Significance
Compounds known as “maturation inhibitors” are potentially important treatments for HIV-1 infections. These compounds strongly inhibit the conversion of immature HIV-1 virions to the mature, infectious state by blocking an essential step in the cleavage of the Gag polyprotein of immature virions into its individual protein subunits. We show that the mechanism of action of the prototype maturation inhibitor bevirimat involves surprisingly subtle changes in molecular structure and molecular motions at the critical cleavage site within the Gag polyprotein lattice of the immature state. Results from these experiments, in which a variety of solid-state nuclear magnetic resonance techniques are applied to virus-like particles prepared in vitro, may contribute to the further development of effective maturation inhibitors.
Keywords: HIV-1 Gag, bevirimat, solid-state NMR, HIV-1 maturation
Abstract
HIV-1 maturation involves conversion of the immature Gag polyprotein lattice, which lines the inner surface of the viral membrane, to the mature capsid protein (CA) lattice, which encloses the viral RNA. Maturation inhibitors such as bevirimat (BVM) bind within six-helix bundles, formed by a segment that spans the junction between the CA and spacer peptide 1 (SP1) subunits of Gag, and interfere with cleavage between CA and SP1 catalyzed by the HIV-1 protease (PR). We report solid-state NMR (ssNMR) measurements on spherical virus-like particles (VLPs), facilitated by segmental isotopic labeling, that provide information about effects of BVM on the structure and dynamics of CA–SP1 junction helices in the immature lattice. Although BVM strongly blocks PR-catalyzed CA–SP1 cleavage in VLPs and blocks conversion of VLPs to tubular CA assemblies, 15N and 13C ssNMR chemical shifts of segmentally labeled VLPs with and without BVM are very similar, indicating that interaction with BVM does not alter the six-helix bundle structure appreciably. Only the 15N chemical shift of A280 (the first residue of SP1) changes significantly, consistent with BVM binding to an internal ring of hydrophobic side chains of L279 residues. Measurements of transverse 15N spin relaxation rates reveal a reduction in the amplitudes and/or timescales of backbone N–H bond motions, corresponding to a rigidification of the six-helix bundles. Overall, our data show that inhibition of HIV-1 maturation by BVM involves changes in structure and dynamics that are surprisingly subtle, but still sufficient to produce a large effect on CA–SP1 cleavage.
HIV-1 virions emerge from infected host cells in an immature state, in which the inner leaflet of the viral membrane is partially covered by a two-dimensional (2D) “immature” protein lattice formed by the Gag polyprotein (1, 2). In the subsequent maturation process, Gag is cleaved into its separate protein subunits by the HIV-1 protease (PR). Gag cleavage proceeds preferentially in a specific sequence (3–5), beginning with cleavage between spacer peptide 1 (SP1) and nucleocapsid protein (NC) segments, continuing with cleavage between matrix (MA) and capsid (CA) protein segments and between spacer peptide 2 (SP2) and p6 segments, and concluding with cleavage between CA and SP1 segments and between NC and spacer peptide 2 (SP2) segments. The final cleavage between CA and SP1 releases CA proteins, which then self-assemble to form the “mature” protein lattice of the capsid shell that encloses the viral RNA within mature HIV-1 (6). Formation of the CA lattice within a mature, infectious virion does not occur until CA–SP1 cleavage (7). Therefore, inhibition of CA–SP1 cleavage by “maturation inhibitor” compounds is a promising strategy for treatment of HIV-1 infections (8–10).
Although disassembly of the immature protein lattice depends on cleavage between CA and SP1, uncleaved CA–SP1 spontaneously self-assembles into the mature (not the immature) protein lattice in vitro, with the C-terminal 13-residue portion of CA and the entire 14-residue SP1 segment being dynamically disordered in this mature lattice (11). Thus, the immature protein lattice that exists within HIV-1 before the final cleavage step by PR is apparently a thermodynamically metastable state. To explain these observations, it was proposed (12–16) and subsequently demonstrated (17–20) that a protein segment around the CA–SP1 junction forms α-helices in the immature Gag lattice, which in turn form six-helix bundles that act as an energetic or steric barrier to disassembly of the immature lattice and subsequent assembly of the mature lattice.
The first maturation inhibitor to be investigated in detail was bevirimat (BVM), which was shown to prevent replication of infectious virions by interfering with CA–SP1 cleavage (8, 9). Experimental data from cryogenic electron tomography (cryo-ET) of immature HIV-1 and virus-like particles (VLPs) (17) and from electron diffraction of crystals formed by the C-terminal domain of CA with SP1 (CACTD–SP1) (20) show that BVM binds within six-helix bundles, but these data do not have sufficient resolution to reveal atomic-level details of the BVM binding site or to allow an atomic-level assessment of changes in CA–SP1 conformation, structural ordering, or dynamics that may arise from interactions with BVM.
In this paper, we use solid-state NMR (ssNMR) to investigate effects of BVM on the structure and dynamics of the CA–SP1 junction in the immature HIV-1 protein lattice. Experiments are performed on VLPs formed by the HIV-1 Gag construct ΔMA–CA–SP1–NC, which includes residues 1 to 15 and 100 to 132 of MA and the entire CA, SP1, and NC sequences (Fig. 1A). The PR cleavage site is between L279 and A280. Previous work has shown that ΔMA–CA–SP1–NC VLPs contain the full immature lattice and are an appropriate model system for immature HIV-1 (2, 13, 17). For ssNMR, we prepare VLPs that are 15N,13C-labeled only in the C-terminal portion of ΔMA–CA–SP1–NC, beginning with residue 266 of this construct (or residue 218 of the CA subunits). Segmental labeling eliminates ssNMR signal contributions from the unlabeled N-terminal portion, which includes structurally ordered segments of CA that would otherwise have strong ssNMR signals and would therefore complicate the interpretation of the data.
