Abstract
Background
Nuclear genome or family mitochondrial screening system has become the hot focus of studies into essential hypertension. The role of mitochondrial DNA (mtDNA) in sporadic Chinese patients with hypertension has not been fully understood. The study was to evaluate the associations of mtDNA mutations with maternally inherited essential hypertensive subjects in China.
Methods
From June 2009 to June 2016, a total of 800 gender-matched Chinese patients with maternally inherited essential hypertension (MIEH) and control group were 1:1 enrolled in this case-control study. Genomic DNA was extracted from each person’s peripheral blood cells. The main mtDNA locations for MIEH were screened with oligodeoxynucleotides 3777-4679 bp, analyzed and compared with the updated consensus Cambridge Sequence. Pathogenic mtDNA mutations were identified from the mitochondrial map.
Results
MIEH subjects presented significantly higher values than those of control group in abdominal circumference (AC), waist circumference (WC), body mass index (BMI), fasting blood glucose (FBG), triglyceride (TG), low-density lipoprotein cholesterol (LDL) and renal function (P < 0.05). MIEH subjects carried more amino acid changes and coding sequence variants (P < 0.01) than control group. The allele frequencies of the eight single nucleotide polymorphisms (SNPs) were significantly different between the two groups, including m.3970 C > T, m.4048G > A, m.4071C > T, m.4086C > T, m. 4164A > G and m.4248 T > C in ND1 gene, and m.4386 T > C and m.4394C > T in tRNAGln gene(P < 0.001). Fifty-five homoplasmic or heteroplasmic mutations were detected in 5 genes: ND1, tRNAIle, tRNAMet, tRNAGln and ND2 gene. The ND1 gene was the main mutation site, where the most mtDNA mutation was m.3970 C > T.
Conclusions
The mtDNA mutations were involved in the process of MIEH. We identified mitochondrial genetic characteristics in MIEH patients in China. The present research serves as a solid foundation for further detailed research on the association between MIEH and mitochondrial dysfunction, and their causal relationship in Chinese and other populations with a similar lifestyle.
Keywords: Mitochondria, DNA, Mutation, Essential hypertension, Maternal inheritance
Background
Essential hypertension (EH) is a common cardiovascular disorder, influencing about 1 billion people around the world [1]. It is generally believed that the interaction between genic and environmental factors affected EH, which may be caused by single-gene defects or multifactorial conditions [2]. Maternally inherited essential hypertension (MIEH) is EH that is consistent with the pattern of maternal inheritance [3]. Mitochondrial DNA (mtDNA) can cause mitochondrial diseases, which are transmitted from the mother exclusively. MtDNA mutations were marked in several pathogenic disorders including mitochondrial myopathy, stroke-like attacks, encephalopathy and maternally hereditary diabetes [4]. In addition, mutations in mtDNA have also been observed to play a role in the pathogenesis of MIEH [5].
The repair and protection systems of mtDNA are less efficient compared to that of nuclear DNA [6]. MtDNA mutations have been linked to MIEH through modifying several functional tRNAs [7]. In particular, a previous study identified that the mutation may reduce the steady-state level of mt-tRNAGlnm.4375C > T and subsequently cause the mitochondrial dysfunction that is responsible for hypertension [8]. So far, the mechanism of mtDNA mutations in MIEH has not been completely elucidated, especially on the interplay between mtDNA mutations and other risk factors, such as the development of blood pressure, nuclear genes, and environmental conditions [9]. While people are more concerned about the role of the nuclear genome [10], investigating the role of mtDNA sequence alteration may help to understand the genetic pathogenesis of MIEH.
Mitochondrial genes in 3777–4679 region were proposed to be hot spots for mutations associated with hypertension as described previously [11]. In order to better understand the pathogenic mechanisms underlying MIEH, we studied clinical and genetic evidence to investigate the association between the mtDNA mutations in 3777–4679 region and MIEH. In this study, we focused on the Han Chinese population, as there is a limited amount of study on this racial group and they might be overlooked for lacking medical knowledge and regular examinations [12].
Methods
Subjects
The current case-control study was based on 400 unrelated patients with MIEH and 400 healthy control individuals in the Jiangsu Province of China. The MIEH patients were recruited according to the following inclusion criteria:
in-patients or outpatients who have undergone regular medical check-up at the Department of Cardiology in Northern Jiangsu People’s Hospital from June 2009 to June 2016;
more than 18 years old;
with a diagnosis of primary hypertension;
diagnosed with MIEH on the basis of the maternal transmission of EH within generations, which was transmitted by the mother or her relatives, rather than by the father.
Participants were excluded if they were diagnosed as follows:
secondary hypertension (e.g. aortic coarctation, renal arterial stenosis, hyperaldosteronism, and pheochromocytoma);
congenital cardiovascular disease;
organic valve diseases.
Another 400 gender-matched healthy individuals were recruited to the current study as controls. Furthermore, the controls were unrelated healthy subjects from the same area who received annual examination in physical examination center of Northern Jiangsu People’s Hospital. They were collected randomly from the physical examination list. The control group included the following criteria:
systolic blood pressure (SBP) of < 130 mmHg and diastolic blood pressure (DBP) of < 85 mmHg.
no personal or family history of hypertension.
Hypertension in one or both biologic parents was considered to be a positive family history of EH. All subjects in the study were interviewed to identify both personal and family medical histories of clinical abnormalities. Verbal Informed Consent, medical history, clinical assessment and genetic analysis were obtained from each individual under protocols involved in the study. Verbal consent is that the mtDNA analysis is used only for diagnosis, not for treatment. It was of no harm to anyone. The protocol was implemented in accordance with the Declaration of Helsinki and approved by the ethics committee of the institutional review board at the Northern Jiangsu People’s Hospital.
