Significance
Glaucoma is a leading cause of irreversible blindness worldwide. A major risk factor is increased intraocular pressure (IOP), and all current treatment strategies for slowing disease progression center on IOP reduction. Increased IOP generally results from reduced drainage of aqueous humor through two routes in the anterior segment of the eye called the conventional and uveoscleral outflow pathways. We use high-throughput single-cell RNA sequencing to identify cell types within these pathways and the genes that they express both in humans and in four model species frequently used to study IOP regulation and glaucoma.
Keywords: trabecular meshwork, ciliary muscle, macaque, pig, mouse
Abstract
Increased intraocular pressure (IOP) represents a major risk factor for glaucoma, a prevalent eye disease characterized by death of retinal ganglion cells; lowering IOP is the only proven treatment strategy to delay disease progression. The main determinant of IOP is the equilibrium between production and drainage of aqueous humor, with compromised drainage generally viewed as the primary contributor to dangerous IOP elevations. Drainage occurs through two pathways in the anterior segment of the eye called conventional and uveoscleral. To gain insights into the cell types that comprise these pathways, we used high-throughput single-cell RNA sequencing (scRNAseq). From ∼24,000 single-cell transcriptomes, we identified 19 cell types with molecular markers for each and used histological methods to localize each type. We then performed similar analyses on four organisms used for experimental studies of IOP dynamics and glaucoma: cynomolgus macaque (Macaca fascicularis), rhesus macaque (Macaca mulatta), pig (Sus scrofa), and mouse (Mus musculus). Many human cell types had counterparts in these models, but differences in cell types and gene expression were evident. Finally, we identified the cell types that express genes implicated in glaucoma in all five species. Together, our results provide foundations for investigating the pathogenesis of glaucoma and for using model systems to assess mechanisms and potential interventions.
Glaucoma, a leading cause of irreversible blindness worldwide (1), results from loss of retinal ganglion cells (RGCs), which carry information about the visual world from the eye to the rest of the brain (2). Of the major risk factors, including age, race, and family history, the only modifiable one is intraocular pressure (IOP). Indeed, lowering IOP remains the only proven treatment strategy to delay disease progression, with several pharmacological and surgical approaches in widespread clinical use (2). However, therapeutic advances have been limited by incomplete understanding of the tissues that regulate IOP and the means by which increased IOP leads to RGC loss. We address the first of these issues here.
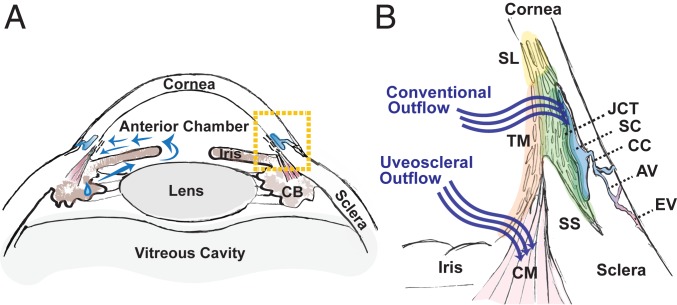
There are two major compartments in the eye, an anterior segment containing aqueous humor (AH) and a posterior segment containing vitreous humor. The primary determinant of IOP is the equilibrium between the production and drainage of AH. AH is produced by the ciliary body, circulates within the anterior chamber, and is then drained through one of two pathways, conventional (trabecular) or uveoscleral (3) (Fig. 1). In the conventional pathway, AH exits the anterior chamber through a lattice-like biological filter called the trabecular meshwork (TM), which is composed of collagenous beams lined with specialized TM cells. It then passes through a region of cells embedded within a denser extracellular matrix called the juxtacanalicular tissue (JCT) prior to being conveyed into a specialized vessel called Schlemm canal (SC). From SC, AH exits the eye through a network of collector channels (CCs) continuous with the venous system. The remaining AH exits the anterior chamber via the uveoscleral pathway, draining through the interstices of the ciliary muscle (CM) and ultimately exiting the eye via the suprachoroidal space and the sclera. In principle, increased IOP could result from either excessive production or insufficient drainage of AH, but in practice, the latter is more commonly implicated (4–7).
Fig. 1.
Human AH outflow pathways. (A) Diagram of the anterior segment of the human eye, which includes the cornea, iris, ciliary body (CB), and lens. AH is secreted by the CB and circulates (blue arrows) within the anterior chamber prior to draining from the eye through one of two pathways located within the iridocorneal angle delineated by boxed area. (B) Enlarged view of the iridocorneal angle (boxed area in A) highlighting two outflow pathways for AH. In the conventional pathway, AH traverses the TM, first through the uveal meshwork (orange highlight), then the corneoscleral meshwork (light green highlight), and finally, the JCT (dark green highlight) prior to entering SC. AH exits the SC via CCs that empty into aqueous veins (AVs) that themselves merge with episcleral veins (EVs). Nonfiltering TM is located at the insert region (yellow highlight), referred to as Schwalbe line (SL), which abuts the corneal endothelium. In the uveoscleral pathway, AH exits via the interstices of the CM. SS, scleral spur.
The cellular composition of the AH outflow pathways has been studied histologically (8, 9), but because of their small size, intricate architecture, and tightly invested tissues, it is impractical to dissect individual components for molecular analysis. Therefore, most studies of gene expression in these tissues have relied on bulk measurements of tissue or cultured cells (10–13). It has, therefore, been difficult to assess gene expression patterns of individual cell types. To circumvent this limitation, we used high-throughput single-cell RNA sequencing (scRNAseq) (SI Appendix, Fig. S1A). We profiled ∼24,000 single cells from adult human TM and surrounding tissues, applied computational methods to cluster the cells based on their transcriptomes, and used histological methods to match the molecularly defined clusters to specific cell types. We identified 19 cell types and defined molecular markers for each of them.
We then used this cell atlas in two ways. First, we assessed expression of genes that have been implicated in glaucoma either as causal genes with Mendelian inheritance or as susceptibility loci identified in genome-wide association studies (GWAS) (14–22), and we compared expression levels in cell types of the outflow pathways with those in RGCs and retinal glia (23). We found that while many genes were preferentially expressed in the anterior segment, others were expressed predominantly in the retina.
Second, we used the human atlas as a foundation for assessing four animal models frequently used to study AH outflow pathways and glaucoma—cynomolgus macaque (Macaca fascicularis), rhesus macaque (Macaca mulatta), pig (Sus scrofa), and mouse (Mus musculus) (24–26). The utility of these models for disease research depends in large part on the extent to which their cell types and patterns of gene expression correspond to those in humans. We show that broad cell classes were generally conserved across all five species and could be localized to expected areas within the eye but that some cell types within classes were less well conserved. In some cases, molecular markers specific for human cell types were either absent or much less specific for cell types within other species. Disease genes in humans, which often demonstrated highly specific expression in outflow pathway cell types, mapped well in macaque species but less reliably in the pig and mouse. These results may help guide tests of therapeutic strategies in these models.
Results
Cell Atlas of Human Trabecular Meshwork and Aqueous Outflow Structures.
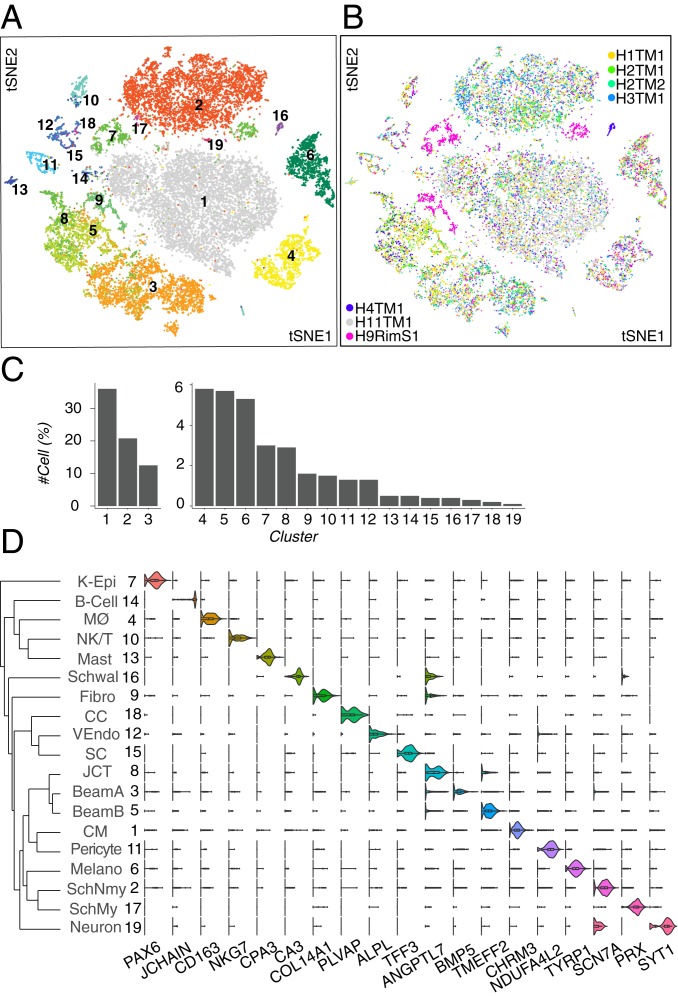
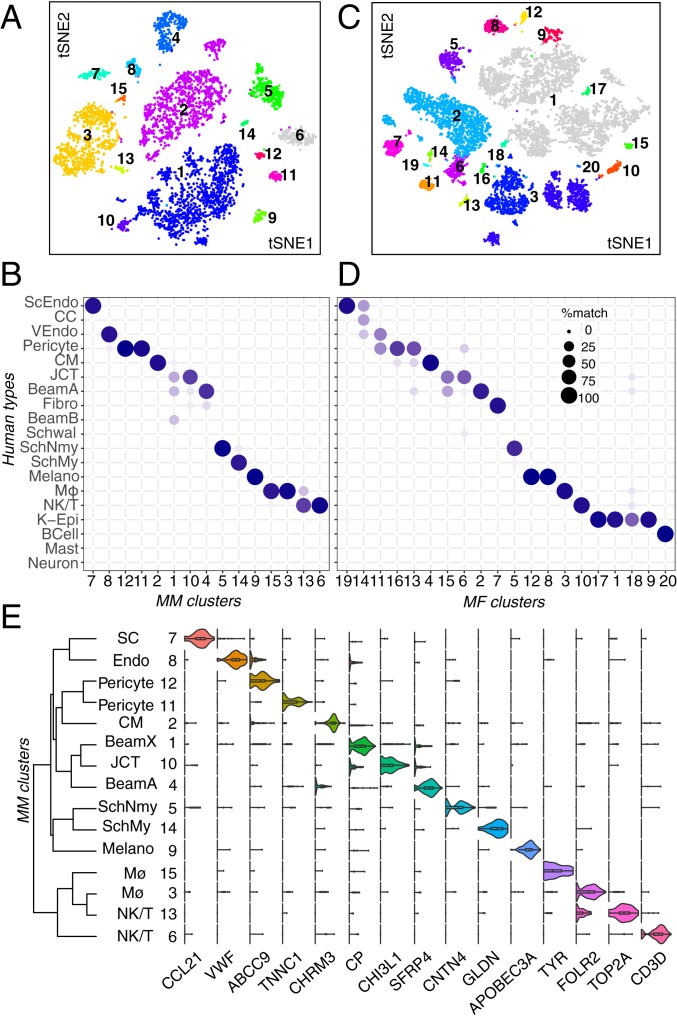
We used a droplet-based method (27) to obtain 24,023 high-quality single-cell transcriptomes from human TM tissue dissected from seven eyes of six postmortem donors (SI Appendix, Fig. S1B and Table S1). Computational analysis (SI Appendix) divided these cells into 19 clusters (Fig. 2A), ranging in frequency from 0.1 to 36% of cells profiled (Fig. 2C). Cells in nearly all clusters were obtained from all individuals (Fig. 2B) (the outlying clusters, C7 and C9, are discussed below). We identified differentially expressed genes for each cluster (Fig. 2D) and then used in situ hybridization and immunohistochemistry to relate the molecularly defined clusters to cell types defined by classical criteria of morphology and location.