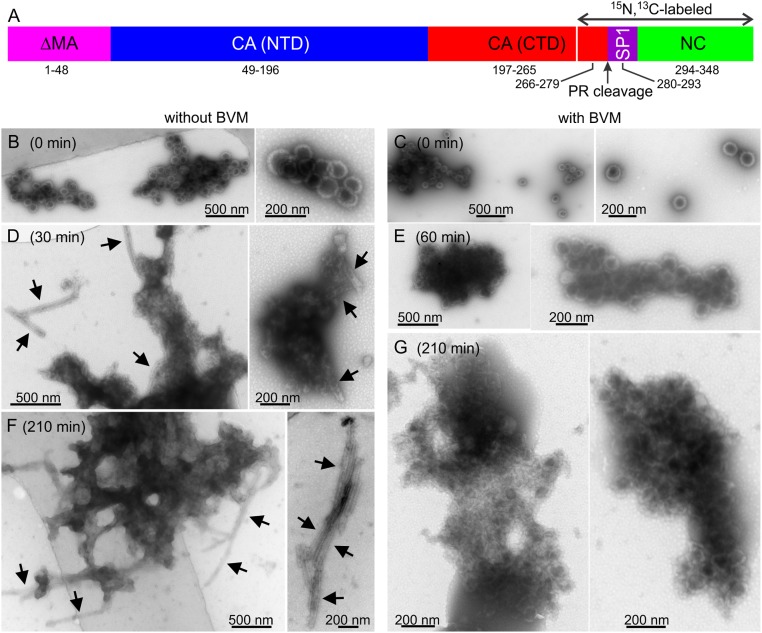
Fig. 1.
(A) Schematic representation of the ΔMA–CA–SP1–NC construct, showing the PR cleavage site and the isotopically labeled segment. (B–G) TEM images of negatively stained ΔMA–CA–SP1–NC VLPs, prepared without BVM (B, D, and F) or with BVM (C, E, and G). Images were recorded after incubation with PR for 0 min (B and C), 30 min (D), 60 min (E), or 210 min (F and G). The arrows in D and F indicate tubular assemblies, formed by CA after complete cleavage of ΔMA–CA–SP1–NC by PR. Such tubular assemblies are not observed in E and G, indicating that BVM inhibits complete cleavage. Segmentally labeled ΔMA–CA–SP1–NC was used in these PR cleavage experiments.
As presented below, by comparing data for VLPs with and without BVM, we observe changes in certain ssNMR chemical shifts and signal intensities upon BVM binding. These changes suggest subtle modifications of the six-helix bundle but do not indicate major structural changes. Additional measurements of 15N spin relaxation times indicate an overall rigidification of the CA–SP1 junction helices, with the amplitudes and/or correlation times of backbone amide N–H bond motions being reduced upon BVM binding. The overall conclusion from our data is that maturation inhibitors such as BVM induce relatively subtle changes in the structure and dynamics of the CA–SP1 junction helices in the protein lattice of immature HIV-1, but that these changes are sufficient to inhibit CA–SP1 cleavage strongly.
Results
Inhibition of CA–SP1 Cleavage in VLPs with BVM.
Fig. 1 compares transmission electron microscopy (TEM) images of VLPs with and without BVM after various periods of incubation with PR. Initially, both samples have the expected hollow spherical morphology of the immature HIV-1 protein lattice (Fig. 1 B and C). VLP diameters are ∼110 ± 20 nm for both samples, and no morphological differences are observed. After only 30 min of incubation with PR, VLPs without BVM are clearly disrupted and tubular assemblies are observed (Fig. 1D). These tubular assemblies are morphologically identical to HIV-1 CA tubes, which are known to contain the mature HIV-1 protein lattice (21, 22). Tubular assemblies are more prevalent after 210 min of incubation with PR, coexisting with apparently amorphous aggregates (Fig. 1F). Hollow spheres are no longer observed. In contrast, VLPs with BVM retain their initial spherical morphologies after 60 min of incubation with PR (Fig. 1E). After 210 min of incubation with PR, the morphology of VLPs with BVM is clearly distorted, possibly due to cleavage at SP1–NC and/or MA–CA junctions, and distorted spheres coexist with apparently amorphous aggregates. However, no tubular assemblies are observed (Fig. 1G). After 48 h of incubation with PR, VLPs without BVM are more fully converted to tubular assemblies (SI Appendix, Fig. S1A). VLPs with BVM are primarily converted to amorphous aggregates after 48 h, coexisting with a minor population of tubular assemblies that constitute less than 5% of the total proteinaceous material in the TEM images (SI Appendix, Fig. S1B).
Data in Fig. 1 and SI Appendix, Fig. S1 are consistent with the known inhibition of CA–SP1 cleavage by BVM within the immature HIV-1 protein lattice (8, 9). In addition, analysis of the VLP/PR reaction mixtures by sodium dodecyl sulfate/polyacrylamide gel electrophoresis (SDS/PAGE) shows nearly complete cleavage of ΔMA–CA–SP1–NC after 120 min in the absence of BVM, but substantial protection of CA–SP1 in the presence of BVM (SI Appendix, Fig. S2). The resistance of our VLPs with BVM to PR-catalyzed cleavage validates our procedure for incorporating BVM into ΔMA–CA–SP1–NC VLPs.
Importantly, as shown in SI Appendix, Fig. S3, BVM does not directly inhibit PR and does not protect unassembled HIV-1 Gag from PR-catalyzed cleavage at the CA–SP1 junction (8). The effect of BVM depends on the immature lattice structure.
Changes in ssNMR Spectra of VLPs Induced by BVM.
For ssNMR measurements, segmentally 15N,13C-labeled ΔMA–CA–SP1–NC was produced with a split-intein–based method (SI Appendix, Fig. S4 and SI Methods), similar to the method we have used for segmental labeling of CA in earlier work (23). The labeled segment begins at C266 (the C-terminal end of helix H11 of CA) and includes all of SP1 and NC. Most of the structurally ordered segments of CA are unlabeled, thereby simplifying the ssNMR spectra and allowing us to focus on the CA–SP1 junction.