Data collection
Body mass index (BMI) refers to a person’s body mass in kilograms divided by height in square meters (kg/m2). Patients reporting cigarette use within 1 year prior to examination were considered as smokers. Blood pressure was measured by an experienced physician who was blinded to the study according to the criteria of the World Health Organization (WHO) [13]. Three measurements of systolic and diastolic blood pressure were taken and the mean value was used as the measurement. According to the 2010 Chinese Hypertension Management, hypertension was diagnosed as follows [14]: the SBP > 140 mmHg and/or DBP > 90 mmHg measured three times on different days or a history of hypertension with current antihypertensive medications. All participants also underwent laboratory tests on hypertension risk factors. 12 h after fasting, lipid profile, fasting blood glucose (FBG), and kidney function test were performed by an automatic biochemistry analyzer (Hitach 7600DDP, Japan).
Mitochondrial DNA analysis
Genomic DNA was extracted from each person’s peripheral blood using standard protocols [15]. MtDNA was isolated by Promega Wizard Genomic DNA Purification Kit (Madison, WI, USA). The main chromosome locations for hypertension as described previously [16] were screened using oligodeoxynucleotides 3777-4679 bp. The mitochondrial tRNAIle gene was amplified by Polymerase chain reaction (PCR) using the primer sequences as follows: forward: 5′- TGGCTCCTTTAACCTCTCCA-3′ and reverse: 5′- AAGGATTATGGATGCGGTTG -3′. PCR cycle program was carried out in a 9700 Thermocycler (Perkin-Elmer Applied Biosystems, Norwalk, USA). Each fragment had been purified and sequenced by ABI 3730 Sequence Analysis software (Applied Biosystems, Inc., Foster City, CA, USA) using the BigDye Terminator v1.1 kit (ABI Company, Carlsbad, CA, USA), and subsequently SeqWeb program GAP (GCG) was analyzed and compared with the updated consensus Cambridge Sequence [17, 18]. Pathogenic mitochondrial variants were identified from the human mitochondrial genome database (https://www.mitomap.org/MITOMAP) [19].
Statistical analysis
Statistical analysis was performed using R and SPSS software (version 22.0; SPSS Inc., Chicago, IL, USA). Continuous variables were tested for normal distribution by Kolmogorov-Smirnov test and then expressed by mean ± standard deviation (SD). The relationship between potential continuous and discrete factors and MIEH were analyzed with Student’s t-test and Fisher’s exact t-test. A P-value ≤0.05 was considered to be statistically significant.
Results
Clinical evaluation of baseline characteristics.
Table 1 shows the main characteristics of all participants. In the present study, age and SBP did not differ between the two groups. There were significant differences in BMI, waist circumference (WC), abdominal circumference (AC), total cholesterol (TC), triglycerides (TG), low-density lipoprotein cholesterol (LDL), FBG, uric acid (UA), creatinine (CR) as well as blood urea nitrogen (BUN) (P < 0.05) between the two groups.
Table 1.
Comparison of baseline clinical data between the MIEH and control groups
| Subjects | MIEH group | Control group | P-value |
|---|---|---|---|
| gender (M/F) | 400 (202/198) | 400 (197/203) | 1 |
| age at test (years) | 68.65 ± 8.34 | 65.36 ± 6.75 | 1.38e-9 |
| age at onset (years) | 48.56 ± 6.7 | NA | |
| SBP (mmHg) | 148.5 ± 19.8 | 145.6 ± 18.6 | 0.033 |
| DBP (mmHg) | 94.8 ± 8.9 | 88.4 ± 12.5 | 3.72e-16 |
| BMI (kg/m2) | 25.90 ± 3.60 | 23.52 ± 3.03 | 1.11e-22 |
| WC (cm) | 86.90 ± 10.78 | 78.08 ± 8.71 | 8.52e-34 |
| AC (cm) | 89.50 ± 11.15 | 80.28 ± 7.79 | 2.02e-37 |
| Alcohol, n (%) | 100(25) | 41(10) | 5.13e-8 |
| Smoking, n(%) | 90(23) | 52(13) | 5.84e-4 |
| TG (mmol/L) | 1.86 ± 1.20 | 1.37 ± 0.88 | 8.65e-11 |
| TC (mmol/L) | 4.58 ± 1.96 | 4.25 ± 1.78 | 0.0129 |
| LDL (mmol/L) | 2.63 ± 1.24 | 2.02 ± 1.39 | 1.04e-10 |
| FBG (mmol/L) | 5.19 ± 2.18 | 4.33 ± 1.84 | 2.54e-09 |
| UA (umol/L) | 368.04 ± 127.28 | 323.39 ± 78.92 | 4.02e-09 |
| Cr (ummol/L) | 105.39 ± 33.71 | 87.38 ± 30.71 | 9.40e-15 |
| BUN (mmol/L) | 5.76 ± 2.08 | 4.84 ± 1.79 | 3.86e-11 |
Abbreviations: F female; M male. SBP Systolic blood pressure; DBP Diastolic blood pressure; BMI Body mass index; WC waist circumference; AC abdomen circumference; TG triglyceride; TC total cholesterol; LDL low-density lipoprotein cholesterol; FBG fasting blood glucose; UA uric acid; Cr creatinine; BUN blood urea nitrogen; *: A P value < 0.05 was marked by a star
mtDNA analysis
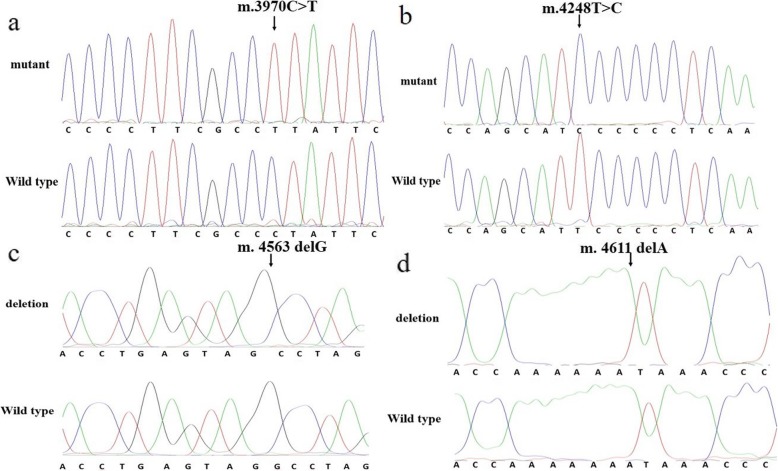
The distribution of mutations in the mtDNA 3777–4679 bp of all participants is shown in Fig. 1. Table 2 shows a comparison of the frequency of mtDNA sequence analyses in the 400 balanced cases and controls. MtDNA analysis revealed 55 mutation sites in the 400 MIEH subjects (Table 3). The ND1 gene was the main mutation site, where the highest mutation frequency was m.3970 C > T (Fig. 2a).
