Significance
Members of the CsrA/RsmA family of global posttranscriptional regulators play critical roles in the control of virulence in a variety of bacteria. These regulators often function to inhibit the translation of target mRNA species and have been thought to act by binding full-length transcripts. Here we show that in the opportunistic pathogen Pseudomonas aeruginosa, RsmA associates with hundreds of transcripts as they are being synthesized by RNA polymerase, some of which are also targeted by another global posttranscriptional regulator called Hfq. Our findings identify which transcripts in P. aeruginosa are targeted by RsmA, reveal connectivity between RsmA and Hfq in this organism, and suggest that the cotranscriptional level of regulation may be an important one for this class of regulator.
Keywords: posttranscriptional regulator, RNA binding proteins, Hfq
Abstract
In the opportunistic pathogen Pseudomonas aeruginosa, RsmA is an RNA-binding protein that plays critical roles in the control of virulence, interbacterial interactions, and biofilm formation. Although RsmA is thought to exert its regulatory effects by binding full-length transcripts, the extent to which RsmA binds nascent transcripts has not been addressed. Moreover, which transcripts are direct targets of this key posttranscriptional regulator is largely unknown. Using chromatin immunoprecipitation coupled with high-throughput DNA sequencing, with cells grown in the presence and absence of the RNA polymerase inhibitor rifampicin, we identify hundreds of nascent transcripts that RsmA associates with in P. aeruginosa. We also find that the RNA chaperone Hfq targets a subset of those nascent transcripts that RsmA associates with and that the two RNA-binding proteins can exert regulatory effects on common targets. Our findings establish that RsmA associates with many transcripts as they are being synthesized in P. aeruginosa, identify the transcripts targeted by RsmA, and suggest that RsmA and Hfq may act in a combinatorial fashion on certain transcripts. The binding of posttranscriptional regulators to nascent transcripts may be commonplace in bacteria where distinct regulators can function alone or in concert to achieve control over the translation of transcripts as soon as they emerge from RNA polymerase.
The CsrA/RsmA family of posttranscriptional regulators play prominent roles in the control of virulence gene expression in a variety of pathogenic bacteria (1–3). Members of this family recognize a core sequence of GGA in target RNA species that is present in the loop of a stem–loop structure (4, 5). CsrA/RsmA orthologs typically act to repress the translation of target mRNA species, with the target GGA sequence residing within, or in close proximity to, the Shine-Dalgarno sequence (1). However, members of this family can also exert stimulatory effects on translation (6). Because the recognition sequence for CsrA/RsmA orthologs is small, must be present within a specific structure, and because each monomer within the CsrA/RsmA dimer can interact with a separate target sequence, predicting which transcripts are directly targeted by these posttranscriptional regulators can be challenging. Target transcripts that are bound by members of the CsrA/RsmA family have therefore typically been identified through RNA coimmunoprecipitation approaches (3, 6, 7).
Transcription and translation can be coupled in bacteria with RNA polymerase (RNAP) making direct contacts with the translation machinery (8–10). Thus, from a regulatory standpoint, it would make sense that posttranscriptional regulators that act to modulate translation would access their targets on transcripts as soon as they emerge from RNAP. Although CsrA/RsmA orthologs have been proposed to act on full-length transcripts (11), there is at least one example in which CsrA is thought to exert its regulatory effects by targeting a nascent transcript (12). However, the extent to which any member of the CsrA/RsmA family binds target transcripts as they are being made by RNAP is not known.
In the opportunistic pathogen Pseudomonas aeruginosa, where RsmA is thought to play an important role in virulence by controlling the switch between acute and chronic infection states (13, 14), we use chromatin immunoprecipitation coupled with high-throughput DNA sequencing (ChIP-seq) with cells grown in the presence and absence of rifampicin to identify hundreds of nascent transcripts that RsmA associates with. Furthermore, we present evidence that in P. aeruginosa RsmA associates with many nascent transcripts that are targeted by a second global posttranscriptional regulator called Hfq. Our findings identify potential regulatory targets of RsmA in P. aeruginosa, have implications for how RsmA and its orthologs may exert their regulatory effects, and lend support to the notion that the binding of posttranscriptional regulators to nascent transcripts may be commonplace in bacteria.
Results
RsmA Acts Cotranscriptionally on over 500 Transcripts in P. aeruginosa.
To determine the extent to which RsmA associates with nascent transcripts in P. aeruginosa, we used ChIP with cells grown in the presence or absence of the RNA polymerase inhibitor rifampicin followed by high-throughput DNA sequencing, an approach we refer to as ChIPPAR-seq (15). The rationale behind this approach is that when bound to a nascent transcript RsmA may be in sufficient proximity to the DNA to be cross-linked to it in a ChIP assay (Fig. 1A) (15). In addition, any cross-linking between RsmA and the DNA would be expected to be prevented by treatment of cells with rifampicin prior to the addition of the cross-linker (Fig. 1A) (15). To facilitate this analysis, we created a strain of PAO1 that synthesized RsmA with a C-terminal vesicular stomatitis virus glycoprotein-G (VSV-G) epitope tag (RsmA-V) from the endogenous rsmA locus. ChIPPAR-seq of RsmA-V in cells grown to midlog (OD600 of 0.4) in lysogeny broth indicated that under these growth conditions RsmA-V associated with 557 different regions of the chromosome, with the vast majority of these associations being sensitive to treatment with rifampicin (Fig. 1B and SI Appendix, Fig. S1A and Table S1) (16), and often occurring within proximity of the translation start site of an associated gene (SI Appendix, Fig. S1B). Similar findings were obtained when the association of RsmA-V with specific genomic regions was tested by ChIPPAR and quantitative PCR (ChIPPAR-qPCR) (Fig. 1C). Furthermore, treatment of cells with rifampicin did not alter the abundance of RsmA-V (Fig. 1D). Our ChIPPAR-seq findings establish that RsmA associates with hundreds of genomic regions in logarithmically growing cells in a manner that is sensitive to treatment with rifampicin, indicating that RsmA associates with hundreds of nascent transcripts in P. aeruginosa.
Fig. 1.
RsmA associates with target RNAs cotranscriptionally. (A) Schematic of ChIPPAR approach. (B) RsmA-V enrichment profiles at the indicated genomic regions in cells cultured to midlog phase prior to rifampicin treatment (−rif), or following rifampicin treatment (+rif). PAO1 WT cells, which do not synthesize RsmA-V, served as the mock IP control (indicated mock). Genomic location in kilobases is provided at the top of each of the two panels. Significantly enriched peaks are indicated by dark gray boxes below the read density plot (in green), red lines within these dark gray boxes indicate site of maximum enrichment. Annotated genes are indicated in yellow in the middle of each panel. RNA-seq normalized reads from WT cells (indicated WT) and ΔrsmAF mutant cells (indicated ΔrsmAF) are provided below the annotated genes. Reads mapping to the plus strand (+ strand) and minus strand (− strand) are indicated. (C) Fold-enrichment of RsmA-V at the indicated loci as determined by ChIPPAR-qPCR. Fold-enrichment of RsmA-V at the indicated loci in cells prior to rifampicin treatment (−rif), or following rifampicin treatment (+rif). (D) Western blot analysis of RsmA-V abundance in cells of the indicated strains to assess the effect of rifampicin treatment (+rif) on RsmA-V abundance. Cells analyzed were PAO1 RsmA-V (indicated RsmA-V). WT PAO1 cells (indicated WT), that do not synthesize RsmA-V were included as a negative control. The α-subunit of RNAP (RNAP-α) served as a loading control. (E) Enrichment of RsmA-V (green bars) and RsmA(R44A)-V (orange bars) at the indicated loci as determined by ChIP-qPCR. (F) Western blot analysis of RsmA-V and RsmA(R44A)-V abundance. Cells analyzed were PAO1 RsmA-V (indicated RsmA-V) and PAO1 RsmA(R44A)-V (indicated RsmA(R44A)-V). WT PAO1 cells (indicated WT), that do not synthesize RsmA-V were included as a negative control. RNAP-α served as a loading control. Throughout the text, error bars represent 1 SD. For C–F, experiments were repeated at least twice and data from biological triplicate samples of a representative experiment are shown. Where indicated, statistical significance was assessed via two-sample t test; *P ≤ 0.05; **P ≤ 0.005.
