Significance
Bacteria have evolved intricate regulatory networks to coordinate their metabolism with internal and external signals of their status. The regulatory phosphotransferase systems (PTSs) constitute a key part of these intricate circuits, with their signal transduction cascades participating in multiple regulatory functions. Although two major systems have been described as being involved in regulating carbon and nitrogen pools, there is very little information on their physiological role in vivo under real-time conditions. In this work we demonstrate the role of PTS as an integrated system, widely conserved in proteobacteria, acting as a complex biological sensor-actuator device enabling bacterial cells to posttranslationally alter bacterial physiology and balance carbon and nitrogen availability.
Keywords: bacterial metabolism, regulatory network, nitrogen, plant–host interactions
Abstract
The nitrogen-related phosphotransferase system (PTSNtr) of Rhizobium leguminosarum bv. viciae 3841 transfers phosphate from PEP via PtsP and NPr to two output regulators, ManX and PtsN. ManX controls central carbon metabolism via the tricarboxylic acid (TCA) cycle, while PtsN controls nitrogen uptake, exopolysaccharide production, and potassium homeostasis, each of which is critical for cellular adaptation and survival. Cellular nitrogen status modulates phosphorylation when glutamine, an abundant amino acid when nitrogen is available, binds to the GAF sensory domain of PtsP, preventing PtsP phosphorylation and subsequent modification of ManX and PtsN. Under nitrogen-rich, carbon-limiting conditions, unphosphorylated ManX stimulates the TCA cycle and carbon oxidation, while unphosphorylated PtsN stimulates potassium uptake. The effects are reversed with the phosphorylation of ManX and PtsN, occurring under nitrogen-limiting, carbon-rich conditions; phosphorylated PtsN triggers uptake and nitrogen metabolism, the TCA cycle and carbon oxidation are decreased, while carbon-storage polymers such as surface polysaccharide are increased. Deleting the GAF domain from PtsP makes cells “blind” to the cellular nitrogen status. PTSNtr constitutes a switch through which carbon and nitrogen metabolism are rapidly, and reversibly, regulated by protein:protein interactions. PTSNtr is widely conserved in proteobacteria, highlighting its global importance.
Maintaining proper intracellular carbon and nitrogen levels is crucial in cell physiology to maximize nutrient utilization and cell growth. Among bacterial regulatory circuits, the well-conserved and ubiquitous PII family proteins (i.e., glnB, glnK, and nifI products) play a major role in coordinating nitrogen metabolism by signal transduction via posttranslational modifications. Among other targets, GlnK controls the activity of the ammonium transporter AmtB; GlnB regulates the expression and activity of glutamine synthetase as well as the NtrBC transcriptional cascade regulating the Ntr operon, and NifI interacts with nitrogenase, posttranslationally inactivating this enzyme in response to ammonium (1). Upon nitrogen starvation NtrC also up-regulates the gene encoding RelA, which synthesizes guanosine tetraphosphate (ppGpp), the effector molecule of the bacterial stringent response (2). Besides transcriptional and posttranslational regulation, small RNAs (sRNAs) have also been identified as a third level of regulation for fine tuning the nitrogen network (3).
In proteobacteria, PII proteins are uridylylated under nitrogen limitation and rapidly deuridylylated under N-excess (4). This switch is mediated by GlnD, an uridylyltransferase/uridylyl-removing enzyme that senses glutamine/α-ketoglutarate (the major metabolic signals reflecting the nitrogen and carbon status of the cell) and ATP/ADP ratios (reflecting the cellular energy status) (4, 5). α-Ketoglutarate is a key intermediate of the tricarboxylic acid (TCA) cycle and the major carbon skeleton for nitrogen assimilation (4, 6). As the α-ketoglutarate pool responds within minutes to a change in extracellular nitrogen availability (7), bacteria have therefore evolved a variety of regulatory mechanisms to sense this effector molecule. The accumulation of α-ketoglutarate under nitrogen (N)-limiting conditions also inhibits carbohydrate uptake in Escherichia coli by binding to the first component of the carbohydrate-phosphoenolpyruvate phosphotransferase system (PTS) (8).
In gram-negative bacteria there are two common PTS variants: the carbohydrate-PTS, which coordinates carbohydrate transport (9, 10), and the nitrogen-related PTS (PTSNtr), a signal transduction cascade with various regulatory roles (11). Most proteobacteria implement an integrated PTS comprising PTSNtr, encoded by the genes ptsP (EINtr), ptsO (NPr), and ptsN (EIIANtr), and an EIIA component remaining from the carbohydrate-PTS, manX (EIIAMan). The importance of PTSNtr is suggested by its wide conservation in α-, β-, and γ-proteobacteria (12, 13) and, because ptsN is often found contiguous with rpoN, which codes for the nitrogen-responsive RNA polymerase σ54 sigma factor (11, 12). PtsP contains a GAF domain homologous to the sensory domain of the NifA protein, which binds small molecules at its N terminus (14). In E. coli, glutamine and α-ketoglutarate control phosphorylation of PTSNtr through allosteric binding to the GAF domain of PtsP. PtsP acquires high-energy phosphate from phosphoenolpyruvate (PEP), which then phosphorylates the small carrier protein NPr (or HPr in the carbohydrate-PTS) on a conserved histidine residue (15). In E. coli and Rhizobium leguminosarum the GAF domain was reported to be dispensable for PEP-dependent PtsP autophosphorylation (16, 17).
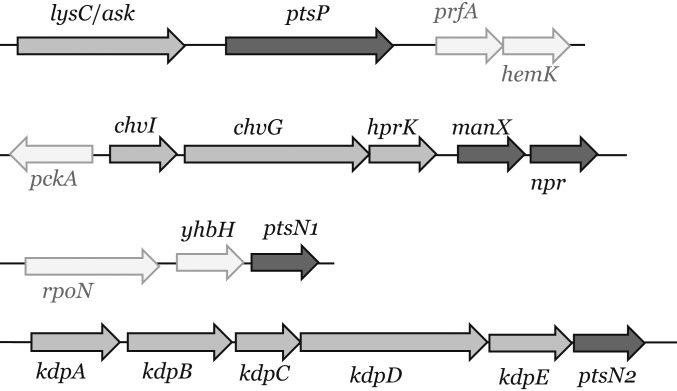
Extensive previous analyses have shown that carbohydrate-PTS transport systems are the exception rather than the rule in bacteria. Most well-studied α- and β-proteobacteria, Chlamydiae and Planctomycetes, do not contain the PTS permease proteins (EIIB and EIIC) responsible for sugar translocation (9, 13, 18, 19), although there are exceptions, such as E. coli. The ultimate acceptors of Pi from NPr∼P are therefore ManX and PtsN (EIIA components), with both proteins having an exclusively regulatory role. It is also striking that α-proteobacteria mainly contain PTSNtr components (18) organized in three operons (Fig. 1). In cluster one, aspartokinase (lysC or ask) is located upstream of ptsP, while in the second cluster, npr is located downstream of manX, encoding the EIIAMan homolog. Further upstream are hprK, the two-component regulatory system chvI/chvG, and phosphoenolpyruvate carboxykinase (pckA). HPrK is a kinase/phosphatase common in gram-positive bacteria, but absent in many gram-negative bacteria, including E. coli (12). It is a major regulator of carbon catabolite repression in firmicutes, where PTSNtr genes are absent (20). Interestingly, proteobacteria of the α-subdivision possess a truncated HPrK missing the N-terminal domain (about 130 amino acids) (19, 21). ptsN is present in the third gene cluster downstream of yhbH, which codes for a putative σ54-modulating protein that can associate with ribosomes. Intriguingly, in 10% of the genomes (most of them rhizobial) there is a second copy of ptsN, always located downstream of the high-affinity potassium (K+) transport system kdpABCDE.
Fig. 1.
Schematic genetic organization of the PTS components in R. leguminosarum bv. viciae 3841. In dark, PTS components; in gray, relevant genes interacting with PTS; and in white, other neighboring genes.
PTS systems, via their EIIA components, exert their regulatory role in numerous bacteria at the protein:protein level (9), coordinating essential processes for cell survival such as K+ homeostasis and phosphate starvation, ATP-binding cassette (ABC) transporters, or central metabolic enzymes, including pyruvate dehydrogenase (PDH) and α-ketoglutarate dehydrogenase (α-KGDH) (22–26). Although EIIA proteins can alter transcription, their regulation is usually exerted by binding to histidine kinases such as KdpD or PhoR (24, 25). The sheer range of regulatory roles assigned to PTS that have been studied independently in different bacteria has made it difficult to understand how phosphorylation coordinated by NPr acts at the global level to control responses in bacterial cells. Therefore, we characterized the entire regulatory network of the integrated PTS in Rhizobium leguminosarum bv. viciae 3841 (Rlv3841) which, as a model α-proteobacterium, lacks the carbohydrate-PTS and sugar translocation components (EIIB and EIIC). Rlv3841 also has a second ptsN copy named ptsN2 and is located on plasmid pRL11 (pRL110376) (23) (Fig. 1). This highly conserved operon arrangement highlights the relevance of the PTSNtr branch in α-proteobacteria and emphasizes the importance of using a model bacterium to unravel the mechanism of action of PTS. We show in this study that this integrated PTS system is another major posttranscriptional regulator, with its two output regulators, PtsN and ManX, acting reciprocally to integrate carbon and nitrogen metabolism signals at the cellular level.
Results and Discussion
PtsN Phosphorylation Is Essential for ABC Transport Activation.
The PTSNtr phosphorelay starts with PEP as the high-energy phosphate donor to PtsP on His367. This phosphate is then transferred to NPr on His17 and PtsN on His66 (15). PtsN (EIIANtr) mediates the effects of PTS in Rlv3841 on the activation of ABC transporters and K+ uptake (17, 23). Mature bacteroids become symbiotic auxotrophs and their host plant must provide branched-chain amino acids to support their development and persistence (27). Active amino acid transport is therefore essential for nitrogen fixation in pea nodules; mutations of the main ABC-type broad-specificity amino acid uptake systems (Aap and Bra) led to severely N-starved plants (27, 28). In both Rlv3841 and E. coli, nonphosphorylated PtsN interacts with KdpD, the sensor kinase that activates the transcription of the high-affinity K+ transporter KdpABC via the response regulator KdpE (23, 24). However, EI (PtsP) phosphorylation on His367 and NPr on His17 are required for ABC transport activation, suggesting that PtsN∼P is the active species (17).