Fig. 1.
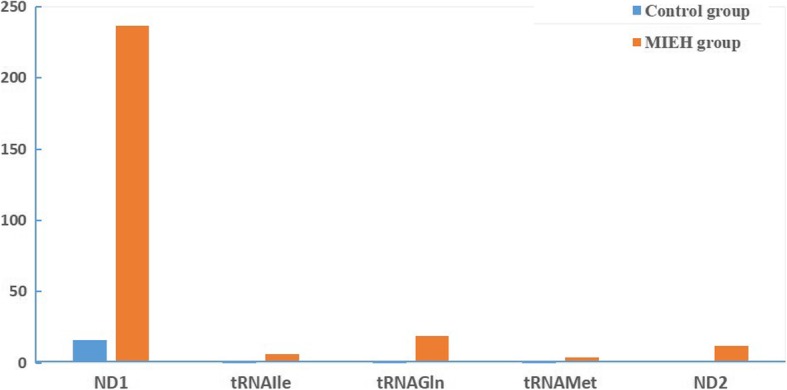
Distribution histogram of the number of variants in mtDNA 3777–4679 bp among 5 genes
Table 2.
Distribution of mtDNA sequence analyses at positions 3777–4679
| Gene | Position | Length | Control group (n(%)) | MIEH group (n(%)) | Fisher’s exact P value |
|---|---|---|---|---|---|
| ND1 | 3777–4262 | 486 | 16 | 237 | < 2.2e-16 |
| tRNAIle | 4263–4331 | 69 | 0 | 6 | 0.0307 |
| tRNAGln | 4329–4400 | 72 | 0 | 19 | 3.065e-06 |
| tRNAMet | 4402–4469 | 68 | 0 | 4 | 0.1241 |
| ND2 | 4470–4679 | 210 | 1 | 12 | 0.003185 |
Table 3.
Mutation sites of mtDNA in MIEH individuals and controls
| Site of mutation | Gene | Replacement | Homoplasmic/heteroplasmic variants | Number of mutations(n) | P value | Previously reporteda | Change of Amino acid | |
|---|---|---|---|---|---|---|---|---|
| (MIEH) | (Controls) | |||||||
| 3902 | ND1 | A to T | heteroplasmic | 1 | 0 | 1 | NO | non-syn;D-V |
| 3905 | ND1 | T to A | heteroplasmic | 1 | 0 | 1 | NO | Non-syn: L-H |
| 3915 | ND1 | G to A | homoplasmic | 6 | 1 | 0.1234 | YES | Syn: G-G |
| 3918 | ND1 | A to G | homoplasmic | 3 | 1 | 0.6241 | YES | Syn: E-E |
| 3941 | ND1 | A to G | homoplasmic | 1 | 0 | 1 | NO | Non-syn: N-S |
| 3948 | ND1 | A to G | homoplasmic | 1 | 0 | 1 | YES | Syn:E-E |
| 3970 | ND1 | C to T | homoplasmic | 67 | 5 | 4.72e-16 | YES | Syn:L-L |
| 4017 | ND1 | C to T | homoplasmic | 1 | 0 | 1 | YES | Syn:L-L |
| 4025 | ND1 | C to T | homoplasmic | 3 | 1 | 0.6241 | YES | Non-syn: T-M |
| 4038 | ND1 | A to G | homoplasmic | 2 | 0 | 0.4994 | YES | Syn:G-G |
| 4047 | ND1 | T to C | homoplasmic | 1 | 0 | 1 | YES | Syn:Y-Y |
| 4048 | ND1 | G to A | homoplasmic | 20 | 2 | 9.93e-05 | YES | Non-syn: D-N |
| 4071 | ND1 | C to T | homoplasmic | 24 | 2 | 7.77e-06 | YES | Syn:Y-Y |
| 4083 | ND1 | T to C | homoplasmic | 1 | 0 | 1 | YES | Syn:F-F |
| 4086 | ND1 | C to T | homoplasmic | 19 | 1 | 3.29e-05 | YES | Syn:V-V |
| 4092 | ND1 | G to A | homoplasmic | 1 | 0 | 1 | YES | Syn:K-K |
| 4093 | ND1 | A to G | homoplasmic | 1 | 0 | 1 | YES | Non-syn: T-A |
| 4104 | ND1 | A to G | homoplasmic | 1 | 0 | 1 | YES | Syn:L-L |
| 4113 | ND1 | G to A | homoplasmic | 1 | 0 | 1 | YES | Syn:L-L |
| 4116 | ND1 | C to T | homoplasmic | 1 | 0 | 1 | YES | Syn:F-F |
| 4117 | ND1 | T to C | homoplasmic | 1 | 0 | 1 | YES | Syn:L-L |
| 4129 | ND1 | A to G | homoplasmic | 3 | 0 | 0.2491 | YES | Non-syn: T-A |
| 4131 | ND1 | A to T | heteroplasmic | 1 | 0 | 1 | NO | Syn:T-T |
