Significance
SF3B1 is the most commonly mutated splicing factor in cancers, and SF3B1 mutations result in aberrant 3′ splice site usage during splicing of a subset of introns. Here, we utilized an unbiased computational biology approach to determine whether cancer-associated mutations in any other genes produce the same pattern of aberrant splicing as do SF3B1 mutations. We found that, while rare, the most frequently mutated gene that causes such splicing defects is SUGP1. This is striking because previous biochemical studies indicated that loss of SF3B1 interaction with SUGP1 underlies the effects of SF3B1 mutations on splicing. Our findings thus establish the cancer relevance of the biochemical link between SF3B1 and SUGP1 and also identify SUGP1 as a cancer-associated gene.
Keywords: SUGP1, SF3B1, G patch, spliceosome
Abstract
The gene encoding the core spliceosomal protein SF3B1 is the most frequently mutated gene encoding a splicing factor in a variety of hematologic malignancies and solid tumors. SF3B1 mutations induce use of cryptic 3′ splice sites (3′ss), and these splicing errors contribute to tumorigenesis. However, it is unclear how widespread this type of cryptic 3′ss usage is in cancers and what is the full spectrum of genetic mutations that cause such missplicing. To address this issue, we performed an unbiased pan-cancer analysis to identify genetic alterations that lead to the same aberrant splicing as observed with SF3B1 mutations. This analysis identified multiple mutations in another spliceosomal gene, SUGP1, that correlated with significant usage of cryptic 3′ss known to be utilized in mutant SF3B1 expressing cells. Remarkably, this is consistent with recent biochemical studies that identified a defective interaction between mutant SF3B1 and SUGP1 as the molecular defect responsible for cryptic 3′ss usage. Experimental validation revealed that five different SUGP1 mutations completely or partially recapitulated the 3′ss defects. Our analysis suggests that SUGP1 mutations in cancers can induce missplicing identical or similar to that observed in mutant SF3B1 cancers.
Frequent mutations in genes encoding splicing factors (SF) have been identified across a variety of hematologic malignancies and solid tumors, highlighting the importance of aberrant splicing to cancerogenesis (1). The most commonly mutated spliceosomal gene, SF3B1, is subject to heterozygous mutations at very specific residues in patients with myelodysplastic syndromes (2, 3), chronic lymphocytic leukemia (4, 5), uveal melanoma (6), breast invasive carcinoma, and skin cutaneous melanoma (1). Previous analyses have shown that SF3B1 mutations promote the usage of upstream branchpoints during the splicing reaction, resulting in the use of cryptic upstream 3′ splice sites (3′ss) (7, 8). SF3B1 encodes a core component of the U2 small nuclear ribonucleoprotein (snRNP) complex of the spliceosome and is involved in early stages of splicing (9). The major spliceosome consists of five snRNP complexes (U1, U2, U4, U5, and U6) and more than 150 proteins, many of which may have direct or indirect physical interactions with SF3B1 during spliceosome assembly (1).
Given this complexity, it was not until recently that the mechanism by which SF3B1 mutations affect splicing was elucidated. We showed that several hot spot mutations, clustered in a domain of the protein consisting of multiple HEAT repeats, disrupt interaction of SF3B1 with another SF, a poorly studied spliceosomal protein called SUGP1 (SURP and G-patch domain containing 1). Supporting the importance of this observation, we also found that siRNA-mediated depletion of SUGP1 recapitulated the splicing defects caused by SF3B1 mutations, while SUGP1 overexpression partially rescued splicing in cells expressing mutant SF3B1. Additionally, overexpression of a SUGP1 derivative with a mutation in the G patch, a domain thought to bind to and activate DEAH-box RNA helicases (10), also resulted in the same splicing defects observed in SF3B1 mutant cells (11).
In addition to SF3B1, other SF-encoding genes have also been found to be mutated in hematologic malignancies, e.g., U2AF1, SRSF2, and ZRSR2. However, these SF gene mutations do not share common alterations in splicing (1, 3), suggesting that different splicing patterns may contribute to different phenotypes of cancers. Because SF3B1 is the most frequently mutated splicing gene, the splicing defects caused by mutant SF3B1 may be the most important to tumorigenesis. To determine if this splicing pattern is widespread in cancer, and what is the full spectrum of genetic mutations that cause such missplicing, we investigated in a pan-cancer manner if different cancer-associated genetic alterations are associated with 3′ cryptic splicing events in a similar fashion as brought about by SF3B1 mutations.
Large-scale sequencing projects in cancer genomics and transcriptomics, e.g., The Cancer Genome Atlas Program (TCGA), provide a unique resource for linking genomics to transcriptomic effects across different tumor types. One recent study performed a systematic analysis of the alternative splicing landscape across all TCGA cancer patients. This analysis identified numerous neojunctions not typically found in normal samples (12). Detection of cancer-specific alternative splicing events was performed on each sample in an unbiased way, regardless of any SF mutations. Coupling the alternative splicing detection analysis with well-annotated somatic variant profiles on matched TCGA samples (13) enables the efficient identification of altered splicing associated genetic lesions in all types of tumors. In this study, we utilized a computational biology approach to identify functional variants that mimic 3′ missplicing behavior characterized previously with SF3B1 mutations (8, 11). Remarkably, although rare, we found that multiple, distinct somatic mutations in SUGP1 are associated with similar aberrant splicing events as detected in samples harboring SF3B1 mutations. Experimental validation using HEK293T cells transiently expressing the SUGP1 mutant proteins confirmed that these somatic variants could completely or partially recapitulate known splicing changes induced by mutant SF3B1. Our analysis has enhanced our understanding of the missplicing brought about by SF3B1 mutations and highlights the importance of the SF3B1–SUGP1 interaction.