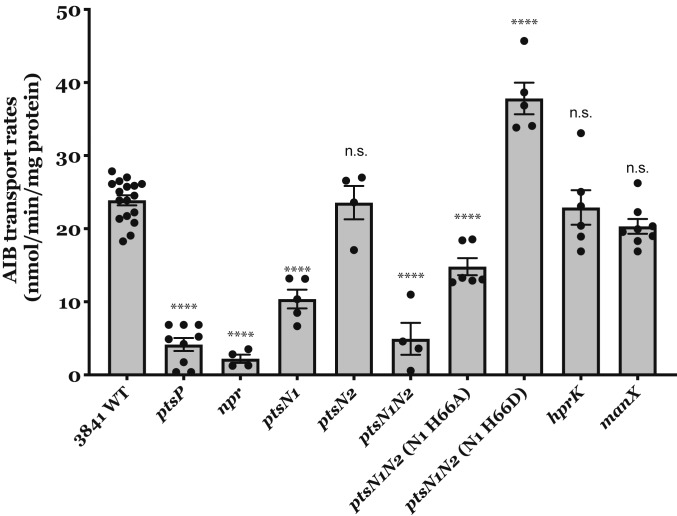
By examining the complete PTS network, we now demonstrate that a manX mutant is not significantly affected in transport, while confirming that PtsN, as well as PtsP and NPr (required for phosphoryl-group transfer), are essential for transport activation (Fig. 2). Amino acid transport was measured with α-aminoisobutyric acid (AIB) because, like glutamine, it is transported exclusively by Aap and Bra, but cannot be metabolized (29). As previously seen, the ptsN1 mutant (LMB271) had a large reduction in transport, whereas the ptsN2 mutant (RU4193) maintained wild-type levels (23). The essentiality of the PTSNtr branch in transport activation was also shown for glucose (SI Appendix, Fig. S1). These results agree with our initial work on PTS in Rlv3841, where we already measured AIB transport (taken up with very similar kinetics to glutamate), γ‐amino butyric acid (GABA), δ‐aminolevulinic acid (ALA), glucose, and myo‐inositol (23). These substrates, taken up by ABC transporters, were all greatly reduced in the ptsP mutant compared with wild-type Rlv3841, whereas this was not the case for succinate transport, driven by the proton-coupled transporter DctA.
Fig. 2.
Transport rates by PTS mutants. Standard rates were obtained from cultures grown on UMS with 10 mM glucose and 10 mM NH4Cl. Rlv3841 wild type, ptsP (PtsP107), npr (AA031), ptsN1 (LMB271), ptsN2 (RU4391), ptsN1N2 (AA047), ptsN1N2 (N1 H66A, OPS1102), ptsN1N2 (N1 H66D, OPS1104), hprK (AA081), and manX (LMB692). All rates are expressed in nmol min−1 mg protein−1. Data are averages (±SEM) from at least three independent cultures analyzed by one-way ANOVA with Dunnett’s posttest for multiple comparisons (****) P < 0.0001 and n.s., not significant.
To confirm the role of phosphorylated PtsN1 in activating ABC transporters, we replaced the chromosomal ptsN1::ΩSpec insertion with a permanent phosphorylation (H66D, strain OPS1104) or nonphosphorylation mimic (H66A, strain OPS1102) in the double ptsN1N2 mutant (AA047). The PtsN1 H66D phosphomimic (OPS1104) significantly increased AIB transport rates above the level observed with the native protein. While H66A shows a partial activation, it does not restore transport rates to wild-type level (Fig. 2). This is consistent with PtsN1∼P being required for full activation of a wide range of ATP‐dependent ABC transport systems in R. leguminosarum.
Exopolysaccharide Secretion Is Regulated by PtsN via Interaction with the ChvI/ChvG System.
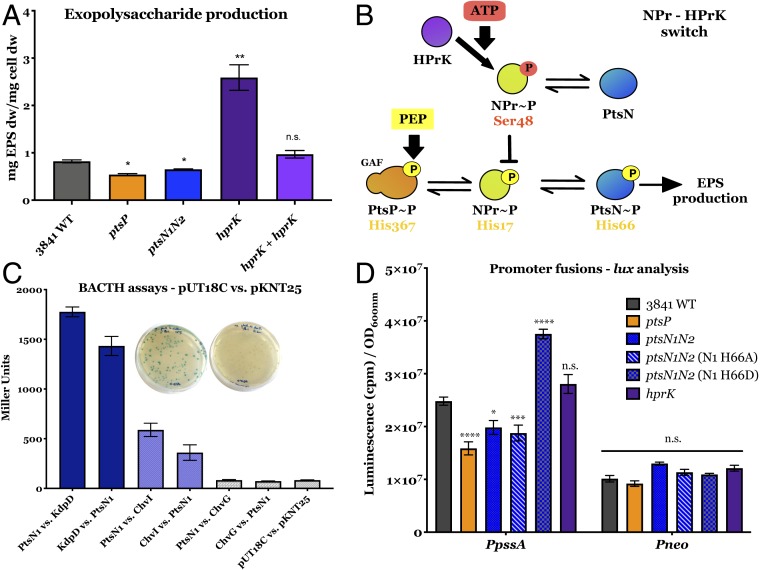
Extracellular polysaccharides are carbon-based surface polymers indispensable for the invasion of a large number of host plants which form indeterminate nodules (30). We had previously shown that a functional PTS is required for normal production of exopolysaccharide (EPS) and therefore a mucoid surface phenotype in R. leguminosarum that produces a high-molecular-weight acidic EPS (17, 23). When grown on agar plates, we now show that mutants that disrupt the phosphorylation cascade of PTSNtr (ptsP, npr, or ptsN1N2) have a dry surface compared to wild type (SI Appendix, Fig. S2). Total EPS was quantified in Rlv3841 and several PTS mutants (Fig. 3A), showing a reduction of 35% in PtsP107 (ptsP::Tn5) and 21% in AA047 (ptsN1::ΩSpec; ΔptsN2). Since ptsN1N2 and mutants in the phosphotransfer cascade have a dry surface, PtsN∼P is likely to be required for EPS production (Fig. 3B). Plasmid complementation of AA047 with either wild-type ptsN or ptsN H66A (permanent nonphosphorylated version) showed that only complementation with ptsN gives a mucoid surface phenotype (SI Appendix, Fig. S2), indicating that PtsN∼P is the likely species that activates both ABC transport and EPS production in Rlv3841.
Fig. 3.
EPS secretion and regulation. (A) EPS production measurements for Rlv3841 wild type, ptsP (PtsP107), ptsN1N2 (AA047), hprK (AA081), and hprK + hprK (AA088 complemented strain). Data are averages (±SEM) from at least four independent cultures analyzed by one-way ANOVA with Dunnett’s posttest for multiple comparisons (*) P < 0.01, (**) P < 0.01 and n.s., not significant. All rates are expressed in mg EPS dry weight mg−1 cell dry weight. (B) Npr-HPrK switch model. PTSNtr components are phosphorylated on histidine residues, initially (PtsP) from PEP as indicated, with PtsN∼P inducing EPS production. Conversely, HPrK phosphorylates NPr on Ser48 from ATP, blocking subsequent phosphorylation of PtsN. (C) BACTH assays of the interaction of PtsN with the ChvI/ChvG system. Interactions expressed in Miller units of LacZ activity. Plates show blue colonies indicating a positive interaction (PtsN1-ChvI) and white colonies indicating a negative interaction (PtsN1-ChvG). PtsN1-KdpD and KdpD-PtsN1 are positive controls. Data are averages (±SEM) from at least three independent cultures. (D) Quantification of promoter activity for PpssA compared to the constitutive promoter nptII (Pneo) in Rlv3841 wild type, ptsP (PtsP107), ptsN1N2 (AA047), ptsN1N2 (N1 H66A, OPS1102), ptsN1N2 (N1 H66D, OPS1104), and hprK (AA081). All rates are expressed in counts per minute (cpm). Data are averages (±SEM) from at least five independent cultures. Statistical analyses are indicated following the level of significance, with (****) as P value <0.0001, (***) as P value <0.001 and n.s., not significant.
The gene encoding NPr is located close to those that encode a two-component global regulatory system, ChvI/ChvG, which modifies the cell surface (Fig. 1); the homologous system in Sinorhizobium meliloti, ChvI/ExoS, controls succinoglycan and galactoglucan production (31, 32). R. leguminosarum synthesizes only an acidic EPS and a chvG mutation was shown to decrease its production, among other pleiotropic effects (33). We investigated the potential link between PTS and ChvI/ChvG using bacterial two-hybrid (BACTH) analysis. PtsN1 and the response regulator ChvI interacted strongly (Fig. 3C), while PtsN1 and the membrane sensor kinase ChvG did not. This interaction suggests that the effect of PTS on EPS is mediated by ChvI, which acts at the transcriptional level (34). A plasmid-borne lux fusion to the promoter of pssA, the gene responsible for the polymerization of EPS (30), was conjugated into the ptsP mutant (dry phenotype) as well as in the double mutant ptsN1N2 and its derivatives with the different versions of ptsN1 (H66A, dry; H66D, mucoid). Increased expression of pssA in the ptsN1 H66D strain and decreased expression in ptsN1N2 and ptsN1 H66A (Fig. 3D) is consistent with PtsN∼P stimulating EPS production via the interaction with the ChvI regulator.