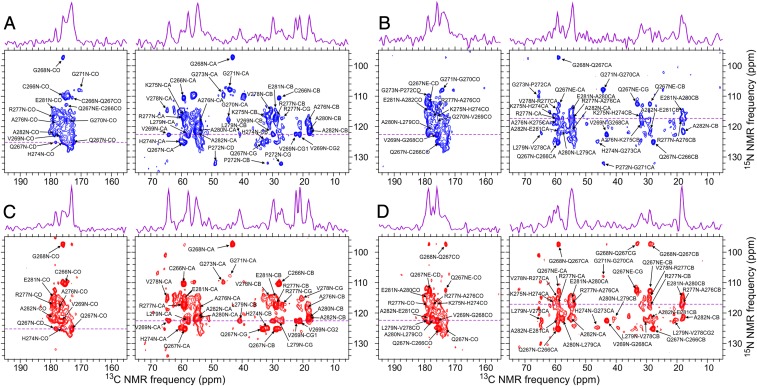
To investigate structural changes induced by BVM binding, 2D ssNMR spectra were acquired under identical conditions for segmentally labeled VLPs with and without BVM. 2D NCACX and NCOCX spectra of both samples are shown in Fig. 2. Site-specific assignments of ssNMR signals were obtained from these 2D spectra, as well as from three-dimensional (3D) NCACX and NCOCX spectra of VLPs without BVM (SI Appendix, Fig. S6). For VLPs without BVM, 15N and 13C chemical shift assignments were obtained for residues 266 to 282, i.e., from the beginning of the labeled segment in CA through the first three residues of SP1, with no unassigned gaps. These assignments are shown in Table 1, along with predicted backbone torsion angles from the TALOS-N program (24). The predicted torsion angles indicate a helical conformation in residues 274 to 281. The CA–SP1 junction helix may extend past residue 281 (Discussion), but the absence of chemical shift assignments beyond residue 282 precludes a prediction.
Fig. 2.
The 2D NCACX (A and C) and NCOCX (B and D) ssNMR spectra of segmentally 15N,13C-labeled VLPs that were prepared without (A and B) or with (C and D) BVM. Site-specific assignments of cross-peak signals for residues 266 to 282 are indicated, with numbering based on the ΔMA–CA–SP1–NC sequence. One-dimensional slices at dashed purple lines are shown above the 2D spectra. Contour levels increase by successive factors of 1.3.
Table 1.
NMR chemical shift assignments and backbone torsion angle predictions from TALOS-N
| Residue | N | CO | CA | CB | CG | CD, CE | Side-chain N | ϕ | ψ |
| C266 | 110.5 | 175.1 | 59.2 | 27.8 | |||||
| 110.4 | 175.2 | 59.1 | 27.9 | ||||||
| Q267 | 125.2 | 173.0 | 58.9 | 28.9 | 33.2 | 178.4 | 58 ± 6 | 39 ± 7 | |
| 125.2 | 173.1 | 58.8 | 28.8 | 33.2 | 178.4 | 112.7 | −60 ± 6 | −33 ± 8 | |
| G268 | 97.3 | 175.9 | 43.1 | 83 ± 17 | 0 ± 24 | ||||
| 97.3 | 175.9 | 43.1 | −88 ± 9 | 1 ± 9 | |||||
| V269 | 122.6 | 176.2 | 64.0 | 30.0 | 22.3, 21.1 | −68 ± 8 | 135 ± 10 | ||
| 122.6 | 176.4 | 64.2 | 29.7 | 22.3, 20.9 | −64 ± 7 | 136 ± 8 | |||
| G270 | 118.1 | 174.1 | 44.4 | 84 ± 13 | 1 ± 9 | ||||
| 174.0 | 44.2 | 90 ± 12 | −4 ± 13 | ||||||
| G271 | 107.9 | 170.7 | 44.1 | 145 ± 49 | 180 ± 13 | ||||
| 107.9 | 43.9 | ||||||||
| P272 | 132.1 | 178.6 | 65.4 | 31.5 | 27.3 | 48.8 | −57 ± 6 | 143 ± 8 | |
| 31.2 | 27.0 | 49.0 | −63 ± 6 | 148 ± 8 | |||||
| G273 | 109.8 | 46.2 | 94 ± 16 | −4 ± 17 | |||||
| 109.8 | 46.2 | −69 ± 10 | 154 ± 18 | ||||||
| H274 | 124.9 | 176.0 | 60.2 | 31.8 | 137.0 | 117.4 | −65 ± 9 | −36 ± 13 | |
| 124.9 | 176.1 | 60.3 | 31.7 | 137.4 | 117.4 | −58 ± 5 | −38 ± 8 | ||
| K275 | 116.6 | 59.5 | 31.6 | 24.3 | 28.6, 41.4 | −69 ± 8 | −36 ± 10 | ||
| 116.8 | 32.8 | 41.4 | −63 ± 6 | −39 ± 6 | |||||
| A276 | 118.7 | 177.5 | 54.3 | 18.4 | −65 ± 5 | −32 ± 10 | |||
| 118.4 | 177.5 | 54.4 | 18.5 | −67 ± 6 | −36 ± 5 | ||||
| R277 | 117.2 | 178.5 | 59.3 | 28.9 | 27.0 | 42.6, 158.9 | 71.8, 70.8, 83.3 | −60 ± 25 | −34 ± 20 |
| 117.3 | 178.9 | 59.2 | 29.0 | 26.8 | 42.7, 159.0 | 71.9, 70.6, 83.4 | −66 ± 5 | −38 ± 5 | |
| V278 | 114.7 | 64.7 | 30.6 | 20.9 | −66 ± 6 | −31 ± 9 | |||
| 115.1 | 178.8 | 64.6 | 30.4 | 20.8, 20.5 | −65 ± 4 | −37 ± 4 | |||
| L279 | 122.5 | 177.3 | 57.6 | 40.7 | −65 ± 6 | −33 ± 9 | |||
| 122.5 | 177.3 | 57.6 | 40.8 | 28.0 | −67 ± 5 | −36 ± 6 | |||
| A280 | 121.0 | 178.5 | 54.8 | 18.0 | −64 ± 7 | −29 ± 10 | |||
| 121.8 | 178.7 | 54.8 | 18.1 | −65 ± 6 | −34 ± 7 | ||||
| E281 | 115.0 | 178.6 | 58.9 | 28.8 | −71 ± 11 | −23 ± 16 | |||
| 115.2 | 178.8 | 59.0 | 28.8 | −63 ± 6 | −34 ± 5 | ||||
| A282 | 121.7 | 178.8 | 54.7 | 17.9 | |||||
| 121.6 | 179.0 | 54.9 | 18.1 |
15N and 13C chemical shifts are in parts per million relative to liquid ammonia and DSS, respectively. Predicted torsion angles ϕ and ψ are in degrees, with uncertainties reported by TALOS-N. For each residue, the upper row contains values for VLPs without BVM. The lower row contains values for VLPs with BVM. Numbering of residues is based on the ΔMA–CA–SP1–NC sequence.
