Significance
Viomycin and the related antibiotic capreomycin have been the main drugs effective against tuberculosis, inhibiting the ribosomes in the pathogenic bacteria. Inhibition is most likely caused by blocking the translocation step of protein synthesis. Viomycin has been shown to stabilize the ribosome in a state of intersubunit rotation, resembling the hybrid-state intermediate of translocation, which may explain its mode of action. Previous structural studies have identified a single viomycin binding site in the ribosome in a nonrotated state. In this paper, four additional viomycin binding sites have been identifed, three of which bind exclusively to the rotated state of the ribosome and are likely responsible for blocking translocation.
Keywords: ribosome, translocation, viomycin
Abstract
Viomycin, an antibiotic that has been used to fight tuberculosis infections, is believed to block the translocation step of protein synthesis by inhibiting ribosomal subunit dissociation and trapping the ribosome in an intermediate state of intersubunit rotation. The mechanism by which viomycin stabilizes this state remains unexplained. To address this, we have determined cryo-EM and X-ray crystal structures of Escherichia coli 70S ribosome complexes trapped in a rotated state by viomycin. The 3.8-Å resolution cryo-EM structure reveals a ribosome trapped in the hybrid state with 8.6° intersubunit rotation and 5.3° rotation of the 30S subunit head domain, bearing a single P/E state transfer RNA (tRNA). We identify five different binding sites for viomycin, four of which have not been previously described. To resolve the details of their binding interactions, we solved the 3.1-Å crystal structure of a viomycin-bound ribosome complex, revealing that all five viomycins bind to ribosomal RNA. One of these (Vio1) corresponds to the single viomycin that was previously identified in a complex with a nonrotated classical-state ribosome. Three of the newly observed binding sites (Vio3, Vio4, and Vio5) are clustered at intersubunit bridges, consistent with the ability of viomycin to inhibit subunit dissociation. We propose that one or more of these same three viomycins induce intersubunit rotation by selectively binding the rotated state of the ribosome at dynamic elements of 16S and 23S rRNA, thus, blocking conformational changes associated with molecular movements that are required for translocation.
The increasing occurrence of resistance of pathogenic bacteria to antibiotic agents has become a major health concern in recent years. One main reason for the emergence of antibiotic resistance is that pathogens can undergo mutation of their target structures to decrease their affinity for the drug. Studies of the modes of action of known antibiotics and the structures of their binding sites can, thus, facilitate the development of more effective antibiotics. Viomycin (SI Appendix, Fig. S1) is a ribosome-targeting cyclic polypeptide antibiotic widely used in the treatment of tuberculosis (1). Understanding the mechanism by which it inhibits protein synthesis may provide the basis for creating new drugs to address the problem of viomycin resistance.
Viomycin has been shown to inhibit ribosome translocation (2–4) and dissociation of the 70S ribosome into its 30S and 50S ribosomal subunits (5). The coupled movement of messenger RNA (mRNA) and tRNAs during translocation is accompanied by large-scale conformational changes of the ribosome, including rotational movements of the 30S subunit and its head domain (6–12). Combined fluorescence resonance energy transfer (FRET) and chemical probing experiments (13) have shown that binding of viomycin can trap the ribosome in a state in which the 30S subunit is rotated with the tRNAs bound in P/E and A/P states, resembling the intermediate hybrid state of translocation (14). The mechanism by which viomycin induces intersubunit rotation and hybrid-state tRNA binding remains largely unknown.
Previous cryo-EM and X-ray crystallographic studies have captured ribosome complexes in different rotated conformations in the presence of elongation factor EF-G (6–10, 15–17), ribosome recycling factor (RRF) (18), or release factor RF3 (19) either in the presence (6–10, 16, 17, 19) or in the absence (15, 18, 20) of antibiotics, including viomycin (8, 16, 17, 19). However, these studies have not addressed the question of how binding of viomycin is able to stabilize a rotated conformation of the ribosome in the absence of translation factors. To date, only a single structural study describing a ribosomal binding site for viomycin bound in the absence of other factors has been reported (21). Ribosomes in that complex contained a single-bound viomycin molecule but were shown to be in the nonrotated classical state; thus, its functional significance remains unclear with regard to how viomycin influences the large-scale dynamics of the ribosome. For this reason, we determined the cryo-EM and X-ray structures of viomycin-bound 70S ribosome complexes. In both cases, a total of five viomycins were bound to ribosomes trapped in conformations with extensive intersubunit rotation. The binding sites for the three of the four newly observed viomycins are found at intersubunit bridges containing dynamic ribosomal RNA elements that undergo conformational changes in the rotated state, explaining how viomycin induces intersubunit rotation and inhibits dissociation of the ribosomal subunits.