| 4134 | ND1 | A to T | heteroplasmic | 1 | 0 | 1 | NO | syn: A-A |
| 4135 | ND1 | T to C | homoplasmic | 1 | 0 | 1 | YES | Non-syn: Y-H |
| 4136 | ND1 | A to G | homoplasmic | 1 | 0 | 1 | YES | Non-syn: Y-H |
| 4140 | ND1 | C to T | homoplasmic | 1 | 0 | 1 | YES | Syn:P-P |
| 4161 | ND1 | C to T | homoplasmic | 1 | 0 | 1 | YES | Syn:L-L |
| 4164 | ND1 | A to G | homoplasmic | 14 | 1 | 8.83e-4 | YES | Syn:M-M |
| 4170 | ND1 | C to T | homoplasmic | 1 | 0 | 1 | YES | Syn:L-L |
| 4176 | ND1 | A to G | homoplasmic | 2 | 0 | 0.4994 | YES | Syn:W-W |
| 4200 | ND1 | A to T | heteroplasmic | 2 | 0 | 0.4994 | YES | Syn:L-L |
| 4203 | ND1 | A to G | homoplasmic | 4 | 0 | 0.1241 | YES | Syn:A-A |
| 4216 | ND1 | T to C | homoplasmic | 4 | 0 | 0.1241 | YES | Non-syn: Y-H |
| 4227 | ND1 | A to G | homoplasmic | 1 | 0 | 1 | YES | syn: M-M |
| 4233 | ND1 | T to C | homoplasmic | 1 | 0 | 1 | YES | Non-syn: I-T |
| 4245 | ND1 | C to T | homoplasmic | 2 | 0 | 0.4994 | YES | Syn:S-S |
| 4248 | ND1 | T to C | homoplasmic | 39 | 2 | 3.25e-10 | YES | Syn:I-I |
| 4254 | ND1 | T to C | homoplasmic | 1 | 0 | 1 | YES | Syn:P-P |
| 4314 | tRNAIle | T to C | homoplasmic | 4 | 0 | 0.1241 | YES | tRNA |
| 4317 | tRNAIle | A to G | homoplasmic | 2 | 0 | 0.4994 | YES | tRNA |
| 4336 | tRNAGln | T to C | homoplasmic | 1 | 0 | 1 | YES | tRNA |
| 4386 | tRNAGln | T to C | homoplasmic | 8 | 0 | 0.0075 | YES | tRNA |
| 4394 | tRNAGln | C to T | homoplasmic | 10 | 0 | 0.0018 | YES | tRNA |
| 4401 | tRNAMet | A to G | homoplasmic | 1 | 0 | 1 | NO | tRNA |
| 4435 | tRNAMet | A to G | homoplasmic | 1 | 0 | 1 | YES | tRNA |
| 4452 | tRNAMet | T to C | homoplasmic | 1 | 0 | 1 | YES | tRNA |
| 4457 | tRNAMet | C to A | heteroplasmic | 1 | 0 | 1 | NO | tRNA |
| 4491 | ND2 | G to A | homoplasmic | 4 | 1 | 0.3734 | YES | Non-syn: V-I |
| 4515 | ND2 | G to C | heteroplasmic | 1 | 0 | 1 | NO | Non-syn: G-R |
| 4535 | ND2 | A to C | heteroplasmic | 2 | 0 | 0.4994 | NO | Syn:L-L |
| 4562 | ND2 | A to G | homoplasmic | 2 | 0 | 0.4994 | YES | Syn:V-V |
| 4563 | ND2 | G to T | heteroplasmic | 1 | 0 | 1 | NO | Syn:G-C |
| 4563 | ND2 | delG | 3 | 0 | 0.2491 | YES | ||
| 4574 | ND2 | A to T | heteroplasmic | 1 | 0 | 1 | NO | Syn:M-M |
| 4576 | ND2 | delA | 1 | 0 | 1 | YES | ||
| 4580 | ND2 | G to A | homoplasmic | 1 | 0 | 1 | YES | Syn:M-M |
| 4611 | ND2 | delA | 10 | 0 | 0.0018 | YES | ||
| 4612 | ND2 | delT | 2 | 0 | 0.4994 | YES | ||
Fig. 2.