The similarity of cross-peak positions in 2D spectra allowed us to transfer the assignments for VLPs without BVM to VLPs with BVM. Superpositions of the 2D NCACX and NCOCX spectra (SI Appendix, Fig. S7) indicate the similarity of these spectra. The 2D 13C–13C ssNMR spectra of both samples were also obtained and are also similar aside from differences in overall signal-to-noise ratios (SI Appendix, Fig. S8).
The 2D NCACX and NCOCX spectra also contain unassigned signals, which are poorly resolved and/or relatively weak. These signals are attributable to C-terminal portions of the SP1 segment and the NC segment. The NC segment was not observed to form an ordered array in cryo-ET measurements by Schur et al. (17) or in earlier electron microscopy studies of the immature protein lattice (2, 7). Apparently, the NC segment is dynamically disordered in the immature lattice. Nonetheless, weak signals from the NC segment are expected if nuclear spin polarization transfers in this segment are attenuated by molecular motions but not fully quenched. Attenuated polarization transfers can occur, for example, if motions in the NC segment are anisotropic (leading to reduced but nonzero motionally averaged nuclear magnetic dipole–dipole couplings) or if the motions are isotropic but slow (i.e., timescales greater than 100 μs). The 2D 13C–13C spectra (SI Appendix, Fig. S8) show stronger signals from residues outside the 266 to 282 range, including threonine and isoleucine signals that may arise from residues 291 to 294, presumably because the 2D 13C–13C spectra do not involve 15N–13C cross-polarization steps that are most sensitive to molecular motions.
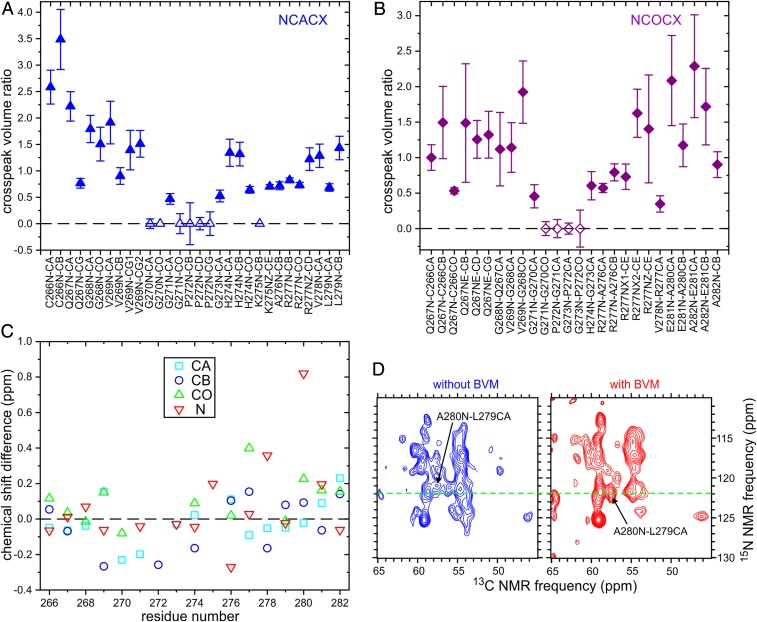
Significant differences in relative cross-peak intensities were observed for certain residues. In particular, signals from residues 270 to 273 are weak or undetectable in 2D NCACX and NCOCX spectra of VLPs with BVM, whereas signals from residues 266 to 269 are relatively strong compared with the corresponding signals in 2D spectra of VLPs without BVM (Fig. 3 A and B). These differences in signal intensities most likely reflect differences in amplitudes or timescales of local backbone motions (see below).
Fig. 3.
Ratios of cross-peak volumes in 2D NCACX (A) and NCOCX (B) ssNMR spectra of segmentally labeled VLPs prepared with BVM to the corresponding cross-peak volumes in 2D spectra of VLPs prepared without BVM. Error bars are estimated from the signal-to-noise ratios in the 2D spectra. Cross-peak volumes for VLPs with BVM are scaled to make the average cross-peak volume ratio equal to 1.0. Hollow symbols indicate that the cross-peak was below the noise level in the 2D spectrum of VLPs with BVM. (C) 13C and 15N chemical shift differences between VLPs with BVM and VLPs without BVM, for Cα, Cβ, CO, and backbone N sites. (D) Portions of 2D NCOCX spectra that show the difference in 15N chemical shift for A280. Contour levels increase by successive factors of 1.2.
For the most part, chemical shift differences between VLP sample with and without BVM are within the uncertainties of the chemical shift measurements (typically ±0.2 ppm) and cannot be considered significant. A significant difference in the chemical shifts of assigned signals is observed only for the backbone amide 15N site of A280 (Fig. 3 C and D).