Results and Discussion
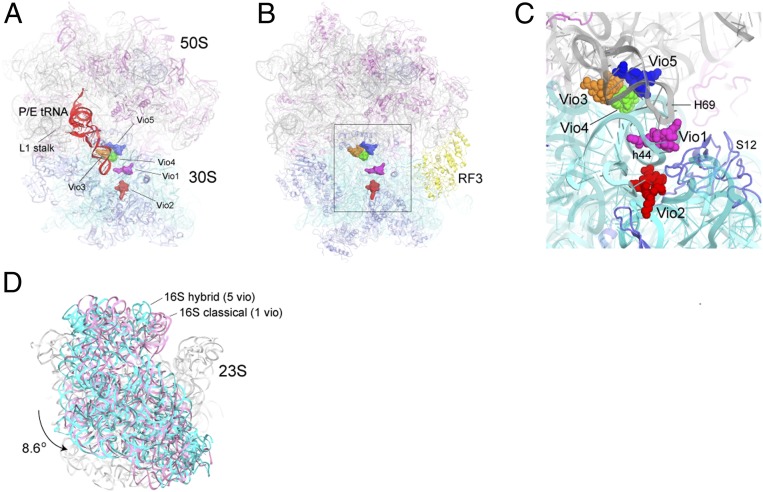
To characterize the effects of viomycin on the overall conformation of the ribosome, we first determined the 3.8-Å cryo-EM structure (Fig. 1A and SI Appendix, Figs. S2–S4) of a viomycin complex with E. coli 70S ribosomes containing mRNA and a deacylated initiator tRNAfMet (SI Appendix, Table S1). In the absence of viomycin, 70S ribosome complexes containing a bound initiator tRNA occupy the classical nonrotated state (13–23). In our viomycin-containing cryo-EM structure, the single main class of tRNA-containing particles obtained by 3D classification showed a rotated hybrid-state ribosome containing density for mRNA, a single tRNA bound in the P/E hybrid state, and five copies of viomycin (Fig. 1A and SI Appendix, Figs. S5 and S6). The cryo-EM structure, therefore, reveals the conformational changes that are unambiguously induced by the binding of viomycin itself. However, we also sought a higher-resolution description of the viomycin binding interactions. For this purpose, we revisited our previous X-ray crystallographic data to solve a crystal structure of viomycin bound to a rotated state of the E. coli 70S ribosome in the presence of release factor RF3 (Fig. 1 B and C) at an improved resolution of 3.1 Å using homogeneous diffraction data obtained from a single crystal (SI Appendix, Table S2). Although the presence of RF3 influences the magnitudes of rotation of the 30S subunit body and head domains, unbiased simulated-annealing Fo−Fc omit maps confirm the presence of the same five viomycin binding sites seen by cryo-EM (SI Appendix, Fig. S7) with occupancies ranging from 0.88 to 1.0 (SI Appendix, Figs. S8 and S9). Their interactions with dynamic features of ribosomal RNA suggest structural explanations for how viomycin inhibits subunit dissociation (Fig. 2) and induces intersubunit rotation and hybrid-state binding of tRNA (Fig. 1D) and, thus, inhibits translocation. Our results are consistent with reports of viomycin-resistance mutations that are indicative of multiple viomycin binding sites (24, 25) as well as single-molecule FRET titration experiments which suggest that multiple viomycins bind and work synergistically to inhibit translocation (26).
Fig. 1.
Overall structures of viomycin-containing complexes. (A) Cryo-EM structure of the E. coli 70S ribosome in a complex with viomycin, mRNA, and a single P/E tRNA (red) in the hybrid state. The positions of the five bound viomycin molecules are indicated. (B and C) X-ray crystal structure of a viomycin-containing complex of the E. coli 70S ribosome bound with release factor RF3 (yellow), showing the positions of the five viomycin molecules and their surroundings in the ribosome, including 16S rRNA (cyan), 23S rRNA (gray), and ribosomal protein S12 (blue). (D) The 30S subunit in the cryo-EM structure shown in A (cyan) has undergone an 8.6° counterclockwise rotation relative to its orientation in the classical-state ribosome containing a single bound viomycin (pink) (21).
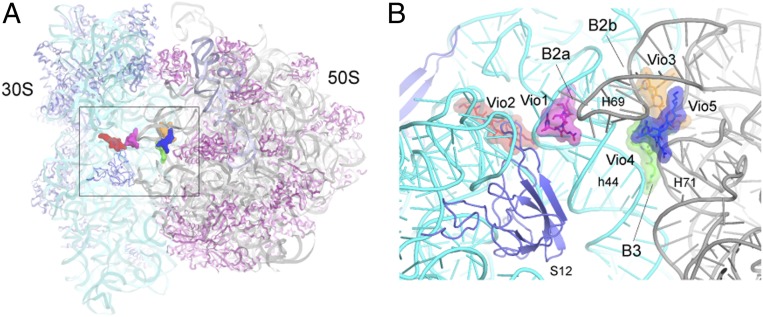
Fig. 2.
Positions of the viomycins relative to intersubunit bridges. (A) Overall view of the crystal structure of the 70S ribosome showing the positions of the five viomycins concentrated around the subunit interface. (B) Closeup view of the structure in A, showing the positions of the viomycins relative to structural features of the ribosome, including helices h44 of 16S rRNA and H69 and H71 of 23S rRNA and ribosomal protein S12. Three of the viomycins, Vio1, Vio3, and Vio4 bind at intersubunit bridges B2a, B2b, and B3.