Fig. 2.
Cell types and gene expression in the human outflow pathways. (A) Clustering of 24,023 single-cell expression profiles from human TM and associated structures visualized by t-distributed stochastic neighbor embedding (tSNE). Arbitrary colors are used to distinguish clusters deemed to be distinct by unsupervised analysis. Clusters were numbered according to relative size, with 1 being the largest. (B) tSNE plot shown in A but with cells colored by sample of origin. Note that corneal epithelial cells (C7) and fibroblasts (C9) were derived primarily from the rim sample (H9). (C) Frequency of each cell type; numbering is the same as in A and D. (D) Violin plots showing expression of genes selectively expressed by cells of each type. The dendrogram in Left shows transcriptional relationships among cell types. Fibro, fibroblast; K-Epi, corneal epithelium; Melano, melanocyte; MØ, macrophage; SchMy, myelinating Schwann cell; SchNmy, nonmyelinating Schwann cell; Schwal, Schwalbe line; VEndo, vascular endothelium.
Conventional pathway.
The conventional pathway includes the TM, SC, CCs and closely associated structures (Fig. 1B). The TM includes an anterior, nonfiltering, insert region and a more posterior filtering region, which itself is divided into an inner part composed of “beam cells” through which AH first filters and an outer part composed of JCT cells residing adjacent to SC. These structures modulate outflow resistance through cell–cell interactions as well as through the generation and degradation of extracellular matrix (5, 7, 28–30).
We identified eight cell types within the conventional pathway, three of which corresponded to filtering TM cells. They were characterized and localized as follows.
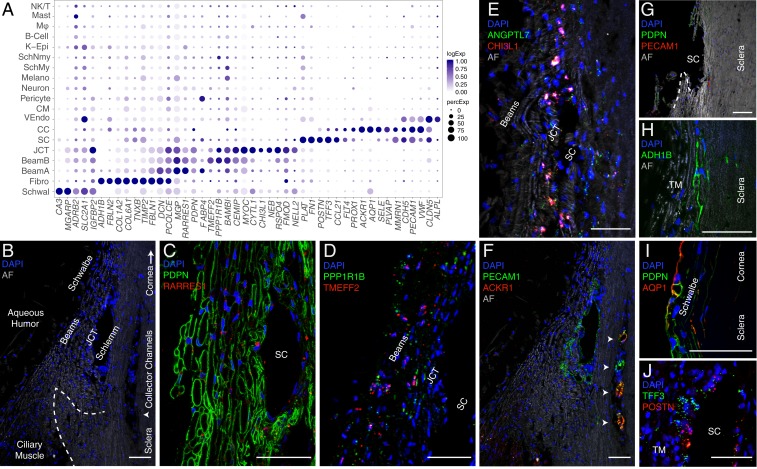
Clusters 3, 5, and 8 demonstrated high expression levels of MYOC, MGP, and PDPN (Fig. 3A), markers previously associated with TM, as well as other genes, such as RARRES1 (7, 31, 32). We used immunostaining for PDPN and RARRES1 to confirm that these genes were expressed by cells encasing the trabecular beams (Fig. 3 B and C and SI Appendix, Fig. S2 K and L). Clusters 3 and 5 could be distinguished by preferential expression of FABP4 and TMEFF2, respectively, each of which marked a subset of beam cells, which we call Beam A and Beam B (Fig. 3D and SI Appendix, Fig. S2 A, B, and J). The FABP4+ Beam A cells and TMEFF2+ Beam B cells were intermingled but with a tendency for the latter to be closer to the JCT. Cluster 8 could be distinguished from C3 and C5 by selective expression of CHI3L1 and ANGPTL7, both previously suggested to be potential markers for a TM cell subpopulation (13). In situ hybridization demonstrated localization of cells expressing these genes predominantly to the JCT (Fig. 3E and SI Appendix, Fig. S2I). Other differentially expressed genes in this cluster (C8) included RSPO4, FMOD, and NELL2. Thus, our transcriptomic method identifies three cell types comprising the filtering TM: two types of beam cells and a distinct JCT cell.
Fig. 3.
Cells of the human conventional outflow pathway. (A) Dot plot showing genes selectively expressed in cells of the conventional outflow pathway. In this and subsequent figures, the size of each circle is proportional to the percentage of cells expressing the gene, and the shade intensity depicts the average normalized transcript count in expressing cells. Fibro, fibroblast; K-Epi, corneal epithelium; Melano, melanocyte; MØ, macrophage; SchMy, myelinating Schwann cell; SchNmy, nonmyelinating Schwann cell; Schwal, Schwalbe line; VEndo, vascular endothelium. (B) Labeled meridional section of a human corneoscleral rim visualized using autofluorescence (AF) demonstrates outflow anatomy at the iridocorneal angle visualized after removal of iris and ciliary body. The dashed line indicates scleral spur and arrowhead indicates region of CCs. (C) TM (beam and JCT) cells immunostained for PDPN (green) and RARRES1 (red). (D) Fluorescent RNA in situ hybridization against TMEFF2 (red) and PPPR1B1 (green) highlights beam cells. (E) Fluorescent RNA in situ hybridization against ANGPTL7 (green) and CHI3L1 (red) highlights cells in the JCT. (F) Immunostaining for PECAM1/CD31 (green) highlights SC, CCs (arrowheads), and CM capillaries, while ACKR1/DARC (red) costains only the CCs. The same field as in B. (G) Corneoscleral rim section demonstrates the tissue left behind after TM dissection. Very few cells in this region are positive for PDPN (green) or PECAM1/CD31 (red), indicating successful removal of relevant structures (TM and SC) during dissection protocol. (H) Scleral fibroblasts identified in the corneoscleral rim collection immunostained for ADH1B. (I) Schwalbe line cells at the junction of TM and corneal endothelium costain with PDPN (green) and AQP1 (red). (J) Fluorescent RNA in situ hybridization against POSTN (red) and TFF3 (green) highlights SC endothelium. DAPI, 4′,6-diamidino-2-phenylindole (nuclear stain). (Scale bars: 50 µm.)
A fourth cluster (C16) corresponded to cells of Schwalbe line, which is located in the “insert” region of the anterior nonfiltering meshwork abutting the peripheral corneal endothelium. This cluster had differentially expressed genes associated with corneal endothelium (e.g., CA3, MGARP, SLC11A2, TGFBI), but some cells shared markers associated with beam cells (e.g., MYOC, IGFBP2, NELL2, PTGDS). The candidate TM marker AQP1 was preferentially expressed in these nonfiltering TM cells, consistent with the observation that deletion of AQP1 in mice did not affect outflow facility (Fig. 3I) (33). Although it has been suggested that Schwalbe line contains TM precursors (34), we detected no differentially expressed genes suggestive of “stemness” (35) in this cluster; if present, they may have been too rare to be detected.
SC endothelial cells (C15) expressed canonical endothelial markers PECAM-1 (CD31), vascular endothelial-cadherin (CDH5), and CLDN5 as well as the hemostasis genes PLAT and VWF (Fig. 3A). This cluster also selectively expressed the lymphatic endothelial cell markers CCL21 and FLT4 (VEGFR3), consistent with the notion that SC is a modified lymphatic vessel (36–38). Other lymphatic endothelial markers expressed in SC at lower levels were PROX1 and LYVE1. Fibronectin-1 (FN1), a Transforming Growth Factor-beta–inducible extracellular matrix (ECM) gene implicated in glaucoma pathogenesis, was differentially expressed in SC endothelium as was MMRN1 (39, 40). We used the markers POSTN and TFF3 (Fig. 3J) to distinguish endothelium of the outflow tract from the vascular endothelium in the CM, which shared many endothelial markers (discussed later). Some intermingled cells in SC expressed PECAM-1 (CD31) and the vascular endothelial marker ALPL (Fig. 4C), raising the possibility that this structure contains two cell types.