RsmA is a member of the CsrA family of proteins that bind RNA as a dimer and recognize sites with the consensus sequence CAnGGAyG, with the critical GGA core sequence centered in the loop region of a stem–loop structure (4, 17). ChIP with a mutant version of RsmA-V containing amino acid substitution R44A [RsmA(R44A)-V] that is defective for RNA binding because it lacks a key residue involved in recognition of the GGA core sequence (18), resulted in significantly less enrichment than RsmA-V at two chromosomal regions (Fig. 1E), even though the RsmA(R44A)-V mutant protein was at least as abundant as the WT version of the protein (Fig. 1F). Thus, the surface of RsmA involved in sequence-specific binding to RNA is required for the association of RsmA-V with nascent transcripts as determined by ChIP.
Each monomer of an RsmA dimer is in principle capable of interacting with a distinct GGA motif in a target transcript (19). Indeed, in Escherichia coli, CsrA is thought to exert control over the translation of certain transcripts by making simultaneous contact with two distinct GGA motifs (20–22). To determine whether GGA motifs present within a target nascent transcript are required in order to detect the association of RsmA by ChIP, we created a mutant strain of P. aeruginosa in which two GGA motifs within the PA3483 transcript were mutated (one motif within and one motif downstream of the predicted Shine-Dalgarno sequence). The qPCR analyses depicted in SI Appendix, Fig. S2 indicate that mutation of these GGA sites specifically abolished the association of RsmA-V with the PA3483 nascent transcript. Our findings with the RsmA(R44A) mutant and the PA3483 mutant transcript support the idea that the enrichment of specific genomic regions through ChIP with RsmA-V reflect the association of RsmA-V with transcripts that are in the process of being synthesized by RNA polymerase.
ChIPPAR-Seq Identifies Potential Direct Regulatory Targets of RsmA.
In P. aeruginosa only a small number of direct targets of RsmA are currently known (5, 7, 23–25). Nevertheless, the majority of these transcripts are among our ChIPPAR-seq dataset, including tssA1 (i.e. PA0082) and fha1 (PA0081), which encode a component and a regulator, respectively, of one of the three distinct type VI secretion systems (T6SS) in P. aeruginosa (Fig. 1B and SI Appendix, Table S1) (23). Indeed, we found that RsmA targets (i.e. binds, either directly or indirectly) transcripts from all three of the major T6SS gene clusters (H1, H2, and H3), as well as many additional transcripts specifying T6SS effector and effector immunity proteins (Fig. 1B and SI Appendix, Fig. S3 and Table S1), thus providing insight into how RsmA may exert direct control over all three of these systems (26).
RsmA has been shown to influence the abundance of hundreds of transcripts in P. aeruginosa (23). Akin to E. coli CsrA, it is thought that interaction of RsmA with target transcripts typically acts to inhibit their translation, resulting in a concomitant reduction in transcript abundance because translation of a mRNA species tends to increase its stability (6, 27, 28). To better define those genes whose expression is influenced by RsmA under the same experimental conditions as were used for our ChIPPAR-seq analyses, we performed RNA sequencing (RNA-seq) to compare gene expression in WT cells to that in cells of a ΔrsmA ΔrsmF double mutant (ΔrsmAF) grown to midlog phase in LB. For these experiments, we used cells that lacked both RsmA and RsmF (a second RsmA family member in P. aeruginosa) (24, 29), as we were concerned that the effects of RsmA on gene expression would be masked through the effects of the ΔrsmA mutation on the abundance of RsmF (also called RsmN) (24, 29). RsmA and RsmF have both overlapping and distinct targets, and although RsmA is the most prominent of these two regulators, we found that the abundance of RsmF increased dramatically in cells that lacked RsmA (SI Appendix, Fig. S4), an effect that has been reported previously to obscure RsmA’s regulatory roles (7, 24). Use of the ΔrsmAF double mutant in our experiments obviated this concern. Consistent with previous findings, we found that RsmA controlled the expression of hundreds of genes in P. aeruginosa (SI Appendix, Table S2) (22). Comparison of RsmA-regulated transcripts from our transcriptomic studies with our ChIPPAR-seq results suggests that many RsmA-regulated transcripts are controlled directly by RsmA (SI Appendix, Fig. S5 and Table S2). It should also be noted that the interaction between RsmA and a particular transcript could still influence the translation of that transcript without necessarily influencing its abundance. Our ChIPPAR-seq findings not only indicate that RsmA acts on hundreds of nascent transcripts in P. aeruginosa but also provide a compendium of the potential direct regulatory targets of RsmA in this organism. We also note that our RNA-seq studies allow a comparison of the RsmA enrichment signal by ChIPPAR-seq at a specific chromosomal location with total transcript abundance (SI Appendix, Table S1). This comparison provides an initial indication of whether the amount of RsmA enrichment at a specific chromosomal location might be explained simply by the level of expression of the corresponding gene.
RsmA Associates with Nascent Transcripts Targeted by Hfq.
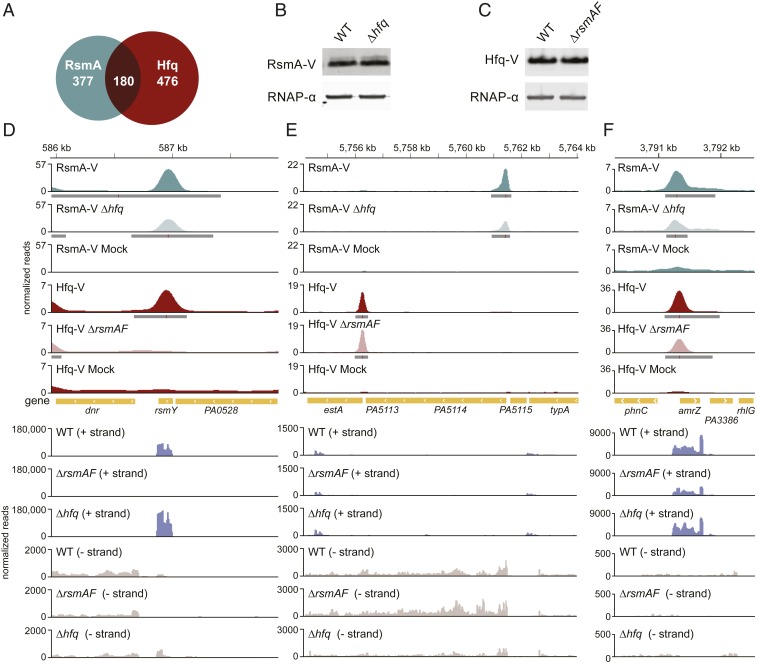
The RNA chaperone Hfq is a prominent posttranscriptional regulator in many bacteria that is best known for its ability to facilitate the base-pairing between small regulatory RNAs (sRNAs) and their target mRNA species, an activity that often culminates in translational repression of the target mRNA (30–33). However, recent work suggests that in some instances this small hexameric RNA-binding protein can act as a translational repressor independently of sRNAs (34–36; reviewed in ref. 37). In P. aeruginosa Hfq is an important posttranscriptional regulator whose activities contribute to the virulence of the organism (38). Using ChIPPAR-seq we have shown previously that Hfq associates with over 600 nascent transcripts in P. aeruginosa (15). A striking finding from our ChIPPAR-seq analyses is that many of the nascent transcripts associated with RsmA were also associated with Hfq; of the 557 nascent transcripts associated with RsmA, 180 are also associated with Hfq (Fig. 2A and SI Appendix, Table S3). Notably, it is not the case that those transcripts that are most highly enriched in the Hfq dataset are necessarily the same as those that are most highly enriched in the RsmA dataset. Indeed, although there is significant overlap between the transcripts targeted by RsmA and those targeted by Hfq, RsmA does not detectably associate with most of the transcripts that Hfq associates with (Fig. 2A).
