Significance
Most studies of pesticide resistance have focused on the identification and functional analysis of resistance genes, but few studies have considered the signaling pathways involved in their regulation. In this work, we discovered that overexpression of a P450 that confers resistance to neonicotinoid insecticides in whitefly is trans-regulated by the transcription factor CREB. Further studies demonstrated that exposure to neonicotinoid insecticides activates a key pathway involved in the cellular response to extracellular signals, the MAPK signaling pathway, that activates CREB by phosphorylation. CREB then binds to a specific site on the promoter of CYP6CM1 resulting in its increased expression. These findings reveal mechanisms underlying the regulation of P450-mediated pesticide resistance and also provide a potential target for pest control.
Keywords: cytochrome P450, CREB, MAPK, neonicotinoid insecticide, insecticide resistance
Abstract
The evolution of insect resistance to pesticides poses a continuing threat to agriculture and human health. While much is known about the proximate molecular and biochemical mechanisms that confer resistance, far less is known about the regulation of the specific genes/gene families involved, particularly by trans-acting factors such as signal-regulated transcription factors. Here we resolve in fine detail the trans-regulation of CYP6CM1, a cytochrome P450 that confers resistance to neonicotinoid insecticides in the whitefly Bemisia tabaci, by the mitogen-activated protein kinase (MAPK)-directed activation of the transcription factor cAMP-response element binding protein (CREB). Reporter gene assays were used to identify the putative promoter of CYP6CM1, but no consistent polymorphisms were observed in the promoter of a resistant strain of B. tabaci (imidacloprid-resistant, IMR), which overexpresses this gene, compared to a susceptible strain (imidacloprid-susceptible, IMS). Investigation of potential trans-acting factors using in vitro and in vivo assays demonstrated that the bZIP transcription factor CREB directly regulates CYP6CM1 expression by binding to a cAMP-response element (CRE)-like site in the promoter of this gene. CREB is overexpressed in the IMR strain, and inhibitor, luciferase, and RNA interference assays revealed that a signaling pathway of MAPKs mediates the activation of CREB, and thus the increased expression of CYP6CM1, by phosphorylation-mediated signal transduction. Collectively, these results provide mechanistic insights into the regulation of xenobiotic responses in insects and implicate both the MAPK-signaling pathway and a transcription factor in the development of pesticide resistance.
Insect resistance to synthetic insecticides is an ongoing challenge to sustainable pest management while also an exceptional model system to study adaptive evolution. Work on this topic has provided a range of insights into the genomic and transcriptomic changes that occur in response to strong selective pressures. One such molecular alteration frequently implicated in resistant insect populations is increased production of metabolic enzymes that detoxify or sequester insecticides before they reach the target site. A key enzyme system in this regard is insect cytochrome P450s, which play a central role in the metabolism of a wide range of natural and synthetic xenobiotics including insecticides (1, 2). The increased production of P450s in insecticide-resistant insects has been shown to most commonly result from gene duplication/amplification or from mutations in cis-acting promoter sequences (3–6). In contrast, evidence for trans-acting factors regulating resistance-associated P450s comes from just a handful of studies (7–10).
Control of gene expression in trans is most commonly mediated by transcription factors, and regulation of genes involved in the metabolism of xenobiotics has been shown to involve several superfamilies, including basic leucine zipper (bZIP), basic-helix–loop–helix/Per-ARNT-Sim, and nuclear receptors (10–12). Well-characterized examples in mammals are the human steroid and xenobiotic receptor and the constitutive androstane receptor (CAR), which appear to function in a partially redundant manner in regulating a range of phase I and phase II detoxification enzymes (13). In insects, much less is known about the transcription factors that regulate the response to xenobiotics with much of our understanding stemming from work on Drosophila melanogaster. In this species, the nuclear receptor DHR96 and the bZIP factor Cap n collar isoform C (CncC) have been shown to play key roles in the regulation of xenobiotic responses, in part by modulating P450 transcription (14, 15). Much less is known about the role of transcription factors in the xenobiotic response of nonmodel insect species; however, recent work has also implicated CncC and the bZIP factor small muscle aponeurosis fibromatosis (maf) in the overexpression of P450 genes involved in insecticide resistance in several crop pest species and disease vectors (7–9). Taken together, these studies suggest that members of the bZIP superfamily may play a central role in regulating the transcriptional response of insects to xenobiotics. A further member of this superfamily, CREB, is involved in wide-ranging processes including glucose homeostasis, growth-factor–dependent cell survival, nervous system development, learning, memory, synaptic plasticity, and drug adaptive responses (16). In insects, CREB has also been shown to play a role in diverse processes including long-term memory formation, waking and rest homeostasis, starvation resistance, age-dependent labor division, and environment-induced behavioral plasticity (17–21). However, the role of CREB in regulating detoxification enzymes in the context of insect resistance to insecticides has never been explored.
Transcription factors such as CREB may themselves be regulated by activity-inducible kinases such as mitogen-activated protein kinase (MAPKs), components of signaling cascades that respond to extracellular stimuli such as environmental stress. MAPKs have been extensively studied in both vertebrates (22) and invertebrates, including D. melanogaster, Caenorhabditis elegans, and Anopheles gambiae (23–25). Three classical kinase subfamilies have been described: extracellular signal-regulated kinases (ERKs), c-Jun N-terminal kinases (JNKs), and p38 kinases (22). To date, the role of the MAPK-signaling pathway in the regulation of P450 genes associated with insecticide resistance is essentially unknown; however, a MAP4K4 gene has recently been shown to alter the expression of genes involved in resistance to Bacillus thuringiensis Cry1Ac toxin in the diamondback moth Plutella xylostella (26).
The sweet potato whitefly Bemisia tabaci is a highly destructive pest of many protected and field crops worldwide that causes damage via direct feeding and as a vector of more than 100 plant viruses (27). Control of B. tabaci has relied largely on the use of synthetic insecticides with members of the neonicotinoid class, such as imidacloprid, particularly widely used. As a result, imidacloprid resistance has been widely reported in the two most economically important species of the B. tabaci complex from a range of geographical origins worldwide and linked to the overexpression of a single P450 gene, CYP6CM1 (28, 29). Subsequent functional analyses have demonstrated the capacity of the encoded enzyme to catalyze the hydroxylation of imidacloprid and other insecticides (30, 31). Despite the importance and global distribution of this mechanism, the molecular basis for CYP6CM1 overexpression in neonicotinoid-resistant strains has never been elucidated. In the present study, we investigated the factors that regulate the expression of CYP6CM1 and lead to the development of resistance to imidacloprid in B. tabaci.
