Significance
Arc is a neural immediate early gene involved in synaptic downscaling and is robustly induced by prolonged wakefulness in rodent brains. However, it remains unclear if and how Arc is involved in sleep regulation. Here we find that Arc is important for inducing multiple homeostatic responses induced by sleep deprivation at behavioral and molecular levels: (1) rebound of sleep time; (2) expression of a subset of sleep deprivation-induced genes; and (3) synaptic glutamate receptor expression. In sleep-deprived wild-type brains, Arc protein levels are significantly increased in the nucleus, cytoplasm, and synapse, suggesting multiple roles for Arc depending on its subcellular location. These findings provide the functional evidence for the role of Arc in homeostatic sleep regulation.
Keywords: sleep homeostasis, nuclear translocation, activity-regulated cytoskeleton-associated protein, GluA1, immediate early gene
Abstract
The activity-regulated cytoskeleton-associated protein (Arc) gene is a neural immediate early gene that is involved in synaptic downscaling and is robustly induced by prolonged wakefulness in rodent brains. Converging evidence has led to the hypothesis that wakefulness potentiates, and sleep reduces, synaptic strengthening. This suggests a potential role for Arc in these and other sleep-related processes. However, the role of Arc in sleep remains unknown. Here, we demonstrated that Arc is important for the induction of multiple behavioral and molecular responses associated with sleep homeostasis. Arc knockout (KO) mice displayed increased time spent in rapid eye movement (REM) sleep under baseline conditions and marked attenuation of sleep rebound to both 4 h of total sleep deprivation (SD) and selective REM deprivation. At the molecular level, the following homeostatic sleep responses to 4-h SD were all blunted in Arc KO mice: increase of α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor GluA1 and its phosphorylation in synaptoneurosomes; induction of a subset of SD-response genes; and suppression of the GluA1 messenger RNA in the cortex. In wild-type brains, SD increased Arc protein expression in multiple subcellular locations, including the nucleus, cytoplasm, and synapse, which is reversed in part by recovery sleep. Arc is critical for these behavioral and multiple molecular responses to SD, thus providing a multifunctional role for Arc in the maintenance of sleep homeostasis, which may be attributed by the sleep/wake-associated changes in subcellular location of Arc.
Introduction
Sleep is homeostatically regulated. A hypothetical substrate of sleep need accumulates in the central nervous system during waking and resolves during sleep (1). Although the molecules or genes involved in sleep need remain unclear, electroencephalography (EEG) delta power (0.5–4 Hz) in non–rapid eye movement (NREM) sleep is well accepted as the most reliable marker of sleep need (1). In response to sleep loss, homeostatic sleep regulation induces sleep rebound and elevated delta power in NREM sleep to compensate for the loss (1). A broad range of molecular events associated with energy metabolism, synaptic plasticity, and macromolecule biosynthesis also occur in response to sleep loss to maintain sleep homeostasis (2). These events are the three major theoretical reasons for sleep, which are supported by the up-regulation of key genes or proteins associated with those events in the brain (2). Currently, based on several lines of evidence, the theoretical model of the relationship between sleep and synaptic plasticity has been developed as the synaptic homeostasis hypothesis (SHY, ref. 3). The SHY posits that synaptic down-regulation is a critical functional characteristic of sleep: the experience during waking increases synaptic strength, and sleep eliminates its excess (3). In sleep-deprived rodent brains, increases in spine number and volume (3, 4), in axon–spine interface (4), in total and phosphorylated GluA1 α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor levels in synaptoneurosomes (5), in synaptic strength (3, 6), and in phosphorylation levels of a subset of synaptic proteins (7) have been reported. In addition to the correlation between these genes/proteins and sleep need, recent omics studies reported an increase in the expression of several immediate early genes in rodent brains exposed to prolonged waking conditions (8–10). However, the direct function of these genes that underlie sleep regulation remains to be elucidated.
Arc, one of such neuronal immediate early genes, is dramatically induced in sleep-deprived brains; it is up-regulated during both forced and spontaneous waking and down-regulated during sleep in the brain (8, 11–13), especially in the cortex and hippocampus (14), where EEG delta and theta oscillations are most robustly measured, respectively. EEG delta power is highest during NREM sleep, while EEG theta power is greatest during REM sleep. These previous reports suggest a potential association between the Arc gene and sleep homeostasis; however, Arc has been considered merely as a sleep marker, because of the poor evidence regarding its function in sleep (11, 13), such as Arc’s involvement in sleep architecture, sleep-related gene expression, sleep-related synaptic scaling, or sleep homeostasis.
In contrast, many studies have implicated Arc as a central player in the synaptic plasticity involved in learning and memory processes (15, 16). The expression of the Arc messenger RNA (mRNA) is tightly regulated by neuronal activity (17, 18). The mRNA is transported to dendrites and translated to the encoded protein locally (18, 19). At the synapse, Arc is involved in long-term potentiation (LTP) consolidation (20), synaptic number and morphology (21), and late-phase LTP (22), which mediates synaptic scaling. Thus, Arc seems to be deeply involved in synaptic strengthening. However, studies have demonstrated that Arc exists in the cytoplasm of excitatory neurons in silent synapses (23) and down-regulates synaptic strength by reducing spine number and increasing AMPA receptor trafficking (16, 24). In addition, neural activities also regulate Arc nuclear shuttling (25). In nuclei, Arc establishes a complex with promyelocytic leukemia protein nuclear bodies (PML-NBs) as a polymer of a transcriptional cofactor (26, 27); in turn, this complex down-regulates the GluA1 mRNA presumably via an epigenetic mechanism (25, 28). Consequently, nuclear Arc reduces GluA1 mRNA translation (25). Thus, currently, it is widely accepted that synaptic down-regulation is the most important function of Arc. However, these opposing findings of Arc suggest the possibility that Arc may contribute to sleep/wake-associated synaptic homeostasis bidirectionally, or in either direction of synaptic upscaling or weakening, as proposed by the SHY.
In this study, to investigate the mechanisms underlying the role of Arc in sleep homeostasis, we first evaluated the sleep phenotype in constitutive Arc knockout (KO) mice (29) under baseline conditions, as well as in response to continuous 4-h total sleep deprivation (SD) and selective REM deprivation. Subsequently, we assessed Arc involvement in the associated molecular events that occur in response to SD: SD-induced sleep-related gene expression, synaptic GluA1 expression, and Arc nuclear distribution within the context of the homeostatic sleep response. We report that, in the absence of Arc, all of the sleep homeostatic responses described above were disrupted at both the behavioral and molecular levels.