Results
Identification of Alternative 3′ss Splicing-Associated Genetic Variants.
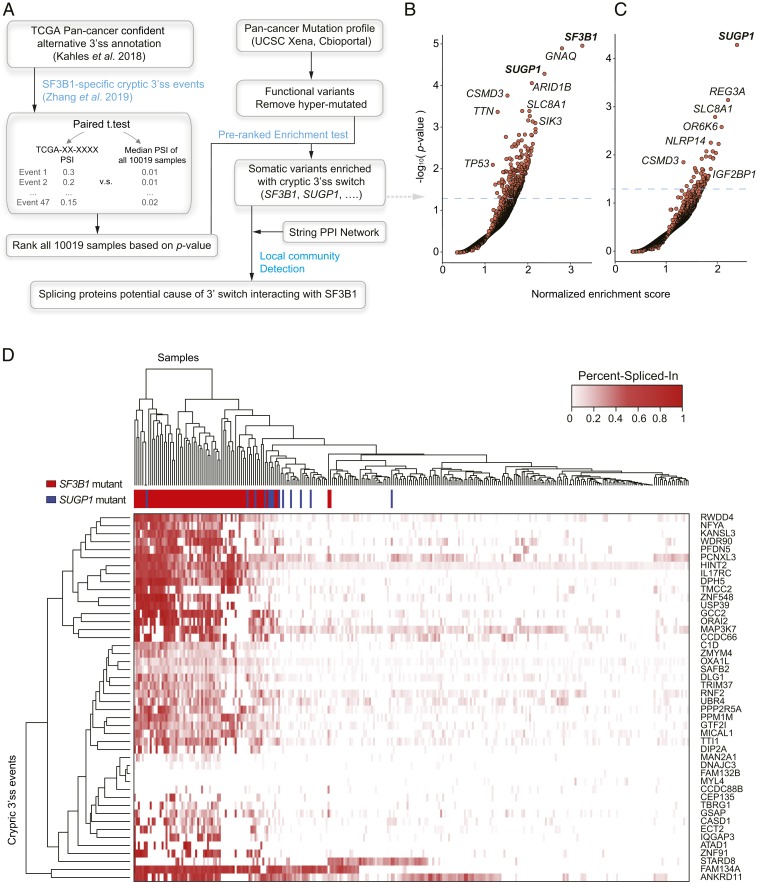
In light of the growing evidence that mutations in genes encoding SFs, especially SF3B1, can play significant roles in a number of cancers, we wished to obtain a better understanding of how SF3B1 mutations alter splicing, and whether cancer-associated mutations in other SF genes can induce this pattern of missplicing. To this end, we performed a pan-cancer analysis using computational approaches that enable the identification and quantification of alternative 3′ss utilization associated with specific somatic variants. For this analysis, we adopted the cryptic 3′ss annotations from a previous comprehensive study that analyzed the alternative splicing landscape across all TCGA cancer patients (12). To capture authentic 3′ss changes caused by SF3B1 hotspot mutations, we narrowed down the list of alternative 3′ss events to 47 high confidence events, based on findings from our recent work (11) (Dataset S1). Of all 10,019 samples in TCGA, 36 of the 47 events had a median value of at least 10 reads at the canonical junction, while 9 events had a median value of at least 1 read at the cryptic site (SI Appendix, Fig. S1 A and B). To evaluate the cryptic 3′ss usage of each of the TCGA samples, we applied a paired t test between the percent-spliced-in (PSI) values of these 47 events against the background missplicing rate (Materials and Methods). Using these results with the somatic mutation reports on the matched TCGA samples, we identified the samples showing abundant 3′ss missplicing, as well as which mutated genes are enriched in those samples (Fig. 1A and Materials and Methods). As expected, we found SF3B1 to be the top enriched gene, which provides a positive control of our pipeline’s validity (Fig. 1B, SI Appendix, Fig. S1C, and Dataset S2). Besides SF3B1, the top mutated genes were GNAQ, SUGP1, and ARID1B (Fig. 1B). We suspected that the presence of GNAQ mutations was due to its cooccurrence with SF3B1 mutations, especially in uveal melanoma (P = 0.03 from cBioportal query), which is a main source of the SF3B1 hotspot R625 mutation among the TCGA cohort.
Fig. 1.
Identification of genetic variants associated with cryptic 3′ss usage. (A) Flowchart of the computational pipeline (see details in Materials and Methods). (B and C) Half-volcano plot representations of genes positively correlated with cryptic 3′ss usage. The normalized enrichment score (horizontal axis) and −log10 P value (vertical axis) were derived from preranked enrichment test. Analysis was performed using all TCGA samples (B) and SF3B1 nonmutated samples (C). (D) Hierarchical clustering using Euclidean distance and heatmap analysis of the usage of 47 cryptic 3′ss events between top-ranked SF3B1 mutant, SUGP1 mutant and WT samples. Rows and columns represent cryptic 3′ss events and samples, respectively. Values in the matrix represent raw PSI.