In an earlier ssNMR study from our laboratory (19), chemical shifts were reported for residues 266 to 289 in uniformly 15N,13C-labeled VLPs without BVM. For residues 266 to 273, agreement with values in Table 1 is good, after correcting for differences in chemical shift references. Chemical shifts for residues 274 to 289 in the earlier study may be partially incorrect, illustrating the difficulty of making unambiguous assignments from congested 3D ssNMR spectra of a high–molecular-weight, noncrystalline protein assembly without segmental labeling.
Changes in 15N Transverse Relaxation Rates Induced by BVM.
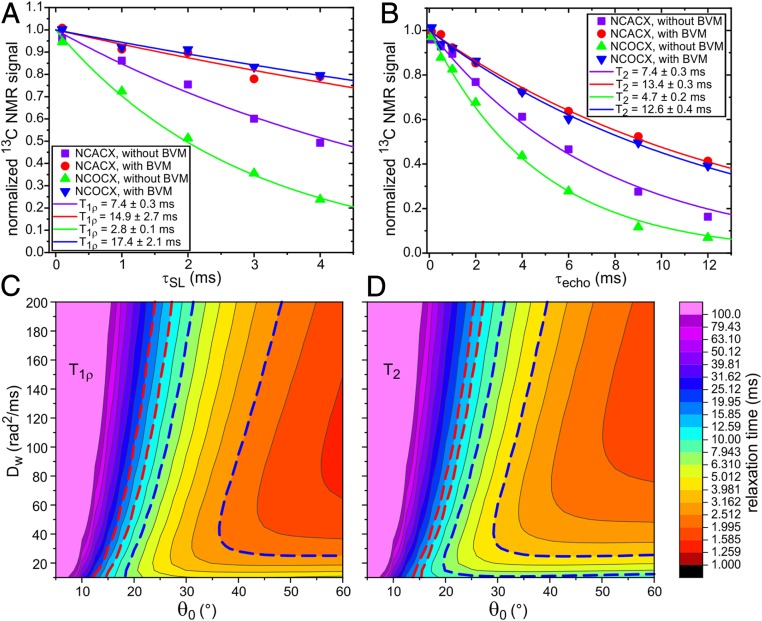
To further characterize dynamical differences, we measured transverse spin relaxation times T1ρ and T2 for backbone amide 15N sites, using 15N spin-locking and spin echo techniques. 15N relaxation times were detected through 1D 13C ssNMR signals after 15N–13Cα or 15N–13CO polarization transfer, followed by `13C–13C polarization transfer (i.e., NCACX or NCOCX detection). Radiofrequency pulse sequences for these measurements are shown in SI Appendix, Fig. S9 A and B. The decays of the 15N magnetization due to transverse relaxation are plotted in Fig. 4 A and B and are fit with single-exponential functions to yield the T1ρ and T2 values. For VLPs with BVM, T1ρ is in the 12- to 19-ms range, while T2 is in the 12- to 14-ms range. For VLPs without BVM, T1ρ is in the 3- to 8-ms range, while T2 is in the 4- to 8-ms range.
Fig. 4.
Measurements of 15N T1ρ (A) and T2 (B) relaxation times in segmentally labeled VLPs prepared with and without BVM, detected through 13C ssNMR signals after NCACX or NCOCX polarization transfers. Pulse sequences for these measurements are shown in SI Appendix, Fig. S9 A and B. Color-coded solid lines are single-exponential fits to the experimental data points, yielding the indicated T1ρ and T2 values. Error limits are SEs reported by the fitting routine. (C and D) Calculated dependences of 15N T1ρ and T2 on the amplitude and rate of N–H bond motion, using theoretical expressions for transverse spin relaxation due to diffusion in a cone with half-angle θ0 and orientational diffusion constant Dw. The dashed red lines enclose the ranges of parameter values that are consistent with experimental measurements on VLPs with BVM. The dashed blue lines enclose the ranges of parameter values that are consistent with experimental measurements on VLPs without BVM.
To interpret the relaxation data, we used a simple “diffusion-in-a-cone” model, i.e., a model in which transverse spin relaxation is driven by fluctuations in one-bond 15N–1H dipole–dipole couplings due to random diffusional motion within a cone with half-angle θ0 and with rotational diffusion constant Dw. As shown in SI Appendix, this model leads to theoretical expressions for T1ρ and T2 that depend on θ0, Dw, and the strengths of 15N spin-locking and 1H decoupling fields employed in the experimental measurements (40 and 92 kHz, respectively). Expressions for T1ρ and T2 also depend on the N–H distance, which we take to be 1.04 Å (25). Fig. 4 C and D show plots of calculated T1ρ and T2 values as functions of θ0 and Dw. A given value of the relaxation time can result from a continuum of values of the two parameters. Nonetheless, the ranges of parameter values that produce the experimentally observed relaxation times for VLPs with and without BVM are distinct and nonoverlapping. Combinations of θ0 and Dw that are consistent with data in Fig. 4 A and B are indicated by regions within dashed red or dashed blue lines in Fig. 4 C and D. Although it is possible that θ0 has the same value in both samples (e.g., θ = 25°), the values of Dw would then be necessarily different, with Dw ∼ 160 rad2/ms for VLPs with BVM and Dw < 80 rad2/ms for VLPs without BVM. Alternatively, if Dw has the same value in both samples (e.g., Dw = 100 rad2/ms), the values of θ0 would then be necessarily different, with θ0 ∼ 21° for VLPs with BVM and θ0 ∼ 30 ± 3° for VLPs without BVM. Within the context of the diffusion-in-a-cone model, binding of BVM causes either a reduction in the amplitude of backbone N–H bond motions or a reduction in the timescale of these motions.
For unrestricted, isotropic rotational diffusion, the orientational correlation time τc that appears in expressions for spin relaxation is related to the diffusion constant by (26). For diffusion-in-a-cone, three different correlation times τm (with m = 1, 2, or 3) describe the decays of the three terms Cm in the relevant correlation function (27). For θ = 25°, the m = 1 component makes the largest contribution to transverse spin relaxation. Values of τ1 are ∼0.34 and 0.91 μs for Dw = 160 rad2/ms and Dw = 60 rad2/ms, respectively (SI Appendix).