Fig. 2.
RsmA associates with nascent transcripts that Hfq binds. (A) Venn diagram depicting overlap between nascent RNAs identified here as associated with RsmA-V by ChIPPAR-seq and nascent RNAs identified previously as being associated with Hfq (15). The significance of the overlap (P = 1.79 × 10−45) was determined by calculating the hypergeometric distribution function in R (39). (B) Western blot analysis of RsmA-V abundance in the presence and absence of Hfq. Cells analyzed were PAO1 RsmA-V (indicated WT), and PAO1 Δhfq RsmA-V (indicated Δhfq). RNAP-α served as a loading control. (C) Western blot analysis of Hfq-V abundance in the presence and absence of RsmA. Cells analyzed were PAO1 Hfq-V (indicated WT), and PAO1 ΔrsmAF Hfq-V (indicated ΔrsmAF). RNAP-α subunit was used as a loading control. (D–F) ChIP-seq enrichment profiles at the indicated genomic regions for RsmA-V in the presence and absence of Hfq, and for Hfq-V in the presence and absence of RsmA. Cells analyzed were PAO1 RsmA-V (indicated RsmA-V), PAO1 Δhfq RsmA-V (indicated RsmA-V Δhfq), PAO1 Hfq-V (indicated Hfq-V), and PAO1 ΔrsmAF Hfq-V (indicated Hfq-V ΔrsmAF). PAO1 WT cells, which do not synthesize RsmA-V or Hfq-V, served as the mock IP control (indicated RsmA-V mock or Hfq-V mock). Genomic location in kilobases is provided at the top of each panel. Significantly enriched peaks are indicated by dark gray boxes below the read density plot (in shades of green for RsmA-V and in shades of red for Hfq-V), red lines within these dark gray boxes indicate site of maximum enrichment. Annotated genes are indicated in yellow in the middle of each panel. RNA-seq normalized reads from WT cells (indicated WT), ΔrsmAF mutant cells (indicated ΔrsmAF), and Δhfq mutant cells (indicated Δhfq) are provided below the annotated genes. Reads mapping to the plus strand (+ strand) and minus strand (− strand) are indicated.
Having first confirmed that Hfq and RsmA do not influence one another’s abundance (Fig. 2 B and C), we next sought to determine to what extent Hfq and RsmA influence each other’s ability to associate with nascent transcripts. Specifically, we determined the effects of Hfq on the ability of RsmA to associate with nascent transcripts by performing ChIP-seq with RsmA-V in otherwise WT cells and in cells lacking Hfq (Δhfq mutant cells) grown to midlog phase in LB. In addition, to determine the effects of RsmA on the ability of Hfq to associate with nascent transcripts, we performed ChIP-seq with Hfq-V in otherwise WT and ΔrsmAF mutant cells grown to midlog phase in LB. For these latter experiments we used cells that again lacked both RsmA and RsmF, as we were concerned that the effects of RsmA on the ability of Hfq to associate with target nascent transcripts would be masked through the effects of the ΔrsmA mutation on the abundance of RsmF (24, 29) (SI Appendix, Fig. S4).
Our ChIP-seq analyses revealed that for the majority of nascent transcripts that appear to be targeted by both RsmA and Hfq, neither regulator is absolutely required for the association of the other with the nascent transcript (Fig. 2 and SI Appendix, Tables S4 and S5). Nevertheless, Hfq and RsmA may influence the association of one another with a subset of nascent transcripts. For example, we found that the association of RsmA-V with the PA4421 nascent transcript appeared to be reduced in cells of the Δhfq mutant when compared to WT, and conversely that the association of Hfq-V with the PA4421 nascent transcript appeared to be reduced in ΔrsmAF mutant cells when compared to WT (SI Appendix, Fig. S6). RNA-seq revealed that the abundance of the PA4421 transcript is essentially unaltered in cells of the ΔrsmAF and Δhfq mutants when compared to WT cells (SI Appendix, Fig. S6), suggesting that the effects of Hfq and RsmA on one another’s association with the PA4421 nascent transcript are unlikely to be explained by differences in PA4421 transcription or transcript stability. Taken together, these findings raise the possibility that RsmA and Hfq may bind cooperatively to the PA4421 nascent transcript. However, we note that there are situations in which the effect of one regulator on the apparent ability of the other to associate with a particular nascent transcript may be explained instead by the effect of the regulator on the transcription of the nascent transcript (i.e. a scenario in which the loss of one regulator limits the amount of nascent transcript that is available for interaction with the other). An example of this type of situation is evident in the case of rsmY, which encodes a small regulatory RNA that acts as a molecular sponge for RsmA and also binds Hfq (40, 41). RsmA is known to positively regulate the transcription of rsmY through an unknown mechanism (42, 43). We found that Hfq-V did not appear to detectably associate with the RsmY nascent transcript in cells of the ΔrsmAF double mutant (Fig. 2D). This finding can be fully explained by an indirect effect of RsmA on the transcription of rsmY; that is, there is simply less RsmY nascent transcript available for interaction with Hfq in cells lacking RsmA (Fig. 2D) (42, 43). In support of this notion, our RNA-seq studies indicate that the abundance of RsmY is dramatically reduced in cells of the ΔrsmAF double mutant when compared to WT (Fig. 2D).
Control of a Polycistronic Transcript by RsmA and Hfq.
For nascent transcripts that we defined as being targeted by both RsmA and Hfq through our ChIPPAR-seq analyses (Fig. 2A), the enrichment peaks for RsmA and Hfq were in close proximity to one another (see, for example, Fig. 2D). However, because bacterial mRNA is often polycistronic, we posited there may be polycistronic transcripts targeted by both RsmA and Hfq in which the regions of the transcript targeted by each regulator are well separated from each other; such transcripts would not be called as targeted by both regulators in our analyses. One example of a nascent polycistronic transcript of this type specifies three genes, PA5114, PA5113, and estA (PA5112); EstA is an esterase involved in rhamnolipid maturation, biofilm development, quorum sensing, and virulence (44), whereas PA5113 and PA5114 are hypothetical proteins of unknown function. Our RNA-seq and ChIPPAR-seq studies suggest that this polycistronic transcript begins upstream of PA5114 and ends downstream of estA, with RsmA associated with the PA5114 portion and Hfq associated with the estA portion of the transcript (Fig. 2E, and illustrated schematically in Fig. 3A). To begin to determine whether this putative PA5114-PA5113-estA polycistronic transcript was subject to control by RsmA and Hfq, we first constructed derivatives of PAO1 WT, ΔrsmAF double mutant, and Δhfq mutant cells in which the native copy of PA5114 specified PA5114 with five copies of the Myc epitope-tag at its C terminus (PA5114-Myc). Quantitative Western blotting in each of these strains indicated that the abundance of PA5114-Myc was higher in cells of the ΔrsmAF double mutant when compared to WT, whereas the abundance of PA5114-Myc was similar in both WT and Δhfq mutant cells (Fig. 3 B and C). Thus, RsmA, but not Hfq, exerts a negative regulatory effect on the abundance of PA5114. In addition, we found that RsmA, but not Hfq, negatively regulates the expression of a PA5114 translational lacZ reporter fusion, but that neither RsmA nor Hfq appreciably control the expression of a PA5114 promoter-lacZ fusion (Fig. 3 D and E). Taken together with our evidence that RsmA associates with the PA5114 nascent transcript, these findings suggest that RsmA inhibits translation of PA5114 by binding directly to the PA5114 mRNA.