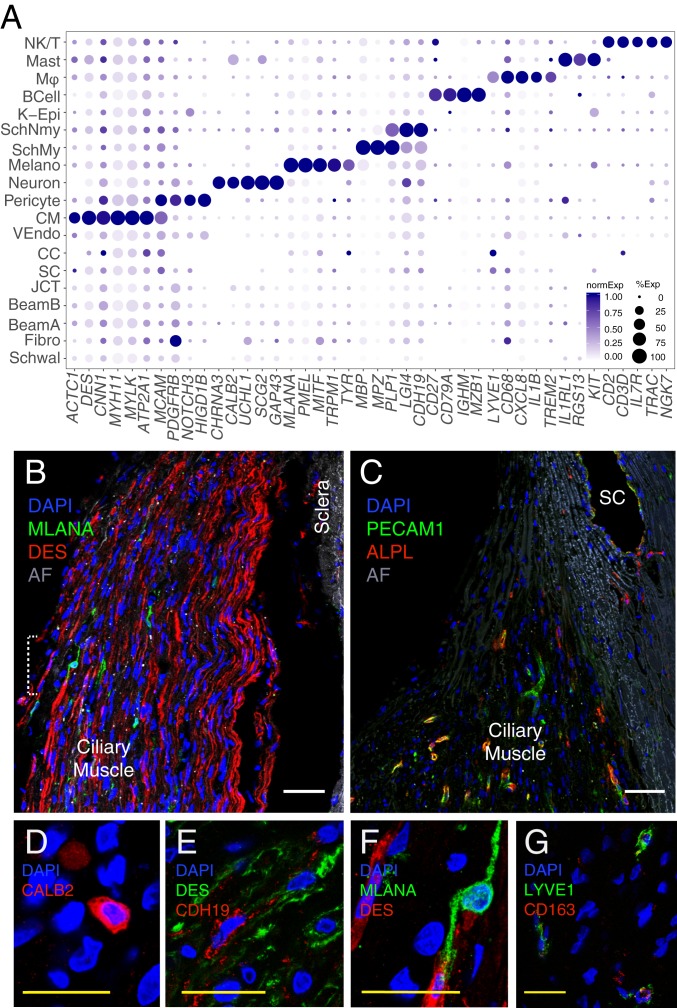
Fig. 4.
Cells of the human uveoscleral pathway. (A) Dot plot showing genes selectively expressed in cells of the uveoscleral outflow pathway. Fibro, fibroblast; K-Epi, corneal epithelium; Melano, melanocyte; MØ, macrophage; SchMy, myelinating Schwann cell; SchNmy, nonmyelinating Schwann cell; Schwal, Schwalbe line; VEndo, vascular endothelium. (B) Smooth muscle cells immunostained for DES (red) and melanocytes immunostained for MLANA (green) in CM. (C) Capillaries in the CM immunostained for PECAM1 (green) and ALPL (red). Occasional PECAM1+ALPL+ staining was also noted in SC, suggesting that this structure contains more than one cell type. (D) Immunostaining for CALB2 (red) highlights intrinsic neurons of the CM. (E) Immunostaining for LYVE1 (green) and CD27 (red) identifies macrophages in the TM. (F) Schwann cells in the CM immunostained for CDH19 (red) amid CM cells stained for DES (green). (G) Higher magnification of the area bracketed in B demonstrates an MLANA+ melanocyte (green). AF, autofluorescence; DAPI, 4′,6-diamidino-2-phenylindole (nuclear stain). (Scale bars: B and C, 50 µm; D–G, 25 µm.)
Because blunt dissection of the TM may have left behind components of the conventional outflow structures such as the outer wall of SC, we collected a corneoscleral rim sample, which included all adjacent tissues after blunt dissection (Fig. 3G). This sample contained an additional vascular cell type (C18) and a fibroblastic type (C9) that were poorly represented in the other samples. Using the differentially expressed gene ACKR1 (DARC), we identified C18 as the endothelium of the CCs downstream from SC (Fig. 3F). ACKR1 has been identified as a marker to distinguish venular from nonvenular endothelial cells, which is consistent with the pre-/perivenular location of the channels (41). Other differentially expressed genes for CC cells include SELE, SELP, and COL15A1. The other cluster, C9, consisted of matrix fibroblasts. Immunostaining for the selectively expressed gene ADH1B showed that C9 cells were present within the sclera located adjacent to the TM and outer wall of SC (Fig. 3H and SI Appendix, Fig. S2H). This cluster was distinguished from TM cell types through its higher level of expression of FBLN2, TIMP2, TNXB, and multiple collagen genes (COL1A2, COL6A1, COL6A2, COL6A3, COL14A1), markers consistent with previous single-cell studies on fibroblasts in other organ systems (42). However, it also shared a set of ECM and complement genes with TM cells (C3, -5, -8), including DCN, PCOLCE, FBLN1, MFAP4, SERPING1, C1S, and C1R, suggesting that these neighboring cells play overlapping roles.
Finally, the corneoscleral rim sample included cells with transcriptomic signatures marking them as corneal epithelium (C7) based on expression of previously described markers, such as AQP3, AQP5, and KRT5 (43). These cells will be described elsewhere.
Uveoscleral pathway.
AH that does not exit the eye through the conventional pathway instead exits via the uveoscleral pathway, draining through the interstices of the CM. Seven clusters in our dataset comprised components of this pathway. The largest cluster (C1) corresponded to CM cells (44). It expressed markers classically associated with well-differentiated smooth muscle, including DES, CNN1, MYH11, MYLK, and ACTC1 (Fig. 4A). We used the differentially expressed genes DES and CHRM3 to localize this cluster histologically to the longitudinal CM (Fig. 4B and SI Appendix, Fig. S2C). The high expression levels of ATP2A1/SERCA1, a marker of type II fast twitch skeletal muscle, suggest that CM may also have characteristics of skeletal muscle, although it lacks expression of other classical skeletal muscle markers, including MYH1, -2, -4, and -7.
Six additional cell types were found within the CM. One, vascular endothelium (C12), was associated with intramuscular capillaries. This cluster shared canonical endothelial markers with SC and CCs but was distinguished from the latter by the presence of ALPL (Fig. 4C) and absence of lymphatic markers. Similarly, PLVAP, while present in SC and CCs, was not expressed in CM capillary endothelium, consistent with observations that these cells do not have pores (39, 45).
C11 comprises contractile pericytes, which express canonical markers PDGFRB, MCAM/CD146, and NOTCH3 as well as HIGD1B (46). These cells were labeled by immunostaining against NDUFA4L2 and were wrapped around small vessels (SI Appendix, Fig. S2D).
A small neuronal cluster (C19) expressing CHRNA3, UCHL1, SCG2, and GAP43 was identified and localized to the CM by immunostaining against CALB2 (aka calretinin) and ELAVL4 (aka HuD) (Fig. 4D and SI Appendix, Fig. S2E). These cells presumably correspond to the sparse neurons described in histological and ultrastructural studies (47, 48).
Two clusters, C2 and C17, expressed markers characteristic of Schwann cells, consistent with prior electron microscopic studies showing that Schwann cells ensheath axons within CM (45). Markers common to both clusters included PLP1 and LGI4. The clusters were distinguished by differential expression of CDH19 in C2 and a set of genes that encode myelin components, such as MBP, MPZ, and PMP2, in C17. Immunostaining for CDH19 marked these cells within the CM (Fig. 4E). We conclude that C2 and C17 represent nonmyelinating and myelinating Schwann cells, respectively.
Finally, C6 comprised uveal melanocytes, demonstrating expression of canonical markers MLANA, PMEL, MITF, TRPM1, and TYR. Using the marker MLANA, we localized these cells to the CM (Fig. 4F).
Immune cells.
Our dataset also included four types of immune cells: B cells, Natural Killer (NK)/T cells, mast cells, and macrophages. The macrophages (C4) were CD163+ and LYVE1+ (49) and localized predominantly to the TM (Fig. 4G). They also expressed CD68, CD14, CCL3, CCL4, CXCL8, IL1B, TREM2, and MS4A genes, all of which have been associated with macrophages in other tissues. Mast cells (C13) were localized to the TM using the marker IL1RL1 and also expressed CPA3, RGS13, and KIT (SI Appendix, Fig. S2F). B cells (C14), characterized by expression of CD27, CD79A, IGHM, IGKC, MZB1, and JCHAIN, were found in only one donor sample but were identified histologically in tissues from other donors using the marker CD27 (SI Appendix, Fig. S2G). NK/T cells (c10) were identified by differential expression of the genes CD2, CD3D, IL7R, TRAC, GZMA, GZMB, and NKG7.
Cell Type-Specific Expression Patterns of Glaucoma-Associated Disease Genes.
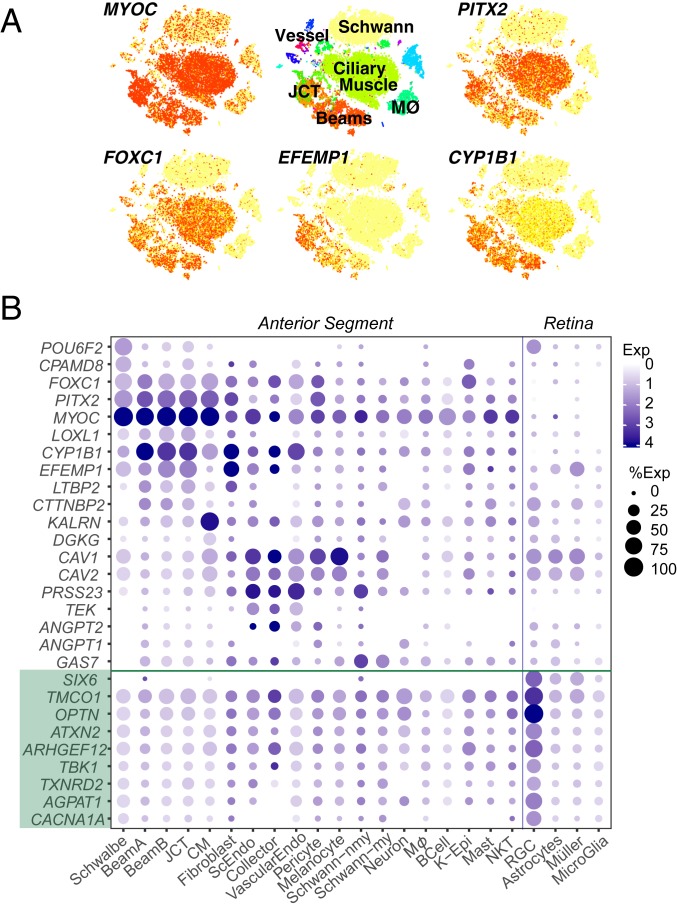
To improve understanding of how genes associated with glaucoma contribute to disease pathogenesis, we mapped their expression by the 19 cell types in our atlas. We included both known monogenic causes (Mendelian genes) and genes implicated as risk factors in GWAS (14–22). Mendelian genes assessed were ANGPT1, ANGPT2, CPAMD8, CYP1B1, FOXC1, LOXL1, LTBP2, MYOC, OPTN, PITX2, TEK (TIE2), and TBK1. Well-established GWAS loci genes included TMCO1, TXNRD2, CAV1, CAV2, ARHGEF12, and ATXN2. We also screened an additional 462 genes listed in the National Human Genome Research Institute–European Bioinformatics Institute (NHGRI-EBI) GWAS Catalog for notable expression patterns (50). Overall, 189 genes were expressed in a minimum of 10% of cells in any given class at a minimum expression level of 0.5. Examples are shown in Fig. 5, with a full list in SI Appendix, Fig. S3.