Results
Sleep Phenotype and Homeostasis in Arc KO Mice.
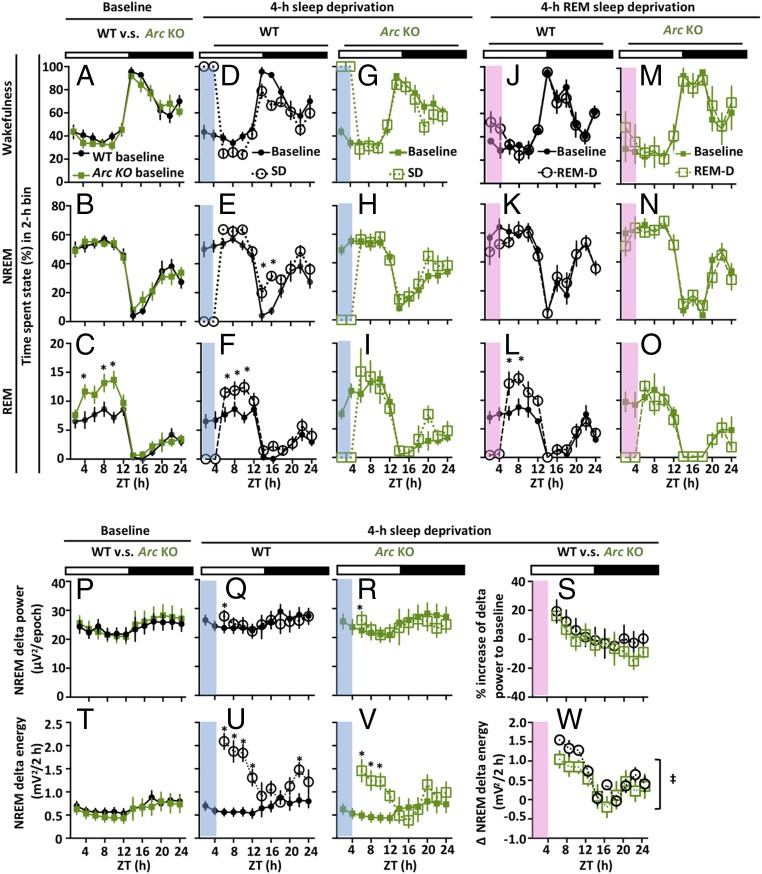
At baseline, Arc KO mice showed a normal diurnal sleep pattern over 24 h (Fig. 1 A–C, P, and T), yet exhibited an increased total time in REM sleep in the light phase due to increased REM sleep episode numbers compared to wild-type (WT) mice (Fig. 1C and SI Appendix, Fig. S1 B, Left). In contrast, there were no differences between the genotypes in the parameters associated with baseline NREM sleep: total time spent in NREM sleep (Fig. 1B), NREM sleep episode frequency (see SI Appendix, Fig. S1A), and 24-h delta power distribution (Fig. 1P). Next, to examine the homeostatic sleep response, continuous SD was employed from ZT0–4, and recovery sleep responses were observed during the recovery period, from ZT4–24. In Arc KO mice, SD induced no appreciable rebound in NREM or REM sleep amount (Fig. 1 H and I). Interestingly, however, they exhibited an apparently normal increase in NREM delta power following 4-h SD (Fig. 1R). Overall, a lower level of NREM delta energy was detected in Arc KO mice compared to WT (Fig. 1W). In addition, after 4 h of selective REM deprivation, the animals did not show detectable REM sleep rebound (Fig. 1O); the REM episode number during the recovery was decreased rather than increased in ZT4–12 (SI Appendix, Fig. S2). Overall, Arc KO mice had significant attenuation in the homeostatic sleep response to both total SD and selective REM deprivation.
Fig. 1.
Sleep phenotype and behavioral homeostatic sleep response in Arc KO mice. (A–C) Percentages of time spent in wakefulness (A), NREM sleep (B), and REM sleep (C) across 24 h baseline in WT (black, n = 8) and Arc KO (green, n = 7) mice. (D–I) Percentage of time spent in states at baseline (solid) and after SD (dashed) in response to 4 h of SD (blue shading) for WT (open circle, n = 8) and Arc KO (open square, n = 7) mice. (J–O) Baseline and response to 4-h selective REM deprivation (REM-D, pink shading) in states in WT (n = 4) and Arc KO (n = 4) mice. (P–R and T–V) Delta power (Top) and NREM delta energy (Bottom) of baseline (P and T) and 4-h SD (Q, R, U, and V). (S and W) Direct comparisons between genotypes of changes in delta power (S) and NREM delta energy (W) following 4-h SD relative to control condition. (W) During recovery sleep, changes in the delta energy in Arc KO mice were significantly lower than those detected in WT mice (F [1, 116] = 5.60, P = 0.020, two-way ANOVA). All averaged values were plotted in 2-h intervals. *P < 0.05 compared with time-matched baseline by Sidak’s multiple-comparisons test following two-way ANOVA; ‡P < 0.05 compared between genotypes by two-way ANOVA.
Attenuation of Synaptoneurosome GluA1 Expression in Response to 4 h of Total Sleep Deprivation in Arc KO Mice.