a. Identification of the m.3970C > T mutation in the mitochondrial ND1 gene. Arrow indicates the position of the gene mutation. b. Identification of the m.4248 T > C mutation in the mitochondrial ND1 gene. Arrow indicates the position of the gene mutation. c. Identification of the m.4563 delG mutation. Arrow indicates the position of the deletion mutation. d. Identification of the m.4611delA mutations. Arrow indicates the position of the deletion mutation
These results showed that the mtDNA in the MIEH group had more variations than the control group. The allele frequencies of eight single nucleotide polymorphisms (SNPs) were significantly (P < 0.001) different between the two groups, including m.3970 C > T, m.4048G > A, m.4071C > T, m.4086C > T, m.4164A > G and m.4248 T > C (Fig. 2b) in ND1 gene, and m.4386 T > C and m.4394C > T in tRNAGln gene. Fifty-five heteroplasmic or homoplasmic mutations were detected in 5 genes: ND1, tRNAIle, tRNAMet, tRNAGln and ND2 gene. We found 45 homoplasmic mutations in 267 subjects of MIEH. Ten heteroplasmic mutations were found in 11 MIEH subjects. MIEH subjects carried more amino acid changes and coding sequence variants (P < 0.01) compared with normotensive (NT) individuals. An interesting observation in MIEH patients was that we found 4 deletions: m.4563 delG (Fig. 2c), m.4576 delA, m.4611 delA (Fig. 2d) and m.4612 delT mutations in MIEH group patients and the site of highest deletion frequency (10/400 = 0.025) was m.4611 delA mutation in 10 MIEH subjects. The results suggested that mtDNA mutations were positively correlated with MIEH.
Discussion
The purpose of this study was to investigate mutations in mtDNA 3777–4679 region of the Chinese MIEH population. The results showed that the group of MIEH had more mtDNA variations, which mainly located at ND1 site and the highest mutation site was m.3970C > T. Mitochondrial dysfunction is an important factor in cardiovascular disorders [20]. As far as we know, this is one of the first large-scale Han Chinese population-based studies about and the potential role of mtDNA in MIEH. The systematic screen of the association between MIEH and mtDNA mutation is not only essential to further our understanding of the specific mechanism of the mutation in disease etiology, but can also improve the diagnosis and treatment of hypertension.
In an attempt to determine whether mtDNA was involved in the biochemical indicators of the MIEH individuals, we compared and analyzed the biochemical abnormalities of all the participants. Clinical evaluation of all the participants suggested that MIEH subjects presented significantly higher values than those of control group in BMI, WC, AC, LDL, TG, FBG and renal function. Notably, MIEH individuals were obese or overweight in comparison to NT group. High BMI for EH was reported as a strong predictor of hypertension in a multivariate analysis [21]. Therefore, a normal body weight (BMI 18.5–24.9 kg/m2) should be recommended for the prevention and management of MIEH [22]. These factors might contribute to the occurrence and development of MIEH, or they might occur as a result of the progress of MIEH, which results in the damage of the target organ.
Many studies have depicted the function of inherited mtDNA mutations in MIEH family, such as the analysis of maternally transmitted hypertension in a large Han Chinese cohort [23]. Here, we investigated the mtDNA 3777–4679 region using PCR amplification and the sequence analysis. Current results showed that eight SNPs were significantly different between the MIEH and the control groups: m.3970C > T, m.4048G > A, m.4071C > T, m.4086C > T, m.4164A > G and m.4248 T > C in ND1 gene, m.4386 T > C and m.4394C > T in tRNAGln gene. ND1 gene is a hotspot for mutations linked to MIEH. Notably, the impaired synthesis of ND1 (subunits of OXPHOS complex I) may be specifically responsible for the decreased activities of complex I [24]. Mitochondrial encoded complex I is a crucial component in the respiratory chain. The ND1 variants in complex I early in the OXPHOS process, which may contribute to the production of reactive oxygen species (ROS) and may participate in key functional development processes of EH [25].
Mutations of mtDNA may lead to disease, and the significant determinant of their clinical manifestation are susceptible to associated with mutations arising in mtDNA [26]. In this study, unlike mild elevation of total cellular ROS production, an increase in the cells carrying the m.4248 T > C and m.3970 C > T mutations suggested that the defective mitochondria are the major producers of ROS. In turn, the increased levels of cytosolic ROS may produce damage to mitochondrial proteins, nucleic acids, stimulating a forward feeding loop of mitochondrial ROS generation and aggravated cell damage [27]. The reduced levels of mitochondrial proteins were observed in cell lines carrying hypertension associated homoplasmic tRNAGlnm.4386 T > C and m. 4394 C > T mutations belonging to mitochondrial haplogroup M. Ancestral variations in human mtDNA define population-specific mtDNA lineages or haplogroups. Importantly, these were used to trace the origins of different races and provide a foundation for mitochondria based evolutionary medicine [28]. The association between mtDNA haplogroup/SNPs and hypertension has been attributed to a significant alteration in the structure of tRNA and may decrease the steady-state levels of tRNA and oxygen consumption rates that may affect protein levels [29]. Failing to keep the balance of oxygen consumption and production induces mitochondrial dysfunction which was implicated in hypertension pathophysiology, as mentioned earlier [30]. Therefore, mtDNA mutations are promising novel biomarkers for the early detection, prevention, and management of MIEH.