Overall Structures
We prepared complexes for cryo-EM reconstruction using E. coli 70S ribosomes containing viomycin, a 27-nucleotide mRNA, and tRNAfMet bound initially to the classical P site using conditions similar to those used in a previous FRET study showing viomycin-induced intersubunit rotation (13) (SI Appendix, Supplementary Methods). Data were collected on a Titan Krios transmission cryoelectron microscope (FEI) operating at 300 kV and equipped with a K2 Summit direct electron detector (Gatan Inc.), generating 2,300 dose-fractionated image stacks that were corrected for beam-induced motion. After semiautomatic particle extraction, a total of 132,064 particles was imaged (SI Appendix, Fig. S2). Subsequent unsupervised 3D classification resulted in two classes of 70S ribosomes, distinguished by the presence or absence of tRNA (SI Appendix, Fig. S2 and Table S1). Cryo-EM reconstruction of the tRNA-containing class using a dataset composed of 103,009 particles revealed a structure with 8.6° intersubunit rotation and 5.3° rotation of the 30S subunit head domain, the deacylated tRNA in the P/E hybrid state with its 3′-CCA end bound to the 50S E site, and its elbow contacting the L1 stalk (Fig. 1 A and D). Importantly, the absence of any bound translation factor or crystal lattice effects in our cryo-EM structure confirms that the source of these large-scale rotational events can be ascribed directly to the presence of viomycin.
Viomycin Binding Sites
The 3.8 Å cryo-EM map revealed density for five viomycin molecules (SI Appendix, Fig. S6), four of which have not been previously described. Vio1 corresponds to the single viomycin bound to a classical state 70S complex previously reported by Stanley et al. (21) with some minor conformational differences as described below (see Fig. 4). No density attributable to Vio3, Vio4, or Vio5 can be seen in electron density maps deposited by Stanley et al. (21) (PDB accession no. 3KNH) (SI Appendix, Fig. S10), although we note some density corresponding to the position of Vio2 (SI Appendix, Fig. S10B). Following the preliminary fit to our cryo-EM map, we reexamined our previous viomycin-containing structure of release factor RF3 bound to the 70S ribosome (19). This complex, such as our cryo-EM complex, shows extensive (6.7°) intersubunit rotation. This rotation could be attributed to either viomycin or RF3, which has also been shown to induce intersubunit rotation (19, 27). After calculating an unbiased simulated-annealing 3.1-Å resolution omit map (SI Appendix, Figs. S8 and S9), we were able to unambiguously place and model the five copies of bound viomycin, all of which bind to elements of ribosomal RNA in the same positions as seen in the cryo-EM structure (Fig. 1 A–C and SI Appendix, Fig. S7). Vio3, Vio4, and Vio5, such as Vio1, are all located at intersubunit bridges (Figs. 1C and 2) providing new evidence for the mechanism of inhibition of subunit dissociation by viomycin (5).
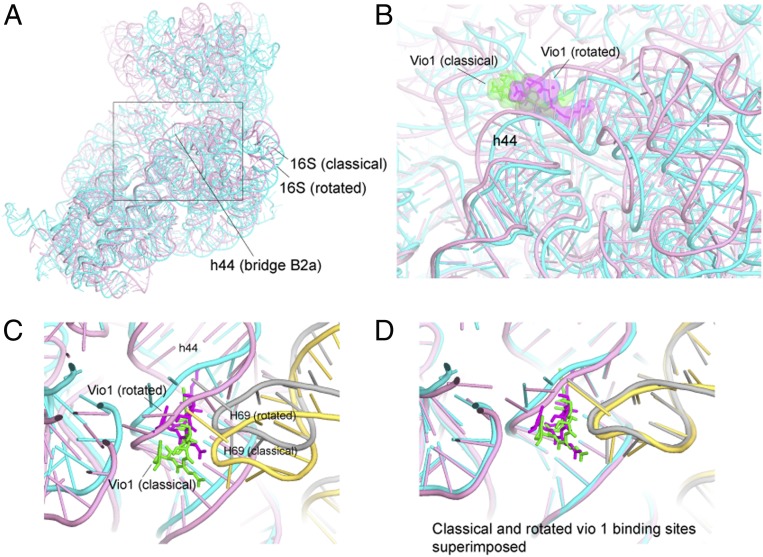
Fig. 4.
Vio1 binds to classical and rotated states. (A) Overall view of 16S rRNA in the classical (pink) and rotated (cyan) states as aligned on the core of 23S rRNA. (B) Movement of the Vio1 binding site between the classical and the rotated states. (C) H69 of 23S rRNA moves synchronously with h44 at the Vio1-binding site. (D) Alignment on 16S rRNA shows that the conformation of the Vio1-binding site is preserved upon intersubunit rotation. Vio1 is shown in green (classical) and magenta (rotated); 23S rRNA is shown in yellow (classical) and gray (rotated).