In summary, while the first defined role assigned to dephosphorylated PtsN was to control K+ homeostasis by direct binding to KdpD in R. leguminosarum (23), as it does in E. coli (23, 24), we have now confirmed that PtsN∼P regulates Aap and Bra and most likely a wide range of ABC transporters. We have also demonstrated the direct interaction of PtsN with ChvI (Fig. 3C), suggesting that the surface phenotype of PTS mutants is mediated through this global regulator of EPS production. The chvIG operon encodes a two-component regulatory system involved in virulence or symbiosis [BvrR/BvrS in Brucella spp (35), ChvI/ChvG in Agrobacterium tumefaciens (36), and ChvI/ExoS in S. meliloti (32)]. Cross-talk between this system and PTSNtr had been proposed in Brucella melitensis, suggesting that PTS communicates the metabolic state of the cell to the virulence gene virB, by phosphorylating or interacting with the BvrR/S system (26). In rhizobia, this two-component regulatory system has already been described as a master transcriptional regulator of EPS (31, 32, 37). Indeed, based on the genetic proximity of chvGI to hprK and the opposite phenotypes between these mutants in terms of growth (chvG and chvI mutants are unable to grow on complex media, while the hprK mutant is able to) and EPS production (chvG and chvI show reduced EPS, hprK increased EPS), PTS was suggested to play a role in the dephosphorylation of ExoS/ChvI in S. meliloti (32). This suggestion fits our model, where HPrK phosphorylation of Ser48 on NPr reduces phosphorylation on its His17, resulting in reduced EPS (Fig. 3B). The BACTH data point toward PtsN∼P binding to ChvI and stimulating EPS production at the transcriptional level, a hypothesis reinforced after showing that pssA is transcriptionally up-regulated when PtsN1 is present as a permanent phosphomimic (strain OPS1104, PtsN1 H66D, Fig. 3D). This differential role of PtsN driven by its phosphorylation status suggests that a phosphorylation switch on PtsN might operate to activate one system while inhibiting another. Given that Rlv3841, along with most α-proteobacteria, has a second EIIA component (ManX) as part of the integrated PtsP-NPr phosphorylation cascade, there may be a more comprehensive switch operating to control a wide range of ABC transporters and metabolism in bacteria.
Expression of manX Is Independent of the chvI Promoter.
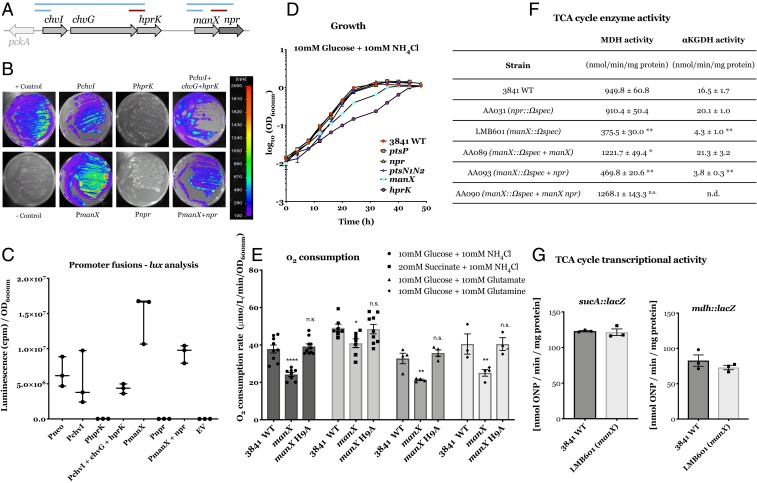
To investigate the role of ManX in R. leguminosarum, we first analyzed its operon structure. manX and npr genes are colocated with chvI, chvG, and hprK, potentially as part of an operon (Fig. 1). Lux-fusion analysis showed that manX and npr are transcribed together, as are chvI, chvG, and hprK (Fig. 4 A–C). The carbohydrate-PTS system is known to be regulated by changes in carbon source in other bacterial species, where it controls their uptake (38). While the manX promoter showed small but significant differences in expression in minimal medium after 22 h but not after 18 h of growth, the main difference was the increased expression in rich medium (tryptone-yeast [TY]) for both the chvI and manX promoters (SI Appendix, Fig. S3). We have therefore confirmed that manX and npr are transcribed from a single promoter in front of manX, independently of the chvI-chvG-hprK operon, with this genomic region conserved across different host-invading bacteria and characteristic of the Rhizobiales (39).
Fig. 4.
Promoter activity analysis, growth phenotype, and TCA enzymatic activity and regulation. (A) Genetic organization of chvI/chvG/hprk and manX/npr operons. In blue, lines indicate regions with promoter activity and in red, those without. (B) Qualitative luminescence imaging showing the activity of the six different potential promoter regions fused to the luxCDABE cassette: PchvI (pOPS0298) encompasses the region immediately upstream of the chvI operon, and PchvI+chvG+hprK (pOPS0313) contains the same region with the chvI, chvG, and hprK genes included. PmanX (pOPS0606) and PmanX+npr (pOPS0296) contain the putative promoter-encoding region in front of manX and this region with the entire manX operon included, respectively. Regions with potential promoters for npr (Pnpr) (pOPS0605) and hprK (PhprK) (pOPS0603) were also included for analysis. (Scale bar on the Right, in counts per minute [cpm].) (C) Quantification of promoter activity from the regions selected for testing. The constitutive Pneo promoter was used as a positive control on the plasmid pIJ11282 and the empty vector pJI11268 (EV) as a negative control. All rates are expressed in cpm. Data are averages (±SEM) from three independent cultures. (D) Growth curves on UMS supplemented with 10 mM glucose + 10 mM NH4Cl for strains wild-type Rlv3841, ptsP (ptsP107), npr (AA031), ptsN1N2 (AA047), manX (LMB692), and hprK (AA081). (E) O2 consumption assay on UMS supplemented with 10 mM glucose + 10 mM NH4Cl, 20 mM succinate + 10 mM NH4Cl, 10 mM glutamate + 10 mM glucose, or 10 mM glutamine + 10 mM glucose. Strains tested are ptsP (ptsP107), npr (AA031), ptsN1N2 (AA047), manX (LMB692), hprK (AA081), and manX H9A (OPS1012). O2 consumption rates are expressed in μmol L−1 min−1 OD600nm-1. Data are averages (±SEM) from at least three independent cultures; two-way ANOVA with Dunnett's posttest for multiple comparisons (*) P < 0.05 (**) P < 0.01 (****) P < 0.0001 and n.s., not significant. (F) MDH and α-KGDH, alpha-ketoglutarate dehydrogenase activities for Rlv3841 wild-type, npr (AA031), manX (LMB601), and complemented strains AA089, AA093, and AA090. Values expressed as nmol−1 min−1 mg protein. Data are averages (±SEM) from three independent cultures. Statistical analyses are indicated following the level of significance, (*) P value <0.01, (**) P value <0.001, and n.s., not significant; n.d., not determined. (G) Transcriptional activity assays in Rlv3841 (black) and LMB601 (manX::ΩSpec, gray) by using transcriptional LacZ fusions in the mdh-sucCDAB operon located on two different cosmids, pRU3068 sucA::lacZ and pRU3070 mdh::lacZ. Data are averages (±SEM) from three independent cultures. Values expressed as nmol ONP min−1 mg protein−1. No significant differences were observed.
ManX Is Required for the Activation of the TCA Cycle.
S. meliloti manX mutants have reduced growth on several carbon sources and altered succinate-mediated catabolite repression (40). Previous work in other systems had already reported direct binding of PTS proteins to enzymatic components (26, 41). This suggested that ManX regulation might be due to direct binding to TCA cycle enzymes. To investigate ManX in Rlv3841, a markerless in-frame manX mutant (LMB692) was generated to prevent polar effects on npr. LMB692 showed reduced growth rates in all of the conditions tested (Fig. 4D and SI Appendix, Table S1), whereas this was not the case for mutants in components needed for phosphorylation (ptsP, npr), nor ptsN1N2. This defect in growth was further investigated by measuring oxygen (O2) consumption, which indicates reductant generated by the TCA cycle feeding directly into the respiratory chain in free-living cultures of the obligate aerobe R. leguminosarum. The manX mutant had a reduced O2 consumption rate, regardless of the carbon or nitrogen source tested (Fig. 4E).
The contiguous arrangement of manX and npr in Rlv3841 suggested the possibility of polar effects between the genes. Although LMB692 is a markerless in-frame mutant, we tested for polarity effects by introducing manX and npr genes individually in LMB692 (manX mutant) and AA031 (npr mutant) under their native promoters on the low-copy plasmid pRK415. AA031 complemented by npr recovered the mucoid colony morphology of the wild-type strain. Likewise, complementation of LMB692 with manX, but not npr, restored growth and O2 consumption to wild-type levels (SI Appendix, Fig. S4). Thus, as expected, the markerless mutation in manX is not polar on npr, indicating that the effect on O2 consumption is exclusively due to the absence of ManX.
It had already been demonstrated by a yeast two-hybrid approach in the α-proteobacterium B. melitensis that ManX interacts with SucA, the E1 component of α-KGDH (26). We therefore analyzed the two halves of the TCA cycle by measuring α-KGDH and malate dehydrogenase (MDH) enzyme activities in Rlv3841, AA031 (npr::ΩSpec), and LMB601 (manX::ΩSpec). Both enzyme activities were reduced by 70 and 60%, respectively, in the manX mutant (Fig. 4F), whereas in the npr mutant, α-KGDH and MDH activities were similar to wild-type levels. LMB601 (manX::ΩSpec) was complemented for enzyme activity by plasmid-borne manX, but not npr. Thus, ManX alone is required for the full activation of these two TCA cycle enzymes. The reduction of TCA cycle activity, as measured by O2 consumption (Fig. 4E) and enzyme activity (Fig. 4F), explains the high generation times of the manX mutant (SI Appendix, Table S1).
The structural genes for MDH (mdh) and α-KGDH (sucCDAB) are organized in the same cluster of genes in R. leguminosarum (42). Transcriptional down-regulation could therefore explain a concerted reduction in activity of α-KGDH and MDH. This cluster of genes has a promoter in front of mdh, suggesting an operon, but there is also a second promoter in front of sucA. We compared their transcriptional activity in LMB601 (manX::ΩSpec) and wild-type strains using transcriptional Tn5-lacZ fusions located in mdh or sucA in two different cosmids containing the entire mdh-sucABCD cluster, pRU3070 Pmdh::lacZ and pRU3068 PsucA::lacZ (42). Both LMB601 and Rlv3841 showed identical levels of lacZ expression for mdh and sucA fusions (Fig. 4G). Therefore, down-regulation of mdh-sucCDAB transcription does not account for the reduced enzyme activities, suggesting that this effect is due to posttranscriptional regulation and agrees with the demonstrated protein:protein interaction between ManX and SucA in B. melitensis (26). We therefore suggest that ManX interacts with these TCA cycle enzymes directly or indirectly to regulate their activity in Rlv3841. There are other established protein:protein interactions of EIIA proteins with enzymes. In Salmonella typhimurium phosphorylated PtsN tightly binds to GlmS (D-glucosamine-6-phosphate synthase) inhibiting the enzyme (43), while in Pseudomonas putida unphosphorylated PtsN inhibits pyruvate dehydrogenase (41). In R. leguminosarum the unphosphorylatable manX mutant (H10A) shows growth at wild-type levels (Fig. 4D) and activates respiration associated with the TCA cycle regardless of the carbon or nitrogen source (Fig. 4E), indicating that nonphosphorylated ManX acts on the TCA cycle.