Results
Imidacloprid Resistance in the Imidacloprid-Resistant Strain of B. tabaci Is Conferred by Overexpression of CYP6CM1 Not by Mutation of the Nicotinic Acetylcholine Receptor.
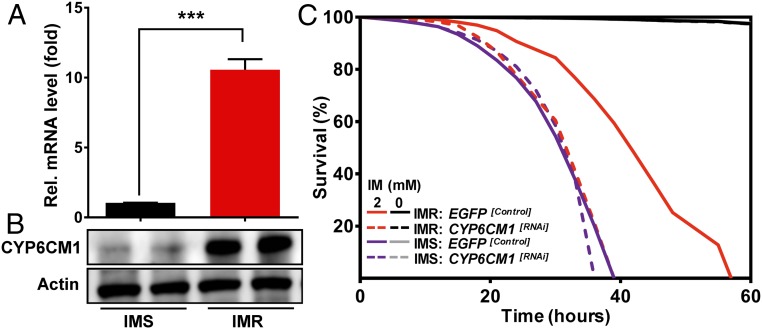
A strain of B. tabaci MED (Mediterranean) collected in the field was split into two cultures and maintained under selection with 2.0 mM imidacloprid for more than 30 generations to form the imidacloprid-resistant (IMR) strain or left unexposed to insecticides to form the imidacloprid-susceptible (IMS) strain. At the time of this study, the IMR strain exhibited a 23-fold resistance to imidacloprid in comparison to the IMS strain (SI Appendix, Table S2). To examine if the resistance of the IMR strain was associated with mutation of the imidacloprid target site, the nicotinic acetylcholine receptor (nAChR), nine nAChR subunit genes (eight α-subunits and one β-subunit genes) present in the genome of B. tabaci were cloned and sequenced from the IMR and IMS strains. No nonsynonymous mutations were observed in the sequences obtained that consistently distinguish the two strains (Dataset S4); thus target-site insensitivity plays no role in the resistance of the IMR strain. As detailed in the Introduction, the primary mechanism of metabolic resistance to imidacloprid in B. tabaci worldwide is enhanced expression of the P450 CYP6CM1. qPCR was therefore used to assess the expression of CYP6CM1 in the two strains, and it revealed 10-fold greater expression of this P450 gene in the IMR strain than in the IMS strain (Fig. 1A). To confirm that overexpression of CYP6CM1 translated to altered expression of the encoded enzyme, we used a rabbit polyclonal antibody raised against a synthetic peptide of CYP6CM1 in Western blot to show that the CYP6CM1 protein is expressed at higher levels in every developmental stage of the IMR strain than in the IMS strain (Fig. 1B and SI Appendix, Fig. S3E). To demonstrate the causal role of CYP6CM1 in the imidacloprid resistance of the IMR strain, RNA interference (RNAi) was used to knock down its expression, and the effect of this on the life span of imidacloprid-treated adults of the IMR strain was examined. After 48 h of feeding on a diet containing double-stranded RNA (dsRNA) specific for CYP6CM1, the messenger RNA (mRNA) levels of this gene decreased by 49.7% (SI Appendix, Fig. S3A). Knockdown of CYP6CM1 significantly decreased the life span of IMR adult whiteflies, relative to the control (fed EGFP dsRNA), when adults were treated with imidacloprid at 0.4 mM (median survival: −23 h; P = 2.05 × 10−147; Fig. 1C) or at 2.0 mM (median survival: −9 h; P = 1.44 × 10−52; SI Appendix, Figs. S6A and S8A and Dataset S1). In contrast, no significant shift was seen in the sensitivity of the IMS strain after CYP6CM1 knockdown. Together, these results demonstrate that reducing the CYP6CM1 mRNA level is sufficient to decrease the resistance of the IMR strain to imidacloprid.
Fig. 1.
CYP6CM1 contributes to imidacloprid (IM) resistance in the B. tabaci strain IMR. (A) Relative expression of CYP6CM1 mRNA (A) and protein (B) in the imidacloprid-resistant strain IMR and the susceptible strain IMS as determined by qPCR and Western blot, respectively (qPCR: n = 3, mean ± SE; ***P < 0.001, two-tailed Student’s t test). Actin was used as a loading control in Western blot. (C) Life span of adults of the IMR and IMS strains exposed to 2.0 mM imidacloprid (2) or no imidacloprid (0) after RNAi knockdown of CYP6CM1. Adults fed on dsEGFP were used as a negative control.
CYP6CM1 promoter analysis.
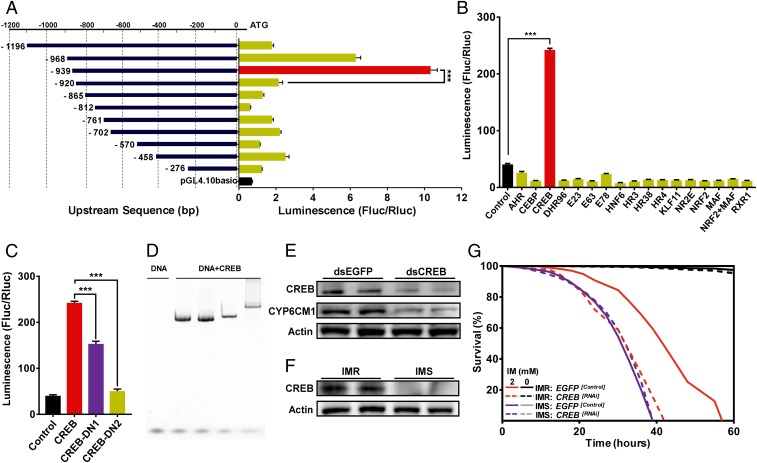
To identify the putative promoter of CYP6CM1, a series of constructs comprising 11 different regions upstream of the gene (−235 to +1, −458 to +1, −570 to +1, −702 to +1, −761 to +1, −812 to +1, −865 to +1, −920/ to +1, −939 to +1, −968 to +1, and −1,196 to +1) were created, and the ability of each of these to drive expression of a reporter gene was assessed using dual luciferase reporter assays. The construct containing the region −939 to +1 (pGL4.10-CYP6CM1−939 to +1) showed the most pronounced expression (14.3-fold higher than the reference reporter plasmid) of the reporter, suggesting that the promoter is located in this region, with particularly important elements located between −920 and −939 bp (Fig. 2A).
Fig. 2.