The Vio1 molecule is bound with full occupancy adjacent to intersubunit bridge B2a, which is formed between the loop of helix H69 of 23S rRNA and the base of helix h44 of 16S rRNA near the decoding site of the 30S subunit (see Fig. 4). Vio1 contacts the base and phosphate groups of A1493 of 16S rRNA and Thr40 of protein S12 in the decoding site (Fig. 3A). The binding of Vio1 closely resembles that of the single viomycin bound to a nonrotated classical-state ribosome (21), although its position is shifted toward the ribosomal E site due to repositioning of bridge B2a in the rotated state (Fig. 4 A–C). Bridge B2a is preserved upon intersubunit rotation by movement of helix H69 to follow the lateral movement of helix h44 (Fig. 4D). Thus, Vio1 precisely follows the movement of h44 and H69, shifting by ∼6 Å from its classical-state position (21). Since Vio1 binds to both rotated and nonrotated ribosomes, it is unlikely to play a role in promoting intersubunit rotation.
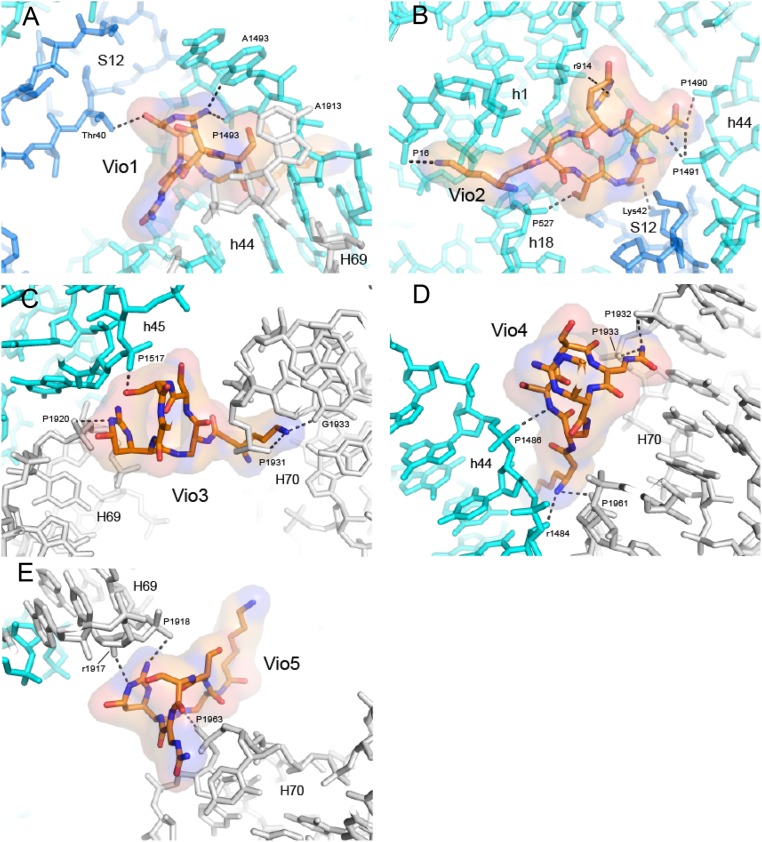
Fig. 3.
Molecular Interactions between the viomycin and the ribosome. (A) Vio1 contacts helix h44 of 16S rRNA, H69 of 23S rRNA, and Thr40 of ribosomal protein S12 at intersubunit bridge B2a, similar to the single viomycin observed for the nonrotated ribosome complex (21). (B) Vio2 binds exclusively to the 30S subunit, contacting h1, h18, and h44 in 16S rRNA and Lys42 and 43 of protein S12. (C) Vio3 binds at intersubunit bridge B2b, contacting h45 in 16S rRNA and H69 and H70 of 23S rRNA. (D) Vio4 binds at intersubunit bridge B3, near the axis of intersubunit rotation, contacting h44 in 16S rRNA and H70 in 23S rRNA. (E) Vio5 binds between H69 and H70 of 23S rRNA, likely restricting movement of H69 during intersubunit rotation. The viomycin structures are shown in all-atom and transparent surface rendering. The 16S rRNA is shown in cyan, 23S rRNA in gray, and S12 in blue. RNA phosphate moieties are labeled by P, and riboses are labeled by r.
In the previously reported viomycin-containing structure (21), the observed nonrotated state was likely favored by a combination of the presence of an initiator tRNA bound to the P site and crystallization of the complex in a crystal form that stabilizes the ribosome in the classical state (28). Our single-particle cryo-EM reconstruction shows that viomycin can stabilize the rotated state in the absence of restraints imposed by a crystal lattice (Fig. 1D), in agreement with ensemble FRET experiments which showed conversion of ribosomes to the rotated state in solution (13). In our crystal structure, we were able to obtain a crystal form that is compatible with the rotated form of the ribosome, which was also favored by the presence of RF3, which itself induces intersubunit rotation in solution (27).
Vio2 (occupancy = 0.92) binds exclusively to the 30S subunit (Fig. 2B), contacting helices h1, h18, h27, and h44 of 16S rRNA and Lys42 and Lys43 of ribosomal protein S12 (Fig. 3B). Along with Vio1, Vio2 is one of the two viomycins that interact with a ribosomal protein. A possible role for Vio2 is suggested by studies which showed that the mutations K43R or K87Q in S12 confer viomycin resistance to Mycobacterium smegmatis (24). The Vio2 binding site overlaps with that of streptomycin (29), explaining how viomycin competes for binding with streptomycin (30) (SI Appendix, Fig. S11). Besides its well-known implication in translational accuracy, streptomycin resistance, and streptomycin binding, protein S12 has also been proposed to function as a control element for ribosomal translocation (31). Electron density can be seen for Vio2 in the structure of the nonrotated viomycin-containing complex (21) (SI Appendix, Fig. S10B). Since the conformation of the binding site for Vio2 is virtually identical in the rotated and nonrotated structures, Vio2 is unlikely to influence intersubunit rotation.