The Phosphorylation Switch through NPr Controls ManX and PtsN Activity.
It was reported from in vitro experiments that B. melitensis NPr also phosphorylates ManX on His9, similarly to PtsN (26). An hprK homolog upstream of manX and npr increases the complexity of PTS-mediated regulation by interfering in this phosphoryl transfer pathway (Figs. 1 and 3B). HPr proteins like NPr have been shown to be phosphorylated on two different residues: His17 and Ser48 (44). In S. meliloti and B. melitensis, phosphorylation of NPr on the serine residue by HPrK slows down or prevents phosphorylation on the histidine by PtsP (26, 45). Thus, histidine phosphorylation of ManX and PtsN should increase in an hprK mutant. Accordingly, we hypothesized that NPr phosphorylation on Ser48 by HPrK would raise the amount of dephospho-EIIAs (ManX and PtsN), while an hprK mutation would show hyperphosphorylation of EIIAs (PtsN and ManX). Insertion of an ΩSpec marker cassette into hprK (strain AA081) led to colonies that were hypermucoid and grew extremely slowly on TY and universal minimal salts (UMS) media. As observed in S. meliloti (45), AA081 (hprK::ΩSpec) showed a large increase in EPS, which was restored to wild-type levels in the complemented strain AA088 (hprK::ΩSpec + hprK) (Fig. 3A and SI Appendix, Fig. S2). The hypermucoid phenotype is consistent with PtsN∼P promoting EPS synthesis (Fig. 3B).
In Ralstonia eutropha the elevated histidine-phosphorylated NPr formed in the hprK mutant negatively affects growth of this organism (46). Consistent with this, an hprK mutant (strain AA081), unable to prevent the phosphorylation of ManX or PtsN, has the most defective growth of all of the mutants tested, with the longest mean generation time (SI Appendix, Table S1 and Fig. 4D) and excess EPS production. These phenotypes are as expected for hyperphosphorylated PtsN, i.e., a wide range of ABC transporters fully active and EPS production up-regulated, while the absence of dephosphorylated ManX suppresses TCA cycle activity. As in Rlv3841, deletion of hprK in S. meliloti also causes a strong growth defect on minimal medium and up-regulation of EPS production (45). Absent in gram-negative enteric bacteria, the HPrK/P regulator is encoded in the genomes of many proteobacteria (19). However, it has a shorter sequence than its counterpart in gram-positive bacteria, lacking about 130 amino acids including an N-terminal region important for the phosphatase activity of HPrK/P (21, 47). This indicates that HPrK in proteobacteria might not be able to efficiently dephosphorylate P∼Ser∼NPr (26, 45), highlighting the importance of the phosphorylation switch coordinated by NPr in these organisms. Indeed, HPrK was shown to be essential in R. eutropha due to the PTS phosphorelay imbalance caused by an elevated amount of P∼His∼NPr (46). We note that protein SixA, a well-conserved protein found in proteobacteria, actinobacteria, and cyanobacteria (48), has recently been reported as a phosphohistidine phosphatase in E. coli acting on NPr (49). It might play an additional role in dephosphorylating P∼His∼NPr.
As seen above, the lack of effect of ptsP and npr mutations on the TCA cycle enzyme activity compared with the strong reduction observed in the manX mutant suggests that ManX in its nonphosphorylated form is needed for full TCA activity. Accordingly, the hyperproduction of EPS and activation of a wide range of ABC transporters (due to PtsN∼P) combined with a lower TCA cycle enzyme activity (elevated ManX∼P depletes nonphosphorylated ManX) would explain why the hprK mutant (AA081) shows the highest generation times of all strains tested (SI Appendix, Table S1). Thus, ManX should activate the TCA cycle in its nonphosphorylated version and, indeed, a genomic manX H9A mutant (OPS1012) restored the growth rate under all conditions tested (SI Appendix, Table S1). O2 consumption in minimal medium with different carbon and nitrogen sources was restored to wild-type levels in the manX H9A strain (Fig. 4E), confirming that nonphosphorylated ManX activates the TCA cycle in Rlv3841 as measured by carbon oxidation. Therefore, while PtsN∼P activates uptake and carbon storage into the EPS layer, carbon catabolism via the TCA cycle will be down-regulated by reduced unphosphorylated ManX (depleted by the formation of ManX∼P). Consequently, PTSNtr acts as a central switch coordinating metabolism of both carbon and nitrogen.
In Vivo Experiments Show PTSNtr Is Regulated by Nitrogen Availability.
The complete carbohydrate-PTS senses the presence or absence of carbohydrates in the medium related to the intracellular PEP/pyruvate ratio (50), increasing the ratio of nonphosphorylated/phosphorylated PTS components with the intake of carbohydrates (38). In the case of the PTSNtr system, the N-terminal GAF sensory domain modulates its autophosphorylation (16). In E. coli PtsP is regulated by its GAF domain binding glutamine and α-ketoglutarate (16), whereas in S. meliloti, only glutamine binds the GAF domain of PtsP in vitro and inhibits its phosphorylation (51). In S. typhimurium, PtsN was also reported to be dephosphorylated in response to nitrogen excess and rapidly degraded by Lon protease upon depletion of cellular amino sugars (43). In Rlv3841, PtsP lacking its GAF domain activates ABC transport (17) and complements the dry surface phenotype of PtsP107 (ptsP mutant), suggesting that the ligand binding to GAF negatively regulates PTS phosphorylation. Overall, while in vitro protein work and mutant complementation studies suggest a role of PtsP in nitrogen signaling (8, 16, 51, 52), in vivo physiological evidence has been lacking. To develop definitive in vivo assays, wild-type cells were grown in minimal medium (UMS) supplemented with either succinate (20 mM) or glucose (10 mM) as carbon sources, and either 10 mM NH4Cl or 10 mM glutamine for N-rich conditions, or 10 mM glutamate and 0.5 to 1 mM NH4Cl as N-limiting conditions. While ammonia either diffuses freely across the membrane or is incorporated via the AmtB permease, glutamate and glutamine are transported actively into cells by the ABC systems Aap and Bra (53).
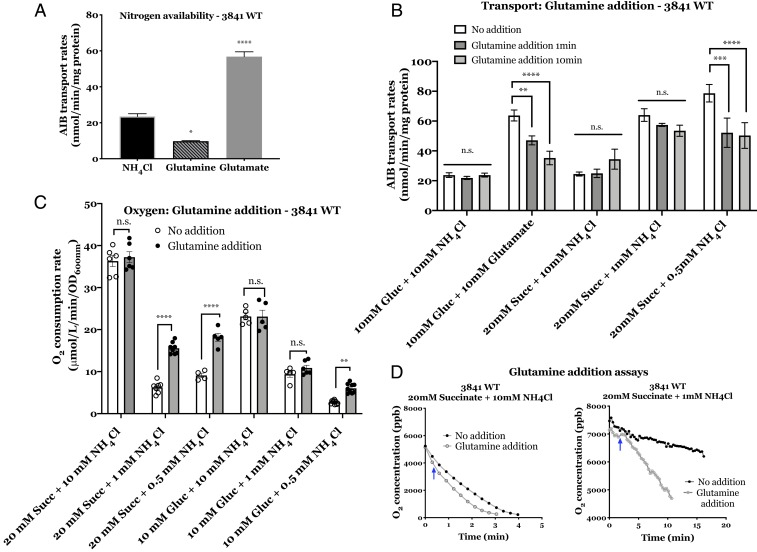
As expected, Rlv3841 cultures grown overnight on glutamine (N-rich) had decreased amino acid transport rates via Aap and Bra relative to Rlv3841 cultures grown on glutamate (N-limiting) (Fig. 5A). This decrease would currently be attributed to transcriptional regulation by NtrC in most bacteria, including Rlv3841 (54). However, we reasoned that it might be due to PtsP regulation, with glutamine signaling via PTS and unrelated to NtrC-dependent transcription. To test this, cells grown on glucose or succinate with glutamate (10 mM) or different concentrations of NH4Cl (10, 1, and 0.5 mM) were subsequently harvested and resuspended without carbon or nitrogen sources. Cells were then incubated in glutamine (100 μM), for 1 or 10 min, harvested, and washed to remove extracellular glutamine, which sterically inhibits AIB transport. Just 1 min after glutamine exposure, AIB transport was significantly decreased in N-limiting conditions (10 mM glucose + 10 mM glutamate, 20 mM succinate + 0.5 mM NH4Cl; Fig. 5B). The speed of the response in this in vivo test of amino acid uptake indicates a rapid protein:protein interaction inhibiting uptake via Aap and Bra. This is consistent with glutamine binding the GAF domain of PtsP to reduce its phosphorylation and subsequently that of PtsN and ManX (16, 51, 52). Reduced PtsN∼P explains the decreased amino acid uptake (Fig. 2) and critically predicts that increased dephosphorylated ManX should activate the TCA cycle and O2 consumption. O2 consumption was therefore measured in liquid cultures growing in exponential phase on succinate or glucose with different NH4Cl concentrations (10, 1, and 0.5 mM). The initial rates of O2 consumption were much higher under N-rich conditions (10 mM NH4Cl) than N-limiting conditions (1 and 0.5 mM NH4Cl), showing that the activity of the TCA cycle in Rlv3841 is reduced when cells are N-starved (Fig. 5 C and D). The O2 consumption rate did not increase significantly when glutamine was added to Rlv3841 grown under N-rich conditions (Fig. 5C). However, the rate increased immediately when glutamine was added to cells grown under N-limiting conditions. We observed an increased O2 consumption when glutamine was added to cells growing on 20 mM succinate with 1 mM and 0.5 mM NH4Cl, whereas when glucose was the carbon source, the increase was only observed when glutamine was added at the lowest NH4Cl concentration, 0.5 mM NH4Cl. This indicates that cells were more severely N-deprived on succinate. Thus, as predicted, the TCA cycle responds dramatically to an up-shift in nitrogen status (i.e., glutamine), indicating that it is immediately and tightly regulated by nitrogen availability. As a control, we carried out both transport and O2 consumption assays adding minimal medium or water and glutamate in place of glutamine to N-limiting conditions (SI Appendix, Figs. S5 and S6). Whereas no significant effect was observed with the addition of plain media or water, there was a small increase when glutamate was added that can be accounted for by the rapid conversion of glutamate to glutamine (7), consistent with the role of glutamine in PTS regulation.