Given the abundance of SF3B1 mutations, we repeated the above analysis after removing all samples with SF3B1 mutations. Using this approach, we found that SUGP1 became the lone highly enriched gene (Fig. 1C, SI Appendix, Fig. S1D, and Dataset S3). We next examined the top-ranked TCGA samples, based on their cryptic 3′ss usage, and found 9 cases with SUPG1 mutations (SF3B1 WT) within the top 150, with 1 in the top 50, along with 48 cases with SF3B1 mutations (SI Appendix, Fig. S2 and Dataset S4). Notably, we also detected mutations in a number of known cancer-associated drivers that are significantly negatively correlated with cryptic 3′ss usage (SI Appendix, Figs. S1 C and D and S3). These include EGFR, IDH1, BRAF, and ATRX, which are frequently found in brain tumors as well as in other tumor types (14). The significance of this finding is unknown.
Subsequently, in order to obtain a global view of misusage of the 47 3′ss events, we performed unsupervised hierarchical clustering on the top-ranked SF3B1 mutant, SUGP1 mutant, and WT samples (Fig. 1D, SI Appendix, Fig. S4A, and Materials and Methods). Overall, SF3B1 and SUGP1 mutants were clustered closer together and with significantly higher PSI than in WT samples, although there were a few exceptions (Fig. 1D). This observation suggests a relative similarity between SF3B1 and SUGP1 mutants in terms of cryptic 3′ss selection. Additionally, we found widespread occurrence of weak cryptic 3′ss usage in WT samples. This phenomenon was heavily biased by particular events, e.g., MAP3K7, a well-known target of mutant SF3B1 (Fig. 1D). This fact indicates that many of the cryptic 3′ss are inherently active to some degree and not entirely dependent on an SF3B1 or SUGP1 mutation. In addition, we applied a principal component analysis to the same data matrix of aberrant 3′ss usage, which generated similar results as did the clustering analysis (SI Appendix, Fig. S4B).
Mutations in SUGP1 and SF3B1 Are Positively Associated with Cryptic 3′ss Switch.
Next, we focus our analysis into variants in SFs, as they are likely more functionally relevant to cryptic 3′ss usage. Thus, we would like to know if there exist somatic variants in the spliceosome genes encoding spliceosomal proteins, particularly those ones with known protein–protein interaction with SF3B1, that are positively associated with cryptic 3′ss switch. For this purpose, we used a computational approach to detect local network communities from one protein–protein interaction network, node weighted by cryptic 3′ss usage (15) (Materials and Methods). This procedure was conducted in order to identify a dense interconnected group of spliceosomal proteins that may contribute to cryptic 3′ss usage by interacting with SF3B1. We found both SUGP1 and SF3B1 are central nodes in the top network community enriched for cryptic 3′ss usage (SI Appendix, Fig. S4C), indicating that direct physical interactions generate similar splicing effects.
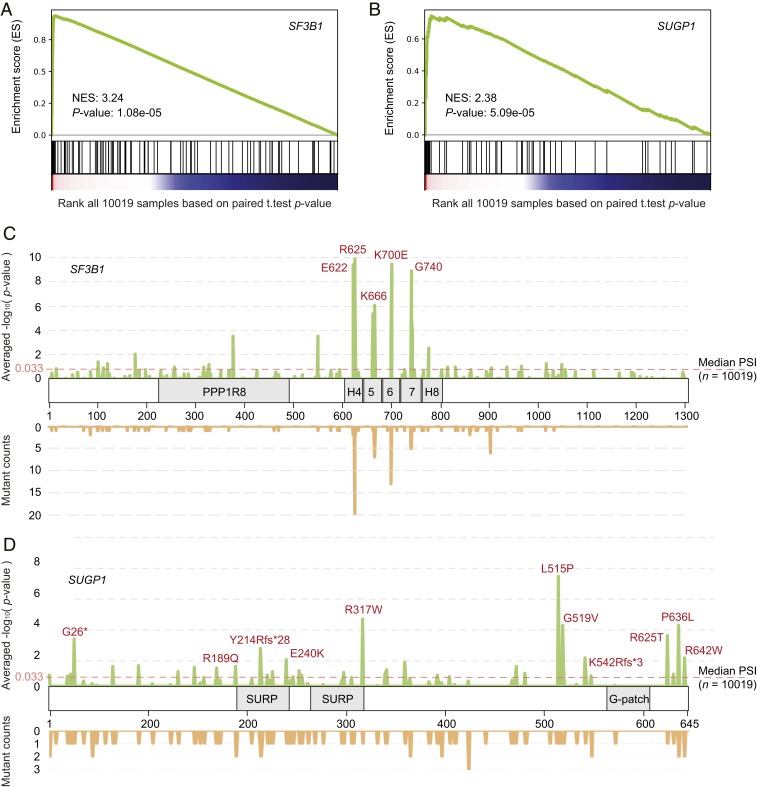
We next determined the locations of missplicing-associated variants on the domain structure of SUGP1. This may computationally predict which part of SUGP1 protein potentially binds to SF3B1 or other spliceosomal proteins to influence 3′ss selection. To this end, we first examined the preranked enrichment curves of SF3B1 and SUGP1 (Materials and Methods) and found that, although many cases gather at the very beginning of the list, there exist pervasive mutations randomly distributed across the entire TCGA database (Fig. 2 A and B). This analysis indicates that not all variants in SUGP1 and SF3B1 are associated with significant missplicing. Next, to determine which mutations lead to missplicing, we examined the extent of cryptic 3′ss usage for each mutated amino acid (Fig. 2 C and D). As expected, recurrent hotspot mutations (including K700, R625, E622, K666, and G740) in SF3B1’s HEAT repeats were strongly associated with 3′ss missplicing (Fig. 2C). Notably, association of the most commonly mutated residue, K700, appeared slightly lower than R625 (Fig. 2C). This observation very likely reflects the absence of many hematologic malignancies that are dominated by SF3B1 K700E, such as CLL and MDS, from the TCGA samples. With SUGP1, among its highly missplicing related amino acids, we found five positions of single-nucleotide variants surrounding the G-patch domain, L515P, G519V, R625T, P636L, and R642W (Fig. 2D). This localization is intriguing given our previous study showing that expression of a SUGP1 derivative mutated in the G patch led to cryptic 3′ss usage similar to mutant SF3B1 (11). However, we did not find any somatic mutations within the actual G-patch domain, which may reflect the low frequency of SUGP1 variants in TCGA samples.