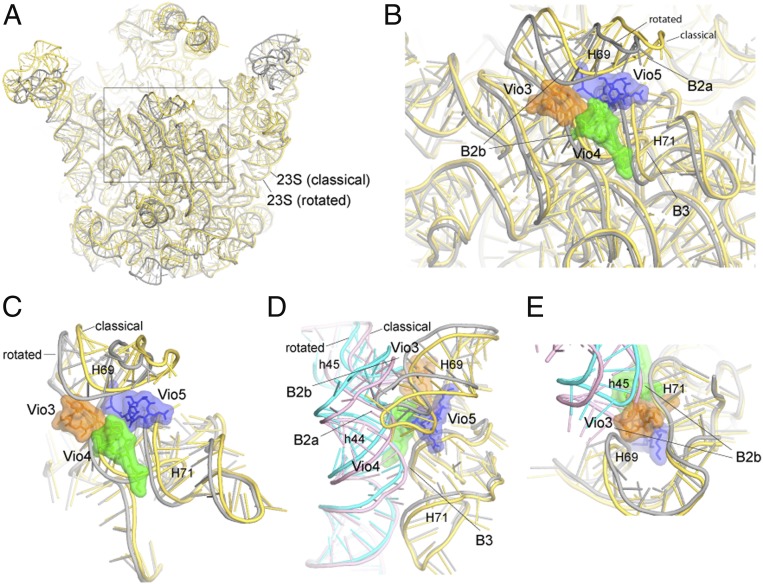
Vio3, Vio4, and Vio5 form a triple cluster (SI Appendix, Fig. S12) in the subunit interface at intersubunit bridges B2b and B3 at the convergence of helices h44 and h45 of 16S rRNA with helices H69, H70, and H71 of 23S rRNA (Fig. 2 A and B). Electron density for Vio3 (occupancy = 0.88) (SI Appendix, Fig. S9A), is located at the subunit interface in contact with bridge B2b connecting the hairpin loops of helices h45 and H69 with helix H70 (Figs. 2B and 3C). Comparison with ribosomes in the nonrotated state shows that bridge B2b undergoes a ∼5-Å movement during intersubunit rotation (15, 18, 19). In our structure, contact between Vio3, H70 and the loops of h45 and H69 would interfere with bridge B2b rearrangements that normally accompany its transition from the rotated to the nonrotated state (Fig. 5E). Mutations in the TlyA gene in B. smegmatis, which encodes a 2′-O-methyltransferase that is specific for ribose methylation at C1409 in h44 of 16S rRNA and C1920 in H69 of 23S rRNA, confer capreomycin and viomycin resistance (32, 33). This finding suggests that 2′-O-methylation of ribose 1920 could confer viomycin sensitivity by promoting binding of Vio3, which contacts the 23S rRNA backbone at position 1920 (Fig. 3C). Ribose 1409 of 16S rRNA does not directly contact any of the viomycins in our structure; however, Vio1 makes a close approach (within ∼4.6 Å) to the 2′-OH of ribose 1409, raising the possibility that they could make contact in the 2′-O-methylated ribosome (SI Appendix, Fig. S13).
Fig. 5.
Binding of Vio3, Vio4, and Vio5 interferes with conformational changes associated with intersubunit rotation. (A) Superimposition of 23S rRNA in the classical (yellow) and rotated (gray) states. (B) Vio3, Vio4, and Vio5 cluster at the site of maximum movement of 23S rRNA components of intersubunit bridges B2b and B3. (C) Rotated view of B. (D) Rotational movement of 16S rRNA and compensating conformational changes in 23S rRNA. (E) Vio3 connects h45 and H69 to H70; in the classical state, h45 would create a steric clash with Vio3. In addition, both Vio3 and Vio5 (C) bridge H69 to H70, interfering with movement of H69.
Vio4 binds to bridge B3 (occupancy = 0.92), which is formed between helices h44 of 16S rRNA and H71 of 23S rRNA (Fig. 2B and SI Appendix, Fig. S14). Vio4 contacts both components of bridge B3; on one side, it contacts positions 1,484 and 1,485 in helix h44 of 16S rRNA and on the other side positions 1,932 and 1,933 in H70 and 1,961 and 1,962 in H71 of 23S rRNA (Figs. 3D, 5 B and C, and SI Appendix, Fig. S14). In addition to its role in subunit association, bridge B3 coincides with the axis of intersubunit rotation (34). Binding of Vio4 to bridge B3 at C1484 of 16S rRNA could, thus, stabilize the rotated state of the ribosome by directly interfering with back rotation of the 30S subunit.