Fig. 5.
Human disease genes. Cell type-specific expression of several genes implicated in glaucoma as illustrated by t-distributed stochastic neighbor embedding (A; arranged as in Fig. 2A) and a dot plot (B). B also includes data from a retinal cell atlas, showing expression in RGCs and three types of retinal glia. Green shading highlights genes that genetics studies suggest may confer additional IOP-independent glaucoma risk; many of these genes are also associated with high IOP. K-Epi, corneal epithelium; MØ, macrophage; ScEndo, Schlemm canal endothelium; Schwann-my, myelinating Schwann cell; Schwann-nmy, nonmyelinating Schwann cell; VascularEndo, vascular endothelium.
Most genes implicated in juvenile glaucoma and high IOP (e.g., MYOC, FOXC1, PITX2, CYP1B1) were expressed strongly by all three TM cell types (Beam A, Beam B, and JCT), with no evidence for selective expression by any one of the three (Fig. 5 and SI Appendix, Fig. S2 K and L). Notably, they were all also expressed at high levels in CM. EFEMP1 exhibited robust cell type-specific expression in beam cells and JCT; in contrast to the first group, it was not present in CM and was expressed differentially among the TM types: JCT > Beam B > Beam A. In contrast, some genes demonstrated stronger expression in non-TM cell types than in TM, including GAS7 (CM, Schwann cells), KALRN (CM), PRSS23 (SC, CC, vessel endothelium, and Schwann cells), and CAV1/CAV2 (all three endothelial types, CM, pericytes, and melanocytes). Thus, although defects in the TM are clear contributors to elevated IOP, genes that regulate IOP may not act exclusively within the TM.
Because glaucoma risk involves susceptibility of RGCs to degeneration as well as increased IOP, we also interrogated expression in four relevant retinal cell types: RGCs and three glial types with which they interact (astrocytes, Müller glia, and microglia). Data for retinal cells are taken from a human retinal cell atlas (Materials and Methods) (23). We found a distinction between genes associated with primary open-angle glaucoma (POAG) involving elevated IOP and those for which an IOP-independent component may exist. Whereas most genes associated with POAG and/or elevated IOP were expressed in cells of the outflow pathways as discussed above, a subset, including OPTN, ATXN2, TMCO1, and SIX6, were expressed at the highest levels in RGCs (Fig. 5B).
Several susceptibility genes, including CAV1, CAV2, and POU6F2, were expressed at high levels in both the anterior segment and retina. POU6F2 is a transcription factor that has been linked to thinner than average central corneal thickness, a highly heritable trait and also a strong risk factor for the development of POAG (51). The expression of POU6F2 has been detected in human RGCs (52), mouse corneal limbal stem cells, and a subset of mouse RGCs (53). We found preferential expression of POU6F2 in both corneal endothelium (specifically, Schwalbe line cells) and RGCs (Fig. 5B).
Two genes associated with exfoliation glaucoma (XFG) and the related exfoliation syndrome (XFS), LOXL1 and CACNA1A, demonstrated divergent regional expression patterns: the former localized predominantly to TM (beam cells and JCT), whereas the latter localized preferentially to RGCs. This result, consistent with prior investigations (22), offers further insight into XFG as both a primary and a secondary open-angle glaucoma characterized not only by IOP elevation due to TM obstruction but also by an inherent susceptibility to glaucomatous optic neuropathy.
Finally, motivated by literature implicating the complement system, aquaporins, matrix metalloproteinases (MMPs), cytochrome P450 enzymes, and prostaglandins in ocular disease pathogenesis, progression, and treatment (54, 55), we investigated their expression in the anterior segment. We found that many genes encoding complement factors were selectively expressed in the conventional outflow pathway. For example, expression of C1QA, C1QB, and C1QC was observed in resident macrophages, whereas the serine proteases C1R and C1S; the C1 inhibitor SERPING1; and other complement genes CFD, CFH, C3, and C7 were expressed preferentially in both TM cells and scleral fibroblasts (SI Appendix, Fig. S4A). Among MMPs, MMP2 and MMP14 were most predominant (SI Appendix, Fig. S4C); the former was selectively expressed by beam cells and SC endothelium as well as scleral fibroblasts, whereas the latter was expressed mostly in scleral fibroblasts. AQP1 and CYP1B1 were uniquely expressed in outflow cell types among their respective family members (SI Appendix, Fig. S4 B and E). Within the Rho-protein family, RHOB demonstrated the strongest expression among outflow cell types followed by RHOA and RHOC (SI Appendix, Fig. S4D). The prostaglandin genes PTGDS and PTGES3 were robustly expressed in all outflow cell types, whereas PTGS2, PTGR1, and PTGER2 were more selectively expressed in the conventional outflow pathway cells (SI Appendix, Fig. S4F).
Model Species.
Analysis of mechanisms underlying IOP regulation and preclinical studies of therapeutic interventions rely almost entirely on model species. Yet, limited information is available on how closely cells and molecules of the anterior segment in commonly used models resemble those of humans. To address this issue, we profiled cells dissociated from anterior segment tissues of four model species: the rhesus macaque (M. mulatta; 5,158 cells), the cynomolgus macaque (M. fascicularis; 9,155 cells), the common swine (S. scrofa; 6,709 cells), and mouse (M. musculus; 5,067 cells). These species are among the most commonly used for studies on glaucoma: rhesus and cynomolgus monkeys are frequently used for basic studies of primate visual physiology and preclinical tests, respectively (26); porcine anterior segments are used in aqueous outflow studies (24); and the broadest range of genetically modified lines is available in mice (25). We used a machine learning algorithm [XGBoost (56)] (Materials and Methods) to find correspondences of cell types in the model species with those in humans. While significant conservation was noted among certain cell types across species, there were also notable differences.
Macaca mulatta.
Cells collected from anterior chamber angle structures of M. mulatta clustered into 15 types (Fig. 6 A and E). We identified three TM-like clusters in this species, similar to humans (Fig. 6B). One cluster corresponded to human Beam Cell A (mmC4), sharing markers including BMP5 and EDN3; another corresponded to human JCT (mmC10), sharing preferential expression of markers including ANGPTL7 and CHI3L1. The third cluster (mmC1), which we call Beam Cell X, could not be mapped to a single human cluster but instead, shared multiple markers of all human TM types; it was distinguished from mmC4 and mmC10 by its preferential expression of CYP1B1 and MGARP. We identified types mapping 1:1 to human SC (mmC7), vascular endothelium (mmC8), and the uveoscleral outflow pathway, including CM (mmC2), melanocytes (mmC9), nonmyelinating Schwann cells (mmC5), and myelinating Schwann cells (mmC14). Two types of pericytes were present (mmC11, mmC12) compared with one in humans. Among immune cells, macrophage (mmC3, mmC15) and NK/T (mmC6, mmC13) clusters were present. B cells, mast cells, and neurons were not detected, possibly due to their sparsity. Finally, due to a conservative dissection of the TM strip in this species, CC endothelium, corneal epithelium, and Schwalbe line cells were not recovered.
Fig. 6.
Cell types and gene expression in the outflow pathways of two macaque species (M. mulatta, MM; and M. fascicularis, MF). (A) A t-distributed stochastic neighbor embedding (tSNE) plot showing 15 cell types derived from TM and associated structures of M. mulatta. (B) Transcriptional correspondence between human and M. mulatta cell types summarized as a ‘‘confusion matrix.’’ In this and subsequent figures, the size of the circle and its intensity indicate the percentage of cells of a given cluster from the model species (column) assigned to a corresponding human cluster (row) by a classification algorithm trained on the human cells. Fibro, fibroblast; K-Epi, corneal epithelium; Melano, melanocyte; MØ, macrophage; ScEndo, Schlemm canal endothelium; SchMy, myelinating Schwann cell; SchNmy, nonmyelinating Schwann cell; Schwal, Schwalbe line; VEndo, vascular endothelium. (C) A tSNE plot showing 20 cell types derived from TM and associated structures of M. fascicularis. (D) Transcriptional correspondence between human and M. fascicularis shown as in B. (E) Violin plot showing examples of genes selectively expressed by each cell type in M. mulatta.
Macaca fascicularis.
Cells collected from anterior chamber angle structures of M. fascicularis clustered into 19 types (Fig. 6C and SI Appendix, Fig. S5A). We identified three TM-like clusters (mfC2, mfC6, mfC15). Of these, one cluster, mfC2, preferentially expressed BMP5 and EDN3 and could be confidently mapped to human Beam Cell A (Fig. 6D). The other two clusters (mfC6 and mfC15) expressed genes observed in multiple human TM clusters, including MYOC, MGP, and ANGPTL7; overall, mfC6 shared more genes with the human JCT cluster (e.g., FMOD, CEMIP) whereas mfC15 expressed a combination of genes found within the the human beam cell, fibroblast, and Schwalbe line clusters (e.g., ANGPTL5, COL6A3, AQP1, and POU3F3) (SI Appendix, Fig. S5A). Three vascular endothelial clusters were identified (mfC11, mfC14, mf19); these corresponded to the human capillary endothelium, CCs, and SC endothelium, respectively. Other clusters mapped closely to cell types in the human dataset, including CM (mfC4), Schwann cells (mfC5), macrophages (mfC3), B cells (mfC20) and NK/T cells (mfC10), melanocytes (mfC8, mfC12), and pericytes (mfC13, mfC16). No neurons were identified.