Fig. 2.
Mutations in SUGP1 and SF3B1 are positively associated with cryptic 3′ss usage. (A and B) Preranked enrichment analysis indicated that mutations in SF3B1 (A) and SUGP1 (B) are strongly associated with cryptic 3′ss usage. (C and D) Distribution plots of averaged associations with cryptic 3′ss usage (Upper), and variant counts (Lower) for each amino acid of SF3B1 (C) and SUGP1 (D).
Next, we examined whether the mutations with 3′ splicing abnormalities are specific to certain cancer types. To examine this, we made comparisons between SF3B1 mutant, SUGP1 mutant, and WT for each cancer type individually (SI Appendix, Fig. S5A). We found elevated use of cryptic 3′ss in SF3B1 mutants in uveal melanoma (UVM), skin cutaneous melanoma (SKCM), breast cancer (BRCA), and other cancer types, which all harbor the hotspot mutations in the HEAT repeats. By contrast, only SUGP1 mutants from lung adenocarcinoma (LUAD) showed clear differences compared to WT (SI Appendix, Fig. S5A). The interpretation of this tumor type preference for SUGP1 mutation is not clear. In addition, due to the rareness of SUGP1 mutations in human cancer, it is difficult to conclude that LUAD is specifically susceptible to SUGP1 mutations. We also examined the number of expressed canonical junctions of the 47 events analyzed and did not find any significant differences, suggesting that gene expression levels did not influence this preference (SI Appendix, Fig. S5 B and C).
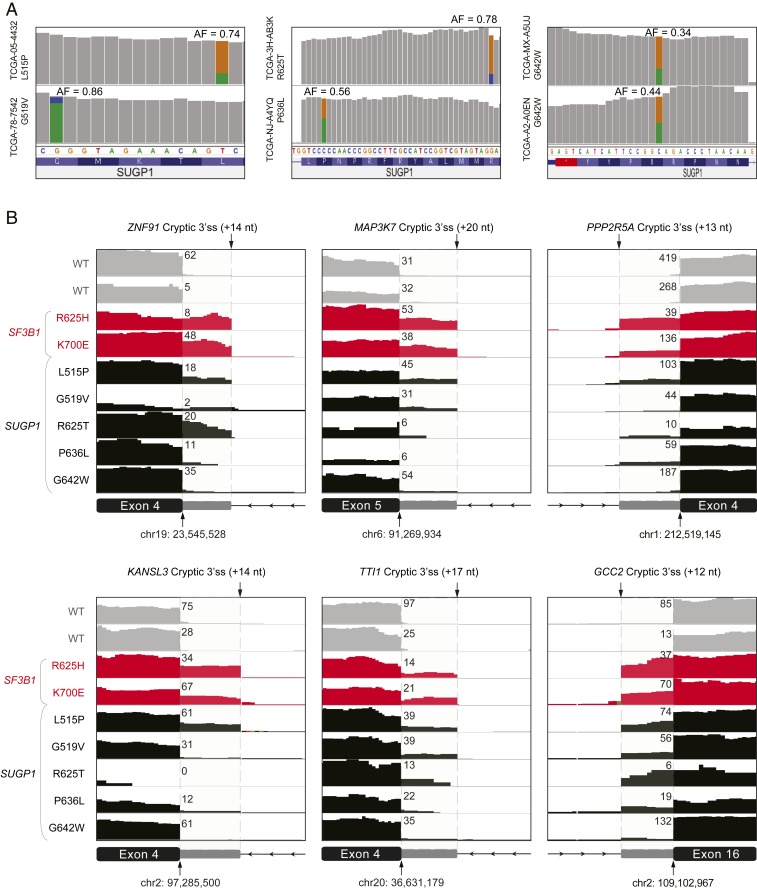
We next examined the raw RNA-sequencing (seq) data of the G patch-associated SUGP1 mutations (Fig. 3). Interestingly, we found three cases of mutations having allele frequency >70% in RNA, which indicates allele-specific expression associated with the mutation. To characterize further the cause of this allele specific expression, we examined possible loss of heterozygosity due to copy number change in the SUGP1 locus (Fig. 3A). We consistently noticed much lower mutation allele frequency in matched DNA-seq as compared to RNA-seq (mutation reports on DNA-seq data obtained from cBioportal query: L515P: 42%, G519V: 41%, R625T: 77%, P636L: 11%, R642W: 19% and 17%), indicating that the allele-specific expression was not due to copy number alterations. Finally, we examined the actual usage of a number of specific cryptic 3′ss events that were all experimentally validated as authentic top targets of SF3B1 mutants (11) (Fig. 3B). We found that the L515P mutation appeared to faithfully recapitulate all these 3′ splicing defects of the SF3B1 hotspot mutations, while other SUGP1 mutations partially reproduced these events, reflecting either low expression or a very low PSI (Fig. 3B).