MtDNA mutations, incorporating point mutations, deletions that affect transcription and translation of mtDNA are implicated in various mitochondrial disorders. Rocha et al. have previously revealed a strong correlation between mitochondrial genetics and respiratory chain deficiency in patients with single, large-scale mtDNA deletion [31]. In line with the previous report, our intriguing observation is that there were also a lot of sporadic single mtDNA deletions in MIEH patients [32]. The mean onset time of hypertension for patients with mtDNA point mutations and deletions was gradually ahead of schedule. Thus, it suggests that mtDNA is probably the molecular cause of this disorder. Deletions arise in the mtDNA of post-mitotic cells in patients with mtDNA maintenance disorders, especially during aging. The impaired mitochondrial function may contribute to increased blood pressure of aging with the accumulated multiple mtDNA deletions later in life. The later onset of clinical symptoms due to mtDNA deletions is likely due to the multiple copies of mtDNA within cells. Therefore, the time for mtDNA deletions exceeded an acritical biochemical threshold in proportion, which is necessary to cause a bioenergetic deficit. The mtDNA deletions are thought to be caused by copying errors or repair, or a combination of the both, with the rate of mtDNA deletion formation likely depends on the disorder, the tissue and mechanism of mtDNA replication [32]. We found that the mtDNA deletions disrupted most of the MT-ND2 genes. This suggests there may be duplications with breakpoints that affect complex I, explaining their isolated complex I defect, or there are other deletions we have not found yet. A possible explanation of mtDNA defects in hypertension is that clonally expanded mtDNA deletions are known to be an underlying cause of mitochondrial OXPHOS deficiency in post-mitotic cells. There are studies demonstrating higher levels of mtDNA deletion heteroplasmy in cytochrome c oxidase deficient cells, ultimately initiating hypertension [33]. Some specific deletions in mitochondria DNA may have an underlying role in Chinese hypertensives due to their dysfunction. How mitochondrial mutations and/or deletions interact synergistically in the phenotypic manifestation of hypertension is the next question, which needs further investigation. However, there is still uncertainty regarding the exact mechanisms by which mtDNA deletions clonally expand in patients with mtDNA maintenance disorders. The present study was the primitive step in evaluating the role of mitochondria in unrelated Chinese hypertension patients.
We observed some limitations in this study. Our result from the single-center with relatively small sample size needs further validation. We studied the mtDNA from peripheral blood, which may not represent the mitochondrial activity in other tissues [34]. Another limitation was the lack of functional experiments. Further investigations should be performed to determine the mitochondrial dysfunctions caused by mtDNA mutations, including assessment of ROS production, ATP production and so on.
Conclusion
The mtDNA mutations were involved in the pathological process of MIEH. Additionally, we identified the mitochondrial genetic characteristics of MIEH patients in Chinese Han population. The investigation of the role of mitochondrial dysfunction in MIEH provides new insights into the understanding and treatment of the disorder. The present research serves as a solid foundation for further detailed research on the association between MIEH and mitochondrial dysfunction, and their causal relationship in Chinese and other populations with a similar lifestyle.
Acknowledgements
We are grateful to Dr. Lei Sun, Dr. Yi Zhang, Dr. Zhengyu Bao and Dr. Jianhua Shen for their valuable advice. We thank the editor and the reviewers whose comments/suggestions helped improve and clarify this manuscript.
Abbreviations
- MtDNA
Mitochondrial DNA
- MIEH
Maternally inherited essential hypertension
- EH
Essential hypertension
- SBP
Systolic blood pressure
- DBP
Diastolic blood pressure
- BMI
Body mass index
- WC
Waist circumference
- AC
Abdominal circumference
Authors’ contributions
YZ, JY and XG carried out the studies, participated in collecting data, and drafted the manuscript. YZ and CX performed the statistical analysis and participated in its design. CX helped to draft the manuscript. All authors read and approved the final manuscript.
Funding
This work was funded by The National Natural Science Foundation of China (81800250) and Yangzhou thirteen five Plan Special Funding for Strengthening Health through Science and Education (ZDRC20181). The funding bodies played no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
The raw data of mitochondrial map reported in this paper have been deposited in the Genome Sequence Archive (Genomics, Proteomics & Bioinformatics 2017) in BIG Data Center (Nucleic Acids Res 2019), Beijing Institute of Genomics (BIG), Chinese Academy of Sciences, under accession numbers CRA002605 that are publicly accessible at https://bigd.big.ac.cn/gsa. The updated consensus Cambridge Sequence and pathogenic mitochondrial variants were identified from the human mitochondrial genome database (https://www.mitomap.org/MITOMAP).
Ethics approval and consent to participate
Verbal informed consent was obtained from the subjects involved in the study. The protocol was conducted in accordance with the Declaration of Helsinki. The procedure with which consent to participate was obtained was approved by the Institutional Review Board of Northern Jiangsu People’s Hospital, Jiangsu Province, China. This study was approved by the Institutional Review Board of Northern Jiangsu People’s Hospital, Jiangsu Province, China.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Ye Zhu, Email: 307971331@qq.com.
Jia You, Email: 976513469@qq.com.
Chao Xu, Email: Chao-Xu@ouhsc.edu.
Xiang Gu, Email: 13773531101@163.com.