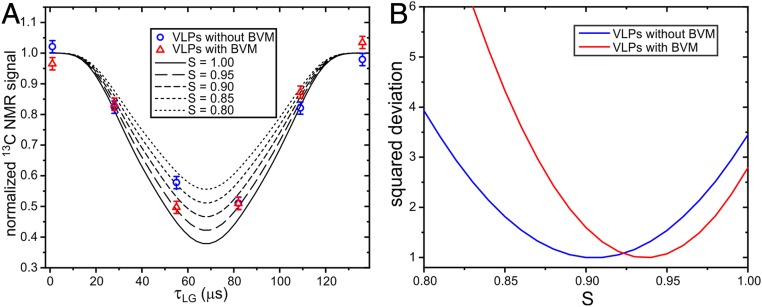
As an additional characterization of backbone dynamics, we performed measurements of motionally averaged 15N–1H dipole–dipole couplings, using the dipolar/shift correlation (DIPSHIFT) method (28–30) shown in SI Appendix, Fig. S9C. In these measurements, 15N spin polarization evolves under the 15N–1H couplings during incremented periods τLG of Lee–Goldburg 1H–1H decoupling. The decay and subsequent refocusing of the 15N polarization within one magic-angle spinning (MAS) rotation period τR is detected through 13C ssNMR signals after 15N–13Cα and 13C–13C polarization transfers. Data for VLPs with and without BVM are plotted in Fig. 5A and compared with numerical simulations for 15N–1H spin pairs. Provided that the motion of the N–H bond direction is rapid compared to the MAS frequency, we expect the DIPSHIFT data in Fig. 5A to show a motionally averaged 15N–1H dipole–dipole coupling that is scaled by , where the overbar indicates averaging over orientations. For diffusion-in-a-cone, . From comparisons with simulations (Fig. 5B), best-fit values of S are 0.94 ± 0.04 and 0.90 ± 0.05 for VLPs with and without BVM, respectively, corresponding to values of θ0 equal to 16 ± 6° and 21 ± 5. Together, the combination of DIPSHIFT and transverse relaxation data indicate that backbone motions in VLPs with BVM have reduced amplitudes and/or reduced correlation times relative to backbone motions in VLPs without BVM.
Fig. 5.
(A) DIPSHIFT measurements of motionally averaged backbone amide 1H–15N dipole–dipole couplings, detected through 13C ssNMR signals after NCACX polarization transfers. Data are shown for segmentally labeled VLPs prepared with and without BVM, scaled so that the average of the data points at τLG = 0.0 and τLG = 136.0 μs is 1.0. The solid and dashed curves are ideal simulations that correspond to 1H–15N bond distances of 1.04/S1/3 Å, where S is the scaling factor that results from N–H bond motions. The pulse sequence for these measurements is shown in SI Appendix, Fig. S9C. (B) Dependence of the total squared deviation between experimental data points and simulations, as a function of the value of S in the simulations. Squared deviations are scaled to make the minimum deviations equal to 1.0.
As shown in Fig. 4 A and B, the NCACX versions of 15N spin-locking and spin echo measurements on VLPs without BVM yield larger values of T1ρ and T2 than do the NCOCX versions. A likely explanation for this observation is that the relaxation times are orientation dependent (as expected from theory), and that different orientations are favored in the 15N–13Cα and 15N–13CO polarization transfers. The fact that 15N–13C cross-polarization times in these experiments (4.0 ms) were comparable to 15N T1ρ values observed for VLPs without BVM may make the orientation-dependent effects more pronounced.
Data points in Figs. 4 A and B and 5 were obtained by integrating 13C ssNMR signals over the full range of aliphatic 13C chemical shifts. The individual 13C spectra are shown in SI Appendix, Figs. S10–S12. To within the precision of these measurements, we did not observed differences in T1ρ or T2 values or in apparent S values when signals were integrated over ranges that correspond to individual peaks in the 13C spectra, indicating that the transverse 15N spin relaxation times and motional amplitudes do not vary significantly within protein segments that contribute to the ssNMR signals.
Discussion
Results from our cleavage assays (Fig. 1 and SI Appendix, Figs. S1 and S2) show that incorporation of BVM into ΔMA–CA–SP1–NC VLPs has a clear effect on the resistance of VLPs to PR-induced disassembly and on conversion of the immature Gag protein lattice of VLPs to the mature protein lattice of tubular CA assemblies. These results are consistent with earlier work (8, 9), which showed that BVM inhibits cleavage at the CA–SP1 junction, thereby inhibiting disruption of the six-helix bundles formed by CA–SP1 junction helices in the immature protein lattice. One might expect there to be major changes in structure and dynamics at the molecular level around the CA–SP1 junction that would underlie this greatly enhanced resistance to cleavage by PR. However, ssNMR data presented above show that effects of BVM on molecular structure and dynamics are rather subtle.
We do not see significant changes in 15N and 13C chemical shifts that would indicate conformational changes between C266 and A282, with the notable exception of the backbone amide 15N chemical shift of A280 (Fig. 3 C and D). Interestingly, PR cleavage would occur between the carbonyl carbon of L279 and the amide nitrogen of A280. The 15N chemical shift change at A280 may then result from binding of BVM near the L279–A280 peptide bond. In the crystal structure of CACTD–SP1 solved by Wagner et al. (18), in which CACTD–SP1 forms hexagonal units that mimic the immature Gag protein lattice (Protein Data Bank ID code 5I4T), this peptide bond is on the inner surface of the junction helix bundle, at the level of an internal ring formed by the side chains of the six copies of L279 within each bundle (L231 in the numbering used by Wagner et al.). This internal hydrophobic ring may be the binding site for BVM. In cryo-ET measurements by Schur et al. (17) and electron diffraction measurements by Purdy et al. (20), density attributable to BVM was observed within junction helix bundles at the level of the L279–A280 peptide bond. No changes in density attributable to the junction helices themselves were reported. Thus, our ssNMR chemical shift data are consistent with these earlier studies.