Fig. 3.
SUGP1 variants completely or partially recapitulate the known 3′ss changes induced by mutant SF3B1. (A) Visualization of top cryptic 3′ss-associated SUGP1 variants using Integrative Genomics Viewer (IGV). RNA-seq bam files of TCGA samples were downloaded from Genomic Data Commons (https://gdc.cancer.gov/). Variant allele frequencies (AF) are added. (B) IGV plots of high-confident 3′ss usage in TCGA samples with SF3B1 mutations, SUGP1 mutations as well as WT. For each track, number of reads using the canonical junction were added.
Experimental Validation of the Use of Cryptic 3′ss by SUGP1 Mutants.
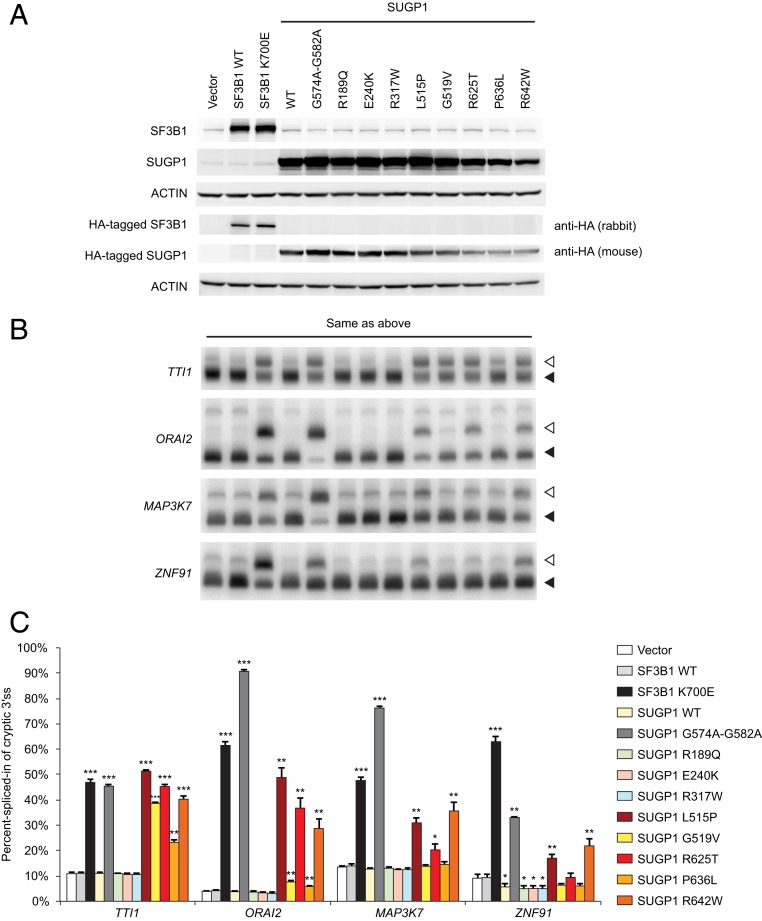
Finally, we wished to validate experimentally the effects of the SUGP1 mutations on cryptic 3′ss use. To do so, we expressed the SUGP1 mutants as well as controls (WT and K700E mutant SF3B1 and WT and the G-patch domain mutant SUGP1) in HEK293T cells to examine directly cryptic 3′ss usage of target gene transcripts. To minimize background from preexisting mRNAs, we used minigenes of four of the mutant SF3B1 targets that we confirmed in our previous study (11). We coexpressed these minigenes with each of the SUGP1 mutants and controls (Fig. 4A) and performed RT-PCR to detect mRNAs produced from the minigenes (Fig. 4 B and C). Consistent with the results in our previous study (11), the positive controls (K700E mutant SF3B1 and the G-patch domain mutant SUGP1) induced robust use of cryptic 3′ss. Two of the SUGP1 mutants identified in this study (L515P and R642W) activated cryptic 3′ss usage in all of the minigene transcripts. R625T switched 3′ss use in three of the four mingenes (not ZNF91), while two other mutants (G519V and P636L) switched 3′ss use of two of the four minigene targets (TTI1 and ORAI2). As mentioned above, all of these five SUGP1 mutations are clustered around (but not within) the G-patch domain. In addition to these 5 SUGP1 mutations, there were 3 other missense mutations that do not cooccur with an SF3B1 mutation among the top 200 samples. These three mutations are located in the two SURP domains of SUGP1 (R189Q and E240K in the first SURP domain and R317W in the second). RT-PCR results showed that these three mutants did not increase cryptic 3′ss usage with any of the four minigenes, and with one minigene (ZNF91), they actually decreased cryptic 3′ss usage slightly when compared to the empty vector control. Because these effects were similar to those of WT SUGP1, we considered these three mutations to be negative. Interestingly, in the top 200 samples, we also identified one nonsense mutation (G26*) and two frame-shift mutations followed by stop codons shortly downstream (Y214Rfs*28 and K542Rfs*3). Although the actual protein levels of SUGP1 in these samples are unknown, it is possible that these three mutations might result in SUGP1 haploinsufficiency. Because we previously showed that reduction in SUGP1 levels can lead to use of cryptic 3′ss (11), these mutations may also activate cryptic 3′ss usage.