The transcription factor CREB regulates CYP6CM1 expression. (A) Identification of the promoter of CYP6CM1 using progressive deletion constructs in dual luciferase reporter assays. S2 cells were cotransfected with pGL4.10 reporter plasmids carrying the indicated promoter regions conjugated to firefly luciferase and a reference reporter plasmid (pGL4.73, containing the hRluc reporter gene and an SV40 early promoter). Fluc/Rluc represents the ratio of Firefly to Renilla luciferase activity (n = 3, mean ± SE; ***P < 0.001, ANOVA with Tukey’s honestly significant difference [HSD] post hoc test). (B) Influence of 16 different B. tabaci transcription factors on expression driven by the promoter of CYP6CM1 in dual luciferase reporter assays. pGL4.10-CYP6CM1−939 to +1 and pGL4.73 were cotransfected into S2 cells with each of the 16 transcription factors in pAC5.1b. Empty pAC5.1b was used as a control (n = 3, mean ± SE; ***P < 0.001, ANOVA with Tukey’s HSD post hoc test). (C) Identification of important functional domains in CREB using dual luciferase reporter assays. pGL4.10-CYP6CM1−939 to +1 and pGL4.73 were cotransfected into S2 cells with pAC5.1b-CREB (CREB) with modified CREB in which the KID domain (CREB-DN1) or the bZIP domain (CERB-DN2) was deleted or with the pAC5.1b empty vector (control) (n = 3, mean ± SE; ***P < 0.001, two-tailed Student’s t test). (D) CREB binds to the upstream promoter sequences of CYP6CM1. EMSA results showing dose-dependent binding of CREB to a CRE-like–containing DNA probe. (E) Expression of CREB and CYP6CM1 protein after RNAi knockdown of CREB in the IMR strain as assessed by Western blot. Actin was used as a loading control. (F) Expression of CREB protein in the IMR and IMS strain as assessed by Western blot. Actin was used as a loading control. (G) Life span of adults of the IMR and IMS strains exposed to 2.0 mM imidacloprid (2) or no imidacloprid (0) after feeding on CREB dsRNA for 48 h. Adults fed on dsEGFP were used as a negative control.
CYP6CM1 overexpression is not associated with polymorphisms in cis-acting promoter elements.
In the first study of CYP6CM1 overexpression in B. tabaci, single-nucleotide polymorphisms (SNPs) were identified in a noncoding region of the gene that associate with imidacloprid resistance, suggesting that CYP6CM1 may be regulated by cis-acting factors (28). To investigate if polymorphisms within the promoter of CYP6CM1 are associated with its overexpression in the IMR strain, the promoter region defined above, along with 6 kb upstream of this region, was PCR amplified and sequenced in six pools of DNA extracted from the IMR and IMS strains. While SNPs were observed at certain positions in the sequences obtained, no consistent polymorphisms were observed between sequences of the two strains (Dataset S2). This finding indicates that the increased expression of CYP6CM1 in the IMR strain does not result from cis-acting elements in the promoter of this gene or in upstream enhancers within 6 kb.
The bZIP transcription factor CREB mediates CYP6CM1 expression.
Transcription factors that sense xenobiotic stress are known as xenobiotic sensors, and we hypothesized that one or more of these factors may be involved in regulating CYP6CM1. Based on an extensive literature search, we selected 16 transcription factors with potential roles in xenobiotic stress response from the recently sequenced genome of B. tabaci (detailed information on these transcription factors is provided in Dataset S3). To explore if any of these regulate CYP6CM1 expression, the 16 transcription factors were individually cloned into the pAC5.1b expression vector and cotransfected into S2 cells with pGL4.10-CYP6CM1S−939 to +1. The successful expression of these transcription factors in S2 cells was confirmed using qPCR (SI Appendix, Fig. S3F). In reporter gene assays, a marked (5.98-fold) and significant (P = 1.49 × 10−7) increase in expression driven by the CYP6CM1 promoter was observed only in combination with the construct expressing the transcription factor CREB (Fig. 2B). In a follow-up experiment, CREB expression also significantly (P = 0.022) increased reporter expression driven by the CYP6CM1 promoter in HEK293T cells (SI Appendix, Fig. S3B).
The full-length gene that encodes CREB in B. tabaci contains an 846-bp open reading frame encoding 282-amino-acid residues with high pairwise amino acid similarity to members of the CREB family from other organisms. The deduced amino acid sequence of the protein includes important conserved domains common to the bZIP family including a basic leucine zipper DNA-binding and dimerization domain (bZIP) and a central kinase-inducible domain (KID) which interacts with protein kinases (SI Appendix, Fig. S1). To investigate the importance of these domains in activating the promoter of CYP6CM1, two mutated forms of CREB—CREB-DN1, which lacks the KID domain, and CREB-DN2, which lacks the bZIP domain—were created and cotransfected into S2 cells with pGL4.10-CYP6CM1−939 to +1. As shown in Fig. 2C, expression of either CREB-DN1 or CREB-DN2 significantly (DN1: P = 1.05 × 10−4; DN2: P = 9.81 × 10−7) reduced the activity of the CYP6CM1 promoter compared to the control (pAC5.1b-CREB). Interestingly, in assays with the CREB-DN1 reporter, gene expression remained significantly (P = 9.43 × 10−5) higher than that of the negative control (pAC5.1b empty vector) whereas expression driven by CREB-DN2 was no different in the negative control. Thus these findings reveal that both the KID and bZIP domains are important for CYP6CM1 promoter activation in vitro with the latter essential.
CREB stimulates target gene expression at promoters that contain cAMP response elements (CREs) (32). These typically appear as either palindromic sequences (TGACGTCA) or half-site sequences (TGACG or CGTCA). Significantly, the promoter region of CYP6CM1 contains a CRE half-site–like sequence TGATTG between positions −930 and −926 bp, which we identified as containing elements of major effect on gene expression. When this site was disrupted, a significant decrease was observed in reporter gene expression driven by the mutant CYP6CM1 promoter, suggesting that this region is important for CREB binding (SI Appendix, Fig. S2). To investigate this further, a DNA probe of the region −920 to −968 bp encompassing this site was produced, and the CREB protein was expressed in Escherichia coli cells and purified. Purified CREB protein and the DNA probe were then used in electrophoretic mobility shift assays (EMSA) to examine protein–nucleic acid interactions. Dose-dependent binding of CREB protein to the CRE-like–containing DNA probe was observed (Fig. 2D), providing strong evidence that CREB enhances CYP6CM1 expression in vitro as a result of its binding to a CRE-like site in the upstream promoter of this gene. To determine whether CREB affects CYP6CM1 gene expression in vivo, RNAi was used to knock down the expression of CREB in adult whiteflies, and the effect of this on the expression of both CREB and CYP6CM1 was examined using qPCR. After 48 h of feeding whiteflies a diet containing dsRNA specific for CREB, the mRNA levels of CREB decreased by 40.2% (SI Appendix, Fig. S3D). Furthermore, a corresponding significant decrease (50.3%) in the transcript and protein levels of CYP6CM1 was observed (Fig. 2E and SI Appendix, Fig. S4D). Taken together, both in vitro and in vivo assays unequivocally demonstrate that CREB directly regulates CYP6CM1 expression.