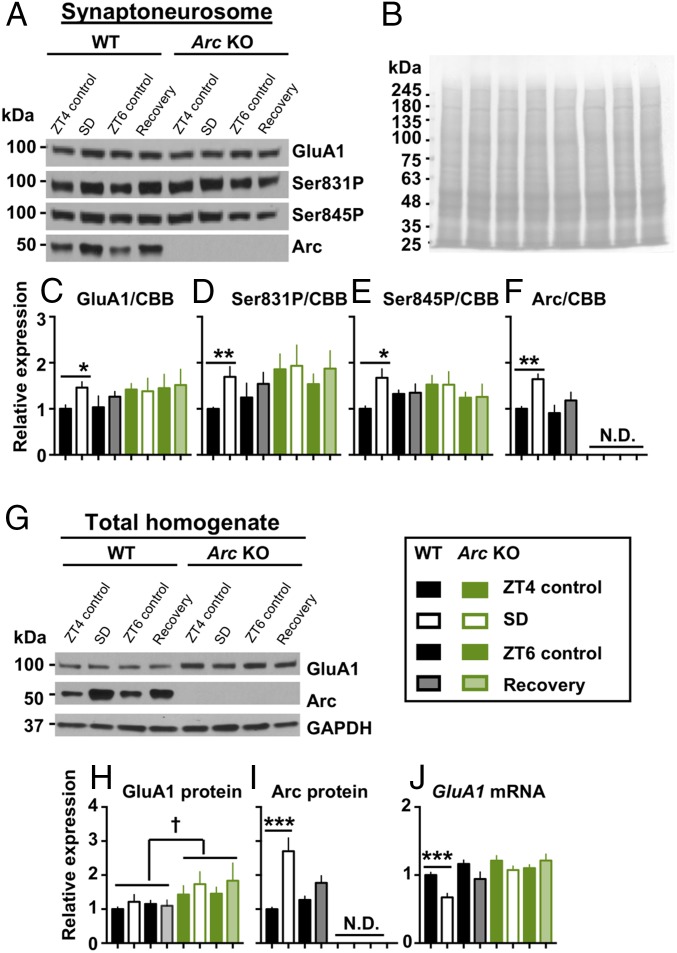
Next, we investigated whether Arc is involved in sleep/wake-associated synaptic homeostasis using an index of GluA1 expression at the synapse and its phosphorylation status, as reported previously (5). We assessed the expression levels of total GluA1 and GluA1 phosphorylated at Ser831 (Ser831P) and Ser845 (Ser845P) across the control, total SD, and recovery conditions. In synaptoneurosomes obtained from the frontal cortex (see SI Appendix, Fig. S3), WT mice showed increased expression of GluA1, Ser831P, and Ser845P during SD compared to the control condition, whereas Arc KO mice brains did not (Fig. 2 C–E and SI Appendix, Fig. S4). Using total homogenates, we confirmed the up-regulation of the Arc protein in the brains of WT animals in the SD condition (Fig. 2I). In total homogenates, GluA1 protein levels were unchanged across sleep/wake conditions within genotypes (Fig. 2H; WT: F [1, 41] = 0.26, P = 0.61; Arc KO: F [1, 24] = 0.8345, P = 0.37, two-way ANOVA); however, as reported previously (16), these levels were higher in Arc KO mice than they were in WT mice across all conditions (Fig. 2H, F [1, 87] = 16.65, P = 0.0001, two-way ANOVA). Interestingly, we found a decrease in GluA1 mRNA levels after 4-h SD in WT mice, but not in Arc KO mice (Fig. 2J). Thus, Arc KO mice seem to lose the ability to maintain synaptic homeostasis that is associated with GluA1 expression by sleep/wake stimuli.
Fig. 2.
Impairment of SD-induced synaptoneurosome GluA1 expression in response to sleep deprivation in Arc KO mice. Representative images of immunoblots (A) regarding GluA1 (C), Ser831P (D), Ser845P (E), and Arc (F) in synaptoneurosomes and a transblotted membrane of whole synaptoneurosomes stained with Coomassie Brilliant Blue (B). Representative immunoblot images (G) of GluA1 (H), Arc (I), and GAPDH in total homogenates. (J). The GluA1 transcript was measured by qRT-PCR and normalized to GAPDH transcript levels. Arc protein expression was not detected (N.D.) in synaptoneurosome (A and F) or total homogenate samples of Arc KO mice (G and I). Protein expression levels of synaptoneurosome (C–F) and total homogenate (H and I) samples of the frontal cortex were normalized to whole synaptoneurosome proteins and GAPDH in total homogenates, respectively. All values were expressed relative to the means of WT control mice at ZT4. *P < 0.05, **P < 0.01, and ***P < 0.001 by Sidak’s multiple-comparisons test following two-way ANOVA; † P < 0.05 by two-way ANOVA between genotypes; N.D., not detected (n = 8–14 in each genotype).
Impairment of the Induction of Sleep-Related Genes in Arc KO Mice.
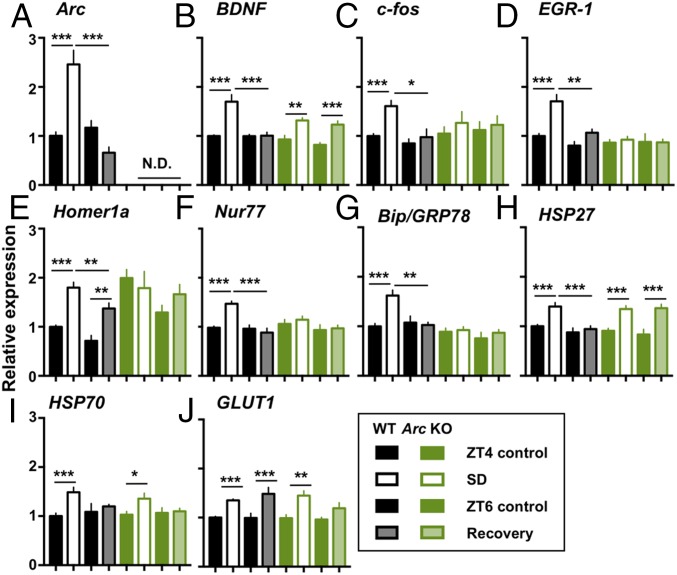
To confirm the importance of Arc in sleep homeostasis, we next tested the involvement of Arc in the expression of genes previously implicated in sleep homeostasis. As the levels of expression of sleep-related genes in the brain vary in response to the homeostatic sleep drive or to the time spent awake, the loss of Arc may affect the expression of those sleep-related immediate early genes (Fig. 3 A–F, ref. 11, 30), the expression of genes associated with macromolecule synthesis (e.g., chaperone proteins, Fig. 3 G–I, refs. 11 and 31), and energy metabolism (glucose transporter 1, GLUT1, Fig. 3J, ref. 11) caused by the disruption of sleep homeostasis observed in Fig. 1. To test the role of Arc as a modulator of the expression of sleep-related genes, we compared the expression patterns of already reported sleep-related genes across sleep/wake conditions in the frontal cortex of WT and Arc KO mice (11, 30, 31). qRT-PCR data showed that, in WT mice, the expression levels of all these genes were significantly increased after 4 h of SD (Fig. 3 A–J) and returned to baseline levels after 2 h of recovery sleep (Fig. 3 A–I), except for homer1a and GLUT1 (Fig. 3 E and J). Importantly, Arc KO mice showed a distinct expression profile, especially regarding activity-regulated genes (Fig. 3 C–F) except for BDNF (Fig. 3B): the expression of c-fos, EGR-1, homer1a, Nur77 (Nr4a1), and a chaperone protein, Bip/GRP78, was not induced after SD and remained at baseline levels after the 2-h recovery sleep (Fig. 3 C–G and SI Appendix, Fig. S5). These results demonstrate that Arc is important for the expression of SD-induced genes, especially activity-regulated genes, after sleep loss.