Fig. 4.
Experimental validation of the use of cryptic 3′ss by SUGP1 mutants. (A) A mixture of four minigenes (TTI1, ORAI2, MAP3K7, and ZNF91) and each of the indicated plasmids were cotransfected to HEK293T cells in six-well plates, followed by Western blotting. SF3B1 WT and SF3B1 K700E, expression plasmids for HA-tagged WT and K700E mutant SF3B1; SUGP1, expression plasmid for HA-tagged WT SUGP1 or each of the SUGP1 mutants as indicated; Vector, empty vector control. (B) Total RNA was extracted from the cells as in A, followed by RT-PCR of cryptic 3′ss (open arrowheads) and the associated canonical 3′ss (filled arrowheads) produced from splicing of the minigene transcripts. (C) Quantification of the RT-PCR products as in B. Error bars represent SDs of the means (n = 3). Unpaired, two-tailed, and unequal variance t tests were performed by comparing each of the expression plasmids to the empty vector control, *P < 0.05, **P < 0.01, and ***P < 0.001 (calculated using Microsoft Excel).
Discussion
Here, we have provided evidence that the gene encoding the spliceosomal protein SUGP1 can harbor cancer-associated mutations that disrupt mRNA splicing in a manner analogous to the more frequently mutated protein SF3B1. Our previous study showed that disease-associated mutations in SF3B1 misregulate splicing by disrupting the interaction of SF3B1 with SUGP1, and further that experimental depletion of SUGP1 could recapitulate the effect of SF3B1 mutations on splicing (11). However, despite the frequent SF3B1 mutations found in hematologic malignancies and certain other cancers, no SUGP1 mutations had been reported in these diseases. In this study, we performed a pan-cancer analysis of misspliced cryptic 3′ss and found that SUGP1 mutations not only do occur in cancers, but also are the top mutations that phenocopy SF3B1 mutations. Our findings confirm the functional link between SF3B1 and SUGP1 demonstrated in our previous study, establish SUGP1 as a potential target for cancer-causing mutations, and also increase the likelihood that SF3B1 mutations contribute to cancer by altering splicing, as opposed to disrupting some other function of the protein.
Five of the SUGP1 mutations induced use of cryptic 3′ss to varying degrees. Importantly, all of these mutations clustered around the G-patch domain. The G patch was first defined in 1999 and is found in a number of proteins throughout eukaryotes. The domain is ∼45 residues, characterized by six nearly invariant Gly residues, and was initially suggested to be involved in RNA binding (16). Subsequent studies, however, have provided considerable evidence that it functions by associating with DEAH box RNA helicases and activating ATP hydrolysis (reviewed in ref. 10). Other G-patch domain proteins have been shown to function in splicing (17, 18), and we believe helicase activation is a critical aspect of SUGP1 function (11). Despite considerable study over 20 y, no G-patch domain structure has been obtained. Thus, we cannot speculate how the five mutations we have identified and characterized here might affect G patch function. However, as mentioned above, it is striking that expression of a SUGP1 derivative with two of the conserved Gly residues mutated to Ala recapitulated the effects of SF3B1 mutants on splicing (11). Thus, we speculate that the SUGP1 cancer mutations affect the ability of the G-patch domain to activate a currently unknown RNA helicase, albeit to differing degrees. The fact that use of cryptic 3′ss of some target minigenes was affected more than others may reflect a substrate specificity of these mutations, although the molecular basis for such specificity is unknown.
Our studies have identified the gene encoding the SF3B1-interacting protein SUGP1 as a target of cancer associated mutations. It is intriguing that genes encoding other proteins involved in branchpoint-3′ss recognition, and that interact with SF3B1 and/or SUGP1, are also mutated in cancers (3, 19). These include genes encoding U2AF1, which recognizes the 3′ss, and, more rarely, U2AF2, which forms a heterodimer with U2AF1, and SF1, which binds the branchpoint early in spliceosome assembly (20, 21). Interestingly, SUGP1 interacts directly with U2AF2 (11) and likely, via its SURP domains, with SF1 (22). Besides these interactions, the prevalence of mutations in genes encoding other proteins in the SF3B1/SUGP1 interactome, e.g., as identified in our community analysis, has not been systematically investigated. One limitation is the likely rarity of such mutations in cancer. Identifying alterations in other splicing components could lead to therapeutic strategies. For example, it has been observed in myelodysplastic syndromes, SF-mutated cells cannot tolerate concurrent mutations in more than one SF (23). Reflecting this synthetic lethal effect, SF3B1-mutated cells may display sensitivity to therapeutic interventions by targeting key splicing partners as noted above. In addition, growing evidence has shown that SF mutations usually promote tumorigenesis through collaboration with established cancer drivers, such as MYC, IDH2, and ATM (24–26). This suggests that it would also be informative to conduct systematic screening exploiting possible functional relationships between SF3B1 and driver mutations.
In summary, our work has identified cancer-associated mutations in the gene encoding the splicing factor SUGP1. Although rare, the effects of these mutations on splicing, activation of cryptic 3′ss, are very similar to those induced by the much more frequently mutated SF3B1. Our findings thus strengthen both the idea that mutant SF3B1-induced splicing errors are important in cancer and also the physiological significance of our previous biochemical data that SF3B1 hotspot mutations result in aberrant splicing by disrupting interaction with SUGP1.
Materials and Methods
Evaluation of Cryptic 3′ss Usage in TCGA Samples.