CREB is overexpressed in the IMR strain, and RNAi knockdown of CREB reduces resistance to imidacloprid.
To explore the expression of CREB in the IMR and IMS strain, we used qPCR and a rabbit polyclonal antibody raised against a synthetic peptide of CREB protein in Western blot to measure mRNA and protein levels, respectively. CREB mRNA levels were 2.9-fold greater in the IMR strain than in the IMS strain (SI Appendix, Fig. S3C) with Western blot confirming that the encoded protein is overexpressed in the IMR strain as a result (Fig. 2F). To further examine the role of CREB overexpression in imidacloprid resistance in the IMR strain, we used RNAi to knockdown the expression of CREB in adult whiteflies and examined the effect of this on the life span of IMR adults that were treated with imidacloprid. Knockdown of CREB significantly decreased the life span of whiteflies relative to the control (fed dsEGFP) when treated with imidacloprid at 0.4 mM (median survival: −14 h; P = 2.37 × 10−52; Fig. 2G) and at 2.0 mM (median survival: −9 h; P = 9.28 × 10−58; SI Appendix, Figs. S6B and S8B and Dataset S1). This finding confirmed that reducing CREB transcript levels (and thereby reducing CYP6CM1 expression) is sufficient to decrease B. tabaci resistance to imidacloprid. Sequencing of the CREB promoter failed to identify consistent polymorphisms in the region 2.2 kb upstream of the translation start site between the IMR and IMS strains (Dataset S2), suggesting that overexpression of CREB results from trans-acting factors.
The ERK- and p38 MAPK-signaling pathways activate CREB transcription.
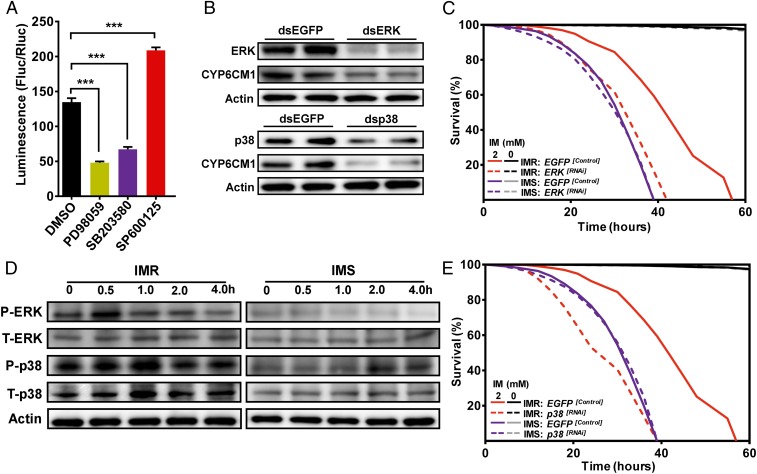
CREB proteins are themselves regulated via phosphorylation by several kinases, including protein kinase A (PKA) (33) and MAPKs (34). To investigate whether PKA and MAPKs activate CREB in vitro, S2 cells were treated with a specific inhibitor of PKA (H89) (35) and with three inhibitors (PD98059, SB203580, and SP600125) (36) of three classical MAPK pathways (ERK, p38, and JNK, respectively) and cotransfected with pGL4.10-CYP6CM1−939 to +1. As shown in SI Appendix, Fig. S4A, reporter gene expression driven by the CYP6CM1 promoter was much greater in cells treated with H89 than in untreated control cells, suggesting that PKA suppresses the expression of CYP6CM1 in this system. In contrast, inhibition of the ERK and p38 pathways significantly (ERK: P = 6.35 × 10−6, p38: P = 4.73 × 10−5) decreased reporter gene expression driven by CYP6CM1−939 to +1, suggesting that these signaling pathways may positively regulate CYP6CM1 expression. Inhibition of the JNK pathway significantly (P = 4.52 × 10−7) increased the expression of CYP6CM1 (Fig. 3A), suggesting a negative regulatory role in CYP6CM1 expression. To examine the effect of inhibition of the ERK and p38 pathways in vivo, PD98059 and SB203580 were fed to IMR adults on an artificial diet, and the effect of this on the life span of IMR adults that were treated with imidacloprid was examined. As shown in SI Appendix, Fig. S4C, both ERK inhibition (median survival: −19 h; P = 5.84 × 10−91) and p38 inhibition (median survival: −16 h; P = 2.27 × 10−45) significantly decreased the resistance of the IMR strain to imidacloprid. The efficacy of these inhibitors was further explored on tomato plants in combination with imidacloprid (0.4 mM) with 100 B. tabaci adults released onto each plant. The life span of adult B. tabaci significantly decreased on plants exposed to 0.4 mM imidacloprid treatment + PD98059 or SB203580 in comparison to plants exposed to imidacloprid without inhibitors (PD98059 treatment—median survival: −3.96 d; P = 8.65 × 10−184; SB203580 treatment—median survival: −26 h; P = 7.46 × 10−91; SI Appendix, Fig. S4E).
Fig. 3.
The MAPK-signaling pathway regulates CYP6CM1 expression and imidacloprid resistance. (A) Expression driven by the promoter of CYP6CM1 in the presence of inhibitors of the ERK, p38, and JNK MAPK pathways. Dual luciferase reporter assays, in which pGL4.10-CYP6CM1−939 to +1 and pGL4.73 were cotransfected into S2 cells with the ERK inhibitor PD98059 (75 μM), the p38 inhibitor SB203580 (53 μM), or the JNK inhibitor SP600125 (45 μM) are shown. Fluc/Rluc represents the ratio of Firefly to Renilla luciferase activity. (n = 3, mean ± SE; ***P < 0.001, ANOVA with Tukey’s HSD post hoc test). (B) Expression of CYP6CM1 protein after knockdown of ERK or p38 in the IMR strain as assessed by Western blot. Actin was used as a loading control. (C) Life span of adult whiteflies of the IMR and IMS strains after feeding on ERK dsRNA for 48 h followed by exposure to 2.0 mM imidacloprid. Adults fed on dsEGFP were used as a negative control. (D) Levels of Total-ERK, Total-p38, and phosphorylated ERK and p38 in IMR and IMS adults assessed by Western blot at four time points after exposure to 2.0 mM imidacloprid. Actin was used as a loading control. (E) Life span of adult whiteflies of the IMR and IMS strains after feeding on p38 dsRNA for 48 h followed by exposure to 2.0 mM imidacloprid (2) or no imidacloprid (0). Adults fed on dsEGFP were used as a negative control.