Vio5 (occupancy = 0.91) bridges helices H69 and H70 at positions 1,917 and 1,918 and 1,962 and 1,963, respectively, of 23S rRNA (Figs. 3E and 5C). Although Vio5 binds exclusively to 23S rRNA, it may interfere with intersubunit rotation by fixing the relative positions of H69 and H70, thus, preventing movement of H69. In addition, Vio5 may also contribute indirectly to inhibition of intersubunit rotation through its close interactions with Vio3 and Vio4 (Figs. 1C, 5 B and C, and SI Appendix, Fig. S12). As described above, intersubunit rotation is accompanied by lateral movement of helix H69 in 23S rRNA. As a component of subunit bridges B2a and B2b, movement of H69 permits relocation of both bridges to cope with rotation of the 30S subunit. Thus, cooperative binding of Vio3, Vio4, and Vio5 to the rotated-state ribosome would inhibit reversal of intersubunit rotation by preventing the repositioning of H69 back to its classical-state position.
There is evidence that neomycin may also influence intersubunit rotation through binding to H69. Addition of neomycin to a tRNA-ribosome complex resulted in a change in FRET efficiency corresponding to partial intersubunit rotation (35). In the same study, neomycin was soaked into single crystals containing both a classical-state ribosome and a hybrid-state ribosome bound to P/E tRNA and RRF (35). The resulting structures showed a neomycin molecule bound to the same position on H69 in both the classical and the hybrid-state complexes at a position distinct from that of Vio3, 4, or 5 (SI Appendix, Fig. S15 A and B). Binding of neomycin to the hybrid-state complex resulted in movement of H69 and the P/E tRNA closer to their classical-state positions (SI Appendix, Fig. S15 A and C). Since intersubunit rotation was constrained by the crystal lattice, it is unclear what the global effect of the H69 conformational change would have been in solution, but the authors inferred that binding of neomycin to H69 stabilizes a partially rotated state, consistent with their FRET measurements (35). However, in another study, cocrystallization of neomycin with 70S ribosomes, EF-G, and two tRNAs showed two neomycins bound near H69 (9) (SI Appendix, Fig. S15D) but at different sites from the one described above. This complex had significant 30S head rotation but very little intersubunit rotation, and it is unclear what role neomycin played in its stabilization versus that of EF-G and the tRNAs. The differences in neomycin binding between these structures show that the presence of ligands that influence ribosome conformation, such as RRF or EF-G, can also influence the mode of antibiotic binding.
The same may be true for viomycin. Here, we show that viomycin binds to the same five binding sites in rotated ribosomes in the presence or absence of RF3 (SI Appendix, Fig. S7). However, electron density maps for previously published crystal structures of complexes containing EF-G bound to vacant rotated ribosomes in the presence of viomycin and RRF (17) show electron density corresponding to Vio1 and Vio2 but not Vio3, Vio4, or Vio5 (SI Appendix, Fig. S16). The absence of Vio3, 4, and 5 in these structures may be related to the presence of RRF, which also binds H69 and so may compete with viomycin binding. Although Vio 3, 4, and 5 are also absent in complexes that were cocrystallized with EF-G and tRNA (8), these complexes show extensive 30S subunit head rotation but very little intersubunit rotation, consistent with our conclusion that Vio3, 4, and 5 bind preferentially to H69 in its rotated conformation.
Conclusions
The existence of multiple binding sites for viomycin is supported by the results of single-molecule FRET studies by Feldman et al. (26). They observed an interesting biphasic dependence of inter-tRNA FRET efficiencies with increasing viomycin concentration that likely reflects the influence of viomycin on hybrid-states formation. Vio1 and Vio2 are unlikely to influence intersubunit rotation. Vio2 binds exclusively to the 30S subunit, while Vio1 binds to both classical and rotated ribosomes as its binding site is preserved upon intersubunit rotation. We, therefore, propose that Vio3, Vio4, and Vio5 are responsible for trapping the ribosome in the rotated state. They form a compact cluster which binds at the subunit interface to mobile 23S rRNA elements of intersubunit bridges that contact the 30S subunit body domain around the axis of 30S rotation. Multiple interactions are formed by the viomycin cluster that fixes the position of H69, preventing its movement relative to H70 that is needed to preserve bridge contacts during rotation of the 30S subunit. In addition, contacts between these viomycins and the bridge elements of 16S rRNA further contribute to interference with intersubunit rotation. Finally, the binding sites for Vio3, Vio4, and Vio5 exist only in the rotated state, thus, trapping the ribosome in the rotated state.
Although it seems at first surprising that binding of a small molecule, such as viomycin, could induce rotation of a >800 KDa macromolecular structure, it is nevertheless clear that the energy barrier for intersubunit rotation is small enough to be triggered by thermal energy alone at 22° (22) or even by the difference in energy between binding an initiator tRNA versus an elongator tRNA to the ribosomal P site. A final question is whether Vio3, Vio4, or Vio5, which are implicated in stabilization of the rotated state of the ribosome are also responsible for antibiotic activity. Some support for this comes from mutations at or near their bindings sites in the loop of H69 of 23S rRNA at A1913U and U1915G as well as deletions of U1915 and A1916, which confer resistance to viomycin and/or capreomycin (32, 36). Ribose methylation of C1920 of 23S rRNA confers sensitivity to viomycin in B. smegmatis (32), implicating Vio3 (which contacts position 1920) in viomycin’s bactericidal action. These findings suggest further directions toward achieving a full understanding of the antibiotic action of viomycin.