We adopted the results from a comprehensive study, which performed a systematic analysis of alternative splicing landscape across all TCGA cancer patients (12). PSI values of all confident alternative 3′ events identified in all TCGA samples were downloaded from the website https://gdc.cancer.gov/about-data/publications/PanCanAtlas-Splicing-2018, with the file name as “merge_graphs_alt_3prime_C2.confirmed.txt.gz”. This file contains 181,915 events across 10,019 TCGA samples. To extract the true aberrant 3′ss switch caused by the mutant SF3B1, we used a list of 47 high-confident targets (PSI difference between mutant vs. WT > 0.2) from one recently published study (11) (Dataset S1). For each of these 47 events, we first calculated the median PSI value across all 10,019 samples as the background missplicing rate. Then, for each sample, a paired t test (one-side test toward greater PSI in the tested sample) was applied between PSI of 47 events against the background missplicing rate (Fig. 1A). Lastly, all samples were ranked based on the ascending order of P value, which indicated the significance level of the abundant 3′ss missplicing usage.
TCGA Pan-Cancer Somatic Mutation Profile.
Somatic mutation profiles of TCGA samples were obtained from UCSC Xena and CBioportal. Mutect2 variant report file “GDC-PANCAN.mutect2_snv.tsv” was downloaded from GDC Pan-Cancer hub of UCSC Xena datasets website. Nonfunctional mutations (for instance: synonymous variant, intron variant, intergenic variant) were removed from the table.
Identification of Alternative Splicing-Associated Genetic Lesions.
We first overlapped the TCGA samples from the mutation table and the 3′ss missplicing table. Then, matched samples were ranked based on the ascending order of previous paired t test P value. Next, a preranked enrichment test was performed using R package “fgsea” (function fgsea, minSize = 50, nperm = 100.000). Here, the preranked list is the TCGA samples described as above. For each gene, the set of mutated samples was tested for enrichment against this preranked list. Both normalized enrichment score and P values from this test were used to measure the association between alternative 3′ss usage and gene mutations. Hypermutated individuals with >500 somatic mutations were removed from this analysis.
Local Network Community Analysis.
STRING interactions were used as the background biological network (https://string-db.org). Only the top 10% of highly confident interactions were kept. The (−1)*log10 P value from the paired t test was used to weigh the nodes of the above network. From the above preranked enrichment analysis, only nodes (genes) with at least 50 mutated TCGA samples were retained for this analysis. R package “igraph” was used to cluster this node weighted network. Specifically, we used the function “cluster_infomap” to detect local network communities with consideration of node weights (15). After the clustering, each gene (node) was uniquely assigned to one community. Then communities were ranked by their weights averaged over all node members.
Heatmap and Hierarchical Clustering.
To explore the relative similarity of cryptic 3′ss usage, an unsupervised hierarchical clustering was performed using the PSI values of the 47 cryptic 3′ss events between SF3B1 mutant (68 top-ranked cases), SUGP1 mutant (13 top-ranked cases), and WT samples (200 randomly selected cases). Missing values in the matrix were imputed as PSI of 0. Command “heatmap.2” from R package “gplots” was adopted for this analysis using Euclidean distance and (1−Pearson correlation)/2 as distance, respectively.
Expression Plasmid Constructs.
The N-terminally HA-tagged WT and K700E mutant SF3B1, and N-terminally HA-tagged WT and G574A-G582A mutant SUGP1 were cloned in p3xFLAG-CMV-14 (Sigma) in our previous study (11). The R189Q, E240K, R317W, L515P, G519V, R625T, P636L, and R642W mutant SUGP1 constructs were generated by site-directed mutagenesis (27).
Minigene Assays.
Minigenes for TTI1, ORAI2, MAP3K7, and ZNF91 were cloned into pcDNA3 (Invitrogen) in our previous study (11). A mixture of these four minigenes (100 ng each) and expression plasmid DNA (2 μg) were cotransfected to HEK293T cells in six-well plates using Lipofectamine 2000 (Thermo Fisher Scientific). At 48 h posttransfection, total RNA was extracted from the transfected cells using TRIzol (Thermo Fisher Scientific), followed by treatment with DNase I (New England Biolabs). RT-PCR was performed as described in our previous study (11). Briefly, 2 μg of DNase-treated total RNA was reverse transcribed using Maxima Reverse Transcriptase (Thermo Scientific) with 50 pmol oligo-dT primer and 0.2 pmol vector-specific reverse primer (5′-TAGAAGGCACAGTCGAGG-3′), followed by PCR reactions containing [α-32P] dCTP. PCR products were resolved in a 6% nondenaturing PAGE, and the gel was then dried and exposed to a phosphor screen. Radioactive signals were scanned by a Typhoon FLA 7000 imager (GE Healthcare) and quantified using ImageQuant (Molecular Dynamics). Primers used in the PCR reactions were as follows: vector-specific forward primer, 5′-TAATACGACTCACTATAGGGAG-3′; ORAI2 reverse, 5′-CTCTCCATCCCATCTCCTTG-3′; TTI1 reverse, 5′-ACATCTGGACGGGTGTCATT-3′; ZNF91 reverse, 5′-CTCTGCTCTGGCCAAAAGTC-3′; and MAP3K7 reverse, 5′-TCCCTGTGAATTAGCGCTTT-3′.
Western Blotting.