Based on results from human, we profiled cells from one sample that included the residual corneoscleral rim in addition to TM strips in hopes of identifying CC endothelium and matrix fibroblasts. Indeed, both additional cell types were recovered in addition to multiple epithelial cell types (not described here). Matrix fibroblasts appeared during the initial unsupervised clustering as mfC7. CC endothelium (mf14) could be identified within the original SC cluster using a supervised approach based on differential expression patterns identified in humans (SI Appendix, Fig. S5B).
Sus scrofa.
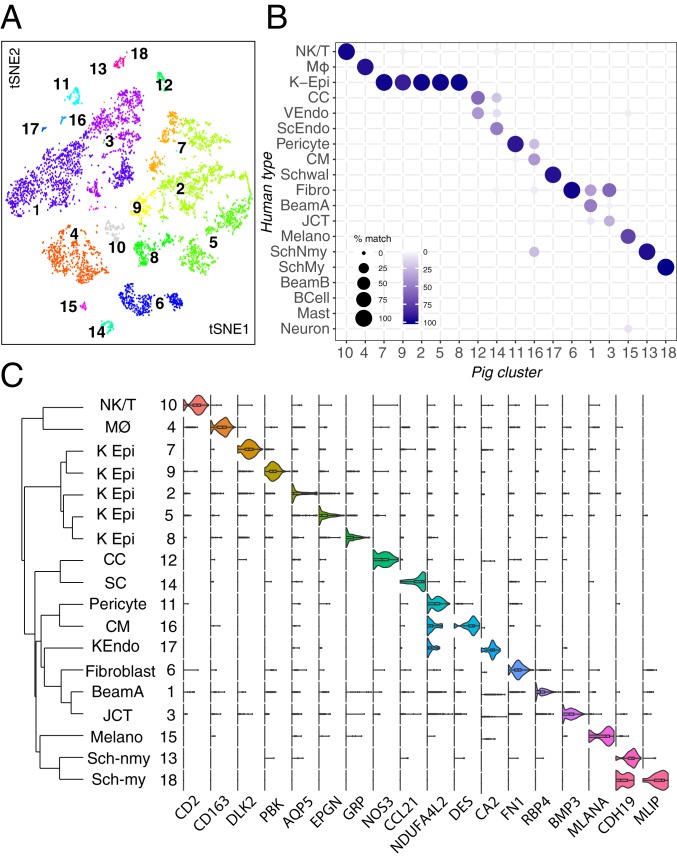
Molecular profiling of porcine anterior segment tissue yielded 18 clusters (Fig. 7A). Two clusters comprised TM cells. One mapped to human Beam Cell A (ssC1), with differential expression of SFRP4 and TMEFF2 in addition to PON1, FMO1, and RARRES1. The other mapped to human JCT (ssC3), selectively expressing CCN3, FMO3, CEMIP, and BMP3 (Fig. 7 B and C). While expression of MYOC was low across all clusters, it was preferentially expressed in ssC1. Both TM clusters expressed CYP1B1, MGP, CFH, and NR2F1.
Fig. 7.
Cell types and gene expression in the outflow pathways of the pig. (A) A t-distributed stochastic neighbor embedding (tSNE) plot showing 18 cell types derived from TM and associated structures of pig. (B) Transcriptional correspondence between human and pig cell types shown as in Fig. 6B. Fibro, fibroblast; K-Epi, corneal epithelium; Melano, melanocyte; MØ, macrophage; SchMy, myelinating Schwann cell; SchNmy, nonmyelinating Schwann cell; Schwal, Schwalbe line; VEndo, vascular endothelium. (C) Violin plot showing examples of genes selectively expressed by each cell type in pig. KEndo, corneal endothelium; K Epi, corneal epithelium; ScEndo, Schlemm canal endothelium; Sch-my, myelinating Schwann cell; Sch-nmy, nonmyelinating Schwann cell.
Of two endothelial clusters identified, one mapped to human SC (ssC14), and the other mapped to capillary endothelium (ssC12). While the pig does not possess a continuous SC and instead, has more discontinuous vessels referred to as an aqueous plexus, cells lining this structure resemble those of human SC. The SC was distinguished from capillary endothelium through differential expression of lymphatic markers CCL21, PROX1, and LYVE1 and was histologically validated with immunostaining against LYVE1 (SI Appendix, Fig. S6A).
A CM cluster (ssC16) was marked by preferential expression of DES and ACTA2, both of which we confirmed with immunostaining; the latter also stained pericytes (SI Appendix, Fig. S6B). NDUFA4L2 was expressed predominantly by pericytes (ssC11) and CM but also, corneal endothelium and to a lesser degree, TM cells (SI Appendix, Fig. S6C). The low yield of CM in our dissection can be attributed to the porcine angle anatomy, which differs from that of primates in having no clear anatomical connection between the CM and conventional drainage structures (57). It is, therefore, easily excluded during dissection.
Other clusters corresponding to human cell types included myelinating (ssC18) and nonmyelinating (ssC13) Schwann cells, melanocytes (ssC15), fibroblasts (ssC6), corneal endothelium (ssC17), macrophages (ssC4), and NK/T cells (ssC10). Five clusters (ssC2, -5, -7 to -9) were derived from ocular surface epithelium due to a permissive dissection technique in which the TM/corneoscleral rim was dissociated. Cell types absent in this collection included mast cells, B cells, and neuronal types.
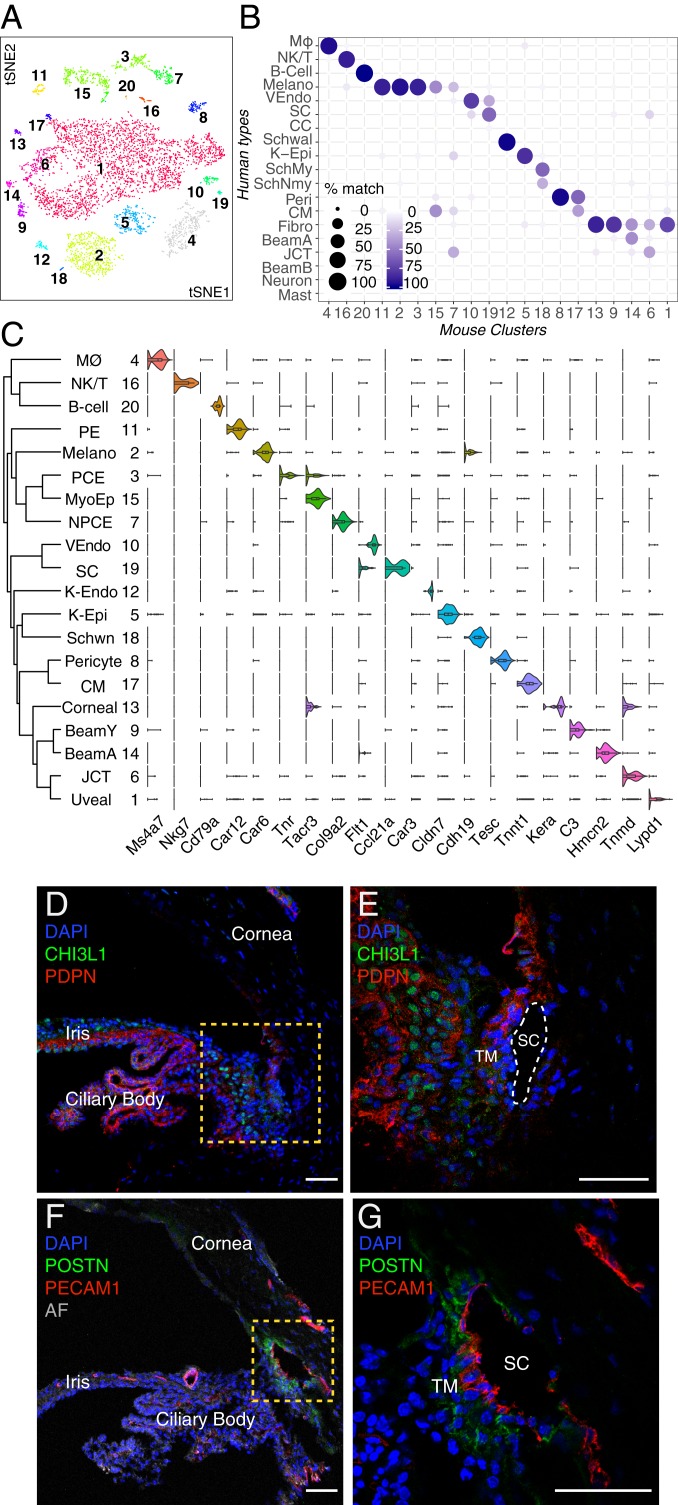
Mus musculus.
The mouse eye is small compared with those of other species profiled, and therefore, we dissociated the entire anterior segment, including cornea, iris, and ciliary body. Presumably for this reason, initial clustering by cell class signatures revealed that 8,766 of 13,833 single-cell profiles corresponded cells of the ocular surface epithelium. We eliminated these cells and reanalyzed the remaining 5,067 cells, yielding 20 clusters (Fig. 8 A and C).
Fig. 8.