Fig. 5.
Effect of glutamine on the PTS regulatory cascade of Rlv3841. (A) Effect of nitrogen availability on AIB transport. Membrane transport of wild-type cells grown on UMS with 10 mM glucose as carbon source and 10 mM NH4Cl, glutamine (N-rich conditions) or glutamate (N-limiting conditions). (B) Effect of glutamine addition on AIB transport. Wild-type cells grown on UMS with 10 mM glucose as carbon source and 10 mM NH4Cl (N-rich conditions) or 10 mM glutamate (N-limiting conditions). Transport rates measured at 1 min and 10 min after glutamine addition. All transport rates are expressed in nmol min−1 mg protein−1. (C) Effect of glutamine addition on O2 consumption of cells grown under different nitrogen conditions. Wild-type cells grown on UMS with 20 mM succinate as carbon source and 10 mM NH4Cl (N-rich conditions) or 1 mM NH4Cl (N-limiting conditions). Values show the rates of O2 consumption expressed in μmol L−1 min−1 OD600nm-1. (D) Effect of glutamine addition on O2 concentration over time. Graphical representation of O2 concentration (ppb) over time (min) for cells with no glutamine added (Top line, filled dots) compared to cells after the addition of glutamine (100 μM) indicated by a blue arrow (Bottom line, empty dots). Conditions are as above. Data are averages (±SEM, at least three independent cultures). Two-way ANOVA with Sidak’s multiple comparisons test (*) P < 0.05, (**) P < 0.01, (***) P < 0.001 (****), P < 0.0001 and n.s. not significant.
GAF-Deletion Mutants Are Blind to Nitrogen Status.
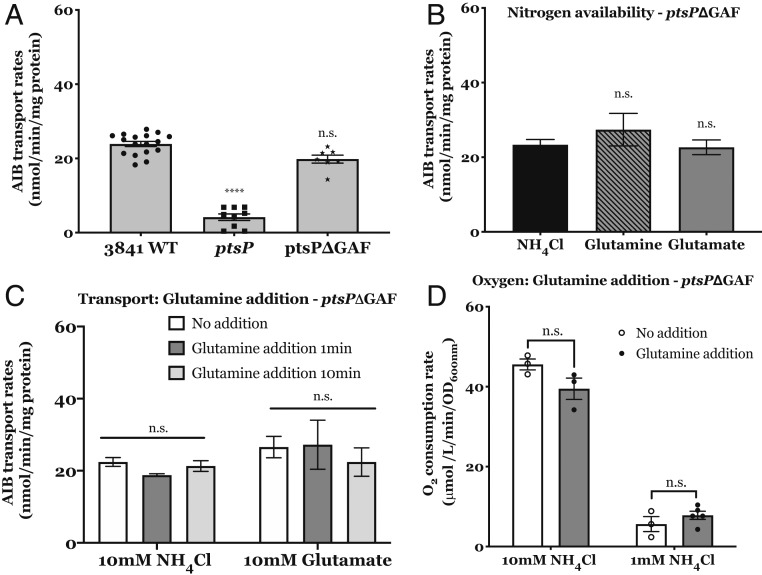
The results of the glutamine addition experiments are consistent with the proposed model of reciprocal regulation of PtsN and ManX. However, we still needed to show that the effects observed were dependent on the presence of the GAF domain binding glutamine to prevent PTS phosphorylation as previously described (16, 51, 52). Therefore, chromosomal PtsP::Tn5 was replaced with a version of PstP lacking the GAF domain, generating ptsPΔGAF (OPS1010), a strain that recovered the wild-type mucoid surface, growth on glutamate, and the ability to transport amino acids (SI Appendix, Table S1 and Fig. 6A), indicating that PtsPΔGAF is a functional protein. Remarkably, OPS1010 no longer responded to the nitrogen status of the growth medium (Fig. 6B). The lack of any significant effect of glutamine addition on amino acid uptake (Fig. 6C) or TCA cycle activity as measured by O2 consumption (Fig. 6D) supports our hypothesis of OPS1010 being nitrogen blind and points toward GAF-mediated nitrogen sensing in Rlv3841. Together, these data support the role of glutamine as the signal inhibiting both PtsP phosphorylation and subsequently, that of PtsN and ManX, down-regulating amino acid uptake and up-regulating TCA cycle activity, respectively. This PtsP-sensing mechanism would allow the cell to balance nitrogen and carbon metabolism.
Fig. 6.
Effect of deletion of the GAF domain on the PTS regulatory cascade. (A) Membrane transport of AIB in wild-type cells (3841 WT), ptsP mutant (PtsP107), and ptsPΔGAF mutant (OPS1010) grown on UMS with 10 mM glucose as the carbon source and 10 mM NH4Cl. (B) Effect of nitrogen availability on AIB transport. Membrane transport of ptsPΔGAF mutant (OPS1010) cells grown on UMS with 10 mM glucose as carbon source and 10 mM NH4Cl, glutamine (N-rich conditions) or glutamate (N-limiting conditions). (C) Effect of glutamine addition on AIB transport. ptsPΔGAF mutant (OPS1010) cells grown on UMS with 10 mM glucose as carbon source and 10 mM NH4Cl (N-rich conditions) or 10 mM glutamate (N-limiting conditions). Transport rates measured at 1 min and 10 min after the glutamine addition. All transport rates are expressed in nmol min−1 mg protein−1. (D) Effect of glutamine addition on O2 consumption. ptsPΔGAF mutant (OPS1010) cells grown on UMS with 20 mM succinate as the carbon source and 10 mM NH4Cl (N-rich conditions) or 1 mM NH4Cl (N-limiting conditions). Values show the rates of O2 consumption expressed in μmol L−1 min−1 OD600nm-1. Data are averages (±SEM, n = 3 independent cultures). Statistical analyses are indicated following the level of significance, with (****) P < 0.0001 and n.s., not significant.
PTS Systems Are Essential for Symbiosis.
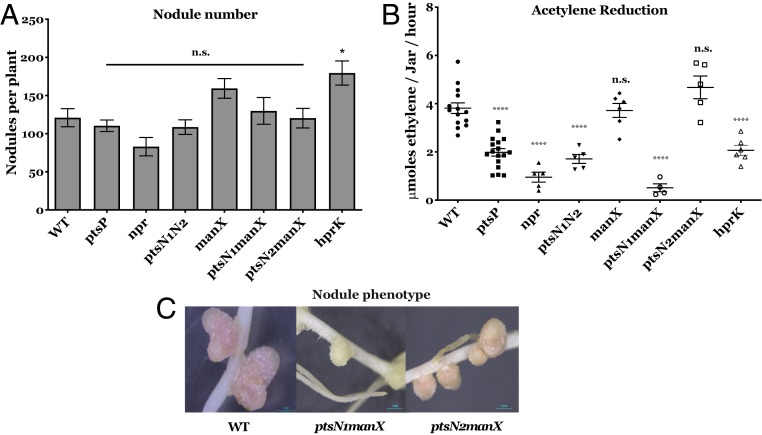
Several rhizobial PTS mutants have symbiotic phenotypes (40, 45, 55). All of the individual PTS mutants derived from Rlv3841 were able to nodulate and fix nitrogen (Fig. 7 A and B), although ptsP, npr, and ptsN1N2 mutants did this at a lower rate and the occurrence of suppressor mutations was observed at a high rate. This suggested that a single blockage through mutation of ptsN or manX does not block nitrogen fixation. However, the double mutant ptsN1manX had white nodules (Fig. 7C) that fixed significantly less nitrogen (Fig. 7B). These data show that either branch of the pathway (PTSNtr or carbohydrate-PTS) is adequate for nitrogen fixation, but disrupting both at the same time blocks it.
Fig. 7.
Symbiotic phenotype of Rlv3841 PTS mutants. (A) Nodule number and (B) acetylene reduction assay carried out 21 d postinoculation (dpi) for Rlv3841 wild type, ptsP (PtsP107), npr (AA031), ptsN1N2 (AA047), manX (LMB692), ptsN1manX (OPS0374), ptsN2manX (OPS0849), and hprK (AA081) mutants. Data are averages (±SEM) from at least three plants inoculated with independent cultures. Statistical analyses are indicated following the level of significance, with (*) P < 0.01, (****) P < 0.0001 and n.s., not significant. (C) Pea nodules formed by wild-type Rlv3841 and double mutants ptsN1manX (OPS0374) and ptsN2manX (OPS0849). (Scale bar, 1 mm.)
In this work, we have determined the physiological consequences of the cross-talk between the two historical PTS branches in rhizobia. Our study provides evidence that this complex biological sensor-actuator device enables bacterial cells to posttranslationally alter physiology to balance carbon and nitrogen availability. By analyzing the PTS components of R. leguminosarum as a single integrated system and identifying the internal signal that activates it, we have been able to assign a role to each of the EIIA regulatory proteins to demonstrate that PTS coordinates the main metabolic cellular functions. The biological relevance of this phosphorylation switch between ManX and PtsN is the ability to balance internal fluxes of carbon and nitrogen. In our model system, the absolute absence of these two output regulators leads to an inefficient symbiosis. Whereas individual mutants are still able to fix nitrogen, potentially due to a partial affinity between EIIAs, the disruption of the phosphotransfer cascade affected negatively the rates of fixation, indicating that the regulatory mechanisms of PTSNtr are essential.