To examine if inhibition of the ERK and p38 pathways increased sensitivity to imidacloprid as a result of their effects on CYP6CM1 gene expression in B. tabaci, the mRNA levels of the ERK and p38 MAPKs were knocked down using RNAi, and the effect of this on the expression of CYP6CM1 was explored using qPCR. As shown in SI Appendix, Fig. S4D, the mRNA transcript levels of ERK and p38 decreased by 47.2 and 41.4%, respectively, after 48 h of feeding on a diet containing ERK and p38 dsRNA (0.5 μg/μL), and knock down of the MAPKs ERK and p38 resulted in a significant decrease (65.7 and 61.9%, respectively) in the expression of CYP6CM1 in the IMR strain. Moreover, a corresponding significant decrease in protein levels of CYP6CM1 was observed (Fig. 3B). Furthermore, knockdown of ERK or p38 significantly decreased the life span of IMR adults treated with 0.4 mM imidacloprid (ERK treatment— median survival: −23 h; P = 1.04 × 10−133; p38 treatment—median survival: −26 h; P = 2.05 × 10−147; Fig. 3 C and E and Dataset S1) and 2.0 mM imidacloprid (ERK treatment—median survival: −9 h; P = 9.22 × 10−31; p38 treatment—median survival: −12 h; P = 1.44 × 10−52; SI Appendix, Figs. S6 C and D and S8 C and D and Dataset S1), demonstrating that a reduction in ERK and p38 expression is sufficient to decrease B. tabaci resistance to imidacloprid. No difference was observed in the expression of ERK in the IMR and IMS strains; however, the expression of p38 was significantly higher in the IMR strain (SI Appendix, Fig. S4B). Both of the MAPKs are phosphorylated in order to activate downstream proteins (analysis of phosphorylation sites is shown in SI Appendix, Fig. S5). To explore if differences were observed in the phosphorylation profile of ERK and p38 protein in the IMR and IMS strains, homophosphorylation antibodies were used in Western blots with the two strains. A significant increase in the level of protein phosphorylation of ERK was observed in the IMR strain compared to the IMS strain (Fig. 3D). Furthermore, when the IMR strain was treated with imidacloprid, the level of protein phosphorylation of ERK increased rapidly (within 0.5 h, Fig. 3D), suggesting that differential phosphorylation, rather than differential expression, of ERK plays a role in the resistance of the IMR strain. Similarly, a dramatic increase in the level of protein phosphorylation of p38 was observed in the IMR strain compared to the IMS strain, and when adult whiteflies were treated with imidacloprid, phosphorylation occurred much more rapidly and reached higher levels in the IMR strain (Fig. 3D), suggesting that both differential phosphorylation and expression of p38 play a role in the resistance of the IMR strain.
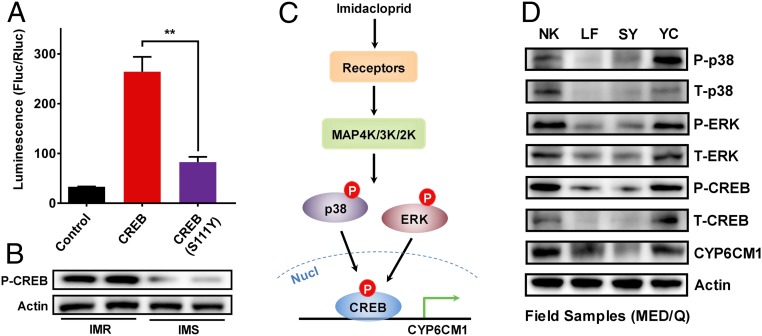
To further explore the role of phosphorylation in resistance, we investigated phosphorylation of CREB itself. In humans, MAPKs activate CREB by phosphorylation of Ser133 which promotes the recruitment of transcriptional coactivators (37). The equivalent site in B. tabaci CREB is Ser111 (RRPSYRK). We first demonstrated the importance of this site on function by mutating site 111 (Ser to Tyr) and cotransfecting S2 cells with the mutant construct and plasmid pGL4.10-CYP6CM1−939 to +1 and examining the effect on luciferase activity. Mutation of CREB at site 111 significantly (P = 0.005) decreased reporter gene expression driven by the promoter of CYP6CM1 (Fig. 4A), demonstrating its functional role. A rabbit polyclonal antibody for the phosphorylated form of B. tabaci CREB protein was then raised and used to determine the phosphorylation profile of this enzyme in the IMR and IMS strain by Western blot. Consistent with the elevated phosphorylation of the upstream MAPKs, we observed higher levels of the phosphorylated form of CREB in the IMR strain, suggesting that resistance results from increased production of the activated (phosphorylated) form of this transcription factor (Fig. 4B).
Fig. 4.
Model of the regulation of CYP6CM1 in B. tabaci. (A) Expression driven by the promoter of CYP6CM1 in the presence of wild type or mutated (Ser111Tyr) CREB. pGL4.10-CYP6CM1−939 to +1 and pGL4.73 plasmids were cotransfected into S2 cells with either CREB or mutated CREB (Ser111Tyr) or the pAC5.1b empty vector (control) and expression assessed using dual luciferase reporter assays (n = 3, mean ± SE; **P < 0.01, two-tailed Student’s t test). (B) Phosphorylation levels of Ser111 in CREB in the IMR and IMS strain as assessed by Western blot. Actin was used as a loading control. (C) A model of the regulation of CYP6CM1 in B. tabaci. In this model, exposure to imidacloprid is detected by an unknown receptor that actives mitogen-activated protein kinases of the MAPK pathway to phosphorylate ERK and p38. These, in turn, phosphorylate CREB, which then activates the expression of CYP6CM1. Activation and overexpression of CREB in the resistant strain increases the production of CYP6CM1 protein, which reduces the toxicity of imidacloprid. (D) Levels of Total-CYP6CM1, Total-CREB, Total-ERK, Total-p38, and phosphorylated CREB, ERK, and p38 in four field strains of B. tabaci MED. Actin was used as a loading control.
Increased phosphorylation of CREB, ERK, and p38 is also observed in imidacloprid-resistant field strains of B. tabaci.
