Significance
The protein corona affects the clinical applications, organ targeting, and safety assessment of nanomaterials, and prediction of the protein corona would be valuable for the design of ideal nanomaterials. However, no methods to predict the protein corona are available. Overcoming the numerous quantitative and qualitative factors influencing corona formation, the present work builds models that precisely predict the functional composition of the protein corona and the cell recognition of nanoparticles (NPs) integrating machine learning and meta-analysis. This workflow provides an effective method to predict the functional composition of the protein corona that determines cell recognition to guide the synthesis and applications of NPs.
Keywords: machine learning, nanotoxicity, nano-bio interface, cellular recognition, protein corona
Abstract
Protein corona formation is critical for the design of ideal and safe nanoparticles (NPs) for nanomedicine, biosensing, organ targeting, and other applications, but methods to quantitatively predict the formation of the protein corona, especially for functional compositions, remain unavailable. The traditional linear regression model performs poorly for the protein corona, as measured by R2 (less than 0.40). Here, the performance with R2 over 0.75 in the prediction of the protein corona was achieved by integrating a machine learning model and meta-analysis. NPs without modification and surface modification were identified as the two most important factors determining protein corona formation. According to experimental verification, the functional protein compositions (e.g., immune proteins, complement proteins, and apolipoproteins) in complex coronas were precisely predicted with good R2 (most over 0.80). Moreover, the method successfully predicted the cellular recognition (e.g., cellular uptake by macrophages and cytokine release) mediated by functional corona proteins. This workflow provides a method to accurately and quantitatively predict the functional composition of the protein corona that determines cellular recognition and nanotoxicity to guide the synthesis and applications of a wide range of NPs by overcoming limitations and uncertainty.
In biological applications, nanoparticles (NPs) interact with numerous proteins and form protein coronas immediately upon administration into blood or contact with the extracellular matrix (1–3). The protein corona reshapes the physicochemical properties (e.g., size, charge, hydrophilicity, and stability) of NPs interfacing with biological systems, thus playing an important role in macrophage uptake, circulation time, immune responses, and cellular recognition of NPs (4, 5). The most conventional approach for analyzing the protein corona involves protein isolation procedures followed by protein identification using mass spectrometry-based proteomics (2, 6). The protein corona composition refers to the relative protein abundance (RPA) accounting for the total proteins in the corona and is an important parameter for describing the protein corona (3). The surface mapping of protein binding sites on the biomolecular corona of NPs was studied using antibody-labeled gold nanoparticles (7). Predicting the composition of the protein corona on a computer instead of via laboratory experiments is cost saving and can predict unknown interactions between biological entities and various NPs. To date, many factors (e.g., NP physicochemical properties, incubation, and separation conditions) have been shown to affect the biological responses (8) and composition of protein coronas (9–14). Therefore, it is difficult to delineate the composition of the protein corona using a general linear regression model or density functional theory (15). Density functional theory requires specific molecular structures, requires much time for calculations of complex systems, and is unable to predict corona formation on NPs accurately and efficiently because of the lack of a specific molecular structure and the coexistence of various proteins (16). The complex relationships between various NPs with protein corona formation and numerous quantitative or qualitative factors limit the application of density functional theory. The general linear regression model cannot handle multivariable problems (15) and will be compared with machine learning in the present work. Furthermore, because of the high heterogeneity between the multidimensional properties of NPs and the protein corona functional components (17), many traditional models (e.g., quantitative structural activity relationship) poorly predict the functional fingerprints of the protein corona (18, 19).
With the robust capability to build models to explain observations through experience, machine learning [e.g., random forest (RF) (20) and neural network] has recently been applied to recognize meaningful complex patterns to control robots (21), predict reproductive responses (19), and predict synthetic reactions (22). RF is a robust machine learning algorithm integrating a decision tree with good learning capability (20). Compared with support vector machines, neural networks, and other machine learning algorithms, RF achieves excellent prediction accuracy on heterogeneous big data with quantitative and qualitative factors (22, 23). Meanwhile, RF could investigate the complex factor-response dependence inside a data-driven model (15, 23) with robust tolerance of heterogeneity. However, the quantitative prediction of the functional compositions of the protein corona by a comprehensive understanding of complex NP–protein binding patterns remains unavailable using machine learning (24). To address these problems, the present work attempts to use a powerful RF model to identify the rules for protein corona formation by associating numerous NP physicochemical properties and distinct experimental conditions with quantitative protein corona compositions (e.g., hydrophily and function). The work aims to develop a general and quantitative prediction of corona formation behaviors on a wide range of (i.e., known and unknown) NPs and experimental conditions.