We speculate that binding of BVM to the internal hydrophobic ring created by L279 side chains prevents transient unfolding of junction helices. Without transient unfolding of junction helices, PR cleavage at the CA–SP1 junction cannot occur, because the cleavage site is not accessible to PR when the six-helix bundle structure is intact (18). We note that the 15N–13Cβ cross-peaks of L279 are stronger in 2D NCACX and NCOCX spectra of VLPs with BVM than in the corresponding spectra of VLPs without BVM (Fig. 2 and SI Appendix, Fig. S13), consistent with structural stabilization. Cross-peaks of V278 and A280 are also somewhat stronger or sharper in spectra of VLPs with BVM (SI Appendix, Fig. S13).
Purdy et al. additionally suggested that the side chain of K275 (K359 in their numbering scheme) interacts electrostatically with the carboxyl group of bound BVM (20). We do not detect differences in K275 side-chain signals in our 2D ssNMR spectra of VLPs with and without BVM. From the existing ssNMR data, we cannot determine the orientation of BVM relative to the junction helices.
Interaction with BVM does not cause an elongation of the junction helices that is detectable in our ssNMR spectra. From 2D and 3D spectra of VLPs with and without BVM (Fig. 2 and SI Appendix, Figs. S6–S8), we can assign ssNMR signals up to, but not beyond, A282. The absence of an effect on the lengths of junction helices is consistent with the binding site for BVM suggested above, since binding to L279 side chains would not be expected to stabilize helical conformations beyond A282. For comparison, junction helices in the CACTD–SP1 crystal structure of Wagner et al. extend to V286. However, residues 284 to 286 in their structural model (Protein Data Bank ID code 5I4T, residues 236 to 238 in their numbering) lack side-chain atoms, presumably reflecting structural disorder, and also lack helix–helix contacts. Density derived from cryo-ET measurements on VLPs with BVM by Schur et al. (Electron Microscopy Data Bank file EMD-4018) apparently also extends to V286 but does not have sufficient resolution to show distinct side chains. Thus, it seems plausible that residues on the C-terminal side of A282 are more disordered and partially dynamic, accounting for their absence from ssNMR spectra.
Cross-peak intensities in 2D NCACX and NCOCX spectra of VLPs with and without BVM are significantly different. Differences are plotted in Fig. 3 A and B and are also obvious in the 2D spectra themselves (Fig. 2 and SI Appendix, Fig. S7). In particular, cross-peaks from G270, G721, and P272 are weak or absent in 2D spectra of VLPs with BVM but are readily observed in 2D spectra of VLPs without BVM. This observation may seem surprising if one expects a general stabilizing effect from binding of BVM. However, the junction helices begin after P272, as indicated by torsion angle predictions from ssNMR chemical shifts (Table 1) and previous structural studies (17–20). Thus, the weak or absent cross-peaks for residues 270 to 272 in 2D spectra of VLPs with BVM indicate a reduction in structural order (or possibly a longer timescale for molecular motions) in the segment that immediately precedes the junction helices, but not in the junction helices themselves.
Measurements of transverse 15N spin relaxation times (Fig. 4) and 15N–1H dipole–dipole couplings (Fig. 5) show that BVM does indeed stabilize the six-helix bundle structure, in the sense that the amplitudes and/or timescales of fluctuations in backbone amide N–H bond directions are reduced in VLPs with BVM. A quantitative analysis based on the diffusion-in-a-cone model (Results) suggests that the amplitude of fluctuations may be reduced by 5 to 10° or the timescale of fluctuations may be reduced by a factor of 3. Variations in 2D cross-peak intensities, discussed in the preceding paragraph, suggest an opposite effect in residues 270 to 272. Because signals from these residues are weak, they do not contribute to the transverse relaxation and dipole–dipole coupling measurements. Thus, BVM appears to stabilize the junction helix structure in VLPs but may destabilize the structure in a short segment that immediately precedes the junction helices.
Finally, experiments described above illustrate the utility of segmental isotopic labeling for enabling ssNMR measurements that probe the structure and dynamics of specific, functionally important segments within large, noncrystalline assemblies formed by high–molecular-weight proteins. In these experiments on VLPs, signals in 2D and 3D triple-resonance ssNMR spectra arise primarily from a specific 17-residue segment of a 348-residue protein, with other segments being either unlabeled or too dynamic to contribute to the ssNMR spectra. The resulting spectral simplifications allowed us to suggest a likely BVM binding site, rule out major conformational changes induced by BVM binding, and detect segment-specific effects on molecular motions. Further developments in segmental labeling methods are likely to expand further the size and complexity of biomolecular systems that are amenable to ssNMR methods.
Materials and Methods
Sample Preparation and ssNMR.
Segmentally labeled ΔMA–CA–SP1–NC with the amino acid sequence in SI Appendix, Fig. S4A was produced from His-tagged fusing constructs, composed of residues 1 to 265 fused to the N-terminal segment of intein DnaE from Nostoc punctiforme and residues 266 to 348 fused to the C-terminal intein segment, as shown in SI Appendix, Fig. S4B. VLPs were prepared under conditions described previously (19). ssNMR measurements were performed at 17.5 T (Figs. 2 and 4 and SI Appendix, Figs. S6–S8, S10, and S11) and 14.1 T (Fig. 5 and SI Appendix, Fig. S12). MAS frequencies were 12.00 and 7.00 kHz, respectively. Sample temperatures in ssNMR measurements were 10 to 12 °C. Full experimental details are given in SI Appendix.
Data Availability.
All 2D and 3D ssNMR spectra are available from Mendeley Data at https://dx.doi.org/10.17632/gtn5jv7vnk.1.
Supplementary Material
Acknowledgments
This work was supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases and by the Intramural AIDS Antiviral Program of the National Institutes of Health.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: All 2D and 3D ssNMR spectra reported in this paper have been deposited in the Mendeley Data repository (https://dx.doi.org/10.17632/gtn5jv7vnk.1).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1917755117/-/DCSupplemental.