References
- 1.Wang L, Dong Z, Lin W, Gao R, Chen C, Xu J. Molecular characterization of a pedigree carrying the hypertensionassociated mitochondrial tRNAGln T4363C mutation. Mol Med Rep. 2017;16(5):6029–6033. doi: 10.3892/mmr.2017.7371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shen Y, Peng X, Wang M, Zheng X, Xu G, Lu L, Xu K, Burstrom B, Burstrom K, Wang J. Family member-based supervision of patients with hypertension: a cluster randomized trial in rural China. J Hum Hypertens. 2017;31(1):29–36. doi: 10.1038/jhh.2016.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qiu Q, Li R, Jiang P, Xue L, Lu Y, Song Y, Han J, Lu Z, Zhi S, Mo JQ, et al. Mitochondrial tRNA mutations are associated with maternally inherited hypertension in two Han Chinese pedigrees. Hum Mutat. 2012;33(8):1285–1293. doi: 10.1002/humu.22109. [DOI] [PubMed] [Google Scholar]
- 4.Schapira AH. Mitochondrial disease. Lancet (London, England) 2006;368(9529):70–82. doi: 10.1016/S0140-6736(06)68970-8. [DOI] [PubMed] [Google Scholar]
- 5.Inaba Y, Chen JA, Bergmann SR. Carotid plaque, compared with carotid intima-media thickness, more accurately predicts coronary artery disease events: a meta-analysis. Atherosclerosis. 2012;220(1):128–133. doi: 10.1016/j.atherosclerosis.2011.06.044. [DOI] [PubMed] [Google Scholar]
- 6.Liu P, Demple B. DNA repair in mammalian mitochondria: much more than we thought? Environ Mol Mutagen. 2010;51(5):417–426. doi: 10.1002/em.20576. [DOI] [PubMed] [Google Scholar]
- 7.Lu Z, Chen H, Meng Y, Wang Y, Xue L, Zhi S, Qiu Q, Yang L, Mo JQ, Guan MX. The tRNAMet 4435A>G mutation in the mitochondrial haplogroup G2a1 is responsible for maternally inherited hypertension in a Chinese pedigree. Eur J Human Genet. 2011;19(11):1181–1186. doi: 10.1038/ejhg.2011.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen H, Sun M, Fan Z, Tong M, Chen G, Li D, Ye J, Yang Y, Zhu Y, Zhu J. Mitochondrial C4375T mutation might be a molecular risk factor in a maternal Chinese hypertensive family under haplotype C. Clin Exp Hypertens. 2018;40(6):518–523. doi: 10.1080/10641963.2017.1403622. [DOI] [PubMed] [Google Scholar]
- 9.Kirichenko TV, Sobenin IA, Khasanova ZB, Orekhova VA, Melnichenko AA, Demakova NA, Grechko AV, Orekhov AN, Ble Castillo JL, Shkurat TP. Data on association of mitochondrial heteroplasmy and cardiovascular risk factors: comparison of samples from Russian and Mexican populations. Data Brief. 2018;18:16–21. doi: 10.1016/j.dib.2018.02.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deng H, Yuan L. Molecular genetics of congenital nuclear cataract. Eur J Med Genet. 2014;57(2–3):113–122. doi: 10.1016/j.ejmg.2013.12.006. [DOI] [PubMed] [Google Scholar]
- 11.Zhu HY, Wang SW, Martin LJ, Liu L, Li YH, Chen R, Wang L, Zhang ML, Benson DW. The role of mitochondrial genome in essential hypertension in a Chinese Han population. Eur J Human Genet. 2009;17(11):1501–1506. doi: 10.1038/ejhg.2009.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han GX, Song Y, Chen L, Zhai YZ, Dong J, Li TS, Zhu HY. The role of mitochondrial DNA mutations in a Han Chinese population on sepsis pathogenesis. Eur Rev Med Pharmacol Sci. 2017;21(14):3247–3252. [PubMed] [Google Scholar]
- 13.Chalmers J, MacMahon S, Mancia G, Whitworth J, Beilin L, Hansson L, Neal B, Rodgers A, Ni Mhurchu C, Clark T. World Health Organization-International Society of Hypertension Guidelines for the management of hypertension. Guidelines sub-committee of the World Health Organization. Clin Exp Hypertension (New York, NY : 1993) 1999;21(5–6):1009–1060. doi: 10.3109/10641969909061028. [DOI] [PubMed] [Google Scholar]
- 14.Liu LS. Chinese guidelines for the management of hypertension. Zhonghua xin xue guan bing za zhi 2011. 2010;39(7):579–615. [PubMed] [Google Scholar]
- 15.Zhu HY, Wang SW, Liu L, Li YH, Chen R, Wang L, Holliman CJ. A mitochondrial mutation A4401G is involved in the pathogenesis of left ventricular hypertrophy in Chinese hypertensives. Eur J Human Genet. 2009;17(2):172–178. doi: 10.1038/ejhg.2008.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Y, Li Y, Gao J, Zhu C, Lan Y, Yang J, Li Z, Guan M, Chen Y. Molecular characterization of a Chinese family carrying a novel C4329A mutation in mitochondrial tRNAIle and tRNAGln genes. BMC Med Genet. 2014;15:84. doi: 10.1186/1471-2350-15-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Echem C, Costa TJD, Oliveira V, Giglio Colli L, Landgraf MA, Rodrigues SF, Franco M, Landgraf RG, Santos-Eichler RA, Bomfim GF, et al. Mitochondrial DNA: a new driver for sex differences in spontaneous hypertension. Pharmacol Res. 2019;144:142–150. doi: 10.1016/j.phrs.2019.04.008. [DOI] [PubMed] [Google Scholar]