The cellular recognition of NPs is well known to determine their applications and adverse effects (3, 5), but the detection and prediction of the cellular recognition of various NPs by experimental methods and traditional models are difficult because of the multidimensional factor-response dependence. The protein corona associates cellular recognition (e.g., cellular uptake by macrophages and immune responses) (3, 25, 26) by presenting key functional motifs interacting with receptors as exposure of critical epitopes (3). For example, clusterin acts as a dyopsonin, increasing the stealth properties of NPs by cloaking them from recognition by macrophages (27). Therefore, predicting the protein corona may be an effective method to predict the biological responses of known and unknown NPs. The present work integrated machine learning and meta-analysis to explore the potential binding patterns of proteins on diverse NPs and then predict the biological effects of NPs based on the functional composition of the protein corona. The construction of a robust and flexible model is crucial for prediction of corona formation on a wide range of NPs and biological responses before experimental efforts, dramatically reducing the cost of experimental efforts. The accurate prediction of the functional composition of the protein corona and the resulting cellular recognition is useful for guiding the design, synthesis, and effective applications of known and unknown NPs.
Results and Discussion
Highly Heterogeneous Data on NPs and Protein Coronas.
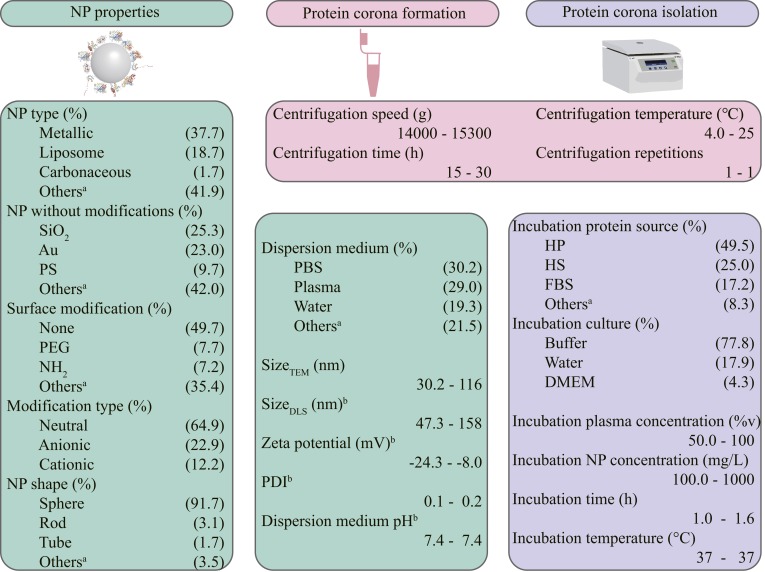
Currently, studies of protein coronas are conducted under specific conditions (e.g., one NP with few properties and one specific exposure pathway), and a data library on protein coronas of various NPs is lacking. Extraction and mining of hidden NP–protein corona-biological response relationships from published evidence with machine learning are urgently needed. Extraction of the data regarding the protein coronas on NPs was performed according to the workflow described in the Methods and SI Appendix. To reduce publication bias and extract information from distinct experimental conditions, strict criteria were applied in the literature extraction and data mining (shown in the Methods) (15, 28). Overall, 652 pieces of data related to the protein coronas on various NPs were mined and analyzed. Eight qualitative factors (NP type, NP shape, NP without modification, surface modification, modification type, dispersion medium, incubation plasma source, and incubation culture) and 13 quantitative factors (size measured by transmission electron microscopy [sizeTEM] and dynamic light scattering [sizeDLS], dispersion medium pH, zeta potential, polydispersity index, incubation plasma concentration, incubation NP concentration, incubation time, incubation temperature, centrifugation speed, centrifugation time, centrifugation temperature, and centrifugation repetitions) were extracted, as listed in Fig. 1. These 21 factors covered the main issues related to protein corona formation on NPs (9, 14, 17, 29). The distribution of the mined multidimensional data in the extracted factors and literature is presented in Fig. 1 and by Krona charts in the SI Appendix, respectively. No particular categories or data from any individual paper supported the dataset with the plentiful hierarchical relationships describing