Fig. 3.
Distinct expression profiles of sleep-related genes in response to sleep deprivation in Arc KO mice. (A–J) Expression of SD-induced genes in WT and Arc KO mice across the control at ZT4 and ZT6, 4 h of SD, and 2 h of recovery sleep conditions. In contrast to WT mice, Arc KO mice exhibited a lower expression profile of SD-related genes in response to SD stimuli (C–G). Samples of the frontal cortex were used for the analysis. Gene expression values were normalized to those of GAPDH and expressed relative to the means of WT control mice at ZT4 (means + standard error of mean). *P < 0.05, **P < 0.01, and ***P < 0.001 compared to the control condition by Sidak’s multiple-comparisons test following two-way ANOVA; N.D., not detected (n = 6–14 in each genotype).
Arc Nuclear Distribution in Association with Sleep Loss.
It has been reported that the activation of excitatory neurons increases the mRNA (18, 19) and protein expression (19, 20) of Arc, as well as its nuclear translocation (25). Then, in the nucleus, Arc down-regulates GluA1 transcription (25). We confirmed an increase in cellular Arc protein and mRNA expression in the frontal cortex of WT mice as previously reported and also found a decrease in GluA1 expression in the brains of WT mice after SD; these changes were abolished in the absence of Arc (Figs. 2 I and J and 3A). These results suggest that SD may lead to the nuclear translocation of Arc (25). Therefore, we investigated the subcellular distribution of Arc protein across sleep/wake conditions in WT mouse brains using immunofluorescence staining.
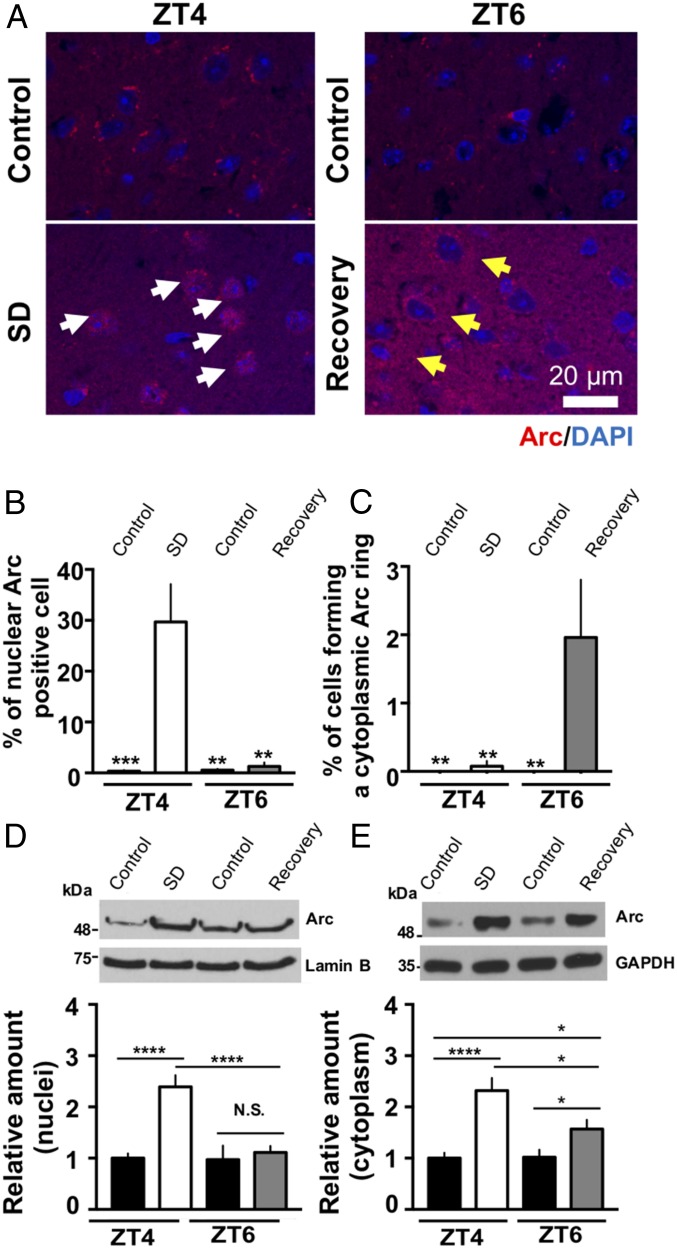
First, we determined the relative number of Arc-positive cells across sleep/wake conditions in the motor cortex of WT animals (Fig. 4 A–C). Qualitatively, we found a unique Arc subcellular distribution that was dependent on sleep/wake conditions, particularly in SD and recovery sleep brains. In 4-h SD brains, Arc accumulated in nuclei (Fig. 4A SD, white arrows, and Fig. 4B). Two hours of recovery sleep following 4 h of SD caused a subset of cells to exhibit the formation of cytoplasmic Arc-positive rings surrounding the nucleus (Fig. 4A Recovery, yellow arrows); however, these rings were rarely observed in the control and SD brains (Fig. 4A Controls, SD, and Fig. 4C). Immunoblotting performed using nuclear fraction samples supported quantitatively the results of the immunofluorescence experiment; in SD brains, the expression of nuclear Arc was increased by nearly 2.5-fold compared to the control condition at ZT4 (Fig. 4D; P = 0.0000007 by Sidak’s test) and returned to baseline levels after 2 h of recovery sleep. Similar to the observed SD-associated increase in nuclear Arc levels, SD induced an up-regulation of cytoplasmic Arc by ∼2.5-fold compared to the control condition at ZT4 (Fig. 4E; P = 0.00009 by Sidak’s test). The expression of cytoplasmic Arc was significantly reduced after 2 h of recovery sleep (Fig. 4E; P = 0.031 by Sidak’s test, SD vs. Recovery). However, in contrast to nuclear Arc, the levels remained higher than the control condition at ZT6 (Fig. 4E; P = 0.026 by Sidak’s test at ZT6). Taken together, these results demonstrate increases in both nuclear and cytoplasmic Arc levels and in the number of nuclear Arc-positive cells associated with sleep loss.