To examine if the key findings on the regulation of resistance in the IMR strain extend to imidacloprid resistant field populations of B. tabaci MED we conducted further analysis on four field samples collected in 2018 (SI Appendix, Table S1). In insecticide bioassays, the NK and YC strains exhibited strong resistance to imidacloprid, the LF strain showed modest resistance, and the SY strain was susceptible to this compound (SI Appendix, Table S2). Western blot was used to estimate the total protein level of CYP6CM1, CREB, ERK, and p38 and the phosphorylation level of CERB, ERK, and p38 (Fig. 4D). The expression of CYP6CM1 protein was higher in the resistant NK and YC strains than in the SY-susceptible strain. In parallel, the level of both total CREB and total P38, and their phosphorylated forms, was greater in the two field-resistance strains than in the susceptible strain. While no difference was seen in the level of total ERK protein in the four field strains, the level of phosphorylated ERK was higher in the NK and YC strains than in both of the other strains. RNAi knockdown of CYP6CM1, CREB, ERK, and p38 in the NK-resistant strain (CYP6CM1 treatment—median survival: −21 h; P = 3.96 × 10−289; CREB treatment—median survival: −6 h; P = 7.63 × 10−67; ERK treatment—median survival: −6 h; P = 3.41 × 10−54; p38 treatment: median survival −6 h, P = 9.76 × 10−37) and YC resistant strain (CYP6CM1 treatment—median survival: −18 h; P = 1.70 × 10−291; CREB treatment—median survival: −15 h; P = 6.06 × 10−279; ERK treatment—median survival: −18 h; P = 5.65 × 10−249; p38 treatment—median survival: −18 h; P = 7.21 × 10−274) significantly decreased the resistance levels to imidacloprid at 2.0 mM (SI Appendix, Fig. S7). In contrast, no significant shift was seen in the sensitivity of the SY strain after these genes were knocked down. Thus the increased expression of CYP6CM1 in field strains of B. tabaci MED also appears to be mediated, at least in part, by CREB and phosphorylation-mediated signal transduction involving the MAPK-signaling pathway.
Discussion
Herbivorous insects have evolved complex defense systems to protect themselves against xenobiotic stressors such as the secondary metabolites produced by their host plants; however, how these toxins are sensed and how this signal is transduced into a physiological response is poorly understood. From an applied perspective such knowledge is important as some of these natural insect defense mechanisms have been co-opted during the evolution of insect resistance to synthetic insecticides. Our data demonstrate that CYP6CM1, a P450 recruited during the evolution of B. tabaci resistance to neonicotinoid insecticides, is regulated by CREB, a transcription factor belonging to the bZIP superfamily. Members of this superfamily have been previously implicated in the regulation of P450 genes involved in resistance to plant allelochemicals and insecticides; however, CREB has never previously been implicated in this role. Indeed, only a single very recent study has previously linked CREB to the regulation of a P450, in this case human CYP2J2, which is highly expressed in the human brain and is responsible for metabolizing endogenous polyunsaturated fatty acids (38, 39). More broadly in mammals, CREB has been shown to play a role in a remarkably diverse range of processes. Of particular relevance to this study is the role of CREB in the response to stressful stimuli including oxidative stress, ischemia, and excitotoxicity, with evidence from knockout mice revealing a role in neuroprotection and neuronal survival (32). Exactly how CREB mediates this activity is not fully understood. Here we demonstrate that CREB performs a neuroprotective function in B. tabaci by up-regulating a key detoxification enzyme. This in turn increases the metabolic conversion of imidacloprid, an agonist of insect nAChRs in neuronal cells, into its less toxic 5-hydroxy form.
Importantly, we show that, while CREB is overexpressed in resistant B. tabaci, a marked increase is also observed in the activated form of this transcription factor as a result of phosphorylation of a key amino acid (Ser111) in the kinase-inducible domain. This finding suggests that additional upstream activators of CREB also play a role in CYP6CM1-mediated insecticide resistance. We confirmed this by demonstrating that two protein kinases, ERK and p38, that are focal points of two different MAPK-signaling pathways, are involved in CYP6CM1-mediated resistance to imidacloprid in B. tabaci. The MAPK-signaling pathway plays a key role in responses to both endogenous and xenobiotic substances (22); increased expression of cytochrome P450 2E1 induces heme oxygenase-1 through the ERK MAPK pathway (40), and p38 regulates the nuclear receptor CAR that activates the CYP2B6 gene in human primary hepatocytes (41). Furthermore, the MAPK p38 pathway has been implicated in insect defense against pore-forming toxins such as Bt Cry toxins (42). We demonstrate that exposure of B. tabaci to imidacloprid results in the phosphorylation of p38 and ERK, and we thus implicate the MAPK-signaling pathway in resistance to synthetic insecticides. MAPKs function in multitiered signaling cascades (43) in which an activated MAP4K phosphorylates and activates a MAP3K, which, in turn, activates a downstream MAP2K, which actives a MAPK (ERK/p38/JNK) that can regulate transcription factors. A MAP4K4 gene was identified in P. xylostella, which is constitutively transcriptionally activated in resistant larvae and alters expression of midgut ALP and ABCC genes leading to resistance to Bt Cry1Ac toxin (26). Furthermore, a MAPK gene was found to be up-regulated in Anopheles stephensi after exposure to pyrethroid for 24 and 48 h (44). In the case of whiteflies, RNA-sequencing analysis of a thiamethoxam-resistant and -susceptible strain of B. tabaci MEAM1 identified the MAPK-signaling pathway as a significantly enriched biological pathway in the differentially expressed genes (45). Together with our results, these findings suggest that the MAPK-signaling pathway may play a previously unappreciated role as a trans-regulator of important insecticide resistance gene effectors in a range of insect species.
Exactly how do the p38 and ERK MAPK pathways and CREB work together to increase the expression of CYP6CM1? We present a model for the regulation of CYP6CM1 in the resistant IMR strain in Fig. 4C and SI Appendix, Fig. S9. In this model, the activation of the MAPK-signaling pathway upon insecticide exposure via an unknown receptor triggers a signaling cascade resulting in the phosphorylation of the MAPKs ERK and p38 by MAPK kinases (MAPKKs); these, in turn, phosphorylate CREB; once phosphorylated, CREB activates the expression of CYP6CM1 by binding to a CRE-like site in the promoter of this gene; as shown previously (28), increased production of CYP6CM1 results in imidacloprid resistance in B. tabaci. Overexpression of CREB plays an additional role in the increased production of CYP6CM1 protein. This model sets out a framework for future studies to identify potential genetic changes at key points in this pathway that result in the constitutive overexpression of CYP6CM1 observed in imidacloprid-resistant strains of B. tabaci.
The receptor that triggers the initiation of the signaling pathway illustrated in Fig. 4C is unknown. However, several previous studies have demonstrated a role for G-protein–coupled receptors (GPCRs) in modulating the expression of P450 genes that lead to resistance (46–48). These previous findings and the fact that GPCRs can activate the conserved MAPK pathway (49, 50) make these receptors candidate upstream activators of the MAPK-signaling pathway associated with imidacloprid-resistant B. tabaci.