NP characteristics. Further details are provided in the SI Appendix. The 40 types of NPs without modification shown in SI Appendix, Fig. S1 included carbonaceous (e.g., multiwalled carbon nanotubes and single-walled carbon nanotubes), metallic (e.g., Ag, Au, and Fe3O4), nonmetallic (e.g., SiO2 and Si), and liposomal (e.g., cholesterol-phosphatidylcholine and thiolated amino-poly[ethylene glycol]; 3 kDa) NPs. The 50 types of surface modifications listed in SI Appendix, Table S1 included anionic (e.g., N-acetyl-l-cysteine and thiolated l-asparagine), cationic (e.g., 11-amino-1-undecanethiol and hexadecyltrimethylammonium bromide), neutral (e.g., carboxymethyl-poly[ethylene glycol]-thiol [5 kDa] and bicyclononyne), common (e.g., carboxyl [COOH] and citrate [CIT]), and rare (e.g., Pluronic F-127 and 16-mercaptohexadecanoic acid) surface ligands. The overall modifications are listed in SI Appendix, Table S1. The enrichment of surface modifications allowed the machine learning model to learn a large number of protein–NP interfaces. The large range of data for quantitative factors (e.g., 30.1 to 115.9 nm for sizeTEM and 100.0 to 1,000.0 mg/L for NP concentration, as listed in Fig. 1) present heterogeneous and complex conditions for protein corona prediction.
Fig. 1.
Overview of the qualitative and quantitative factors (652 data pieces). The superscript “a” represents that the other types of qualitative factors are shown in the SI Appendix. The superscript “b” represents that the factors are measured in the same culture with the same NP concentration The overall dataset is listed in the SI Appendix. The first to third quartiles were used to describe the data distribution. DMEM, Dulbecco’s modified Eagle medium; HP, human plasma; PDI, polydispersity index; PEG, polyethylene glycol; PS, polystyrene.
The limited amount of data and high heterogeneity were the major factors limiting the prediction accuracy of traditional statistical approaches and machine learning models (15, 28). As observed in SI Appendix, Figs. S1 and S2, a portion of the categories of qualitative factors contained limited data (e.g., calcium phosphate, multiwalled carbon nanotubes, and 6-amino-1-hexanethiol), making it difficult to obtain high prediction accuracy from the models regarding these categories. The very large response (73 corona components and 178 selected independent proteins) presented another challenge for obtaining high prediction accuracy from models of complex NP–protein interactions in complex biological environments. The traditional linear regression model cannot easily reveal the complex relationships between multiple (qualitative or quantitative) factors and protein corona compositions. As illustrated in SI Appendix, Fig. S3, the value of the correlation coefficient (R2 < 0.4) indicated poor prediction accuracy and an ambiguous relationship between quantitative factors and protein corona composition, using a linear regression model. To reduce the prediction errors from protein corona composition classifications, two methods were used: the corona compositions were classified by different physicochemical and functional properties (e.g., theoretical isoelectric point [pI], length, molecular weight, grand average of hydropathicity [GRAVY] score and function), and the protein compositions were measured by RPA in the subsequent analysis.
Prediction of the Protein Corona Composition.