Cell types and gene expression in the outflow pathways of the mouse. (A) A t-distributed stochastic neighbor embedding (tSNE) plot showing 20 cell types derived from TM and associated structure of mouse. (B) Transcriptional correspondence between human and mouse cell types shown as in Fig. 6B. Fibro, fibroblast; K-Epi, corneal epithelium; Melano, melanocyte; MØ, macrophage; SchMy, myelinating Schwann cell; SchNmy, nonmyelinating Schwann cell; Schwal, Schwalbe line; VEndo, vascular endothelium. (C) Violin plot showing examples of genes selectively expressed by each cell type in mouse. K-Endo, corneal endothelium; NPCE, nonpigmented ciliary epithelium; PE, pigmented epithelium; Schwn, Schwann cell. (D and E) Pdpn (red) is present in multiple cell types, including pigmented and nonpigmented epithelium of the iris and ciliary body (CB) as well as a subset of TM cells, K-Endo, and K-Epi, whereas Chil1 (green; ortholog to CHI3L1 in humans) stains a different subset of TM cells and to a lesser extent, cells within the CB. (E) Higher magnification of the boxed area in D. (F and G) Immunostaining against Postn (green), a secreted protein, highlights SC and JCT cells. Pecam1 (red) highlights SC as well as vascular endothelial clusters. AF, autofluorescence. DAPI, 4′,6-diamidino-2-phenylindole (nuclear stain). (Scale bars: 50 µm.)
Although the mouse iridocorneal angle is more compact than that of humans, anatomical studies suggest that conventional outflow structures in mice are similar in many respects, with trabecular lamellae, distinctive JCT, and a continuous SC (58). We identified three candidate TM clusters (mC6, mC9, and mC14) through their high levels of Myoc expression. Two of these clusters, mC14 and mC6, mapped preferentially to human Beam Cell A and JCT, respectively, while also demonstrating some ECM expression patterns similar to the human corneoscleral fibroblast cluster (Fig. 8B). The third candidate TM cluster, mC9, despite sharing many markers with fibroblasts (e.g., Pi16, Fbn1, Mfap5, Tnxb, Clec3b), was closely related to mC14 and thus, tentatively named Beam Cell Y. All three TM clusters also expressed Mgp and Pdpn similar to human TM cells; the latter was confirmed histologically with immunostaining as was expression of Chil1 (Chi3l1) in mC6 and mC14 (Fig. 8 D and E and SI Appendix, Fig. S7). Other differentially expressed genes within the mouse JCT cluster included Nell2, Chad, and Tnmd, whereas differentially expressed genes within the mouse beam clusters included Sfrp4 (both mC9 and mC14), Tmeff2 (mC9), and Fmo2 (mC14).
Among other outflow cell types, we identified two separate vessel clusters (mC10, mC19) corresponding to vascular and SC endothelium, respectively. Postn, which was also differentially expressed within the human SC cluster, was also expressed in mouse SC as well as JCT (Fig. 8 F and G).
Other clusters mapping to human cell types included corneal endothelium/Schwalbe line (mC12), smooth muscle (mC17), pericytes (mC8), Schwann cells (mC18), uveal melanocytes (mC2), macrophages (mC4), B cells (mC20), and NK/T cells (mC16) (Fig. 8B). In contrast to our human data, the smooth muscle and Schwann cell clusters were small, which can be attributed in large part to the diminutive murine accommodative apparatus (58).
Owing to the dissection method, the mouse dataset also included a large number of cells derived from iris and ciliary body. We tentatively identified these clusters based on markers identified in previous studies (43, 59); they comprised melanocytes, stromal fibroblasts, pigmented epithelium, and nonpigmented epithelium of the iris and ciliary body.
Conservation of Expression Patterns among Species.
Next, we assessed expression patterns in model species of key genes selectively expressed in cell types of the human AH outflow pathways. In general, conservation was striking.
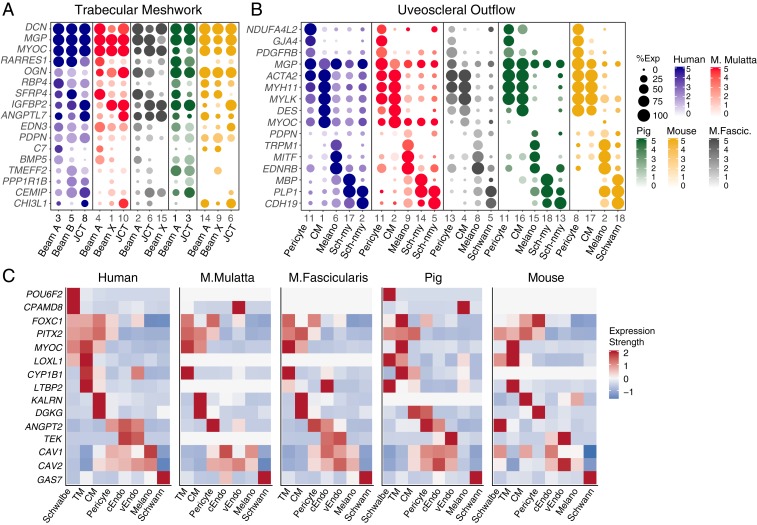
Many markers expressed across all human TM clusters were conserved in other species (Fig. 9A). They included matrix-related genes, such as DCN and MGP, and the retinoic acid-related genes RARRES1 and RBP. However, some differences among species were evident. PDPN, a selective marker for TM cells in humans, was less selective for these cells in other species. For example, it was also found at similar levels in uveal melanocytes in all four nonhuman species. BMP5, expressed in Beam Cell A in human and macaque, was present but less specific in pig and absent in mouse. The human JCT marker, CHI3L1, was preferentially expressed in JCT clusters in monkey and mouse but was notably absent in pig.
Fig. 9.
Comparison of gene expression across species. (A and B) Key genes are shown in dot plots for cell types comprising the TM (A) and uveoscleral outflow pathway (B). (C) Heat map showing expression of genes implicated in human POAG in aqueous outflow cells of humans (replotted from SI Appendix, Fig. S3) and model species. In some cases (i.e. TM, cEndo), multiple cell types from each species have been merged into their broader class to facilitate comparison. cEndo, canal endothelium (includes both Schlemm canal and collector channel clusters where available); Melano, melanocyte; Sch-my, myelinating Schwann cell; Sch-nmy, nonmyelinating Schwann cell; TM, trabecular meshwork (includes beam cells and JCT where available), vEndo, vascular endothelium.
There was also excellent correspondence between markers of other cell types that compose the conventional pathway. Markers of CC, SC, and vascular endothelium markers were generally well conserved among human and macaque (SI Appendix, Fig. S8A). Pig and mouse also shared many markers; however, SC in these species tended to demonstrate more prominent expression of lymphatic markers than primates. Among species in which Schwalbe line/corneal endothelial cells were obtained, there was also good correspondence (SI Appendix, Fig. S8B). Of note, whereas the Schwalbe line cluster in humans includes cells at the junction between cornea and TM, the corresponding clusters in pig and mouse more likely represent a larger proportion of corneal endothelia. Markers of cell types comprising the uveoscleral pathway and immune cells were also very well conserved among species (Fig. 9B and SI Appendix, Fig. S8C).
Finally, we analyzed patterns of disease gene expression in these model species (Fig. 9C). To facilitate comparison, individual TM cell clusters from each species were merged during this analysis, and a threshold was set such that only genes with expression in >10% of cells in at least one cluster were plotted. While some genes were consistently expressed in corresponding cell types across species, others exhibited significant differences. For example, we noted that the classic anterior segment dysgenesis genes FOXC1 and PITX2 were reliably expressed in TM cell types across all species. Similarly, MYOC was also expressed in TM cells across species; however, in this case, subtle differences in other cell types were observed. For example, in human and macaque, MYOC was strongly expressed in both TM and CM, whereas in pig and mouse, it was expressed only in TM. Similarly, MYOC was highly expressed in Schwalbe line cells of human but not in the corresponding corneal endothelial clusters of pig (ssC17) and mouse (ssC12). As mentioned above, this is likely because ssC17 and mC12 represent a larger set of corneal endothelia, not just the peripheral subset. LOXL1, while expressed in TM clusters of human, pig, and mouse, was absent in the TM of both macaque species. KALRN, expressed in human, macaque, and mouse CM, was absent in pig. Finally, CYP1B1, expressed in TM cells across human, macaque, and pig, did not meet threshold expression levels in mouse TM. This is consistent with prior studies, which have reported CYP1B1 complementary DNA but not protein expression in human adult TM samples and negative immunohistochemistry (IHC) staining in mouse TM (60).
Discussion
We used scRNAseq to profile cells composing the AH outflow pathways in humans, generating a cell atlas for these tissues and identifying markers for each cell type. We then used the atlas to localize the expression of genes implicated in glaucoma. These findings offer insights into the molecular architecture of TM cells and highlight potential roles in IOP homeostasis for non-TM cell types in the anterior chamber angle. Finally, we profiled cells in the outflow pathways of four model species—M. mulatta, M. fascicularis, S. scrofa, and M. musculus—providing a foundation for using these models in studies on regulation and dysregulation of IOP.
Technical Issues.
A major challenge in human vision research is that essentially all ocular tissues must be obtained either postmortem or postenucleation. Furthermore, postmortem ocular tissues of the anterior segment suitable for transplantation are understandably prioritized for this purpose over research. In this study, all but one of the postmortem tissues sequenced were obtained within ∼6 h of death from a rapid autopsy program enrolling predominantly oncology patients. While donors had no documented pathological ocular history or clinical evidence of eye disease on examination by an ophthalmologist (T.v.Z), they did have varying degrees of chronic systemic disease and different levels of antemortem exposure to chemotherapeutic and steroid medications. This represents a limitation of our study. Furthermore, lack of documented or physically evident ocular pathology cannot be taken as evidence that none existed. Since we focused primarily on cell type classification in this study rather than quantitative determination of gene expression levels, we believe that the donors’ systemic diseases did not influence the ultimate cell atlas. This was further corroborated in two ways: 1) demonstrating that the same cell types were obtained from all individuals and 2) performing histological validation on tissues obtained from a wider variety of donors through an eye bank as well as on separate rapid autopsy patients.
Human Cell Atlas.
In the human AH outflow pathways, we identified 19 major cell types: 8 belonging to the conventional outflow pathway (including 3 distinct populations within the filtering TM), 7 belonging to the uveoscleral pathway, and 4 representing immune cell populations.
By histological criteria, the filtering TM has been divided into three layers: a uveal layer adjacent to the anterior chamber, a juxtacanalicular layer adjacent to SC, and a corneoscleral layer in between (7, 9). The JCT cells in our dataset clearly localized closest to SC, with the Beam A and B cells localizing to the other two layers. It is tempting to assign Beam A and B to uveal and corneoscleral layers, respectively, but our histological analysis suggests that they are in fact intermingled.