Methods
For cryo-EM structure determination, viomycin was bound to tight-couple 70S ribosomes from E. coli MRE600 containing tRNAfMet and a defined 27-nucleotide mRNA. Cryo-EM data were acquired using an FEI Titan Krios cryotransmission electron microscope equipped with a Gatan K2 Summit direct electron detector. Following motion correction and estimation of contrast transfer function parameters, 13,000 particles were picked using semiautomated particle picking and subjected to 2D classification, 3D classification, and high-resolution refinement. For crystallographic structure determination, crystals of a viomycin-RF3-ribosome complex containing a 27-nucleotide mRNA and tRNAMet were grown via vapor diffusion. Diffraction data were integrated, merged, and scaled, and the structure was solved by molecular replacement using an all-atom model of a ribosome-RF3-GDPNP structure at 3.7-Å for the initial search.
Data Availability.
Coordinates and structure factors for the X-ray crystal structure of viomycin bound to a 70S ribosome containing mRNA, initiator tRNA and RF3 have been deposited with the Protein Data Bank, http://www.wwpdb.org/ (PDB ID code 6LKQ). The Cryo-EM structure of viomycin bound to a 70S ribosome containing mRNA and initiator tRNA has been deposited with the Protein Data Bank, http://www.wwpdb.org/ (PDB ID code EMD-0939).
Supplementary Material
Acknowledgments
We thank the staffs of the beamlines at the Advanced Light Source, the Advanced Photon Source, and the Shanghai Synchrotron Radiation Facility. Research at Zhejiang University was supported by Grants from the Natural Science Foundation of China (31971226), the Zhejiang Natural Science Foundation (LR20C050003), the Fundamental Research Funds for the Central Universities (2018QN81010), the new faculty start-up funds from Zhejiang University, and the Thousand Young Talents Plan of China. Research at the University of California, Santa Cruz was supported by Grant R35-GM118156 from the NIH (to H.F.N.) and funds from the Robert L. Sinsheimer Chair.
Footnotes
The authors declare no competing interest.
Data deposition: Coordinates and structure factors for the X-ray crystal structure of viomycin bound to a 70S ribosome containing mRNA, initiator tRNA, and RF3 have been deposited in the Worldwide Protein Data Bank, http://www.wwpdb.org/ (PDB ID code 6LKQ). The Cryo-EM structure of viomycin bound to a 70S ribosome containing mRNA and initiator tRNA has been deposited in the Worldwide Protein Data Bank, http://www.wwpdb.org/ (PDB ID codes 3KNH and EMD-0939).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2002888117/-/DCSupplemental.
References
- 1.Akbergenov R., et al. , Molecular basis for the selectivity of antituberculosis compounds capreomycin and viomycin. Antimicrob. Agents Chemother. 55, 4712–4717 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liou Y.-F., Tanaka N., Dual actions of viomycin on the ribosomal functions. Biochem. Biophys. Res. Commun. 71, 477–483 (1976). [DOI] [PubMed] [Google Scholar]
- 3.Modolell J., Vázquez D., The inhibition of ribosomal translocation by viomycin. Eur. J. Biochem. 81, 491–497 (1977). [DOI] [PubMed] [Google Scholar]
- 4.Polikanov Y. S., Aleksashin N. A., Beckert B., Wilson D. N., The mechanisms of action of ribosome-targeting peptide antibiotics. Front. Mol. Biosci. 5, 48 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamada T., Nierhaus K. H., Viomycin favours the formation of 70S ribosome couples. Mol. Gen. Genet. 161, 261–265 (1978). [DOI] [PubMed] [Google Scholar]
- 6.Ratje A. H., et al. , Head swivel on the ribosome facilitates translocation by means of intra-subunit tRNA hybrid sites. Nature 468, 713–716 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramrath D. J., et al. , Visualization of two transfer RNAs trapped in transit during elongation factor G-mediated translocation. Proc. Natl. Acad. Sci. U.S.A. 110, 20964–20969 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou J., Lancaster L., Donohue J. P., Noller H. F., Crystal structures of EF-G-ribosome complexes trapped in intermediate states of translocation. Science 340, 1236086 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou J., Lancaster L., Donohue J. P., Noller H. F., How the ribosome hands the A-site tRNA to the P site during EF-G-catalyzed translocation. Science 345, 1188–1191 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frank J., Agrawal R. K., A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature 406, 318–322 (2000). [DOI] [PubMed] [Google Scholar]
- 11.Belardinelli R., et al. , Choreography of molecular movements during ribosome progression along mRNA. Nat. Struct. Mol. Biol. 23, 342–348 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Noller H. F., Lancaster L., Mohan S., Zhou J., Ribosome structural dynamics in translocation: Yet another functional role for ribosomal RNA. Q. Rev. Biophys. 50, e12 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Ermolenko D. N., et al. , The antibiotic viomycin traps the ribosome in an intermediate state of translocation. Nat. Struct. Mol. Biol. 14, 493–497 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Moazed D., Noller H. F., Intermediate states in the movement of transfer RNA in the ribosome. Nature 342, 142–148 (1989). [DOI] [PubMed] [Google Scholar]