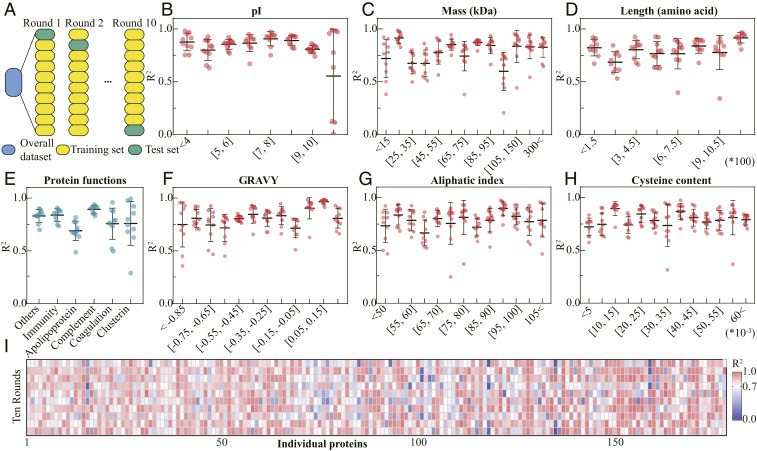
Given the above complex data, a robust RF model with high heterogeneity tolerance was used to explore the physicochemical properties and biological functions of the proteins in the complex protein corona. The identified proteins on NPs were classified by GRAVY score, length, mass, and pI. As data-driven models, machine learning models (e.g., RF) explain observations by learning previous experience. With robust learning capability, machine learning is likely to be overfit with a limited set of training data (15). To avoid overfitting and evaluate prediction accuracy credibly, the model performance was estimated by 10-fold cross-validation. For 10-fold cross-validation analysis, the original dataset was randomly partitioned into 10 folds. Nine folds were used to train the model as the training set, and the remaining one fold evaluated model as the test set. The average R2 and root-mean-square error (RMSE) were applied to measure model performance. Enhancing model complexity may increase accuracy but decrease the generalization ability for variable conditions, and vice versa (30). To balance the model prediction accuracy and the generalization capability of the models, factor selection was applied in the present work. Before selecting factors, the composition prediction models were built using original datasets with 21 overall factors. According to the variable importance (shown in SI Appendix, Fig. S4) and relationships among factors, 10 important and independent factors were selected: NP without modification, surface modification, incubation plasma source sizeTEM, zeta potential, incubation plasma concentration, incubation NP concentration, centrifugation speed, centrifugation time, and centrifugation temperature. In Fig. 2 B–D and F–H, the significantly high R2 (>0.85; P < 0.05) and low RMSE (<8%; shown in SI Appendix, Fig. S5) values of models in different pI value ranges are shown. The results supported that RF learned statistically tighter relationships between corona components classified by the pI value and 10 factors than between corona components classified by other properties (e.g., GRAVY score and mass). According to the high accuracies and tight relationships for the corona components classified by pI, electrostatic interaction could be the most important force determining protein corona formation in NPs (17). Fig. 2 B–I and SI Appendix, Fig. S5 show the good model performance (high R2, mostly >0.75, and low RMSE, mostly below 5%) on the prediction of protein corona composition, even on hundreds of individuals (R2 > 0.7; RMSE < 1). Unlike other models (e.g., linear regression, classification tree, and neural network model) (15), the RF model was suitable for learning the limited and heterogeneous data on the NP–protein coronas and biological responses, and did not exhibit overfitting.
Fig. 2.
Model performance of RF on protein corona compositions classified by physicochemical and functional properties and selected individual proteins. (A) Tenfold cross-validation was utilized to estimate the model prediction accuracy. (B–H) Each point or (I) each box represents one model built from a nine-tenth dataset, estimated by the correlation coefficient (R2). The model performance evaluated by the rootRMSE of physicochemical and functional properties and individual proteins is shown in the SI Appendix. The specific 178 proteins are listed in the SI Appendix.
Inspired by the specific protein adsorption on NPs (27), the present work further predicted the corona patterns of proteins with different biological functions on diverse NPs. The protein corona was divided into apolipoproteins, clusterin, coagulation proteins, complement proteins, immune proteins, and other proteins by biological functions and molecular composition in the UniProt database. As shown in Fig. 2 and SI Appendix, Fig. S5, the analysis obtained high accuracies in predicting the RPA values of various protein corona compositions, mostly with R2 values over 0.7 and RMSE values below 5%. The models with good performance overcame the great heterogeneity in the dataset and offered the possibility of screening the most important factors determining the compositions of the protein corona.