Fig. 4.
Subcellular distribution of Arc across the control, SD, and recovery sleep conditions in the brains of WT mice. (A) Representative images of the motor cortex (bregma ± 100 μm) for each state. Arc and DAPI signals are indicated in red and blue colors, respectively. White and yellow arrows show representative images of Arc accumulation in the nucleus (SD) and a ring formation of cytoplasmic Arc around the nucleus (recovery), respectively. Percentage of nuclear–Arc-positive cells (B) and cells forming a cytoplasmic Arc ring (C) in the control, SD, and recovery condition. Arc-positive cells were expressed as percentages of the total DAPI-stained cell (both neurons and glia) numbers. SD and recovery brains showed significant increases in nuclear Arc-positive cells and cells forming a cytoplasmic ring compared to the other groups, respectively (B: **P < 0.01, ***P < 0.0001 compared to SD, C: **P < 0.01 compared to recovery, Sidak’s multiple-comparisons test, n = 10–15 mice). (D and E) Evaluation of nuclear (D) and cytoplasmic (E) Arc expression by immunoblot. The expression levels of nuclear and cytoplasmic Arc were normalized to those of lamin B and GAPDH, respectively, and expressed relatively. *P < 0.05, ****P < 0.0001 by Sidak’s multiple-comparisons test following two-way ANOVA; N.S., not significant (n = 4–9).
Discussion
In the present study, we demonstrate that Arc KO mice have greatly attenuated homeostatic responses to SD, including the loss of (1) sleep rebound after total SD and selective REM deprivation, (2) SD-induced synaptoneurosome GluA1 expression changes, and (3) induction of a subset of sleep-related genes. In addition, Arc changes subcellular distribution under SD and during recovery sleep. These results are indicative of the multiple roles of Arc in the homeostatic sleep response at the behavioral and molecular levels.
Role of Arc in Sleep Behavior.
The amount of REM sleep is associated with NREM sleep duration (32). A longer NREM sleep duration tends to induce REM sleep. However, Arc KO mice tend to have additional REM sleep episodes at the end of NREM sleep episodes more than WT mice do, resulting in a consequent increase in REM sleep amounts (Fig. 1C) without changes in a mean duration of NREM sleep episodes under baseline conditions (SI Appendix, Fig. S1). Compared with WT animals at baseline, Arc KO mice exhibited increases in REM frequencies in the light phase and transitions between NREM and REM sleep, without changes in REM sleep latency. Thus, Arc KO mice exhibit a unique phenotype of an increased total time spent in REM sleep. At this time, the mechanism for REM sleep regulation through Arc is unclear. It might be related to the phenomenon that “stress” tends to increase REM sleep (33, 34). Further studies are required to elucidate the Arc-mediated regulation of REM sleep.
It is interesting to note that, after a single exposure to 4 h of SD, Arc KO mice showed increased delta power similar to that of WT (Fig. 1 Q–S), but not increased NREM sleep amount, compared with baseline (Fig. 1H). Consequently, NREM delta energy as the integral of delta power density through NREM sleep, which is correlated with a compensation for short-term sleep loss to satisfy sleep need (35), was significantly decreased in Arc KO mice compared to WT animals (Fig. 1W). Also, in contrast to WT, Arc KO mice did not show NREM sleep episode consolidation after 4-h SD (see SI Appendix, Fig. S1F), suggesting that the animals failed to exhibit this additional index of sleep homeostatic response (36). Moreover, other homeostatic responses including increase of synaptic GluA1 expression (Fig. 2 C–E) and sleep-related gene induction (Fig. 3) are also blunted in Arc KO mice. Thus, we speculate that Arc may mediate specific signal(s) through which increased delta power, or sleep need, induces behavioral and molecular changes to maintain normal sleep homeostasis. BDNF has been reported to have a role in the increase of NREM delta power under increased sleep need in humans and rodents (37, 38). Notably, Arc KO mice show an apparently intact increase in BDNF mRNA levels and delta-power rebound after 4-h SD (Figs. 1R and 3B). This suggests that BDNF may induce NREM delta-power rebound either through an Arc-independent pathway, or acting upstream of Arc signaling. It would be interesting to subject Arc KO mice to longer periods of SD; it would also be worth analyzing delta power decay within each NREM episode, which is another index of sleep need (36).
Arc-Mediated Regulation of the Synaptic GluA1 Accumulation.
Our results (Fig. 2 A and C–F) demonstrate that Arc is important for the SD-induced up-regulation of the GluA1 synaptic pool and of GluA1 phosphorylation (3, 39, 40), which is involved in synaptic strengthening (16–19). In addition, due to the increased GluA1 phosphorylation at Ser831 and Ser845 (16–19, 40), GluA1 surface insertion is likely increased during SD, while decreased in recovery sleep in WT but not Arc KO mice (Fig. 2 A, D, and E). Thus, our observations suggest Arc’s involvement in synaptic strengthening and downscaling during SD and recovery sleep, respectively. A recent study of the interaction of Arc with PSD-95 shows that this complex is necessary for several physiological postsynaptic functions (41), consistent with Arc’s involvement in sleep homeostasis affecting postsynaptic function during SD. Alternatively, a substantial amount of SD-induced synaptic Arc accumulation may be required to induce sleep-related synaptic downscaling during recovery sleep (3, 15, 16, 23). Together, although one of the most important roles of Arc is synaptic downscaling via AMPA receptor internalization at inactivated synapses (23), Arc might contribute to sleep/wake-associated synaptic down/upscaling bidirectionally through the regulation of synaptic GluA1.
Arc Involvement in SD-Induced Gene Expression.