In conclusion, we uncover the trans-regulatory pathway involved in the increased expression of a key resistance gene in B. tabaci and implicate both the MAPK-signaling pathway and a transcription factor in the development of resistance to synthetic insecticides. Collectively, these results advance our understanding of the regulation of xenobiotic responses in insects and illustrate the remarkable sophistication of the molecular pathways involved. Finally, from an applied perspective identification of the pivotal genes involved in the regulation of CYP6CM1 also highlight potential targets for control interventions against a global crop pest. In this regard we provide preliminary demonstration of the potential use of inhibitors of these pathways to enhance the effectiveness of imidacloprid applications against B. tabaci.
Materials and Methods
Detailed information on insect strains and cell lines, extraction of DNA and RNA, qRT-PCR, promoter analysis, dual-luciferase reporter assay, RNAi experiments, life-span measurement, Western blot and inhibitor assays, and statistical analysis are described in SI Appendix, SI Materials and Methods.
Data Availability.
All data used in the study are included in the paper and SI Appendix. All protocols are described in SI Appendix, SI Materials and Methods or in the references therein. Plasmids are used in the study are freely available upon request to the corresponding author.
Supplementary Material
Acknowledgments
We thank Murad Ghanim (The Volcani Center) and Prof. Xiaoqiang Yu (University of Missouri, Kansas City) for suggestions that improved the paper. This research was supported by the National Natural Science Foundation of China (grants 31420103919 and 31601664); National Key R&D Program of China (grant 2016YFD0200500); China Agriculture Research System (CARS-24-C-02); the Beijing Key Laboratory for Pest Control and Sustainable Cultivation of Vegetables; and the Science and Technology Innovation Program of the Chinese Academy of Agricultural Sciences (CAAS-ASTIP-IVFCAAS). C.B. received funding from the European Research Council under the European Union’s Horizon 2020 Research and Innovation Programme (grant 646625).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1913603117/-/DCSupplemental.
References
- 1.Li X., Schuler M. A., Berenbaum M. R., Molecular mechanisms of metabolic resistance to synthetic and natural xenobiotics. Annu. Rev. Entomol. 52, 231–253 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Liu N., Insecticide resistance in mosquitoes: Impact, mechanisms, and research directions. Annu. Rev. Entomol. 60, 537–559 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Daborn P. J., et al. , A single p450 allele associated with insecticide resistance in Drosophila. Science 297, 2253–2256 (2002). [DOI] [PubMed] [Google Scholar]
- 4.Li X., Baudry J., Berenbaum M. R., Schuler M. A., Structural and functional divergence of insect CYP6B proteins: From specialist to generalist cytochrome P450. Proc. Natl. Acad. Sci. U.S.A. 101, 2939–2944 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bass C., et al. , Gene amplification and microsatellite polymorphism underlie a recent insect host shift. Proc. Natl. Acad. Sci. U.S.A. 110, 19460–19465 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zimmer C. T., et al. , Neofunctionalization of duplicated P450 genes drives the evolution of insecticide resistance in the brown planthopper. Curr. Biol. 28, 268–274.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kalsi M., Palli S. R., Transcription factor cap n collar C regulates multiple cytochrome P450 genes conferring adaptation to potato plant allelochemicals and resistance to imidacloprid in Leptinotarsa decemlineata (Say). Insect Biochem. Mol. Biol. 83, 1–12 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Kalsi M., Palli S. R., Transcription factors, CncC and Maf, regulate expression of CYP6BQ genes responsible for deltamethrin resistance in Tribolium castaneum. Insect Biochem. Mol. Biol. 65, 47–56 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Ingham V. A., Pignatelli P., Moore J. D., Wagstaff S., Ranson H., The transcription factor Maf-S regulates metabolic resistance to insecticides in the malaria vector Anopheles gambiae. BMC Genomics 18, 669 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilding C. S., Regulating resistance: CncC:Maf, antioxidant response elements and the overexpression of detoxification genes in insecticide resistance. Curr. Opin. Insect Sci. 27, 89–96 (2018). [DOI] [PubMed] [Google Scholar]
- 11.McDonnell C. M., Brown R. P., Berenbaum M. R., Schuler M. A., Conserved regulatory elements in the promoters of two allelochemical-inducible cytochrome P450 genes differentially regulate transcription. Insect Biochem. Mol. Biol. 34, 1129–1139 (2004). [DOI] [PubMed] [Google Scholar]
- 12.Brown R. P., McDonnell C. M., Berenbaum M. R., Schuler M. A., Regulation of an insect cytochrome P450 monooxygenase gene (CYP6B1) by aryl hydrocarbon and xanthotoxin response cascades. Gene 358, 39–52 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Nakata K., et al. , Nuclear receptor-mediated transcriptional regulation in Phase I, II, and III xenobiotic metabolizing systems. Drug Metab. Pharmacokinet. 21, 437–457 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Misra J. R., Horner M. A., Lam G., Thummel C. S., Transcriptional regulation of xenobiotic detoxification in Drosophila. Genes Dev. 25, 1796–1806 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Afschar S., et al. , Nuclear hormone receptor DHR96 mediates the resistance to xenobiotics but not the increased lifespan of insulin-mutant Drosophila. Proc. Natl. Acad. Sci. U.S.A. 113, 1321–1326 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mayr B., Montminy M., Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat. Rev. Mol. Cell Biol. 2, 599–609 (2001). [DOI] [PubMed] [Google Scholar]
- 17.Hendricks J. C., et al. , A non-circadian role for cAMP signaling and CREB activity in Drosophila rest homeostasis. Nat. Neurosci. 4, 1108–1115 (2001). [DOI] [PubMed] [Google Scholar]