The lymphatic marker podoplanin (PDPN/D2-40) emerged as a robust marker for all human TM cell types (C3, C5, and C8), consistent with previous studies (31, 32). Along with CCL2 (a chemoattractant for monocytes) and VCAM1 (a mediator of immune cell migration), there was a marked specificity of PDPN expression among cells of the conventional pathway compared with the uveoscleral pathway, suggesting that the former acts as an immunological “sink” guiding antigen presenting cells and other immune cells toward SC, the venous system, and ultimately, the spleen (36). Consistent with this idea, SC expresses markers of lymphatic vessels (e.g., CCL21 and FLT4/VEGFR3), whereas CCs express venular markers (e.g., ACKR1).
Model Species.
In comparing the cellular composition of human AH outflow pathways with those of commonly used models, we documented excellent correspondence for many cell types across species. CM cells, pericytes, melanocytes, and Schwann cells were present in all species and demonstrated shared expression of canonical markers. Corneal endothelium was identified in pig and mouse but in neither macaque species, likely due to our targeted dissection technique. Two or more types of vascular endothelium, defined as PECAM1+ TIE1+ clusters, were also present in each species and could be assigned either to SC or to other vasculature. Among immune cells, LYVE1+ CD163+ CD68+ macrophages were also present in all species; other immune cells were identified in each species with less correlation, perhaps due to their relatively sparse numbers. Similarly, neurons were limited to the human dataset, most likely due to their low numbers.
On the other hand, while TM cells could be identified in each species, they demonstrated substantial variability across species. Three TM types—two beam cell types and one JCT cell type—were present in all three primates as well as the mouse, but they failed to map 1:1 across species. Beam A and JCT cells were present in these four species, but the second beam type, which we call Beam X in both macaque species and Beam Y in the mouse, demonstrated more unique expression patterns. Moreover, we identified only one beam cell type in pig, although it is possible that further analysis with increased sample size would allow a subdivision of this cluster. Some differentially expressed markers, such as PDPN, RARRES1, CHI3L1, and ANGPTL7, which were selectively expressed by beam and/or JCT cells in human tissue, were either absent in the model species or nonspecific. Others were conserved across species, including MYOC, EDN3, and RBP4. Interestingly, PDPN did not demonstrate the same specificity in any of our model species, suggesting that this may be a human-specific feature.
Glaucoma.
Glaucoma is a phenotypically heterogeneous disease with traits dictated by complex interactions among age, environment, and genes. Examination of cell type-specific expression patterns of both Mendelian genes and GWAS susceptibility loci revealed multiple expression patterns, of which we note four groups. First, several IOP-associated disease genes were selectively expressed by TM cell types (e.g., CYP1B1 and EFEMP1), supporting their potential contribution to function and or/dysfunction of this tissue. Second, others were expressed not only by TM but also by cells of the uveoscleral pathway (e.g., MYOC, PITX2, and FOXC1). Third, a few genes implicated in high IOP mapped selectively to cells of the uveoscleral pathway (e.g., KALRN). Together, these results may indicate an integral contribution of the uveoscleral pathway to disorders of IOP. Fourth, some genes were expressed by RGCs, often in addition to TM cells. For example, TMCO1, while clearly expressed in the anterior segment outflow pathways, also demonstrated robust expression in RGCs, supporting the notion that it may also confer risk for glaucoma development through an IOP-independent mechanism (61).
Although TM cells have been shown to exhibit phagocytic abilities, it is possible that resident macrophages in the conventional outflow pathway also contribute to the phagocytic workload. Their stimulation or recruitment to the TM may serve to lower IOP as has been suggested to occur after selective laser trabeculoplasty (62). Conversely, dysfunctional macrophages may contribute to elevated IOP (63).
Conclusion
In conclusion, we have generated a cell atlas of the trabecular meshwork and surrounding structures constituting the aqueous humor outflow pathways in humans and four model species. We identified markers for each cell type, enabling in-depth study of individual components of this complex tissue and shedding light on features such as the immunological milieu and the lymphatic characteristics of the conventional outflow pathway. Our atlas also provides biological context for susceptibility loci identified in genome-wide association studies. Together, these results may guide further investigation in both humans and model species.
Materials and Methods
Tissue Acquisition and Processing.
Human ocular tissues used for sequencing, immunohistochemistry, and in situ hybridization were collected a median of 6 h postmortem (range 3 to 14 h) (SI Appendix, Table S1). Human tissue used for IHC and in situ hybridization was provided by the Lions Vision Gift, Portland, OR. No ocular disease was reported in any of the human donors, and no abnormalities were noted during microdissection. Nonhuman primate eyes were obtained from macaques 4 to 10 y of age that had reached the end of unrelated studies at supplying institutions. Porcine (S. scrofa) eyes were obtained from a local abattoir. Murine eyes were collected from male and female 12-wk-old CD1 mice obtained from Charles River Laboratories.
Procedures were conducted in compliance with the Association for Research in Vision and Ophthalmology’s Statement for the Use of Animals in Ophthalmic and Vision Research and the guidelines for the care and use of animals and human subjects at Harvard University and Partners Healthcare. Acquisition and use of human tissue were approved by the Human Study Subject Committees (Dana Farber/Harvard Cancer Center Protocol No. 13–416 and Massachusetts Eye and Ear - Non-Human Subjects Research Protocol No. 18–034H). Acquisition and use of nonhuman tissue were approved by the Institutional Animal Care and Use Committee at Harvard University.
RNA Sequencing.
Tissues were digested enzymatically, and single cells were loaded into 10× Chromium Single Cell Chips (27). Single-cell libraries were generated following the manufacturer’s protocol. Libraries were sequenced on Illumina HiSeq 2500. Sequencing data were demultiplexed and aligned using Cellranger software (10× Genomics). Reads were aligned to the following reference genome: human samples, GRCh38; M. mulatta, Mmul8; M. fascicularis, MacFas5 with our augmented transcriptome file (64); pig, Sscrofa11; and mouse, mm10. Subsequent clustering and identification of differentially expressed genes followed methods described by Peng et al. (64) with modifications detailed in SI Appendix, Supplementary Materials and Methods. Transcriptomic similarity between humans and other model species was assessed with a machine learning algorithm XGBoost (56).
Histology.
Corneoscleral wedges were fixed, embedded in tissue freezing medium, and sectioned in a cryostat. SI Appendix, Table S2 shows donor information. Sections were immunostained or processed for in situ hybridization using probes listed in SI Appendix, Table S3. Images were acquired on Zeiss LSM 710 confocal microscopes.
Data Availability.
The accession number for the raw and unprocessed data files reported in this paper is Gene Expression Omnibus accession number GSE146188. Data can be visualized in the Broad Institute’s Single Cell Portal at https://singlecell.broadinstitute.org/single_cell/study/SCP780.
Supplementary Material
Acknowledgments
This work was supported by NIH Grants 5K12EY016335, R21 EY028633, and U01 MH105960; Chan–Zuckerberg Initiative Grant CZF-2019-002459; the Susan Eid Tumor Heterogeneity Initiative; and the Klarman Cell Observatory of the Broad Institute of the Massachusetts Institute of Technology and Harvard. We thank the donors and their families for their generosity. We also thank Michael Do for providing macaque tissue and Carl Romano and Dan Stamer for sharing results of their parallel study prior to submission.
Footnotes
J.R.S. is a consultant at Biogen. All other authors declare no competing interest.
Data deposition: The raw and unprocessed data files reported in this paper have been deposited in the Gene Expression Omnibus (accession no. GSE146188). Data can be visualized in the Broad Institute’s Single Cell Portal at https://singlecell.broadinstitute.org/single_cell/study/SCP780.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2001250117/-/DCSupplemental.