As part of the role of Arc in sleep homeostasis, we examined the expression of sleep-related genes and found that Arc was important for the induction of a subset of those genes: the SD-induced expression of c-fos, EGR-1, Homer1a, Nur77, and Bip/GRP78 was greatly disrupted (Fig. 3 C–G), while that of BDNF, HSP27, HSP70, and GLUT1 was largely unaffected in Arc KO mice (Fig. 3 B and H–J). This result leads to three speculations. First, considering that these disrupted genes are all CREB targets (42–46), the CREB pathway or its associated pathways, including calcium-signaling pathways, may be important for the Arc-mediated induction of sleep-related genes. Indeed, the link between Arc and cellular calcium (47), the involvement of Arc in late-phase LTP (22), and the interaction of Arc and CREB-binding protein (48) have been reported. Second, SD-induced genes that are inhibited by Arc loss are all immediate early genes, except for Bip/Grp78 (Fig. 3 C–G). This suggests that Arc may be an important regulator of molecules that are associated with synaptic plasticity via multiple direct or indirect pathways (42–46). Thus, the disrupted expression of sleep-related immediate early genes may also cause the inhibition of sleep-related response of synaptic GluA1 in Arc KO mice (Fig. 2C). Third, the SD-induced up-regulation of GLUT1 as a metabolism-related gene or HSP27 and HSP70 as chaperone proteins that was normally observed in Arc KO mice (Fig. 3 H–J) suggests that Arc is not crucial for the function of sleep related to energy conservation or macromolecule biosynthesis (2). Taken together, these results suggest that Arc, itself an immediate early gene, is highly involved in the SD-mediated expression of activity-induced immediate early genes in the brain.
Interestingly, the SD-induced expression of BDNF, which is also a target gene of CREB (48), is not affected by the loss of Arc (Fig. 3B). This seems reasonable because Arc is known as a BDNF-induced gene (48). On the other hand, the SD-induced expression of EGR-1 that also induces Arc expression (49), and a target gene of CREB, was inhibited in Arc KO mice (Fig. 3D), which likely indicates additional possibilities. Supporting this, the recent report (50) that KCl or light exposure results in a slower induction of BDNF compared to the Arc or Fos genes suggests that, although BDNF is an immediate early gene that responds to neural activities, the induction process of BDNF may be differentially activated by extracellular or physiological stimuli, supposedly including SD, compared to other SD-induced immediate early genes we examined here.
Sleep Homeostasis-Associated Arc Translocation between the Cytoplasm and Nucleus.
We demonstrated that SD promoted increase in both nuclear and cytoplasmic Arc. Recovery sleep returned nuclear Arc to baseline levels, as assessed by immunoblotting of nuclear proteins together with immunostaining (Fig. 4 A, D, and E). These results fit well with reports that neural activation increases the nuclear import of Arc (25), as the firing rate of glutamatergic neurons increases during waking (51). Notably, cytoplasmic Arc remained higher than baseline levels after 2 h of recovery sleep (Fig. 4E), suggesting the possibility that recovery sleep facilitates the cytoplasmic retranslocation of Arc (25). Cytoplasmic Arc are known to be distributed either to the nonsubmembrane cytoplasm or postsynaptic sites (in the soma, dendrites, and spines) through its binding partners (52). SD-induced cytoplasmic Arc may exert spine F-actin formation and reorganization (52) for wake-associated synaptic upscaling, which fits the SHY well (3). A recent histological study reported a sleep-mediated increase in the nuclear:cytoplasmic ratio of Arc-GFP fusion protein (53), which our biochemical analysis did not positively replicate (Fig. 4 D and E), perhaps reflecting methodological differences.
Interestingly, the cytoplasmic rings of Arc are not observed at ZT4 (Fig. 4C), when total cytoplasmic Arc is high (Fig. 4E). Thus, the rings are not associated with increased total Arc in either the nucleus or the cytoplasm (ZT4), but rather with a decrease of nuclear Arc after recovery sleep (ZT6). The total amount of Arc in the cytoplasm (Fig. 4E) does not necessarily predict its pattern of distribution within the cytoplasm (e.g., the formation of the ring). The number of cells with a visible ring (only about 2% even at ZT6 after the recovery sleep) probably reflects a small subset of those cells that had high Arc in the nucleus at ZT4 (Fig. 4B) and less nuclear Arc after 2 h of recovery at ZT6 (Fig. 4 B and D). The ring may reflect a transient Arc accumulation just outside of the nuclear envelope in the cytoplasm before it has a chance to disperse evenly into the cytoplasm in association with recovery sleep. Understanding the mechanism for the perinuclear accumulation may be an interesting topic for future studies.
The function of nuclear Arc largely remains to be elucidated compared to that of its cytoplasmic counterpart. Accordingly, little is known about the role of the SD-induced nuclear translocation of Arc. However, in vitro studies have demonstrated that, in the nucleus, Arc down-regulates the GluA1 mRNA epigenetically through an association with PML-NBs (26). In the nucleus, PML-NBs recruit histone acetyltransferases (HATs)/histone deacetylases (HDACs) (27), implying the possibility that the complex of Arc and PML-NBs may be involved in the bidirectional regulation of downstream gene expression (25, 26, 28). More than 30% of cells exhibited strong nuclear Arc expression in the brains of WT mice in the SD condition (Fig. 4 A, B, and D), which may be involved in transcriptional events of the epigenetically regulated GluA1 gene. In support of this contention, our results similarly revealed an SD-induced down-regulation of the GluA1 mRNA in WT (Fig. 2J). Moreover, the reduction of nuclear Arc expression level during the recovery sleep (Fig. 4 B and D WT mice) may cause alterations in the binding affinity of PML-NBs to HATs/HDACs, which may reverse GluA1 levels to those of baseline in WT brains (Fig. 2J Recovery). Furthermore, it was reported that the nuclear Arc-mediated down-regulation of the GluA1 mRNA consequently reduces the synthesis of the encoded protein, which was observed 30 h after Arc transfection in an in vitro study (25). However, in 4- or 6-h WT SD brains, GluA1 protein levels in total homogenates were unchanged (Fig. 2H, ref. 5). Supposedly, this time-consuming process of GluA1 mRNA translation may prevent the detection of any changes in GluA1 protein levels in 4-h SD brains, regardless of its mRNA levels (Fig. 2J WT mice). Additional mechanisms for regulating GluA1 translation may also exist in SD brains. Thus, our finding of sleep-related changes in Arc subcellular distribution implies that nuclear Arc may contribute to a mechanism modulating sleep/wake-associated gene expression.