Fig. 3.
Control of a polycistronic transcript by RsmA and Hfq. (A) Schematic of PA5114, PA5113 and estA genomic locus. (Top) In WT cells, the PA5114 promoter (PPA5114) drives expression of the PA5114-PA5113-estA polycistronic transcript and the estA promoter (PestA) drives expression of the estA monocistronic transcript. Promoters are indicated with arrows. RsmA and Hfq are depicted interacting with the PA5114-PA5113-estA polycistronic transcript and Hfq is depicted interacting with the estA monocistronic transcript. (Middle) In PA5114::Ω mutant cells, the PA5114-PA5113-estA polycistronic transcript is no longer made. (Bottom) In PestA* mutant cells, the estA monocistronic transcript is no longer made. (B) Western blot analysis of PA5114-Myc abundance in cells of the indicated strains. Cells analyzed were PAO1 PA5114-Myc (indicated WT + PA5114-Myc), PAO1 ΔrsmAF PA5114-Myc (indicated ΔrsmAF + PA5114-Myc), and PAO1 Δhfq PA5114-Myc (indicated Δhfq + PA5114-Myc). WT PAO1 cells (indicated WT), that do not synthesize PA5114-Myc were included as a negative control. RNAP-α served as a loading control. (C) Quantification of PA5114-Myc abundance from cells in B. (D) β-Galactosidase activity of PAO1 WT, PAO1 ΔrsmAF (ΔrsmAF), and PAO1 Δhfq (Δhfq) mutant cells containing a PA5114-lacZ translational fusion. (E) β-Galactosidase activity of PAO1 WT, PAO1 ΔrsmAF (ΔrsmAF), and PAO1 Δhfq (Δhfq) mutant cells containing a PA5114-lacZ transcriptional fusion. (F) Western blot analysis of EstA-Myc abundance in cells of the indicated strains. Cells analyzed were PAO1 EstA-Myc (indicated WT + EstA-Myc), PAO1 ΔrsmAF EstA-Myc (indicated ΔrsmAF + EstA-Myc), and PAO1 Δhfq EstA-Myc (indicated Δhfq + EstA-Myc). WT PAO1 cells (indicated WT), that do not synthesize EstA-Myc were included as a negative control. RNAP-α served as a loading control. (G) Quantification of EstA-Myc abundance from cells in F. (H) β-Galactosidase activity of PAO1 WT, PAO1 ΔrsmAF (ΔrsmAF), and PAO1 Δhfq (Δhfq) mutant cells containing an estA-lacZ translational fusion. (I) β-Galactosidase activity of PAO1 WT, PAO1 ΔrsmAF (ΔrsmAF), and PAO1 Δhfq (Δhfq) mutant cells containing an estA-lacZ transcriptional fusion. (J) Western blot analysis of EstA-Myc abundance in cells of the indicated strains. Cells analyzed were PAO1 EstA-Myc (indicated WT + EstA-Myc), PAO1 ΔrsmAF EstA-Myc (indicated ΔrsmAF + EstA-Myc), PAO1 PA5114::Ω EstA-Myc (indicated WT + EstA-Myc + PA5114::Ω), and PAO1 PA5114::Ω ΔrsmAF EstA-Myc (indicated ΔrsmAF + EstA-Myc + PA5114::Ω). WT PAO1 cells (indicated WT), that do not synthesize EstA-Myc were included as a negative control. RNAP-α served as a loading control. (K) Quantification of EstA-Myc abundance from cells in J. (L) Western blot analysis of EstA-Myc abundance in cells of the indicated strains. Cells analyzed were PAO1 EstA-Myc (indicated WT + EstA-Myc), PAO1 PestA* EstA-Myc (indicated WT + EstA-Myc + PestA*), and PAO1 Δhfq PestA* EstA-Myc (indicated Δhfq + EstA-Myc + PestA*). WT PAO1 cells (indicated WT), that do not synthesize EstA-Myc were included as a negative control. RNAP-α served as a loading control. (M) Quantification of EstA-Myc abundance from cells in L. Data shown in B, D–F, H–J, and L are from a representative experiment conducted with triplicate biological samples and were repeated independently at least twice. Where indicated, statistical significance was assessed via two-sample t test; *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
Postulating that the PA5114 mRNA species was a polycistronic transcript that also contained estA (i.e. PA5114-PA5113-estA), we next asked whether estA was regulated by RsmA and Hfq. To this end, we constructed derivatives of PAO1 WT, ΔrsmAF double mutant, and Δhfq mutant cells in which the native copy of estA specified EstA with five copies of the Myc epitope-tag at its C terminus (EstA-Myc). The results of quantitative Western blotting indicated that the abundance of EstA-Myc was higher in cells of the ΔrsmAF double mutant, as well as cells of the Δhfq mutant, when compared to otherwise WT cells (Fig. 3 F and G). Thus, both RsmA and Hfq negatively regulate the abundance of EstA.
We reasoned that RsmA influenced the abundance of EstA by binding the PA5114 portion of the PA5114-PA5113-estA polycistronic transcript. However, evidence suggests that estA mRNA species exist that are also monocistronic. Indeed, the translation of estA has been shown previously to be inhibited by Hfq (45), and this prior work placed the estA promoter within a 592-bp portion of DNA lying immediately upstream (45, 46) (represented schematically as PestA in Fig. 3A). Moreover, global RNA-seq analyses are consistent with such a location, placing a possible transcription start site for the estA promoter upstream of the 3′-end of the PA5113 gene (47) (Fig. 3A). Consistent with previous findings (45), we found that the expression of an estA-lacZ translational fusion whose expression was driven by the PestA promoter (within 592 bp of estA) was higher in Δhfq mutant cells compared to WT (Fig. 3H). Moreover, the activity of the PestA promoter was not negatively regulated by Hfq (Fig. 3I). Importantly, RsmA did not influence the expression of an estA-lacZ translational fusion that was under the control of the PestA promoter region only (Fig. 3G). We infer from this that the effect of RsmA on EstA abundance cannot be explained through an effect of RsmA on the translation of estA transcripts initiating from the PestA promoter region.
We next asked whether RsmA influenced the abundance of EstA by binding the PA5114-PA5113-estA polycistronic transcript. We reasoned that if RsmA influenced the abundance of EstA by binding the PA5114-PA5113-estA polycistronic transcript, then in cells that no longer produced this transcript, RsmA would no longer exert a regulatory effect over estA. To test this prediction we introduced a transcription terminator immediately upstream of the PA5114 gene (PA5114::Ω), between the putative promoter (depicted PPA5114) and the predicted translation start, in cells synthesizing EstA-Myc (see schematic in Fig. 3 A, Middle). Consistent with this expectation, quantitative Western blotting revealed that RsmA no longer influenced the abundance of EstA-Myc in cells harboring the transcription terminator immediately upstream of PA5114 (Fig. 3 J and K). Collectively, our findings indicate that RsmA negatively regulates the synthesis of EstA specified by the PA5114-PA5113-estA polycistronic transcript.