References
- 1.Quigley H. A., Broman A. T., The number of people with glaucoma worldwide in 2010 and 2020. Br. J. Ophthalmol. 90, 262–267 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weinreb R. N., Aung T., Medeiros F. A., The pathophysiology and treatment of glaucoma: A review. JAMA 311, 1901–1911 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Costagliola C., et al. , How many aqueous humor outflow pathways are there? Surv. Ophthalmol. 65, 144–170 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Grant W. M., Further studies on facility of flow through the trabecular meshwork. AMA Arch. Opthalmol. 60, 523–533 (1958). [DOI] [PubMed] [Google Scholar]
- 5.Tian B., Geiger B., Epstein D. L., Kaufman P. L., Cytoskeletal involvement in the regulation of aqueous humor outflow. Invest. Ophthalmol. Vis. Sci. 41, 619–623 (2000). [PubMed] [Google Scholar]
- 6.Braunger B. M., Fuchshofer R., Tamm E. R., The aqueous humor outflow pathways in glaucoma: A unifying concept of disease mechanisms and causative treatment. Eur. J. Pharm. Biopharm. 95, 173–181 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Stamer W. D., Clark A. F., The many faces of the trabecular meshwork cell. Exp. Eye Res. 158, 112–123 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alvarado J., Murphy C., Juster R., Trabecular meshwork cellularity in primary open-angle glaucoma and nonglaucomatous normals. Ophthalmology 91, 564–579 (1984). [DOI] [PubMed] [Google Scholar]
- 9.Tamm E. R., The trabecular meshwork outflow pathways: Structural and functional aspects. Exp. Eye Res. 88, 648–655 (2009). [DOI] [PubMed] [Google Scholar]
- 10.Liu Y., et al. , Gene expression profile in human trabecular meshwork from patients with primary open-angle glaucoma. Invest. Ophthalmol. Vis. Sci. 54, 6382–6389 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sathiyanathan P., Tay C. Y., Stanton L. W., Transcriptome analysis for the identification of cellular markers related to trabecular meshwork differentiation. BMC Genomics 18, 383 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carnes M. U., Allingham R. R., Ashley-Koch A., Hauser M. A., Transcriptome analysis of adult and fetal trabecular meshwork, cornea, and ciliary body tissues by RNA sequencing. Exp. Eye Res. 167, 91–99 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Liton P. B., Luna C., Challa P., Epstein D. L., Gonzalez P., Genome-wide expression profile of human trabecular meshwork cultured cells, nonglaucomatous and primary open angle glaucoma tissue. Mol. Vis. 12, 774–790 (2006). [PMC free article] [PubMed] [Google Scholar]
- 14.Wiggs J. L., Pasquale L. R., Genetics of glaucoma. Hum. Mol. Genet. 26, R21–R27 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lewis C. J., et al. , Primary congenital and developmental glaucomas. Hum. Mol. Genet. 26, R28–R36 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choquet H., et al. , A multiethnic genome-wide association study of primary open-angle glaucoma identifies novel risk loci. Nat. Commun. 9, 2278 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao X. R., Huang H., Nannini D. R., Fan F., Kim H., Genome-wide association analyses identify new loci influencing intraocular pressure. Hum. Mol. Genet. 27, 2205–2213 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khawaja A. P., et al. , A common glaucoma-risk variant of SIX6 alters retinal nerve fiber layer and optic disc measures in a European population: The EPIC-Norfolk Eye Study. J. Glaucoma 27, 743–749 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.MacGregor S., et al. , Genome-wide association study of intraocular pressure uncovers new pathways to glaucoma. Nat. Genet. 50, 1067–1071 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Sears N. C., Boese E. A., Miller M. A., Fingert J. H., Mendelian genes in primary open angle glaucoma. Exp. Eye Res. 186, 107702 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Youngblood H., Hauser M. A., Liu Y., Update on the genetics of primary open-angle glaucoma. Exp. Eye Res. 188, 107795 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aung T., et al. , Genetic association study of exfoliation syndrome identifies a protective rare variant at LOXL1 and five new susceptibility loci. Nat. Genet. 49, 993–1004 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan W., et al. , Cell atlas of the human fovea and peripheral retina. bioRxiv:10.1101/2020.02.11.943779 (12 February 2020). [DOI] [PMC free article] [PubMed]
- 24.Bachmann B., Birke M., Kook D., Eichhorn M., Lütjen-Drecoll E., Ultrastructural and biochemical evaluation of the porcine anterior chamber perfusion model. Invest. Ophthalmol. Vis. Sci. 47, 2011–2020 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Fernandes K. A., et al. , Using genetic mouse models to gain insight into glaucoma: Past results and future possibilities. Exp. Eye Res. 141, 42–56 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Picaud S., et al. , The primate model for understanding and restoring vision. Proc. Natl. Acad. Sci. U.S.A. 116, 26280–26287 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng G. X., et al. , Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 14049 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Acott T. S., Kelley M. J., Extracellular matrix in the trabecular meshwork. Exp. Eye Res. 86, 543–561 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Epstein D. L., Rohen J. W., Morphology of the trabecular meshwork and inner-wall endothelium after cationized ferritin perfusion in the monkey eye. Invest. Ophthalmol. Vis. Sci. 32, 160–171 (1991). [PubMed] [Google Scholar]
- 30.Rao P. V., Deng P. F., Kumar J., Epstein D. L., Modulation of aqueous humor outflow facility by the Rho kinase-specific inhibitor Y-27632. Invest. Ophthalmol. Vis. Sci. 42, 1029–1037 (2001). [PubMed] [Google Scholar]
- 31.Birke K., Lütjen-Drecoll E., Kerjaschki D., Birke M. T., Expression of podoplanin and other lymphatic markers in the human anterior eye segment. Invest. Ophthalmol. Vis. Sci. 51, 344–354 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Watanabe Y., Hamanaka T., Takemura T., Murakami A., Involvement of platelet coagulation and inflammation in the endothelium of Schlemm’s canal. Invest. Ophthalmol. Vis. Sci. 51, 277–283 (2010). [DOI] [PubMed] [Google Scholar]
- 33.Zhang D., Vetrivel L., Verkman A. S., Aquaporin deletion in mice reduces intraocular pressure and aqueous fluid production. J. Gen. Physiol. 119, 561–569 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kelley M. J., et al. , Stem cells in the trabecular meshwork: Present and future promises. Exp. Eye Res. 88, 747–751 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yun H., Zhou Y., Wills A., Du Y., Stem cells in the trabecular meshwork for regulating intraocular pressure. J. Ocul. Pharmacol. Ther. 32, 253–260 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aspelund A., et al. , The Schlemm’s canal is a VEGF-C/VEGFR-3-responsive lymphatic-like vessel. J. Clin. Invest. 124, 3975–3986 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomson B. R., et al. , A lymphatic defect causes ocular hypertension and glaucoma in mice. J. Clin. Invest. 124, 4320–4324 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ulvmar M. H., Mäkinen T., Heterogeneity in the lymphatic vascular system and its origin. Cardiovasc. Res. 111, 310–321 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Herrnberger L., Ebner K., Junglas B., Tamm E. R., The role of plasmalemma vesicle-associated protein (PLVAP) in endothelial cells of Schlemm’s canal and ocular capillaries. Exp. Eye Res. 105, 27–33 (2012). [DOI] [PubMed] [Google Scholar]
- 40.Medina-Ortiz W. E., Belmares R., Neubauer S., Wordinger R. J., Clark A. F., Cellular fibronectin expression in human trabecular meshwork and induction by transforming growth factor-β2. Invest. Ophthalmol. Vis. Sci. 54, 6779–6788 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thiriot A., et al. , Differential DARC/ACKR1 expression distinguishes venular from non-venular endothelial cells in murine tissues. BMC Biol. 15, 45 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xie T., et al. , Single-cell deconvolution of fibroblast heterogeneity in mouse pulmonary fibrosis. Cell Rep. 22, 3625–3640 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Diehn J. J., Diehn M., Marmor M. F., Brown P. O., Differential gene expression in anatomical compartments of the human eye. Genome Biol. 6, R74 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wiederholt M., Thieme H., Stumpff F., The regulation of trabecular meshwork and ciliary muscle contractility. Prog. Retin. Eye Res. 19, 271–295 (2000). [DOI] [PubMed] [Google Scholar]
- 45.Ishikawa T., Fine structure of the human cillary muscle. Invest. Ophthalmol. 1, 587–608 (1962). [PubMed] [Google Scholar]
- 46.Barron L., Gharib S. A., Duffield J. S., Lung pericytes and resident fibroblasts: Busy multitaskers. Am. J. Pathol. 186, 2519–2531 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tamm E. R., Flügel-Koch C., Mayer B., Lütjen-Drecoll E., Nerve cells in the human ciliary muscle: Ultrastructural and immunocytochemical characterization. Invest. Ophthalmol. Vis. Sci. 36, 414–426 (1995). [PubMed] [Google Scholar]
- 48.Flügel-Koch C., Neuhuber W. L., Kaufman P. L., Lütjen-Drecoll E., Morphologic indication for proprioception in the human ciliary muscle. Invest. Ophthalmol. Vis. Sci. 50, 5529–5536 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Margeta M. A., Lad E. M., Proia A. D., CD163+ macrophages infiltrate axon bundles of postmortem optic nerves with glaucoma. Graefes Arch. Clin. Exp. Ophthalmol. 256, 2449–2456 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Buniello A., et al. , The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 47, D1005–D1012 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gordon M. O., et al. , The ocular hypertension treatment study: Baseline factors that predict the onset of primary open-angle glaucoma. Arch. Ophthalmol. 120, 714–720 (2002). [DOI] [PubMed] [Google Scholar]
- 52.Zhou H., Yoshioka T., Nathans J., Retina-derived POU-domain factor-1: A complex POU-domain gene implicated in the development of retinal ganglion and amacrine cells. J. Neurosci. 16, 2261–2274 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.King R., et al. ; International Glaucoma Genetics Consortium; NEIGHBORHOOD Consortium , Genomic locus modulating corneal thickness in the mouse identifies POU6F2 as a potential risk of developing glaucoma. PLoS Genet. 14, e1007145 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Clark S. J., Bishop P. N., The eye as a complement dysregulation hotspot. Semin. Immunopathol. 40, 65–74 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pauly D., et al. , Cell-type-specific complement expression in the healthy and diseased retina. Cell Rep. 29, 2835–2848.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen T., Guestrin C., Xgboost: A scalable tree boosting system. arXiv:1603.02754 (10 June 2016).
- 57.McMenamin P. G., Steptoe R. J., Normal anatomy of the aqueous humour outflow system in the domestic pig eye. J. Anat. 178, 65–77 (1991). [PMC free article] [PubMed] [Google Scholar]
- 58.Overby D. R., et al. , The structure of the trabecular meshwork, its connections to the ciliary muscle, and the effect of pilocarpine on outflow facility in mice. Invest. Ophthalmol. Vis. Sci. 55, 3727–3736 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Janssen S. F., et al. , Gene expression and functional annotation of the human ciliary body epithelia. PLoS One 7, e44973 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vasiliou V., Gonzalez F. J., Role of CYP1B1 in glaucoma. Annu. Rev. Pharmacol. Toxicol. 48, 333–358 (2008). [DOI] [PubMed] [Google Scholar]
- 61.Scheetz T. E., et al. , Glaucoma risk alleles in the ocular hypertension treatment study. Ophthalmology 123, 2527–2536 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Alvarado J. A., Katz L. J., Trivedi S., Shifera A. S., Monocyte modulation of aqueous outflow and recruitment to the trabecular meshwork following selective laser trabeculoplasty. Arch. Ophthalmol. 128, 731–737 (2010). [DOI] [PubMed] [Google Scholar]
- 63.Hamanaka T., Takei A., Takemura T., Oritsu M., Pathological study of cases with secondary open-angle glaucoma due to sarcoidosis. Am. J. Ophthalmol. 134, 17–26 (2002). [DOI] [PubMed] [Google Scholar]
- 64.Peng Y. R., et al. , Molecular classification and comparative taxonomics of foveal and peripheral cells in primate retina. Cell 176, 1222–1237.e22 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the raw and unprocessed data files reported in this paper is Gene Expression Omnibus accession number GSE146188. Data can be visualized in the Broad Institute’s Single Cell Portal at https://singlecell.broadinstitute.org/single_cell/study/SCP780.