Screening for priority factors determining corona composition will provide deep insight into the formation mechanisms of protein coronas (4, 26). To evaluate the importance of the factors, two methods were provided by RF to measure factor importance: the percentage of increase in mean square error (MSE) and the mean decrease in node impurity, as shown in SI Appendix, Fig. S6. NP without modification and surface modification were identified as the most important factors dominating the formation of the protein corona. The above results supported the hypothesis that the use of surface modifications and NP without modification are the two main strategies for designing and synthesizing nanocarriers with accurate targeted delivery of cancer therapeutics (26). The surface modifications determined the surface interacting directly with proteins. For NPs without modification, the surfaces of bare NPs adsorbed proteins directly by physicochemical interactions [e.g., electrostatic attraction and hydrophilicity (27, 31)]. However, the specific interactions between surface modifications or NPs without modification and proteins remain unclear. Given the accurate and tight factor-response relationship hidden in the RF models with good performance to comprehensively analyze and compare the influence of two priority factors on corona formation, the well-performed functional composition prediction models were utilized to extract the factor-response dependence on priority factors by the similarity network.
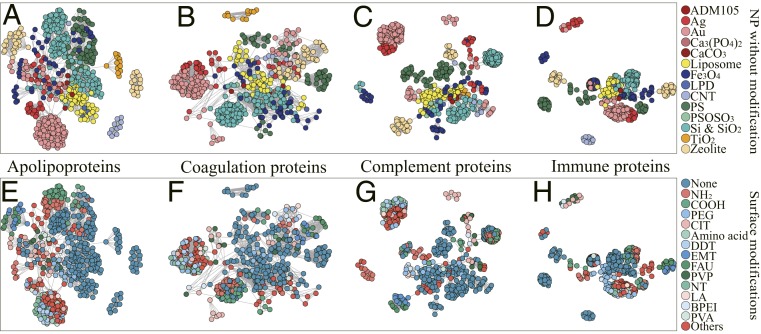
As shown in Fig. 3, the similarity network visualized the heterogeneity distribution of priority factors according to the proximity matrixes from different functional composition models. The proximity matrix quantified the factor-response dependence similarity according to the frequency of two data pieces appearing in the same node of a tree in an RF model. In the similarity network, connected nodes represented the data pieces containing homogeneity of physicochemical properties learned by RF models. Given the factor-response relationships hidden in RF models, the high homogeneity of tight-knit clusters indicated similar factor-response relationships. In addition, the data pieces with tight connections shared similar corona formation patterns. In Fig. 3 and SI Appendix, Figs. S7 and S8, significantly tight-knit clusters with an abundance of nodes associated with five functional proteins were present. With increasing prediction accuracy, the clusters in the model became more tightly grouped, according to the high R2 between the network density and model performance shown in SI Appendix, Fig. S8. The consistent tendency between model performance and clustering density indicated that the models could explore the tighter protein binding patterns of datasets with high homogeneity (e.g., the complement and immune proteins in Fig. 3). Hence, the present work applied the heterogeneity distribution to measure the prediction accuracy distribution of data pieces from models using the similarity network. In various NPs without modification (Fig. 3 A–D), there were clearly tight connections in the clusters of Au, Si, and SiO2 NPs and liposomes. The extensive homogeneity sharing in clusters with individual NPs indicated that the NP without modification played a crucial role in the factor-composition relationships associated with protein functions. However, for the liposomes in Fig. 3, the nodes representing various liposomes that were tightly grouped together indicated that the various liposomes shared the same factor-response dependence or protein binding pattern. Tight-knit connections were also observed for the same modification (e.g., Na88[AlO2]88[SiO2]104 and poly[vinylpyrrolidone] in Fig. 3 E–H). In addition, less distinct boundaries existed between the various surface modifications than between the results of NPs without modification. Therefore, additional methods describing surface modifications [e.g., charge and log P (4, 26)] are necessary to achieve a deeper understanding of the interactions between surface modifications and proteins (27). Given the heterogeneity distribution of priority factors in Fig. 3, the similarity network offers a way to explore the driving force determining the factor–response relationships hidden in RF models and provides a perspective for evaluation of the model performance of priority factors by measuring the heterogeneity contribution from the priority factors. The strong driving forces at the NP–protein interface (5, 31) can be applied to design nanocarriers that adsorb certain proteins to increase the targeting accuracy and mediate biological recognition, as was further confirmed by the following experiments.
Fig. 3.