In summary, the present study demonstrates the importance of Arc in sleep homeostasis in mice. Arc was induced at high levels in the nucleus, cytoplasm, and synapse during SD and was involved in the induction of sleep rebound, sleep-related gene expression, and GluA1 expression in synaptoneurosomes. Arc is essential for homeostatic sleep regulation at the behavioral and molecular levels, in addition to its role in synaptic plasticity.
Materials and Methods
See SI Appendix for detailed materials and methods.
Animals.
The animal study protocols approved by the Institutional Animal Care and Use Committee of University of Texas (UT) Southwestern Medical Center were carried out in strict accordance with the NIH Guide for the Care and Use of Laboratory Animals (54). For animal source and husbandry conditions, refer to SI Appendix, Materials and Methods.
Data Availability.
All data are made available in the paper.
Supplementary Material
Acknowledgments
We thank Dr. Kimberly Huber (UT Southwestern Medical Center) for providing Arc KO mouse breeding pairs. This work was supported by National Institute of Neurological Disorders and Stroke (R01NS103422-01), and Veterans Affairs (1I01BX002646-01A2) (to R.W.G.); Japan Society for the Promotion of Science (JSPS) Grants-in-Aid for Scientific Research (KAKENHI) 26220207 and 17H06095; Japan Science and Technology Agency (JST) Core Research for Evolutionary Science and Technology (CREST) Grant JPMJCR1655; Funding Program for World-Leading Innovative R&D on Science & Technology (FIRST) program from JSPS; the Uehara and Takeda Foundations; and the World Premier International Research Center Initiative program from Japan’s Ministry of Education, Culture, Sports, Science, and Technology (to M.Y.).
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1906840117/-/DCSupplemental.
References
- 1.Achermann P., Borbély A. A., Mathematical models of sleep regulation. Front. Biosci. 8, s683–s693 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Mignot E., Why we sleep: The temporal organization of recovery. PLoS Biol. 6, e106 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tononi G., Cirelli C., Sleep and the price of plasticity: From synaptic and cellular homeostasis to memory consolidation and integration. Neuron 81, 12–34 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Vivo L. et al., Ultrastructural evidence for synaptic scaling across the wake/sleep cycle. Science 355, 507–510 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vyazovskiy V. V., Cirelli C., Pfister-Genskow M., Faraguna U., Tononi G., Molecular and electrophysiological evidence for net synaptic potentiation in wake and depression in sleep. Nat. Neurosci. 11, 200–208 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Diering G. H. et al., Homer1a drives homeostatic scaling-down of excitatory synapses during sleep. Science 355, 511–515 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Z. et al., Quantitative phosphoproteomic analysis of the molecular substrates of sleep need. Nature 558, 435–439 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cirelli C., Tononi G., Differential expression of plasticity-related genes in waking and sleep and their regulation by the noradrenergic system. J. Neurosci. 20, 9187–9194 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cirelli C., Gutierrez C. M., Tononi G., Extensive and divergent effects of sleep and wakefulness on brain gene expression. Neuron 41, 35–43 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Terao A. et al., Gene expression in the rat brain during sleep deprivation and recovery sleep: An Affymetrix GeneChip study. Neuroscience 137, 593–605 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suzuki A., Sinton C. M., Greene R. W., Yanagisawa M., Behavioral and biochemical dissociation of arousal and homeostatic sleep need influenced by prior wakeful experience in mice. Proc. Natl. Acad. Sci. U.S.A. 110, 10288–10293 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taishi P. et al., Conditions that affect sleep alter the expression of molecules associated with synaptic plasticity. Am. J. Physiol. Regul. Integr. Comp. Physiol. 281, R839–R845 (2001). [DOI] [PubMed] [Google Scholar]
- 13.Maret S. et al., Homer1a is a core brain molecular correlate of sleep loss. Proc. Natl. Acad. Sci. U.S.A. 104, 20090–20095 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thompson C. L. et al., Molecular and anatomical signatures of sleep deprivation in the mouse brain. Front. Neurosci. 4, 165 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shepherd J. D., Bear M. F., New views of Arc, a master regulator of synaptic plasticity. Nat. Neurosci. 14, 279–284 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chowdhury S. et al., Arc/Arg3.1 interacts with the endocytic machinery to regulate AMPA receptor trafficking. Neuron 52, 445–459 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Link W. et al., Somatodendritic expression of an immediate early gene is regulated by synaptic activity. Proc. Natl. Acad. Sci. U.S.A. 92, 5734–5738 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Steward O., Wallace C. S., Lyford G. L., Worley P. F., Synaptic activation causes the mRNA for the IEG Arc to localize selectively near activated postsynaptic sites on dendrites. Neuron 21, 741–751 (1998). [DOI] [PubMed] [Google Scholar]
- 19.Lyford G. L. et al., Arc, a growth factor and activity-regulated gene, encodes a novel cytoskeleton-associated protein that is enriched in neuronal dendrites. Neuron 14, 433–445 (1995). [DOI] [PubMed] [Google Scholar]
- 20.Panja D. et al., Novel translational control in Arc-dependent long term potentiation consolidation in vivo. J. Biol. Chem. 284, 31498–31511 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peebles C. L. et al., Arc regulates spine morphology and maintains network stability in vivo. Proc. Natl. Acad. Sci. U.S.A. 107, 18173–18178 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang H. et al., Metabotropic glutamate receptors induce a form of LTP controlled by translation and Arc signaling in the Hippocampus. J. Neurosci. 36, 1723–1729 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okuno H. et al., Inverse synaptic tagging of inactive synapses via dynamic interaction of Arc/Arg3.1 with CaMKIIβ. Cell 149, 886–898 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shepherd J. D. et al., Arc/Arg3.1 mediates homeostatic synaptic scaling of AMPA receptors. Neuron 52, 475–484 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Korb E., Wilkinson C. L., Delgado R. N., Lovero K. L., Finkbeiner S., Arc in the nucleus regulates PML-dependent GluA1 transcription and homeostatic plasticity. Nat. Neurosci. 16, 874–883 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bloomer W. A. C., VanDongen H. M. A., VanDongen A. M. J., Activity-regulated cytoskeleton-associated protein Arc/Arg3.1 binds to spectrin and associates with nuclear promyelocytic leukemia (PML) bodies. Brain Res. 1153, 20–33 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Torok D., Ching R. W., Bazett-Jones D. P., PML nuclear bodies as sites of epigenetic regulation. Front. Biosci. 14, 1325–1336 (2009). [DOI] [PubMed] [Google Scholar]