- 18.Widmer Y. F., et al. , Multiple neurons encode CrebB dependent appetitive long-term memory in the mushroom body circuit. eLife 7, e39196 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gehring K. B., Heufelder K., Kersting I., Eisenhardt D., Abundance of phosphorylated Apis mellifera CREB in the honeybee’s mushroom body inner compact cells varies with age. J. Comp. Neurol. 524, 1165–1180 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Shen R., et al. , Neuronal energy-sensing pathway promotes energy balance by modulating disease tolerance. Proc. Natl. Acad. Sci. U.S.A. 113, E3307–E3314 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hou L., Li B., Ding D., Kang L., Wang X., CREB-B acts as a key mediator of NPF/NO pathway involved in phase-related locomotor plasticity in locusts. PLoS Genet. 15, e1008176 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cargnello M., Roux P. P., Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol. Mol. Biol. Rev. 75, 50–83 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ragab A., et al. , Drosophila Ras/MAPK signalling regulates innate immune responses in immune and intestinal stem cells. EMBO J. 30, 1123–1136 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sakaguchi A., Matsumoto K., Hisamoto N., Roles of MAP kinase cascades in Caenorhabditis elegans. J. Biochem. 136, 7–11 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Horton A. A., et al. , The mitogen-activated protein kinome from Anopheles gambiae: Identification, phylogeny and functional characterization of the ERK, JNK and p38 MAP kinases. BMC Genomics 12, 574 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo Z., et al. , MAPK signaling pathway alters expression of midgut ALP and ABCC genes and causes resistance to Bacillus thuringiensis Cry1Ac toxin in diamondback moth. PLoS Genet. 11, e1005124 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Barro P. J., Liu S. S., Boykin L. M., Dinsdale A. B., Bemisia tabaci: A statement of species status. Annu. Rev. Entomol. 56, 1–19 (2011). [DOI] [PubMed] [Google Scholar]
- 28.Karunker I., et al. , Over-expression of cytochrome P450 CYP6CM1 is associated with high resistance to imidacloprid in the B and Q biotypes of Bemisia tabaci (Hemiptera: Aleyrodidae). Insect Biochem. Mol. Biol. 38, 634–644 (2008). [DOI] [PubMed] [Google Scholar]
- 29.Yang X., et al. , Two cytochrome P450 genes are involved in imidacloprid resistance in field populations of the whitefly, Bemisia tabaci, in China. Pestic. Biochem. Physiol. 107, 343–350 (2013). [DOI] [PubMed] [Google Scholar]
- 30.Karunker I., et al. , Structural model and functional characterization of the Bemisia tabaci CYP6CM1vQ, a cytochrome P450 associated with high levels of imidacloprid resistance. Insect Biochem. Mol. Biol. 39, 697–706 (2009). [DOI] [PubMed] [Google Scholar]
- 31.Nauen R., Vontas J., Kaussmann M., Wölfel K., Pymetrozine is hydroxylated by CYP6CM1, a cytochrome P450 conferring neonicotinoid resistance in Bemisia tabaci. Pest Manag. Sci. 69, 457–461 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Lonze B. E., Ginty D. D., Function and regulation of CREB family transcription factors in the nervous system. Neuron 35, 605–623 (2002). [DOI] [PubMed] [Google Scholar]
- 33.Dash P. K., Karl K. A., Colicos M. A., Prywes R., Kandel E. R., cAMP response element-binding protein is activated by Ca2+/calmodulin- as well as cAMP-dependent protein kinase. Proc. Natl. Acad. Sci. U.S.A. 88, 5061–5065 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xing J., Ginty D. D., Greenberg M. E., Coupling of the RAS-MAPK pathway to gene activation by RSK2, a growth factor-regulated CREB kinase. Science 273, 959–963 (1996). [DOI] [PubMed] [Google Scholar]
- 35.Chijiwa T., et al. , Inhibition of forskolin-induced neurite outgrowth and protein phosphorylation by a newly synthesized selective inhibitor of cyclic AMP-dependent protein kinase, N-[2-(p-bromocinnamylamino)ethyl]-5-isoquinolinesulfonamide (H-89), of PC12D pheochromocytoma cells. J. Biol. Chem. 265, 5267–5272 (1990). [PubMed] [Google Scholar]
- 36.English J. M., Cobb M. H., Pharmacological inhibitors of MAPK pathways. Trends Pharmacol. Sci. 23, 40–45 (2002). [DOI] [PubMed] [Google Scholar]
- 37.Gonzalez G. A., Montminy M. R., Cyclic AMP stimulates somatostatin gene transcription by phosphorylation of CREB at serine 133. Cell 59, 675–680 (1989). [DOI] [PubMed] [Google Scholar]
- 38.Liu M., et al. , Glutamate affects the production of epoxyeicosanoids within the brain: The up-regulation of brain CYP2J through the MAPK-CREB signaling pathway. Toxicology 381, 31–38 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Xu M., Ju W., Hao H., Wang G., Li P., Cytochrome P450 2J2: Distribution, function, regulation, genetic polymorphisms and clinical significance. Drug Metab. Rev. 45, 311–352 (2013). [DOI] [PubMed] [Google Scholar]
- 40.Gong P., Cederbaum A. I., Nieto N., Increased expression of cytochrome P450 2E1 induces heme oxygenase-1 through ERK MAPK pathway. J. Biol. Chem. 278, 29693–29700 (2003). [DOI] [PubMed] [Google Scholar]
- 41.Joannard F., et al. , Role for mitogen-activated protein kinases in phenobarbital-induced expression of cytochrome P450 2B in primary cultures of rat hepatocytes. Toxicol. Lett. 161, 61–72 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Cancino-Rodezno A., et al. , The mitogen-activated protein kinase p38 is involved in insect defense against Cry toxins from Bacillus thuringiensis. Insect Biochem. Mol. Biol. 40, 58–63 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chang L., Karin M., Mammalian MAP kinase signalling cascades. Nature 410, 37–40 (2001). [DOI] [PubMed] [Google Scholar]
- 44.De Marco L., et al. , The choreography of the chemical defensome response to insecticide stress: Insights into the Anopheles stephensi transcriptome using RNA-seq. Sci. Rep. 7, 41312 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang N., et al. , Transcriptome profiling of the whitefly Bemisia tabaci reveals stage-specific gene expression signatures for thiamethoxam resistance. Insect Mol. Biol. 22, 485–496 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li T., Liu L., Zhang L., Liu N., Role of G-protein-coupled receptor-related genes in insecticide resistance of the mosquito, Culex quinquefasciatus. Sci. Rep. 4, 6474 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hill C. A., Sharan S., Watts V. J., Genomics, GPCRs and new targets for the control of insect pests and vectors. Curr. Opin. Insect Sci. 30, 99–106 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Lagerström M. C., Schiöth H. B., Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat. Rev. Drug Discov. 7, 339–357 (2008). [DOI] [PubMed] [Google Scholar]
- 49.Goldsmith Z. G., Dhanasekaran D. N., G protein regulation of MAPK networks. Oncogene 26, 3122–3142 (2007). [DOI] [PubMed] [Google Scholar]
- 50.Wetzker R., Böhmer F. D., Transactivation joins multiple tracks to the ERK/MAPK cascade. Nat. Rev. Mol. Cell Biol. 4, 651–657 (2003). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data used in the study are included in the paper and SI Appendix. All protocols are described in SI Appendix, SI Materials and Methods or in the references therein. Plasmids are used in the study are freely available upon request to the corresponding author.