Conclusion
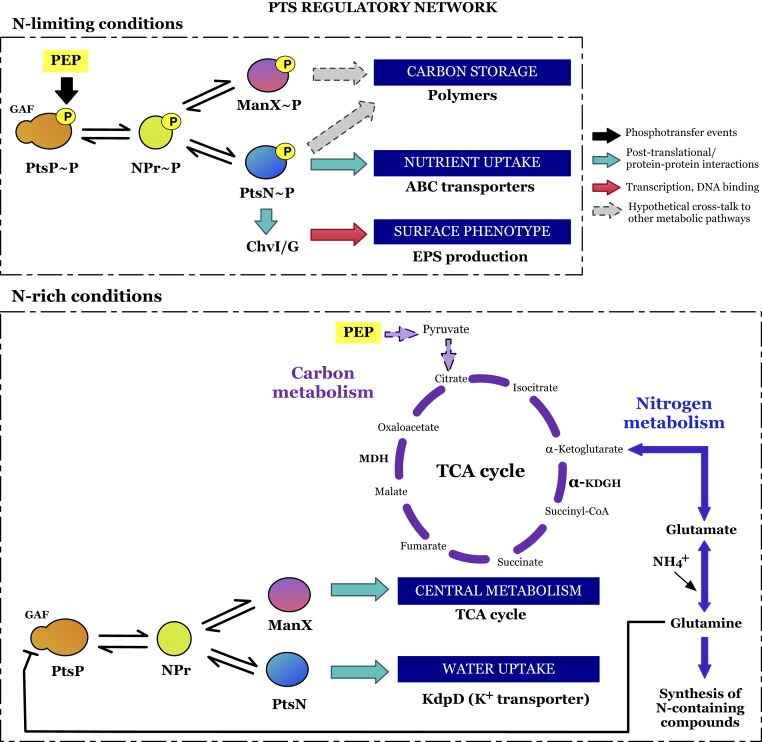
The PTS system has been described as an autonomous biochemical device that modulates a variety of cell functions in response to the intracellular PEP/pyruvate ratio, the N/C ratio, and the intracellular/extracellular fraction of K+ ions (56). However, despite its apparent universal importance in proteobacteria, it was unclear how the nitrogen and carbon balance was globally integrated. We reasoned that to understand PTS at the global level, all of the components needed to be tested in a single system. Furthermore, it was crucial to have single-copy integrated versions of PtsN and ManX that were either wild type or nonphosphorylatable and, in the case of ptsN, to have a functional phosphomimic version. It was also essential to have single chromosomal copies of full-length PtsP and PtsPΔGAF. With these strains as tools it was then possible to design real-time in vivo experiments to test how the components work. To illustrate our findings, we propose a model for an integrated PTS regulatory network in Fig. 8. This two-component EIIA switch modulated by NPr integrates PTSNtr and the carbohydrate-PTS systems as a single PTS system in α-proteobacteria, where glutamine is the signal binding the GAF domain of PtsP. Our results on transport and O2 consumption rates for both wild type and the GAF deletion mutant (Figs. 5 and 6) point toward a model where, as seen in other α-proteobacteria (51, 52), glutamine prevents PtsP phosphorylation by binding to GAF under N-rich conditions. This binding would drive a reduction in amino acid transport (due to a decrease in PtsN∼P) while increasing the activity of the TCA cycle (ManX also dephosphorylated). We hypothesize that carbon catabolism increases to match the nitrogen status of the cell. By contrast, under N-limiting and carbon-rich conditions PtsP∼P would increase as would PtsN∼P and ManX∼P. We predict that PtsN∼P will increase amino acid uptake while the drop in dephosphorylated ManX will reduce the activity of the TCA cycle, increasing nitrogen acquisition while slowing down carbon catabolism. Concomitantly, PtsN∼P stimulates extracellular EPS production (Fig. 3A) but is also likely that either PtsN∼P or ManX∼P increase the production of intracellular carbon storage polymers such as glycogen or polyhydroxybutyrate (PHB). Effectively, when carbon cannot be catabolized because of limiting nitrogen, carbon surplus is stored as polymers. Accordingly, PTS mutants in R. eutropha, P. putida, and Sinorhizobium fredii that cannot phosphorylate EIIA have significantly reduced PHB levels (46, 55, 57). Therefore, this two-component EIIA switch allows bacteria to balance the availability of nitrogen and carbon skeletons and, therefore, nitrogen and carbon metabolism, by means of PTS phosphorylation (Fig. 8) and independently of the Ntr system. The speed of response observed after addition of glutamine also confirmed that the regulation exerted by PTS follows the model of allosteric binding of signaling metabolites and rapid protein posttranslational modifications, allowing regulatory mechanisms to act at different timings relative to nitrogen levels. Thus, PTS constitutes another major regulatory system that is able to interact with both metabolic and transcriptional networks (50), where cross-talk between PTS systems could increase the power and connectivity of signal-response networks (58). Indeed, it was recently revealed that in Caulobacter crescentus and R. eutropha PTS interacts with the global stress stringent response (52, 59), emphasizing its profound importance to bacterial signaling and control. In C. crescentus, this PTSNtr-dependent regulation of (p)ppGpp accumulation upon N-starvation is mediated by direct binding of SpoT to the phosphorylated version of PtsN interfering with its (p)ppGpp hydrolase activity, a regulatory mechanism also conserved in S. meliloti (60).
Fig. 8.
Schematic model for PTS interactions according to nitrogen availability. Under N-limiting conditions, PtsP autophosphorylation via PEP modulates the phosphorylation of NPr and subsequentially, PtsN (EIIANtr) and ManX (carbohydrate-EIIA). Once phosphorylated, PtsN activates ABC transporters and interacts with ChvI to control EPS production, with both phosphorylated EIIAs possibly acting on carbon storage. Under N-rich conditions, intracellular glutamine inhibits PtsP autophosphorylation and, therefore, the upcoming phosphorylation of PTS components. Dephosphorylated PtsN interacts with KdpD, controlling K+ homeostasis, and ManX acts on the TCA cycle. Dual control of PtsN and ManX mediated by the NPr switch is affected by nitrogen availability, balancing carbon metabolism.
Given the broad conservation of this system, the reciprocal regulation of carbon and nitrogen metabolism by an EIIA switch as proposed here is likely to be another global regulatory mechanism, enabling bacterial cells to alter their physiology in real time to balance carbon and nitrogen availability. PTS and PII proteins coordinate posttranslationally many facets of bacterial metabolism by interacting with and regulating the activities of enzymes, transcription factors, and transport proteins. These regulatory mechanisms work in parallel using the glutamine/α-ketoglutarate level to time their responses differentially, reinforcing the importance of coordinating nitrogen and carbon sources as essential elements for all life. Such behavior allows bacteria to efficiently adapt to nutritional adversity and reveals how versatile they are at modifying their metabolism in response to nutrient availability. An attractive line of future investigation would be to model regulatory networks, exploring how bacteria integrate PTS regulation with PII proteins, the stringent response, and Ntr regulation to unravel how nitrogen levels drive these bacterial metabolic connections.
Methods
Bacterial Strains and Growth Conditions.
The bacterial strains and plasmids used in this study are listed in SI Appendix, Table S2. E. coli strains were grown in liquid or solid Luria–Bertani (LB) medium (61) at 37 °C supplemented with appropriate antibiotics (µg ml−1): ampicillin, 100; tetracycline, 10; and kanamycin, 20. R. leguminosarum strains, listed in SI Appendix, Table S2, were grown at 28 °C in TY extract (62) or UMS (63) with appropriate carbon and nitrogen sources at 10 mM unless otherwise stated. Antibiotics were used at the following concentrations (µg ml−1) unless otherwise stated: gentamicin, 20; kanamycin, 20; neomycin, 40; spectinomycin, 50; streptomycin, 500; and tetracycline (2 in UMS, 5 in TY). Mean generation times (MGTs) for Rlv3841-derived strains were obtained from cells grown in 50 mL UMS with the corresponding carbon and nitrogen sources at an initial optical density (OD600nm) of 0.01. OD600nm measurements were taken at 4-h time intervals until growth reached stationary phase from at least three biological replicates. The MGT of Rlv strains was calculated as the number of hours it takes the population to double while in exponential growth phase.
Mutant and Plasmid Construction.
All routine DNA analyses were done using standard protocols (61). The cloning details are described in SI Appendix. Primers used in this work are listed in SI Appendix, Table S3. Conjugations and transductions with bacteriophage RL38 were performed as previously described (64, 65).
Transport Assays.
Transport assays were performed with 25 μM (4.625 kBq) of 14C-labeled solute (53, 66), with cultures grown in UMS with 10 mM glucose and 10 mM NH4Cl unless otherwise specified. Glutamine and glutamate addition experiments were done by adding 5 μL of a 100 μM stock to 5 mL of starved cells. The same procedure was followed by adding 5 μL of rhizobium minimal salts (RMS) medium as control. At two different time points after their addition, 1 min and 10 min, 2 mL of cells were filtered using a Millipore filtration unit with vacuum suction, washed twice, and resuspended in RMS with no carbon or nitrogen sources for subsequent transport assays.
Exopolysaccharide Measurements.
The 250-mL cultures were grown up to an OD600nm of 0.5. Cells were spun down, dried, and weighted. The supernatant was treated with two volumes (500 mL) of cold isopropanol. After precipitation, the EPS was spun down and the pellet dried at 37 °C. Values are expressed in mg/mg dry weight of cells.
Promoter Analysis.
Promoter analyses with lux fusions were done by growing rhizobial strains on UMS agar slopes or liquid cultures (with appropriate antibiotics) following the protocol by Pini et al. (67). Sensitivity of each promoter fusion was measured either with the NightOWL in Vivo Imaging System (Berthold Technologies) with agar plates or the Promega GloMax Microplate Reader with liquid cultures. β-Galactosidase fusions were assayed according to Miller (68), with modifications as described by Poole et al. (69).
O2 Consumption Assays.
