo FK, Wan Sulaiman WA, Inche Mat LN, Mohamed MH. Association of HTRA1 and ARMS2 gene polymorphisms with response to intravitreal ranibizumab among neovascular age-related macular degenerative subjects. Human Genom. 2019;13(1):13. doi: 10.1186/s40246-019-0197-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang X, Hu J, Zhang J, Guan H. Polymorphisms in CFH, HTRA1 and CX3CR1 confer risk to exudative age-related macular degeneration in Han Chinese. Br J Ophthalmol. 2010;94(9):1211–1214. doi: 10.1136/bjo.2009.165811. [DOI] [PubMed] [Google Scholar]
- 72.Amoaku WM, Chakravarthy U, Gale R, Gavin M, Ghanchi F, Gibson J, Harding S, Johnston RL, Kelly SP, Lotery A, et al. Defining response to anti-VEGF therapies in neovascular AMD. Eye (London, England) 2015;29(6):721–731. doi: 10.1038/eye.2015.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cheung GCM, Lai TYY, Gomi F, Ruamviboonsuk P, Koh A, Lee WK. Anti-VEGF Therapy for Neovascular AMD and Polypoidal Choroidal Vasculopathy. Asia-Pac J Ophthalmol (Philadelphia, Pa) 2017;6(6):527–534. doi: 10.22608/APO.2017260. [DOI] [PubMed] [Google Scholar]
- 74.Jabbarpoor Bonyadi MH, Yaseri M, Bonyadi M, Soheilian M, Nikkhah H. Association of combined cigarette smoking and ARMS2/LOC387715 A69S polymorphisms with age-related macular degeneration: a meta-analysis. Ophthalmic Genet. 2017;38(4):308–313. doi: 10.1080/13816810.2016.1237664. [DOI] [PubMed] [Google Scholar]
- 75.Munafo MR, Flint J. Meta-analysis of genetic association studies. Trends Genet. 2004;20(9):439–444. doi: 10.1016/j.tig.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 76.Chen W, Xu W, Tao Q, Liu J, Li X, Gan X, Hu H, Lu Y. Meta-analysis of the association of the HTRA1 polymorphisms with the risk of age-related macular degeneration. Exp Eye Res. 2009;89(3):292–300. doi: 10.1016/j.exer.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 77.Tang NP, Zhou B, Wang B, Yu RB. HTRA1 promoter polymorphism and risk of age-related macular degeneration: a meta-analysis. Ann Epidemiol. 2009;19(10):740–745. doi: 10.1016/j.annepidem.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 78.Narayanan R, Butani V, Boyer DS, Atilano SR, Resende GP, Kim DS, Chakrabarti S, Kuppermann BD, Khatibi N, Chwa M, et al. Complement factor H polymorphism in age-related macular degeneration. Ophthalmology. 2007;114(7):1327–1331. doi: 10.1016/j.ophtha.2006.10.035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Allele Frequency from 1000 Genomes Browser and present study.
Additional file 2: Figure S1. A: Begg’s funnel plot for publication bias test (A-allele vs. G-allele). Each point represents a separate study for the indicated association. B: Egger’s publication bias plot (A-allele vs. G-allele).
Additional file 3: Figure S2. Sensitivity analysis between HTRA1 gene rs11200638 polymorphism and AMD risk (A-allele vs. G-allele).
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files.