Our results indicate that the expression of estA is driven by at least two promoters: PestA that is proximal to estA (Fig. 3A), and PPA5114 that is proximal to PA5114 and drives expression of the PA5114-PA5113-estA polycistronic transcript. To determine whether Hfq could influence the abundance of EstA by binding the PA5114-PA5113-estA polycistronic transcript, we therefore sought to study the effects of Hfq on EstA abundance in cells in which the PestA promoter was inactivated (i.e. in cells in which only the PA5114 promoter drives estA expression) (see schematic in Fig. 3 A, Bottom). Mutation of the putative promoter sequences of the PestA promoter on the PAO1 chromosome resulted in a decrease in the abundance of EstA-Myc (Fig. 3 L and M). Furthermore, EstA-Myc was more abundant in Δhfq mutant cells containing the mutated estA promoter (PestA*) than in otherwise WT (i.e. hfq+) cells containing the PestA* mutant promoter (Fig. 3 L and M). These findings suggest that Hfq can repress the translation of estA specified by the PA5114-PA5113-estA polycistronic transcript. Thus, the PA5114-PA5113-estA polycistronic transcript is a target that is common to both RsmA and Hfq in which the binding sites for RsmA and Hfq are well separated from one another.
Control of AmrZ by RsmA and Hfq.
AmrZ is an important global transcription regulator that controls motility, virulence, and biofilm formation in P. aeruginosa (48, 49). Our ChIP-seq studies indicated that both Hfq and RsmA can bind the amrZ nascent transcript (Fig. 2F; illustrated schematically in Fig. 4A). We therefore asked whether Hfq and RsmA could influence the abundance of AmrZ. To do this, we constructed derivatives of PAO1 WT, ΔrsmAF double mutant, and Δhfq mutant cells that synthesized AmrZ with five copies of the Myc epitope tag at its C terminus (AmrZ-Myc). Consistent with the idea that amrZ might be subject to control by both RsmA and Hfq, quantitative Western blotting revealed that AmrZ-Myc was more abundant in ΔrsmAF and Δhfq mutant cells than in otherwise WT cells (Fig. 4 B and C). RsmA appeared to exert a greater regulatory effect than Hfq (Fig. 4 B and C) and the effects of the ΔrsmAF mutations could be complemented by ectopic expression of rsmA (SI Appendix, Fig. S7).
Fig. 4.
Control of AmrZ abundance by RsmA and Hfq. (A) Schematic of amrZ genomic locus and control of amrZ by RsmA and Hfq. (B) Western blot analysis of AmrZ-Myc abundance in cells of the indicated strains. Cells analyzed were PAO1 AmrZ-Myc (indicated WT + AmrZ-Myc), PAO1 ΔrsmAF AmrZ-Myc (indicated ΔrsmAF + AmrZ-Myc), and PAO1 Δhfq AmrZ-Myc (indicated Δhfq + AmrZ-Myc). WT PAO1 cells (indicated WT), that do not synthesize AmrZ-Myc were included as a negative control. RNAP-α served as a loading control. (C) Quantification of AmrZ-Myc abundance from cells in B. (D) β-Galactosidase activity of PAO1 WT, PAO1 ΔrsmF (ΔrsmF), PAO1 ΔrsmA (ΔrsmA), PAO1 ΔamrZ (ΔamrZ), PAO1 ΔrsmAF (ΔrsmAF), and PAO1 ΔrsmAF ΔamrZ (ΔrsmAF ΔamrZ) mutant cells containing a tssA1-lacZ transcriptional fusion. (E) β-Galactosidase activity of the tssA1-lacZ transcriptional reporter in ΔrsmAF mutant cells harboring an empty vector control (pEV) or a plasmid encoding rsmA (pRsmA). (F) β-Galactosidase activity of PAO1 WT, PAO1 ΔrsmF (ΔrsmF), PAO1 ΔrsmA (ΔrsmA), PAO1 ΔamrZ (ΔamrZ), PAO1 ΔrsmAF (ΔrsmAF), and PAO1 ΔrsmAF ΔamrZ (ΔrsmAF ΔamrZ) mutant cells containing a tssA1-lacZ translational fusion. (G) Model for control of tssA1 transcription and translation by RsmA. Data shown in B and D–F are from a representative experiment conducted with triplicate biological samples and were repeated independently at least twice. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ns, not significant.
It has been shown previously that AmrZ acts as a positive regulator of type VI secretion genes in the H1 cluster in P. aeruginosa, such as tssA1 (26, 49). Consistent with our finding that RsmA influences the abundance of AmrZ (Fig. 4 B and C), we found that expression of a tssA1-lacZ transcriptional reporter fusion was approximately sixfold higher in cells of a ΔrsmAF mutant than in WT (Fig. 4D); this effect of the ΔrsmAF mutations on tssA1 transcription could be complemented by the introduction of a plasmid directing the synthesis of RsmA (Fig. 4E). Importantly, the observed stimulatory effect of the ΔrsmAF mutations on the tssA1-lacZ transcriptional reporter was dependent upon AmrZ (Fig. 4D). These findings support the idea that RsmA acts indirectly to silence the transcription of tssA1 by limiting the abundance of AmrZ.
It is well established that RsmA can bind the tssA1 transcript directly and repress its translation (23, 24). Consistent with this idea, we found that expression of a tssA1-lacZ translational reporter fusion was considerably higher in cells of both a ΔrsmAF double mutant and in cells of a ΔrsmAF ΔamrZ triple mutant when compared to WT (Fig. 4F). Moreover, consistent with the idea that RsmA controls the abundance of AmrZ, which in turn activates the transcription of tssA1, expression of the tssA1-lacZ translational reporter fusion was diminished in cells of the ΔrsmAF ΔamrZ triple mutant when compared to that in cells of the ΔrsmAF double mutant (Fig. 4F). Our findings suggest that RsmA exerts negative control over tssA1 through a two-pronged mechanism, acting at both the translational level (directly) and the transcriptional level (indirectly via its effect on AmrZ) (Fig. 4G).
Discussion
We have found that RsmA associates cotranscriptionally with hundreds of transcripts in P. aeruginosa, and that a subset of these nascent transcripts is targeted by both RsmA and the RNA chaperone Hfq. RsmA influences the abundance of hundreds of RNA species in this organism (23), and our findings indicate which of these RsmA binds, either directly or indirectly.
The widespread targeting of nascent transcripts by RsmA that we observe in P. aeruginosa suggests that some of RsmA’s regulatory effects are exerted cotranscriptionally. Indeed, for bacteria like P. aeruginosa in which transcription and translation are thought to be coupled, the binding of RsmA to nascent mRNAs would allow this regulator to mediate effects on translation at the earliest possible opportunity, before synthesis of the mature transcript was complete. CsrA and its orthologs have been shown to influence the stability of certain target transcripts, which likely results, in many instances, from the destabilizing effect of reducing their translation (6, 20). It will therefore be interesting to determine whether the degradation of transcripts occurs cotranscriptionally in a manner that can be modulated by RsmA. Furthermore, repressing the translation of a nascent mRNA, or restructuring its 5′-untranslated region, could render that transcript an attractive substrate for the transcription termination factor Rho, resulting in its termination. Indeed, the one example from E. coli in which CsrA has been shown to exert a regulatory effect cotranscriptionally involves CsrA binding the pgaA nascent transcript and promoting transcription termination by facilitating Rho loading (12). Conversely, the binding of RsmA to a nascent transcript might prevent Rho from accessing a loading site on that transcript. Thus, the association of RsmA with a nascent transcript could exert a negative or positive regulatory effect at the level of transcription by exploiting regulatory mechanisms that are unique to nascent transcripts (i.e. that operate only during transcription elongation).