The similarity network visualizes heterogeneity of functional corona composition in models. Each node represents a data piece in the prediction models of functional corona compositions. The nodes are colored according to the priority factors, NP without modification (A–D) and surface modification (E–H). For well-performed RF models, the values of connected nodes are more than four times higher than the average in each proximity matrix. Tighter connections in each cluster indicate the higher homogeneity of nodes for the factor-response dependence learned by RF models. In contrast, the sparse connections represent the heterogeneity of nodes in terms of the NP properties and experimental conditions in the cluster. The similarity network of functional composition models for protein clusters is shown in SI Appendix, Fig. S7. The full names of abbreviations are listed in SI Appendix, Table S1.
Prediction of Protein Coronas and Experimental Verification.
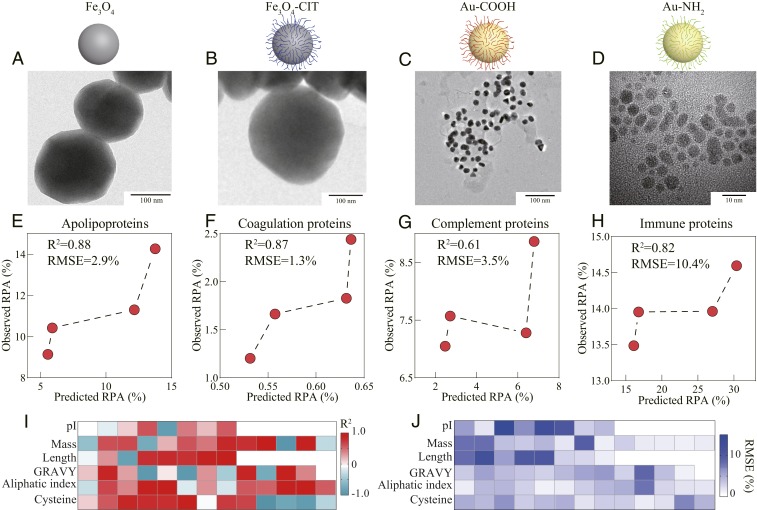
As data-driven models, machine learning models (e.g., RF) could explain observations using available training data with remarkable accuracy (30, 32). However, the factor-response dependence hidden in models requires new experimental data for verification to enable highly accurate generalization regarding corona formation behavior (especially for distinct combinations of NPs without modification and surface modifications). To assess the model quality, corona composition identification was performed by another independent researcher (double-blind test). In the model datasets, 40 types of NPs without surface modifications and 50 types of surface modifications covered the common NPs, and 10-fold cross-validation was utilized to estimate the model prediction accuracy. Given the wide application of and significant cell responses to Au NPs and Fe3O4 NPs in therapy and imaging (17, 33, 34), the protein coronas of Au- and Fe3O4-based NPs were analyzed in the laboratory to further verify the performance of the model. To verify the prediction capacity of the model for NPs, the protein coronas of Fe3O4 and Fe3O4-CIT NPs in the model datasets and Au-NH2 and Au-COOH not in the model datasets were detected. The NPs that were not included in the training set made the performance of the machine learning model challenging and valuable. Moreover, two other frequently used NPs (Ag NPs and TiO2 NPs) were added to verify the prediction of cellular recognition. The characterizations of NPs are presented in Fig. 4A and SI Appendix, Fig. S9. The protein coronas were identified using liquid chromatography tandem mass spectrometry, as shown in Fig. 4 A–F (more details are shown in the SI Appendix). The high R2 confirmed the consistency between observations and predictions regarding the four functional protein compositions on four distinct NPs, especially for apolipoprotein, coagulation, and immune proteins, where the values of R2 were more than 0.8. The high R2 (majority over 0.6) and small RMSEs (majority below 5%) also indicate that the model can predict the physicochemical compositions of protein coronas on various NPs. The present method can predict and evaluate protein corona formation to guide the design of nanocarriers before NPs enter complex biological environments.
Fig. 4.