- 18.Zhu Y, Gu X, Xu C. Mitochondrial DNA 7908-8816 region mutations in maternally inherited essential hypertensive subjects in China. BMC Med Genet. 2018;11(1):89. doi: 10.1186/s12920-018-0408-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ingman M, Gyllensten U. mtDB: human mitochondrial genome database, a resource for population genetics and medical sciences. Nucleic Acids Res. 2006;34(Database issue):D749–D751. doi: 10.1093/nar/gkj010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Samuels DC, Li C, Li B, Song Z, Torstenson E, Boyd Clay H, Rokas A, Thornton-Wells TA, Moore JH, Hughes TM, et al. Recurrent tissue-specific mtDNA mutations are common in humans. PLoS Genet. 2013;9(11):e1003929. doi: 10.1371/journal.pgen.1003929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bruno RM, Pucci G, Rosticci M, Guarino L, Guglielmo C, Agabiti Rosei C, Monticone S, Giavarini A, Lonati C, Torlasco C, et al. Association between lifestyle and systemic arterial hypertension in young adults: a national, survey-based, cross-sectional study. High Blood Press Cardiovasc Prev. 2016;23(1):31–40. doi: 10.1007/s40292-016-0135-6. [DOI] [PubMed] [Google Scholar]
- 22.Chobanian AV, Bakris GL, Black HR, Cushman WC, Green LA, Izzo JL, Jr, Jones DW, Materson BJ, Oparil S, Wright JT, Jr, et al. Seventh report of the Joint National Committee on Prevention, Detection, Evaluation, and Treatment of High Blood Pressure. Hypertension (Dallas, Tex : 1979) 2003;42(6):1206–1252. doi: 10.1161/01.HYP.0000107251.49515.c2. [DOI] [PubMed] [Google Scholar]
- 23.Chen X, Zhang Y, Xu B, Cai Z, Wang L, Tian J, Liu Y, Li Y. The mitochondrial calcium uniporter is involved in mitochondrial calcium cycle dysfunction: underlying mechanism of hypertension associated with mitochondrial tRNA (Ile) A4263G mutation. Int J Biochem Cell Biol. 2016;78:307–314. doi: 10.1016/j.biocel.2016.07.018. [DOI] [PubMed] [Google Scholar]
- 24.Zhou M, Wang M, Xue L, Lin Z, He Q, Shi W, Chen Y, Jin X, Li H, Jiang P, et al. A hypertension-associated mitochondrial DNA mutation alters the tertiary interaction and function of tRNA (Leu (UUR)) J Biol Chem. 2017;292(34):13934–13946. doi: 10.1074/jbc.M117.787028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao Y, Chen X, Li H, Zhu C, Li Y, Liu Y. Mitochondrial genome mutations in 13 subunits of respiratory chain complexes in Chinese Han and Mongolian hypertensive individuals. Mitochondrial DNA A DNA Mapp Seq Anal. 2018;29(7):1090–1099. doi: 10.1080/24701394.2017.1407762. [DOI] [PubMed] [Google Scholar]
- 26.Shimizu A, Mito T, Hayashi C, Ogasawara E, Koba R, Negishi I, Takenaga K, Nakada K, Hayashi J. Transmitochondrial mice as models for primary prevention of diseases caused by mutation in the tRNA (Lys) gene. Proc Natl Acad Sci U S A. 2014;111(8):3104–3109. doi: 10.1073/pnas.1318109111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mohanty K, Dada R, Dada T. Neurodegenerative Eye Disorders: Role of Mitochondrial Dynamics and Genomics. Asia-Pac J Ophthalmology (Philadelphia, Pa) 2016;5(4):293–299. doi: 10.1097/APO.0000000000000203. [DOI] [PubMed] [Google Scholar]
- 28.Zhou H, Nie K, Qiu R, Xiong J, Shao X, Wang B, Shen L, Lyu J, Fang H. Generation and bioenergetic profiles of Cybrids with east Asian mtDNA Haplogroups. Oxidative Med Cell Longev. 2017;2017:1062314. doi: 10.1155/2017/1062314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crescenzo R, Bianco F, Falcone I, Coppola P, Liverini G, Iossa S. Increased hepatic de novo lipogenesis and mitochondrial efficiency in a model of obesity induced by diets rich in fructose. Eur J Nutr. 2013;52(2):537–545. doi: 10.1007/s00394-012-0356-y. [DOI] [PubMed] [Google Scholar]
- 30.Singh A, Pezeshki A, Zapata RC, Yee NJ, Knight CG, Tuor UI, Chelikani PK. Diets enriched in whey or casein improve energy balance and prevent morbidity and renal damage in salt-loaded and high-fat-fed spontaneously hypertensive stroke-prone rats. J Nutr Biochem. 2016;37:47–59. doi: 10.1016/j.jnutbio.2016.07.011. [DOI] [PubMed] [Google Scholar]
- 31.Rocha MC, Rosa HS, Grady JP, Blakely EL, He L, Romain N, Haller RG, Newman J, McFarland R, Ng YS, et al. Pathological mechanisms underlying single large-scale mitochondrial DNA deletions. Ann Neurol. 2018;83(1):115–130. doi: 10.1002/ana.25127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lehmann D, Tuppen HAL, Campbell GE, Alston CL, Lawless C, Rosa HS, Rocha MC, Reeve AK, Nicholls TJ, Deschauer M, et al. Understanding mitochondrial DNA maintenance disorders at the single muscle fibre level. Nucleic Acids Res. 2019. [DOI] [PMC free article] [PubMed]
- 33.Moskalev AA, Aliper AM, Smit-McBride Z, Buzdin A, Zhavoronkov A. Genetics and epigenetics of aging and longevity. Cell cycle (Georgetown, Tex) 2014;13(7):1063–1077. doi: 10.4161/cc.28433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Herbers E, Kekalainen NJ, Hangas A, Pohjoismaki JL, Goffart S. Tissue specific differences in mitochondrial DNA maintenance and expression. Mitochondrion. 2019;44:85–92. doi: 10.1016/j.mito.2018.01.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw data of mitochondrial map reported in this paper have been deposited in the Genome Sequence Archive (Genomics, Proteomics & Bioinformatics 2017) in BIG Data Center (Nucleic Acids Res 2019), Beijing Institute of Genomics (BIG), Chinese Academy of Sciences, under accession numbers CRA002605 that are publicly accessible at https://bigd.big.ac.cn/gsa. The updated consensus Cambridge Sequence and pathogenic mitochondrial variants were identified from the human mitochondrial genome database (https://www.mitomap.org/MITOMAP).