O2 consumption rates were obtained from UMS cultures grown with 10 mM glucose and 10 mM NH4Cl unless otherwise specified to early exponential phase. A 25-mL sealed glass universal, containing an OxyDot was quickly filled completely with the liquid culture, and O2 measurements were taken every 15 s for 1,000 s, or until the O2 level decreased below 1%. These noninvasive measurements were performed with the O2 electrode OxySense 325I System and the data analyzed with the OxySense Gen III software. O2 consumption rates were calculated as the time it takes the population to decrease the O2 concentration by 3%. Glutamine and glutamate addition experiments were done by measuring O2 consumption after injecting 250 μL of a 100-mM stock into the sealed universal. The same procedure was followed by adding 250 μL of water as control.
Enzyme Assays.
Cultures of R. leguminosarum strains were harvested at exponential growth, washed, and resuspended in 10 mL 40 mM Hepes pH 7, containing 1 mM dithiothreitol. Cells were disrupted by two passages on a FastPrep-24 5G ribolyser (MP Biomedicals), following centrifugation. Oxoglutarate dehydrogenase (EC 1.2.4.2) was assayed according to Reeves et al. (70) and malate dehydrogenase (EC 1.1.1.37) by the technique of Saroso et al. (71). The protein concentration of whole cells was determined by the method of Lowry et al. (72), using bovine serum albumin as standard.
BACTH Assays.
Interacting partners on the high-copy pUT18C vectors were transformed into MAE01 (ΔcyaA::ApraR) cells. Single transformants were grown at 37 °C in 5 mL LB to an OD600nm of 0.3 to 0.5 and subsequently transformed with the interaction partners on the low-copy pKNT25 vector, plated onto LB plates with 0.5 mM isopropyl-β-D-1-thiogalactopyranosid (IPTG) and X-gal, and incubated for 2 d at 28 °C. To quantify BACTH interactions, these colonies were grown overnight at 28 °C in 10 mL LB with ampicillin and kanamycin at standard concentrations (73). Overnight cultures were then used to inoculate 5-mL cultures of LB with 1% wt/vol glucose, ampicillin, and kanamycin to an initial OD600nm of 0.1 and grown until they reached an OD600nm of 0.4. Cultures were then induced with 2 mM IPTG and grown for 6 h at 28 °C. A standard β-galactosidase assay (68) was used to quantify the interaction between proteins. KdpD/PtsN1 pairs were used as positive controls (23), while empty vectors, as negative controls.
Plant Growth and Acetylene Reduction.
Pisum sativum cv. Avola seeds were surface sterilized using 95% ethanol and 2% sodium hypochlorite at the time of sowing. Plants were inoculated with 1 × 107 cells of the appropriate rhizobial strain and grown in 1-L beakers filled with sterile medium-grade vermiculite and N-free nutrient solution as previously described (69) in a growth room (16 h light/8 h dark). They were harvested at pea flowering (3 wk) and acetylene reduction was determined as previously described (74).
Statistical Analysis.
All analyses were performed using GraphPad Prism 8 (GraphPad Software). Significant differences between pairs of parameters were determined by Student’s t tests. Comparisons of more than two groups were done by ANOVA followed by multiple comparisons post hoc corrections as indicated in each figure legend. A P value of less than 0.05 was considered as statistically significant.
Data Availability.
The data supporting the findings of the study are available in this article and its SI Appendix.
Supplementary Material
Acknowledgments
This work was supported by the Biotechnology and Biological Sciences Research Council (grants BB/K006134/1, BB/N003608/1, and BB/M011224/1). We thank Frank Sargeant for providing the strain MAE01 for BACTH assays and Alison East for critical reading of this manuscript.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1917471117/-/DCSupplemental.
References
- 1.Huergo L. F., Chandra G., Merrick M., P(II) signal transduction proteins: Nitrogen regulation and beyond. FEMS Microbiol. Rev. 37, 251–283 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Brown D. R., Barton G., Pan Z., Buck M., Wigneshweraraj S., Nitrogen stress response and stringent response are coupled in Escherichia coli. Nat. Commun. 5, 4115 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prasse D., Schmitz R. A., Small RNAs involved in regulation of nitrogen metabolism. Microbiol. Spectr. 6, RWR-0018 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Huergo L. F., Dixon R., The emergence of 2-oxoglutarate as a master regulator metabolite. Microbiol. Mol. Biol. Rev. 79, 419–435 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Radchenko M. V., Thornton J., Merrick M., P(II) signal transduction proteins are ATPases whose activity is regulated by 2-oxoglutarate. Proc. Natl. Acad. Sci. U.S.A. 110, 12948–12953 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schumacher J., et al. , Nitrogen and carbon status are integrated at the transcriptional level by the nitrogen regulator NtrC in vivo. MBio 4, e00881–e13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Radchenko M. V., Thornton J., Merrick M., Control of AmtB-GlnK complex formation by intracellular levels of ATP, ADP, and 2-oxoglutarate. J. Biol. Chem. 285, 31037–31045 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doucette C. D., Schwab D. J., Wingreen N. S., Rabinowitz J. D., α-Ketoglutarate coordinates carbon and nitrogen utilization via enzyme I inhibition. Nat. Chem. Biol. 7, 894–901 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pflüger-Grau K., de Lorenzo V., From the phosphoenolpyruvate phosphotransferase system to selfish metabolism: A story retraced in Pseudomonas putida. FEMS Microbiol. Lett. 356, 144–153 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Deutscher J., et al. , The bacterial phosphoenolpyruvate:carbohydrate phosphotransferase system: Regulation by protein phosphorylation and phosphorylation-dependent protein-protein interactions. Microbiol. Mol. Biol. Rev. 78, 231–256 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Powell B. S., et al. , Novel proteins of the phosphotransferase system encoded within the rpoN operon of Escherichia coli. Enzyme IIANtr affects growth on organic nitrogen and the conditional lethality of an erats mutant. J. Biol. Chem. 270, 4822–4839 (1995). [DOI] [PubMed] [Google Scholar]
- 12.Pflüger-Grau K., Görke B., Regulatory roles of the bacterial nitrogen-related phosphotransferase system. Trends Microbiol. 18, 205–214 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Cases I., Velázquez F., de Lorenzo V., The ancestral role of the phosphoenolpyruvate-carbohydrate phosphotransferase system (PTS) as exposed by comparative genomics. Res. Microbiol. 158, 666–670 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Aravind L., Ponting C. P., The GAF domain: An evolutionary link between diverse phototransducing proteins. Trends Biochem. Sci. 22, 458–459 (1997). [DOI] [PubMed] [Google Scholar]
- 15.Zimmer D. P., et al. , Nitrogen regulatory protein C-controlled genes of Escherichia coli: Scavenging as a defense against nitrogen limitation. Proc. Natl. Acad. Sci. U.S.A. 97, 14674–14679 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee C. R., et al. , Reciprocal regulation of the autophosphorylation of enzyme INtr by glutamine and α-ketoglutarate in Escherichia coli. Mol. Microbiol. 88, 473–485 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Untiet V., et al. , ABC transport is inactivated by the PTS(Ntr) under potassium limitation in Rhizobium leguminosarum 3841. PLoS One 8, e64682 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barabote R. D., Saier M. H. Jr, Comparative genomic analyses of the bacterial phosphotransferase system. Microbiol. Mol. Biol. Rev. 69, 608–634 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boël G., et al. , Transcription regulators potentially controlled by HPr kinase/phosphorylase in Gram-negative bacteria. J. Mol. Microbiol. Biotechnol. 5, 206–215 (2003). [DOI] [PubMed] [Google Scholar]
- 20.Stülke J., Hillen W., Carbon catabolite repression in bacteria. Curr. Opin. Microbiol. 2, 195–201 (1999). [DOI] [PubMed] [Google Scholar]
- 21.Hu K. Y., Saier M. H. Jr, Phylogeny of phosphoryl transfer proteins of the phosphoenolpyruvate-dependent sugar-transporting phosphotransferase system. Res. Microbiol. 153, 405–415 (2002). [DOI] [PubMed] [Google Scholar]
- 22.Lee C. R., Cho S. H., Yoon M. J., Peterkofsky A., Seok Y. J., Escherichia coli enzyme IIANtr regulates the K+ transporter TrkA. Proc. Natl. Acad. Sci. U.S.A. 104, 4124–4129 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prell J., et al. , The PTS(Ntr) system globally regulates ATP-dependent transporters in Rhizobium leguminosarum. Mol. Microbiol. 84, 117–129 (2012). [DOI] [PubMed] [Google Scholar]
- 24.Lüttmann D., et al. , Stimulation of the potassium sensor KdpD kinase activity by interaction with the phosphotransferase protein IIA(Ntr) in Escherichia coli. Mol. Microbiol. 72, 978–994 (2009). [DOI] [PubMed] [Google Scholar]
- 25.Lüttmann D., Göpel Y., Görke B., The phosphotransferase protein EIIA(Ntr) modulates the phosphate starvation response through interaction with histidine kinase PhoR in Escherichia coli. Mol. Microbiol. 86, 96–110 (2012). [DOI] [PubMed] [Google Scholar]
- 26.Dozot M., et al. , Functional characterization of the incomplete phosphotransferase system (PTS) of the intracellular pathogen Brucella melitensis. PLoS One 5, e12679 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prell J., et al. , Legumes regulate Rhizobium bacteroid development and persistence by the supply of branched-chain amino acids. Proc. Natl. Acad. Sci. U.S.A. 106, 12477–12482 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lodwig E. M., et al. , Amino-acid cycling drives nitrogen fixation in the legume-Rhizobium symbiosis. Nature 422, 722–726 (2003). [DOI] [PubMed] [Google Scholar]