Comparison between those nascent transcripts identified here as being bound by RsmA (either directly or indirectly) and those we identified previously as being bound by Hfq (15) revealed extensive overlap between the two. We found that RsmA and Hfq can influence the abundance of proteins specified by common target transcripts. Specifically, we showed that the abundance of EstA was subject to control by both RsmA and Hfq, with each of these regulators associating with distinct portions of a polycistronic transcript that are physically well separated from one another (Fig. 3). Moreover, we showed that the abundance of the transcription regulator AmrZ, which is predicted to be specified by a monocistronic transcript (Fig. 2F), was controlled by both RsmA and Hfq (Fig. 4). Thus, akin to distinct transcription regulators that bind the DNA and influence the transcription of a common gene (50), distinct RNA-binding proteins can act to influence the translation and stability of common target transcripts. By binding the same mRNA species, RsmA and Hfq could work in a combinatorial fashion to control the translation and (or) abundance of that species. This would allow control of the common target transcript to be sensitive to any environmental input that governs the activity of either global posttranscriptional regulator. It is noteworthy that in E. coli, Hfq has been shown to bind a third of the RNA-pairs that are targeted by the RNA-chaperone ProQ, suggesting overlapping or competing roles of these two RNA-binding proteins (51). Hfq may therefore exert its regulatory effects on target transcripts that are shared by a variety of different posttranscriptional regulators.
A final point of note is that RsmA can exert regulatory effects on the same transcript at multiple levels. It has previously been established that both RsmA and AmrZ regulate the expression of various T6SS loci in P. aeruginosa (26). Indeed, prior work indicated that both the transcription and translation of tssA1, the first gene in a structural operon for the H1-T6SS, is repressed by RsmA (23, 24). Although RsmA was shown to bind the tssA1 transcript directly, explaining how it exerted its effects on translation, how RsmA influenced the transcription of tssA1 was not known. Our findings that RsmA associates with the amrZ nascent transcript, that RsmA influences the abundance of AmrZ, and that the effect of RsmA on the expression of a tssA1-lacZ transcriptional reporter is AmrZ-dependent, suggest that RsmA influences the transcription of tssA1 through its effects on AmrZ abundance. These data support a model in which RsmA exerts control over tssA1 through a two-pronged mechanism, involving: 1) RsmA directly repressing the translation of tssA1, and 2) RsmA indirectly influencing the transcription of tssA1 through its effect on the abundance of AmrZ (itself a positive regulator of tssA1 transcription) (Fig. 4G). Our findings further suggest that RsmA may indirectly influence the abundance of other transcripts by influencing the abundance of a variety of transcription regulators (including AmrZ), whose corresponding transcripts are direct targets of RsmA (SI Appendix, Table S1).
Our ChIPPAR approach involved the enrichment of DNA that is cross-linked to RsmA, either directly or indirectly, when this posttranscriptional regulator is bound to nascent transcripts. The rifampicin-sensitive enrichment of DNA we observed through ChIP with RsmA may reflect the cross-linking of RNA-bound RsmA directly to the DNA, or may reflect the cross-linking to the DNA of RNA polymerase that is tethered to an RsmA-bound nascent transcript (this could involve cross-linking of RNA-bound RsmA directly to RNA polymerase that is in turn cross-linked to the DNA) (15). However, ChIP signals from RNA polymerase in transcription elongation complexes are generally low (52), which might support the notion that the ChIP signals we detect through the immunoprecipitation of RsmA reflect the direct cross-linking of RNA-bound RsmA to the DNA. That our RsmA ChIP signals 1) are dependent upon transcription (i.e. are sensitive to treatment of cells with rifampicin), 2) depend on the RNA-binding surface of RsmA, and 3) depend on specific GGA sequences in the target RNA, support the idea that they reflect the binding of RsmA to specific nascent transcripts. However, it is possible that at some locations RsmA could be bound to a target nascent transcript indirectly through its interaction with another RNA-binding protein. We note that because our DNA-enrichment signals following ChIPPAR-seq of RsmA provide no strand-specific information, the accurate assignment of a DNA-enrichment signal to a particular nascent transcript could prove difficult in situations where an enrichment peak is centered between two divergently transcribed genes. However, the RNA-seq data obtained from WT cells can provide another point of reference to ensure that such enrichment peaks are assigned to the appropriate nascent transcripts.
RsmA is a critical regulator of the switch between the acute and chronic stages of infection in P. aeruginosa that influences the abundance of hundreds of different transcripts in this organism. Our ChIPPAR-seq findings indicate which of these are bound by RsmA (whether directly or indirectly). In addition, our data include transcripts that were not known previously to interact with RsmA that may also be subject to control by this posttranscriptional regulator. More generally, our findings that RsmA and Hfq each target hundreds of nascent transcripts suggest that the binding of posttranscriptional regulators to nascent transcripts might be commonplace in P. aeruginosa as well as other bacteria. The cotranscriptional level of regulation may therefore be a particularly important mode of regulation for these types of proteins.
Materials and Methods
Plasmids and Strains.
All bacterial strains, plasmids, and oligonucleotide primers used in this study (SI Appendix, Tables S6–S8, respectively), as well as details of strain and plasmid construction and growth conditions are described in the SI Appendix.
ChIP.
ChIPPAR-seq was performed on biological triplicate samples in PAO1 RsmA-V, as previously described (15), and a detailed protocol is included in SI Appendix. Briefly, cells were diluted from overnight cultures and grown to midlog in 200 mL LB at 37 °C with shaking and 80 mL were collected. The remaining culture was treated with 150 μg/mL rifampicin and returned to the incubator for 30 min before another 80 mL were collected. WT PAO1 was subjected to the same experimental conditions to be used as a mock IP control (referred to as a mock control). ChIP-seq was performed on biological triplicate samples in PAO1 RsmA-V, PAO1 RsmA-V ∆hfq, PAO1 Hfq-V, PAO1 Hfq-V ∆rsmAF, and WT PAO1 (the mock control). ChIP-seq experimental conditions were the same as those for ChIPPAR-seq, with the exception that the cells were not treated with rifampicin. Immediately upon collection, cells were cross-linked with formaldehyde (1% final concentration) for 30 min and then treated with 250 mM glycine to quench the cross-linking reaction. Cells were lysed and DNA was sheared with a Bioruptor water bath sonicator (Diagenode). Lysates were combined with anti–VSV-G beads (Sigma-Aldrich) for IP. After extensive washing and overnight cross-link reversal, DNA was isolated with a PCR purification kit (Qiagen). DNA yields were determined via Nanodrop or Agilent Bioanalyzer. For ChIPPAR- and ChIP-qPCR, triplicate cultures (3 mL) were grown from independent colonies and lysed via a tip sonicator. For ChIP-seq experiments including a ∆hfq mutant strain, overnight cultures were grown in no carbon essential (NCE) growth medium supplemented with 20 mM succinate as the carbon source and subsequently back-diluted into LB before sample collection, as described above.
ChIP-Seq Library Preparation and Sequencing.
Sequencing libraries were prepared using the NEBNext Ultra II DNA Library Prep Kit for Illumina (New England Biolabs) according to the manufacturer’s specifications. Approximately 1 to 40 ng immunoprecipitated DNA was used, and adaptors were diluted 10-fold prior to ligation. AMPure XP beads (Beckman Coulter) were used to purify the libraries, which were subjected to 10 rounds of amplification without size selection. Libraries were sequenced by Elim Biopharmaceuticals on an Illumina HiSeq2500 producing 50-bp paired-end reads.
ChIPPAR-Seq and ChIP-Seq Data Analysis.