References
- 1.Fuller S. D., Wilk T., Gowen B. E., Kräusslich H. G., Vogt V. M., Cryo-electron microscopy reveals ordered domains in the immature HIV-1 particle. Curr. Biol. 7, 729–738 (1997). [DOI] [PubMed] [Google Scholar]
- 2.Briggs J. A. G., et al. , Structure and assembly of immature HIV. Proc. Natl. Acad. Sci. U.S.A. 106, 11090–11095 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mervis R. J., et al. , The gag gene products of human immunodeficiency virus type 1: Alignment within the gag open reading frame, identification of posttranslational modifications, and evidence for alternative gag precursors. J. Virol. 62, 3993–4002 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gowda S. D., Stein B. S., Engleman E. G., Identification of protein intermediates in the processing of the p55 HIV-1 gag precursor in cells infected with recombinant vaccinia virus. J. Biol. Chem. 264, 8459–8462 (1989). [PubMed] [Google Scholar]
- 5.Pettit S. C., et al. , The p2 domain of human immunodeficiency virus type 1 Gag regulates sequential proteolytic processing and is required to produce fully infectious virions. J. Virol. 68, 8017–8027 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ganser-Pornillos B. K., Yeager M., Sundquist W. I., The structural biology of HIV assembly. Curr. Opin. Struct. Biol. 18, 203–217 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keller P. W., Adamson C. S., Heymann J. B., Freed E. O., Steven A. C., HIV-1 maturation inhibitor bevirimat stabilizes the immature Gag lattice. J. Virol. 85, 1420–1428 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li F., et al. , PA-457: A potent HIV inhibitor that disrupts core condensation by targeting a late step in gag processing. Proc. Natl. Acad. Sci. U.S.A. 100, 13555–13560 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou J., et al. , Small-molecule inhibition of human immunodeficiency virus type 1 replication by specific targeting of the final step of virion maturation. J. Virol. 78, 922–929 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Waheed A. A., Freed E. O., HIV type 1 Gag as a target for antiviral therapy. AIDS Res. Hum. Retroviruses 28, 54–75 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han Y., et al. , Magic angle spinning NMR reveals sequence-dependent structural plasticity, dynamics, and the spacer peptide 1 conformation in HIV-1 capsid protein assemblies. J. Am. Chem. Soc. 135, 17793–17803 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Accola M. A., Höglund S., Göttlinger H. G. A., A putative alpha-helical structure which overlaps the capsid-p2 boundary in the human immunodeficiency virus type 1 Gag precursor is crucial for viral particle assembly. J. Virol. 72, 2072–2078 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gross I., et al. , A conformational switch controlling HIV-1 morphogenesis. EMBO J. 19, 103–113 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liang C., et al. , Characterization of a putative alpha-helix across the capsid-SP1 boundary that is critical for the multimerization of human immunodeficiency virus type 1 gag. J. Virol. 76, 11729–11737 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wright E. R., et al. , Electron cryotomography of immature HIV-1 virions reveals the structure of the CA and SP1 Gag shells. EMBO J. 26, 2218–2226 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Datta S. A. K., et al. , On the role of the SP1 domain in HIV-1 particle assembly: A molecular switch? J. Virol. 85, 4111–4121 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schur F. K. M., et al. , An atomic model of HIV-1 capsid-SP1 reveals structures regulating assembly and maturation. Science 353, 506–508 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Wagner J. M., et al. , Crystal structure of an HIV assembly and maturation switch. eLife 5, e17063 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bayro M. J., Ganser-Pornillos B. K., Zadrozny K. K., Yeager M., Tycko R., Helical conformation in the CA-SP1 junction of the immature HIV-1 lattice determined from solid-state NMR of virus-like particles. J. Am. Chem. Soc. 138, 12029–12032 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Purdy M. D., et al. , MicroED structures of HIV-1 Gag CTD-SP1 reveal binding interactions with the maturation inhibitor bevirimat. Proc. Natl. Acad. Sci. U.S.A. 115, 13258–13263 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li S., Hill C. P., Sundquist W. I., Finch J. T., Image reconstructions of helical assemblies of the HIV-1 CA protein. Nature 407, 409–413 (2000). [DOI] [PubMed] [Google Scholar]
- 22.Zhao G., et al. , Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics. Nature 497, 643–646 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gupta S., Tycko R., Segmental isotopic labeling of HIV-1 capsid protein assemblies for solid state NMR. J. Biomol. NMR 70, 103–114 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen Y., Bax A., Protein backbone and sidechain torsion angles predicted from NMR chemical shifts using artificial neural networks. J. Biomol. NMR 56, 227–241 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yao L. S., Vogeli B., Ying J. F., Bax A., NMR determination of amide N-H equilibrium bond length from concerted dipolar coupling measurements. J. Am. Chem. Soc. 130, 16518–16520 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abragam A., The Principles of Nuclear Magnetism (Oxford University Press, Oxford, 1961). [Google Scholar]
- 27.Lipari G., Szabo A., Pade approximants to correlation-functions for restricted rotational diffusion. J. Chem. Phys. 75, 2971–2976 (1981). [Google Scholar]
- 28.Cady S. D., Hong M., Amantadine-induced conformational and dynamical changes of the influenza M2 transmembrane proton channel. Proc. Natl. Acad. Sci. U.S.A. 105, 1483–1488 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu J. X., Yau W. M., Tycko R., Evidence from solid-state NMR for nonhelical conformations in the transmembrane domain of the amyloid precursor protein. Biophys. J. 100, 711–719 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Munowitz M. G., Griffin R. G., Bodenhausen G., Huang T. H., Two-dimensional rotational spin-echo nuclear magnetic-resonance in solids—correlation of chemical-shift and dipolar interactions. J. Am. Chem. Soc. 103, 2529–2533 (1981). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All 2D and 3D ssNMR spectra are available from Mendeley Data at https://dx.doi.org/10.17632/gtn5jv7vnk.1.