- 29.Cheng G., Karunakaran R., East A. K., Munoz-Azcarate O., Poole P. S., Glutathione affects the transport activity of Rhizobium leguminosarum 3841 and is essential for efficient nodulation. FEMS Microbiol. Lett. 364, fnx045 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Skorupska A., Janczarek M., Marczak M., Mazur A., Król J., Rhizobial exopolysaccharides: Genetic control and symbiotic functions. Microb. Cell Fact. 5, 7 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheng H. P., Walker G. C., Succinoglycan production by Rhizobium meliloti is regulated through the ExoS-ChvI two-component regulatory system. J. Bacteriol. 180, 20–26 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bélanger L., Dimmick K. A., Fleming J. S., Charles T. C., Null mutations in Sinorhizobium meliloti exoS and chvI demonstrate the importance of this two-component regulatory system for symbiosis. Mol. Microbiol. 74, 1223–1237 (2009). [DOI] [PubMed] [Google Scholar]
- 33.Vanderlinde E. M., Yost C. K., Mutation of the sensor kinase chvG in Rhizobium leguminosarum negatively impacts cellular metabolism, outer membrane stability, and symbiosis. J. Bacteriol. 194, 768–777 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen E. J., Fisher R. F., Perovich V. M., Sabio E. A., Long S. R., Identification of direct transcriptional target genes of ExoS/ChvI two-component signaling in Sinorhizobium meliloti. J. Bacteriol. 191, 6833–6842 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sola-Landa A., et al. , A two-component regulatory system playing a critical role in plant pathogens and endosymbionts is present in Brucella abortus and controls cell invasion and virulence. Mol. Microbiol. 29, 125–138 (1998). [DOI] [PubMed] [Google Scholar]
- 36.Charles T. C., Nester E. W., A chromosomally encoded two-component sensory transduction system is required for virulence of Agrobacterium tumefaciens. J. Bacteriol. 175, 6614–6625 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang C., et al. , Sinorhizobium meliloti 1021 loss-of-function deletion mutation in chvI and its phenotypic characteristics. Mol. Plant Microbe Interact. 23, 153–160 (2010). [DOI] [PubMed] [Google Scholar]
- 38.Deutscher J., Francke C., Postma P. W., How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol. Mol. Biol. Rev. 70, 939–1031 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heavner M. E., Qiu W. G., Cheng H. P., Phylogenetic co-occurrence of ExoR, ExoS, and ChvI, components of the RSI bacterial invasion switch, suggests a key adaptive mechanism regulating the transition between free-living and host-invading phases in Rhizobiales. PLoS One 10, e0135655 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pinedo C. A., Bringhurst R. M., Gage D. J., Sinorhizobium meliloti mutants lacking phosphotransferase system enzyme HPr or EIIA are altered in diverse processes, including carbon metabolism, cobalt requirements, and succinoglycan production. J. Bacteriol. 190, 2947–2956 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pflüger-Grau K., Chavarría M., de Lorenzo V., The interplay of the EIIA(Ntr) component of the nitrogen-related phosphotransferase system (PTS(Ntr)) of Pseudomonas putida with pyruvate dehydrogenase. Biochim. Biophys. Acta 1810, 995–1005 (2011). [DOI] [PubMed] [Google Scholar]
- 42.Walshaw D. L., Wilkinson A., Mundy M., Smith M., Poole P. S., Regulation of the TCA cycle and the general amino acid permease by overflow metabolism in Rhizobium leguminosarum. Microbiology 143, 2209–2221 (1997). [DOI] [PubMed] [Google Scholar]
- 43.Yoo W., et al. , Fine-tuning of amino sugar homeostasis by EIIANtr in Salmonella typhimurium. Sci. Rep. 6, 33055 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reizer J., Novotny M. J., Hengstenberg W., Saier M. H. Jr, Properties of ATP-dependent protein kinase from Streptococcus pyogenes that phosphorylates a seryl residue in HPr, a phosphocarrier protein of the phosphotransferase system. J. Bacteriol. 160, 333–340 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pinedo C. A., Gage D. J., HPrK regulates succinate-mediated catabolite repression in the gram-negative symbiont Sinorhizobium meliloti. J. Bacteriol. 191, 298–309 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krausse D., et al. , Essential role of the hprK gene in Ralstonia eutropha H16. J. Mol. Microbiol. Biotechnol. 17, 146–152 (2009). [DOI] [PubMed] [Google Scholar]
- 47.Dossonnet V., et al. , Phosphorylation of HPr by the bifunctional HPr Kinase/P-ser-HPr phosphatase from Lactobacillus casei controls catabolite repression and inducer exclusion but not inducer expulsion. J. Bacteriol. 182, 2582–2590 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hakoshima T., Ichihara H., Structure of SixA, a histidine protein phosphatase of the ArcB histidine-containing phosphotransfer domain in Escherichia coli. Methods Enzymol. 422, 288–304 (2007). [DOI] [PubMed] [Google Scholar]
- 49.Schulte J. E., Goulian M., The phosphohistidine phosphatase SixA targets a phosphotransferase system. MBio 9, e01666-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chavarría M., Fuhrer T., Sauer U., Pflüger-Grau K., de Lorenzo V., Cra regulates the cross-talk between the two branches of the phosphoenolpyruvate : Phosphotransferase system of Pseudomonas putida. Environ. Microbiol. 15, 121–132 (2013). [DOI] [PubMed] [Google Scholar]
- 51.Goodwin R. A., Gage D. J., Biochemical characterization of a nitrogen-type phosphotransferase system reveals that enzyme EI(Ntr) integrates carbon and nitrogen signaling in Sinorhizobium meliloti. J. Bacteriol. 196, 1901–1907 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ronneau S., Petit K., De Bolle X., Hallez R., Phosphotransferase-dependent accumulation of (p)ppGpp in response to glutamine deprivation in Caulobacter crescentus. Nat. Commun. 7, 11423 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hosie A. H., Allaway D., Galloway C. S., Dunsby H. A., Poole P. S., Rhizobium leguminosarum has a second general amino acid permease with unusually broad substrate specificity and high similarity to branched-chain amino acid transporters (Bra/LIV) of the ABC family. J. Bacteriol. 184, 4071–4080 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Walshaw D. L., Lowthorpe S., East A., Poole P. S., Distribution of a sub-class of bacterial ABC polar amino acid transporter and identification of an N-terminal region involved in solute specificity. FEBS Lett. 414, 397–401 (1997). [DOI] [PubMed] [Google Scholar]
- 55.Li Y. Z., et al. , Genetic analysis reveals the essential role of nitrogen phosphotransferase system components in Sinorhizobium fredii CCBAU 45436 symbioses with soybean and pigeonpea plants. Appl. Environ. Microbiol. 82, 1305–1315 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chavarría M., Kleijn R. J., Sauer U., Pflüger-Grau K., de Lorenzo V., Regulatory tasks of the phosphoenolpyruvate-phosphotransferase system of Pseudomonas putida in central carbon metabolism. MBio 3, e00028-12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Velázquez F., Pflüger K., Cases I., De Eugenio L. I., de Lorenzo V., The phosphotransferase system formed by PtsP, PtsO, and PtsN proteins controls production of polyhydroxyalkanoates in Pseudomonas putida. J. Bacteriol. 189, 4529–4533 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Silva-Rocha R., de Lorenzo V., Noise and robustness in prokaryotic regulatory networks. Annu. Rev. Microbiol. 64, 257–275 (2010). [DOI] [PubMed] [Google Scholar]
- 59.Karstens K., Zschiedrich C. P., Bowien B., Stülke J., Görke B., Phosphotransferase protein EIIANtr interacts with SpoT, a key enzyme of the stringent response, in Ralstonia eutropha H16. Microbiology 160, 711–722 (2014). [DOI] [PubMed] [Google Scholar]
- 60.Ronneau S., et al. , Regulation of (p)ppGpp hydrolysis by a conserved archetypal regulatory domain. Nucleic Acids Res. 47, 843–854 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sambrook J., Russell D. W., Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Laboratory Press, 2001). [Google Scholar]
- 62.Beringer J. E., R factor transfer in Rhizobium leguminosarum. J. Gen. Microbiol. 84, 188–198 (1974). [DOI] [PubMed] [Google Scholar]
- 63.Wheatley R. M., et al. , Role of O2 in the growth of Rhizobium leguminosarum bv. viciae 3841 on glucose and succinate. J. Bacteriol. 199, e00572-16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Poole P. S., Schofield N. A., Reid C. J., Drew E. M., Walshaw D. L., Identification of chromosomal genes located downstream of dctD that affect the requirement for calcium and the lipopolysaccharide layer of Rhizobium leguminosarum. Microbiology 140, 2797–2809 (1994). [DOI] [PubMed] [Google Scholar]
- 65.Buchanan-Wollaston V., Generalized transduction in Rhizobium leguminosarum. J. Gen. Microbiol. 112, 135–142 (1979). [Google Scholar]
- 66.Poole P. S., Franklin M., Glenn A. R., Dilworth M. J., The transport of L-glutamate by Rhizobium leguminosarum Involves a common amino acid carrier. J. Gen. Microbiol. 131, 1441–1448 (1985). [Google Scholar]
- 67.Pini F., et al. , Bacterial biosensors for in vivo spatiotemporal mapping of root secretion. Plant Physiol. 174, 1289–1306 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Miller J. H., Experiments in Molecular Genetics (Cold Spring Harbor Laboratory, 1972). [Google Scholar]
- 69.Poole P. S., Blyth A., Reid C. J., Walters K., Myoinositol catabolism and catabolite regulation in Rhizobium leguminosarum bv viciae. Microbiol-UK 140, 2787–2795 (1994). [Google Scholar]
- 70.Reeves H. C., Rabin R., Wegener W. S., Ajl S. J., “Assays of enzymes of the tricarboxylic acid and glyoxylate cycles” in Methods in Microbiology, Norris J. R., Ribbons D. W., Eds. (Academic Press, 1971), vol. 6, chap. X, pp. 425–462. [Google Scholar]
- 71.Saroso S., Dilworth M. J., Glenn A. R., The use of activities of carbon catabolic enzymes as a probe for the carbon nutrition of snakebean nodule bacteroids. J. Gen. Microbiol. 132, 243–249 (1986). [Google Scholar]
- 72.Lowry O. H., Rosebrough N. J., Farr A. L., Randall R. J., Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193, 265–275 (1951). [PubMed] [Google Scholar]
- 73.Karimova G., Pidoux J., Ullmann A., Ladant D., A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. Acad. Sci. U.S.A. 95, 5752–5756 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Allaway D., et al. , Identification of alanine dehydrogenase and its role in mixed secretion of ammonium and alanine by pea bacteroids. Mol. Microbiol. 36, 508–515 (2000). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the findings of the study are available in this article and its SI Appendix.