Paired-end sequencing reads were mapped to the PAO1 genome (National Center for Biotechnology Information RefSeq NC_002516) using bowtie2 v2.3.4.3 (53). Only reads corresponding to fragments of 200 bp or less were used. A custom script was used to extract only read 1 from each pair and regions of enrichment were identified using QuEST v2.4 (54). Sequencing reads collected from the appropriate PAO1 mock biological replicates (i.e., IP from WT PAO1 that does not synthesize any VSV-G–tagged protein) were merged and served as the mock control. This mock control data were subsequently used to determine background for each ChIPPAR or ChIP biological replicate. The following criteria were used to identify regions of enrichment (peaks): They are twofold enriched in reads compared to the background, are not present in the mock control, have a positive peak shift and strand correlation, and have a q-value of less than 0.01. Peaks for each immunoprecipitated protein were defined as the maximal region identified in at least two biological replicates. Data were visualized using the Integrative Genomics Viewer (IGV) v2.5.0 (55). Peak analyses used custom scripts and BEDtools, v2.27.1.
qPCR.
qPCR was performed on DNA isolated from ChIPPAR and ChIP experiments using FastStart Essential DNA Green Master (Roche) and a LightCycler 96 (Roche). Primer efficiencies were calculated for each target gene via serial dilutions and melting curve analyses. Data analyses were supported by LightCycler software v1.1.0.1320 (Roche). Relative fold-enrichment indicates the relative abundance of a DNA region-of-interest relative to a negative control region (herein, we use a sequence within the gene PA2155) and the amount of DNA in the input. Specifically, we calculated fold-enrichment = 1.9∆∆Ct, ∆∆Ct = (Ct_ChIPPA2155 – Ct_ChIPtarget) – (Ct_InputPA2155 – Ct_Inputtarget). Reported fold-enrichments are the average of three biological replicates, and error bars denote SD. All data shown are representative of at least two independent experiments.
RNA Isolation.
Triplicate overnight cultures of PAO1, PAO1 ∆rsmAF, and PAO1 ∆hfq were back-diluted in fresh LB medium and grown until midlog phase, at which time, cells from 1 mL of culture were pelleted and resuspended in Tri-Reagent. Total RNA was collected by Direct-zol RNA-Miniprep (Zymo Research) according to the manufacturer’s specifications. RNA was eluted from the column in molecular-grade water.
Generation and Analysis of RNA-Seq Data.
RNA-seq was conducted by the Microbial ‘Omics Core facility at the Broad Institute using a modified version of the RNAtag-Seq protocol (56). Differential expression analysis was conducted with DESeq2 (57). For visualization of the RNA-seq data and coverage plots in the context of genome sequences and gene annotations, the read depths of the ∆rsmAF and ∆hfq datasets were normalized to that of the WT sample (PAO1), converted to bigwig format, and visualized in IGV (55). A detailed description of both the RNAtag-seq methodologies and data analysis steps can be found in SI Appendix.
Western Blot Analyses.
Whole-cell lysates from biological triplicates were separated by SDS/PAGE on either 12% or 4–12% Bis-Tris NuPAGE gels in MES or MOPS running buffer (Thermo Fisher). Proteins were transferred to polyvinylidene fluoride (PVDF) membranes with the XCell-II Blot Module (Thermo Fisher). Membranes were blocked for 1 h or overnight with Odyssey Blocking Buffer diluted 1:5 in PBS (LI-COR). Membranes were probed with anti-VSV-G, anti-Myc, and/or anti-RNA polymerase-α antibodies. Membranes were reblocked and incubated with near-Infrared secondary antibodies, 680LT donkey anti-mouse and 800CW donkey anti-rabbit (LI-COR). Imaging was performed on a LI-COR Odyssey CLx Imager, and fluorescence intensity was quantified using Image Studio software (LI-COR). When quantitative Western blots are shown, protein abundances were calculated as the mean fold-change of the target protein relative to the abundance of RNA polymerase-α subunit for three biological replicates. Error bars represent SD. The three biological replicates for each strain under comparison were analyzed on the same blot.
β-Galactosidase Assays.
Cells were permeabilized with ChCl3 and SDS and assayed for β-galactosidase activity as previously described (58). Values are the average measurements from triplicate cultures of a representative experiment. Error bars represent the SD from the mean. β-Galactosidase experiments were performed at least twice.
Data Availability.
The sequencing data reported in this paper have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) under accession number GSE138338.
Supplementary Material
Acknowledgments
We thank members of the S.L.D. laboratory for discussions; Renate Hellmiss for artwork; and Ann Hochschild for discussions and comments on the manuscript. Bioanalyzer analysis was performed in the Boston Children’s Hospital Intellectual and Developmental Disabilities Research Center Molecular Genetics Core that is supported by National Institutes of Health Award NIH-P30-HD 18655. RNA-sequencing libraries were constructed and sequenced at the Broad Institute of Massachusetts Institute of Technology and Harvard by the Microbial ‘Omics Core and Genomics Platform, respectively. The Microbial ‘Omics Core also provided guidance on experimental design and conducted preliminary analyses for all RNA-sequencing data. This work was supported by National Institutes of Health Grants AI143771 and AI125876 (to S.L.D.). T.K.K. was supported by a Graduate Research Fellowship from the NSF. M.J.G. was supported by a Postdoctoral Research Fellowship from the Cystic Fibrosis Foundation and by National Institutes of Health Training Grant 5T32HD055148-10.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE138338).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1917587117/-/DCSupplemental.
References
- 1.Romeo T., Babitzke P., Global regulation by CsrA and its RNA antagonists. Microbiol. Spectr. 6, RWR-0009-2017 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vakulskas C. A., Potts A. H., Babitzke P., Ahmer B. M., Romeo T., Regulation of bacterial virulence by Csr (Rsm) systems. Microbiol. Mol. Biol. Rev. 79, 193–224 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holmqvist E., et al. , Global RNA recognition patterns of post-transcriptional regulators Hfq and CsrA revealed by UV crosslinking in vivo. EMBO J. 35, 991–1011, 10.15252/embj.201593360 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dubey A. K., Baker C. S., Romeo T., Babitzke P., RNA sequence and secondary structure participate in high-affinity CsrA-RNA interaction. RNA 11, 1579–1587 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schulmeyer K. H., et al. , Primary and secondary sequence structure requirements for recognition and discrimination of target RNAs by Pseudomonas aeruginosa RsmA and RsmF. J. Bacteriol. 198, 2458–2469 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Potts A. H., et al. , Global role of the bacterial post-transcriptional regulator CsrA revealed by integrated transcriptomics. Nat. Commun. 8, 1596 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Romero M., et al. , Genome-wide mapping of the RNA targets of the Pseudomonas aeruginosa riboregulatory protein RsmN. Nucleic Acids Res. 46, 6823–6840 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miller O. L. Jr, Hamkalo B. A., Thomas C. A. Jr, Visualization of bacterial genes in action. Science 169, 392–395 (1970). [DOI] [PubMed] [Google Scholar]
- 9.Kohler R., Mooney R. A., Mills D. J., Landick R., Cramer P., Architecture of a transcribing-translating expressome. Science 356, 194–197 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Demo G., et al. , Structure of RNA polymerase bound to ribosomal 30S subunit. eLife 6, e28560 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holmqvist E., Vogel J., RNA-binding proteins in bacteria. Nat. Rev. Microbiol. 16, 601–615 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Figueroa-Bossi N., et al. , RNA remodeling by bacterial global regulator CsrA promotes Rho-dependent transcription termination. Genes Dev. 28, 1239–1251 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mulcahy H., et al. , Pseudomonas aeruginosa RsmA plays an important role during murine infection by influencing colonization, virulence, persistence, and pulmonary inflammation. Infect. Immun. 76, 632–638 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moscoso J. A., Mikkelsen H., Heeb S., Williams P., Filloux A., The Pseudomonas aeruginosa sensor RetS switches type III and type VI secretion via c-di-GMP signalling. Environ. Microbiol. 13, 3128–3138 (2011). [DOI] [PubMed] [Google Scholar]