Western blotting was performed as described in our previous study (11). Briefly, proteins were resolved by SDS/PAGE and transferred to nitrocellulose membranes (Bio-Rad), followed by immunoblotting with primary and secondary antibodies. Primary antibodies were as follows: anti-ACTIN (Sigma, A2066), anti-SF3B1 (Bethyl Laboratories, A300-996A), anti-SUGP1 (Bethyl Laboratories, A304-675A-M), anti-HA rabbit (Abm, G166), anti-HA mouse (Sigma, H3663). Secondary antibodies were as follows: Donkey anti-Rabbit IgG (LI-COR, 926-68073) and Goat anti-Mouse IgG (LI-COR, 926-32210). Immunofluorescence signals were detected using the ChemiDoc Imaging System (Bio-Rad).
Data Availability.
All data generated in this study are included in the main text, SI Appendix, or Datasets S1–S4.
Supplementary Material
Acknowledgments
This work has been funded by NIH Grants U54 CA193313 (to R.R.) and R35 GM118136 (to J.L.M.) and the Edward P. Evans Foundation (J.L.M.). Z.L. is supported by NIH Grant P01CA087497.
Footnotes
Competing interest statement: G.Y. and Y.S. are coauthors on a 2019 research paper.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1922622117/-/DCSupplemental.
References
- 1.Seiler M., et al. , Somatic mutational landscape of splicing factor genes and their functional consequences across 33 cancer types. Cell Rep. 23, 282–296.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Papaemmanuil E., et al. ; Chronic Myeloid Disorders Working Group of the International Cancer Genome Consortium , Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N. Engl. J. Med. 365, 1384–1395 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yoshida K., et al. , Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 478, 64–69 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Quesada V., Ramsay A. J., Lopez-Otin C., Chronic lymphocytic leukemia with SF3B1 mutation. N. Engl. J. Med. 366, 2530 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Wang L., et al. , SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N. Engl. J. Med. 365, 2497–2506 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Furney S. J., et al. , SF3B1 mutations are associated with alternative splicing in uveal melanoma. Cancer Discov. 3, 1122–1129 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Darman R. B., et al. , Cancer-associated SF3B1 hotspot mutations induce cryptic 3′ splice site selection through use of a different branch point. Cell Rep. 13, 1033–1045 (2015). [DOI] [PubMed] [Google Scholar]
- 8.DeBoever C., et al. , Transcriptome sequencing reveals potential mechanism of cryptic 3′ splice site selection in SF3B1-mutated cancers. PLOS Comput. Biol. 11, e1004105 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alsafadi S., et al. , Cancer-associated SF3B1 mutations affect alternative splicing by promoting alternative branchpoint usage. Nat. Commun. 7, 10615 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robert-Paganin J., Réty S., Leulliot N., Regulation of DEAH/RHA helicases by G-patch proteins. BioMed Res. Int. 2015, 931857 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang J., et al. , Disease-causing mutations in SF3B1 alter splicing by disrupting interaction with SUGP1. Mol. Cell 76, 82–95.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kahles A., et al. , Comprehensive analysis of alternative splicing across tumors from 8,705 patients. Cancer Cell 34, 211–224.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao J., et al. , Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang J., et al. , Clonal evolution of glioblastoma under therapy. Nat. Genet. 48, 768–776 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosvall M., Bergstrom C. T., Maps of random walks on complex networks reveal community structure. Proc. Natl. Acad. Sci. U.S.A. 105, 1118–1123 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aravind L., Koonin E. V., Novel predicted RNA-binding domains associated with the translation machinery. J. Mol. Evol. 48, 291–302 (1999). [DOI] [PubMed] [Google Scholar]
- 17.Niu Z., Jin W., Zhang L., Li X., Tumor suppressor RBM5 directly interacts with the DExD/H-box protein DHX15 and stimulates its helicase activity. FEBS Lett. 586, 977–983 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Warkocki Z., et al. , The G-patch protein Spp2 couples the spliceosome-stimulated ATPase activity of the DEAH-box protein Prp2 to catalytic activation of the spliceosome. Genes Dev. 29, 94–107 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Larsson C. A., Cote G., Quintás-Cardama A., The changing mutational landscape of acute myeloid leukemia and myelodysplastic syndrome. Mol. Cancer Res. 11, 815–827 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berglund J. A., Abovich N., Rosbash M., A cooperative interaction between U2AF65 and mBBP/SF1 facilitates branchpoint region recognition. Genes Dev. 12, 858–867 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruskin B., Zamore P. D., Green M. R., A factor, U2AF, is required for U2 snRNP binding and splicing complex assembly. Cell 52, 207–219 (1988). [DOI] [PubMed] [Google Scholar]
- 22.Crisci A., et al. , Mammalian splicing factor SF1 interacts with SURP domains of U2 snRNP-associated proteins. Nucleic Acids Res. 43, 10456–10473 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee S. C.-W., et al. , Synthetic lethal and convergent biological effects of cancer-associated spliceosomal gene mutations. Cancer Cell 34, 225–241.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsu T. Y.-T., et al. , The spliceosome is a therapeutic vulnerability in MYC-driven cancer. Nature 525, 384–388 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yin S., et al. , A murine model of chronic lymphocytic leukemia based on B cell-restricted expression of Sf3b1 mutation and Atm deletion. Cancer Cell 35, 283–296.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoshimi A., et al. , Coordinated alterations in RNA splicing and epigenetic regulation drive leukaemogenesis. Nature 574, 273–277 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R., Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77, 51–59 (1989). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated in this study are included in the main text, SI Appendix, or Datasets S1–S4.