- 15.Tourigny D. S., Fernández I. S., Kelley A. C., Ramakrishnan V., Elongation factor G bound to the ribosome in an intermediate state of translocation. Science 340, 1235490 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brilot A. F., Korostelev A. A., Ermolenko D. N., Grigorieff N., Structure of the ribosome with elongation factor G trapped in the pretranslocation state. Proc. Natl. Acad. Sci. U.S.A. 110, 20994–20999 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pulk A., Cate J. H., Control of ribosomal subunit rotation by elongation factor G. Science 340, 1235970 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dunkle J. A., et al. , Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science 332, 981–984 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou J., Lancaster L., Trakhanov S., Noller H. F., Crystal structure of release factor RF3 trapped in the GTP state on a rotated conformation of the ribosome. RNA 18, 230–240 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin H., Kelley A. C., Ramakrishnan V., Crystal structure of the hybrid state of ribosome in complex with the guanosine triphosphatase release factor 3. Proc. Natl. Acad. Sci. U.S.A. 108, 15798–15803 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stanley R. E., Blaha G., Grodzicki R. L., Strickler M. D., Steitz T. A., The structures of the anti-tuberculosis antibiotics viomycin and capreomycin bound to the 70S ribosome. Nat. Struct. Mol. Biol. 17, 289–293 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cornish P. V., Ermolenko D. N., Noller H. F., Ha T., Spontaneous intersubunit rotation in single ribosomes. Mol. Cell 30, 578–588 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dorner S., Brunelle J. L., Sharma D., Green R., The hybrid state of tRNA binding is an authentic translation elongation intermediate. Nat. Struct. Mol. Biol. 13, 234–241 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taniguchi H., et al. , Molecular analysis of kanamycin and viomycin resistance in Mycobacterium smegmatis by use of the conjugation system. J. Bacteriol. 179, 4795–4801 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamada T., Mizugichi Y., Nierhaus K. H., Wittmann H. G., Resistance to viomycin conferred by RNA of either ribosomal subunit. Nature 275, 460–461 (1978). [DOI] [PubMed] [Google Scholar]
- 26.Feldman M. B., Terry D. S., Altman R. B., Blanchard S. C., Aminoglycoside activity observed on single pre-translocation ribosome complexes. Nat. Chem. Biol. 6, 54–62 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ermolenko D. N., et al. , Observation of intersubunit movement of the ribosome in solution using FRET. J. Mol. Biol. 370, 530–540 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Selmer M., Gao Y. G., Weixlbaumer A., Ramakrishnan V., Ribosome engineering to promote new crystal forms. Acta Crystallogr. D Biol. Crystallogr. 68, 578–583 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carter A. P., et al. , Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature 407, 340–348 (2000). [DOI] [PubMed] [Google Scholar]
- 30.Masuda K., Yamada T., Effect of viomycin on dihydrostreptomycin binding to bacterial ribosomes. Biochim. Biophys. Acta 435, 333–339 (1976). [DOI] [PubMed] [Google Scholar]
- 31.Cukras A. R., Southworth D. R., Brunelle J. L., Culver G. M., Green R., Ribosomal proteins S12 and S13 function as control elements for translocation of the mRNA:tRNA complex. Mol. Cell 12, 321–328 (2003). [DOI] [PubMed] [Google Scholar]
- 32.Johansen S. K., Maus C. E., Plikaytis B. B., Douthwaite S., Capreomycin binds across the ribosomal subunit interface using tlyA-encoded 2′-O-methylations in 16S and 23S rRNAs. Mol. Cell 23, 173–182 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Maus C. E., Plikaytis B. B., Shinnick T. M., Mutation of tlyA confers capreomycin resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 49, 571–577 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao H., et al. , Study of the structural dynamics of the E coli 70S ribosome using real-space refinement. Cell 113, 789–801 (2003). [DOI] [PubMed] [Google Scholar]
- 35.Wang L., et al. , Allosteric control of the ribosome by small-molecule antibiotics. Nat. Struct. Mol. Biol. 19, 957–963 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Monshupanee T., Gregory S. T., Douthwaite S., Chungjatupornchai W., Dahlberg A. E., Mutations in conserved helix 69 of 23S rRNA of Thermus thermophilus that affect capreomycin resistance but not posttranscriptional modifications. J. Bacteriol. 190, 7754–7761 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Coordinates and structure factors for the X-ray crystal structure of viomycin bound to a 70S ribosome containing mRNA, initiator tRNA and RF3 have been deposited with the Protein Data Bank, http://www.wwpdb.org/ (PDB ID code 6LKQ). The Cryo-EM structure of viomycin bound to a 70S ribosome containing mRNA and initiator tRNA has been deposited with the Protein Data Bank, http://www.wwpdb.org/ (PDB ID code EMD-0939).