- 28.Wee C. L. et al., Nuclear Arc interacts with the histone acetyltransferase Tip60 to modify H4K12 acetylation(1,2,3). eNeuro 1, ENEURO.0019-14.2014 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang K. H. et al., In vivo two-photon imaging reveals a role of arc in enhancing orientation specificity in visual cortex. Cell 126, 389–402 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Terao A., Greco M. A., Davis R. W., Heller H. C., Kilduff T. S., Region-specific changes in immediate early gene expression in response to sleep deprivation and recovery sleep in the mouse brain. Neuroscience 120, 1115–1124 (2003). [DOI] [PubMed] [Google Scholar]
- 31.Terao A. et al., Differential increase in the expression of heat shock protein family members during sleep deprivation and during sleep. Neuroscience 116, 187–200 (2003). [DOI] [PubMed] [Google Scholar]
- 32.Benington J. H., Heller H. C., Does the function of REM sleep concern non-REM sleep or waking? Prog. Neurobiol. 44, 433–449 (1994). [DOI] [PubMed] [Google Scholar]
- 33.Suchecki D., Tiba P. A., Machado R. B., REM sleep rebound as an adaptive response to stressful situations. Front. Neurol. 3, 41 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nollet M. et al., REM sleep’s unique associations with corticosterone regulation, apoptotic pathways, and behavior in chronic stress in mice. Proc. Natl. Acad. Sci. U.S.A. 116, 2733–2742 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim Y., Laposky A. D., Bergmann B. M., Turek F. W., Repeated sleep restriction in rats leads to homeostatic and allostatic responses during recovery sleep. Proc. Natl. Acad. Sci. U.S.A. 104, 10697–10702 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bjorness T. E. et al., An adenosine-mediated glial-neuronal circuit for homeostatic sleep. J. Neurosci. 36, 3709–3721 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Faraguna U., Vyazovskiy V. V., Nelson A. B., Tononi G., Cirelli C., A causal role for brain-derived neurotrophic factor in the homeostatic regulation of sleep. J. Neurosci. 28, 4088–4095 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bachmann V. et al., The BDNF Val66Met polymorphism modulates sleep intensity: EEG frequency- and state-specificity. Sleep (Basel) 35, 335–344 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ehlers M. D., Reinsertion or degradation of AMPA receptors determined by activity-dependent endocytic sorting. Neuron 28, 511–525 (2000). [DOI] [PubMed] [Google Scholar]
- 40.Oh M. C., Derkach V. A., Guire E. S., Soderling T. R., Extrasynaptic membrane trafficking regulated by GluR1 serine 845 phosphorylation primes AMPA receptors for long-term potentiation. J. Biol. Chem. 281, 752–758 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Fernández E. et al., Arc requires PSD95 for assembly into postsynaptic complexes involved with neural dysfunction and intelligence. Cell Rep. 21, 679–691 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dudek S. M., Fields R. D., Somatic action potentials are sufficient for late-phase LTP-related cell signaling. Proc. Natl. Acad. Sci. U.S.A. 99, 3962–3967 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luo S., Baumeister P., Yang S., Abcouwer S. F., Lee A. S., Induction of Grp78/BiP by translational block: Activation of the Grp78 promoter by ATF4 through and upstream ATF/CRE site independent of the endoplasmic reticulum stress elements. J. Biol. Chem. 278, 37375–37385 (2003). [DOI] [PubMed] [Google Scholar]
- 44.Flavell S. W., Greenberg M. E., Signaling mechanisms linking neuronal activity to gene expression and plasticity of the nervous system. Annu. Rev. Neurosci. 31, 563–590 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang G.-C. et al., In vivo regulation of Homer1a expression in the striatum by cocaine. Mol. Pharmacol. 71, 1148–1158 (2007). [DOI] [PubMed] [Google Scholar]
- 46.Chen Y. et al., Activity-induced Nr4a1 regulates spine density and distribution pattern of excitatory synapses in pyramidal neurons. Neuron 83, 431–443 (2014). [DOI] [PubMed] [Google Scholar]
- 47.Vazdarjanova A. et al., Spatial exploration induces ARC, a plasticity-related immediate-early gene, only in calcium/calmodulin-dependent protein kinase II-positive principal excitatory and inhibitory neurons of the rat forebrain. J. Comp. Neurol. 498, 317–329 (2006). [DOI] [PubMed] [Google Scholar]
- 48.Ying S. W. et al., Brain-derived neurotrophic factor induces long-term potentiation in intact adult hippocampus: Requirement for ERK activation coupled to CREB and upregulation of Arc synthesis. J. Neurosci. 22, 1532–1540 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li L., Carter J., Gao X., Whitehead J., Tourtellotte W. G., The neuroplasticity-associated arc gene is a direct transcriptional target of early growth response (Egr) transcription factors. Mol. Cell. Biol. 25, 10286–10300 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ebert D. H. et al., Activity-dependent phosphorylation of MeCP2 threonine 308 regulates interaction with NCoR. Nature 499, 341–345 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hengen K. B., Torrado Pacheco A., McGregor J. N., Van Hooser S. D., Turrigiano G. G., Neuronal firing rate homeostasis is inhibited by sleep and promoted by wake. Cell 165, 180–191 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nikolaienko O., Patil S., Eriksen M. S., Bramham C. R., Arc protein: A flexible hub for synaptic plasticity and cognition. Semin. Cell Dev. Biol. 77, 33–42 (2018). [DOI] [PubMed] [Google Scholar]
- 53.Honjoh S. et al., Higher arc nucleus-to-cytoplasm ratio during sleep in the superficial layers of the mouse cortex. Front. Neural Circuits 11, 60 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.National Research Council , Guide for the Care and Use of Laboratory Animals, (National Academies Press, Washington, DC, ed. 8, 2011). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are made available in the paper.