- 15.Kambara T. K., Ramsey K. M., Dove S. L., Pervasive targeting of nascent transcripts by Hfq. Cell Rep. 23, 1543–1552 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gebhardt M. J., Kambara T. K., Ramsey K. M., Dove S. L., Widespread targeting of nascent transcripts by RsmA in Pseudomonas aeruginosa. National Center for Biotechnology Information Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE138338 Deposited 2 October (2019). [DOI] [PMC free article] [PubMed]
- 17.Schubert M., et al. , Molecular basis of messenger RNA recognition by the specific bacterial repressing clamp RsmA/CsrA. Nat. Struct. Mol. Biol. 14, 807–813 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Kulkarni P. R., et al. , A sequence-based approach for prediction of CsrA/RsmA targets in bacteria with experimental validation in Pseudomonas aeruginosa. Nucleic Acids Res. 42, 6811–6825 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duss O., et al. , Structural basis of the non-coding RNA RsmZ acting as a protein sponge. Nature 509, 588–592 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Baker C. S., Morozov I., Suzuki K., Romeo T., Babitzke P., CsrA regulates glycogen biosynthesis by preventing translation of glgC in Escherichia coli. Mol. Microbiol. 44, 1599–1610 (2002). [DOI] [PubMed] [Google Scholar]
- 21.Dubey A. K., et al. , CsrA regulates translation of the Escherichia coli carbon starvation gene, cstA, by blocking ribosome access to the cstA transcript. J. Bacteriol. 185, 4450–4460 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gutiérrez P., et al. , Solution structure of the carbon storage regulator protein CsrA from Escherichia coli. J. Bacteriol. 187, 3496–3501 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brencic A., Lory S., Determination of the regulon and identification of novel mRNA targets of Pseudomonas aeruginosa RsmA. Mol. Microbiol. 72, 612–632 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marden J. N., et al. , An unusual CsrA family member operates in series with RsmA to amplify posttranscriptional responses in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 110, 15055–15060 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Janssen K. H., Diaz M. R., Gode C. J., Wolfgang M. C., Yahr T. L., RsmV, a small noncoding regulatory RNA in Pseudomonas aeruginosa that sequesters RsmA and RsmF from target mRNAs. J. Bacteriol. 200, e00277-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Allsopp L. P., et al. , RsmA and AmrZ orchestrate the assembly of all three type VI secretion systems in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 114, 7707–7712 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baker C. S., et al. , CsrA inhibits translation initiation of Escherichia coli hfq by binding to a single site overlapping the Shine-Dalgarno sequence. J. Bacteriol. 189, 5472–5481 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu M. Y., Yang H., Romeo T., The product of the pleiotropic Escherichia coli gene csrA modulates glycogen biosynthesis via effects on mRNA stability. J. Bacteriol. 177, 2663–2672 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morris E. R., et al. , Structural rearrangement in an RsmA/CsrA ortholog of Pseudomonas aeruginosa creates a dimeric RNA-binding protein, RsmN. Structure 21, 1659–1671 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chao Y., Vogel J., The role of Hfq in bacterial pathogens. Curr. Opin. Microbiol. 13, 24–33 (2010). [DOI] [PubMed] [Google Scholar]
- 31.Hajnsdorf E., Boni I. V., Multiple activities of RNA-binding proteins S1 and Hfq. Biochimie 94, 1544–1553 (2012). [DOI] [PubMed] [Google Scholar]
- 32.Vogel J., Luisi B. F., Hfq and its constellation of RNA. Nat. Rev. Microbiol. 9, 578–589 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Updegrove T. B., Zhang A., Storz G., Hfq: The flexible RNA matchmaker. Curr. Opin. Microbiol. 30, 133–138 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vecerek B., Moll I., Bläsi U., Translational autocontrol of the Escherichia coli hfq RNA chaperone gene. RNA 11, 976–984 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ellis M. J., Trussler R. S., Haniford D. B., Hfq binds directly to the ribosome-binding site of IS10 transposase mRNA to inhibit translation. Mol. Microbiol. 96, 633–650 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen J., Gottesman S., Hfq links translation repression to stress-induced mutagenesis in E. coli. Genes Dev. 31, 1382–1395 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kavita K., de Mets F., Gottesman S., New aspects of RNA-based regulation by Hfq and its partner sRNAs. Curr. Opin. Microbiol. 42, 53–61 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sonnleitner E., et al. , Reduced virulence of a hfq mutant of Pseudomonas aeruginosa O1. Microb. Pathog. 35, 217–228 (2003). [DOI] [PubMed] [Google Scholar]
- 39.Team R. C., R: A language and environment for statistical computing (R Foundation for Statistical Computing, Vienna, Austria, 2016), https://www.R-project.org/.
- 40.Sonnleitner E., Schuster M., Sorger-Domenigg T., Greenberg E. P., Bläsi U., Hfq-dependent alterations of the transcriptome profile and effects on quorum sensing in Pseudomonas aeruginosa. Mol. Microbiol. 59, 1542–1558 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Sorger-Domenigg T., Sonnleitner E., Kaberdin V. R., Bläsi U., Distinct and overlapping binding sites of Pseudomonas aeruginosa Hfq and RsmA proteins on the non-coding RNA RsmY. Biochem. Biophys. Res. Commun. 352, 769–773 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Kay E., et al. , Two GacA-dependent small RNAs modulate the quorum-sensing response in Pseudomonas aeruginosa. J. Bacteriol. 188, 6026–6033 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Intile P. J., Diaz M. R., Urbanowski M. L., Wolfgang M. C., Yahr T. L., The AlgZR two-component system recalibrates the RsmAYZ posttranscriptional regulatory system to inhibit expression of the Pseudomonas aeruginosa type III secretion system. J. Bacteriol. 196, 357–366 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilhelm S., Gdynia A., Tielen P., Rosenau F., Jaeger K.-E., The autotransporter esterase EstA of Pseudomonas aeruginosa is required for rhamnolipid production, cell motility, and biofilm formation. J. Bacteriol. 189, 6695–6703 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sonnleitner E., Bläsi U., Regulation of Hfq by the RNA CrcZ in Pseudomonas aeruginosa carbon catabolite repression. PLoS Genet. 10, e1004440 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sonnleitner E., et al. , Novel targets of the CbrAB/Crc carbon catabolite control system revealed by transcript abundance in Pseudomonas aeruginosa. PLoS One 7, e44637 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gill E. E., et al. , High-throughput detection of RNA processing in bacteria. BMC Genomics 19, 223 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Waligora E. A., et al. , AmrZ beta-sheet residues are essential for DNA binding and transcriptional control of Pseudomonas aeruginosa virulence genes. J. Bacteriol. 192, 5390–5401 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jones C. J., et al. , ChIP-Seq and RNA-Seq reveal an AmrZ-mediated mechanism for cyclic di-GMP synthesis and biofilm development by Pseudomonas aeruginosa. PLoS Pathog. 10, e1003984 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Browning D. F., Busby S. J., Local and global regulation of transcription initiation in bacteria. Nat. Rev. Microbiol. 14, 638–650 (2016). [DOI] [PubMed] [Google Scholar]
- 51.Melamed S., Adams P. P., Zhang A., Zhang H., Storz G., RNA-RNA interactomes of ProQ and Hfq reveal overlapping and competing roles. Mol. Cell 77, 411–425.e7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mooney R. A., et al. , Regulator trafficking on bacterial transcription units in vivo. Mol. Cell 33, 97–108 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Valouev A., et al. , Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nat. Methods 5, 829–834 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thorvaldsdóttir H., Robinson J. T., Mesirov J. P., Integrative genomics viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 14, 178–192 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shishkin A. A., et al. , Simultaneous generation of many RNA-seq libraries in a single reaction. Nat. Methods 12, 323–325 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dove S. L., Hochschild A., A bacterial two-hybrid system based on transcription activation. Methods Mol. Biol. 261, 231–246 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data reported in this paper have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) under accession number GSE138338.