Experiments evaluating the functional and physicochemical composition predictions of protein coronas. (A–D) TEM micrograph representing the morphology and size of Fe3O4, Fe3O4-CIT, Au-COOH, and Au-NH2 NPs. The observed functional components of the protein coronas on the four NPs were measured by liquid chromatography tandem mass spectrometry. The model prediction accuracy was measured by the correlation coefficient (R2) and between observations and predictions of the functional (E, apolipoproteins; F, coagulation proteins; G, complement proteins; and H, immune proteins) and physicochemical (I and J) compositions of selected NPs. The heatmap visualizes the model performance for compositions classified by physicochemical properties, as measured by R2 (I) and RMSE (J). The model evaluations of clusterin and other proteins are shown in SI Appendix, Fig. S10.
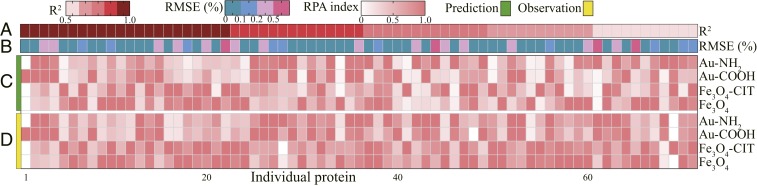
It is worth noting that the corona formation mechanism operates on individual proteins, and proteins with limited RPA values (e.g., clusterin and IgM) play important roles in the biological recognition of NPs (26, 27). According to the good model performance on the functional and physicochemical components of the protein corona, the established model further predicted the individual protein components in the corona. Fig. 5 represents the model performance for individual protein components measured by RMSE and R2 between the observations and predictions. As shown in Fig. 5, the good prediction accuracy was illustrated by the high R2 (>0.5 for 71 overall proteins and >0.7 for over half the proteins) and low RMSE (majority <0.2%). The consistent adsorption tendency of protein coronas on different NPs between the predictions and the observations demonstrated that the present models were powerful enough to learn individual protein binding patterns on NPs, although it was difficult to predict the absolute overall RPA value of individual proteins in the protein corona. The method provided a potential platform for designing targeted nanocarriers and regulating biological responses (1, 5) by predicting and designing corona fingerprints in a complex biological environment before administration.
Fig. 5.
Individual protein composition predictions evaluated by experiments. Correlation coefficients (R2 in A) and RMSE (B) of the predictions (C) and observations (D) evaluating the model performance regarding 71 individuals adsorbed on selected NPs (Fe3O4, Fe3O4-CIT, Au-COOH, and Au-NH2). The RPA values were normalized to the maximum value in each prediction or observation as the RPA index. The model performance and details for proteins are shown in SI Appendix.
Predicting Cellular Recognition of Protein Coronas on NPs.
Fig. 4 verified that the machine learning models could accurately predict the critical functional compositions of the corona and the protein binding patterns of various NPs. The epitopes (rather than merely single protein composition and amount) in the protein corona representing biomolecular recognition motifs played a critical role in cellular recognition, complement activation, macrophage phagocytosis, and immune response by interacting with various receptors (3, 7, 35–37). Models that achieve robust prediction of the functional compositions with epitopes of the protein corona will provide insights into predicting the cellular recognition of NPs by associating the functional composition of the corona with cellular recognition. Predicting the cellular recognition of NPs is useful for guiding the design of ideal nanocarriers (1).
Increasing attention has been paid to the unwanted cellular uptake and inflammatory and immune responses of NPs shielded by protein coronas in biological systems, as these responses are important causes of shortened circulation life and limited nanocarrier targeting efficiency (5). The coronas on NPs labeled with endogenous proteins could be mistaken for exogenous matter (e.g., viruses or lipoproteins) by presenting functional epitopes allowing specific receptor recognition (38). The recognition indexes were applied to measure the relationships of functional corona compositions with immune system recognition (37) by analyzing uptake efficiency, proinflammatory effects, and immune perturbation due to the presence of the corona. The relationships of the recognition index with the predicted functional composition of the protein corona were measured by R2, as shown in SI Appendix, Figs. S11 and S12. In SI Appendix, Fig. S11, tight correlations were present in corona compositions with specific cellular recognition indexes and high R2 values (e.g., R2 = 0.92 between apolipoprotein and TNF-α releases and R2 = −0.80 between complement proteins and cellular uptake). The results also suggested the presence of functional compositions of the corona closely associated with the cellular recognition of different NP surfaces. For example, apolipoprotein and complement proteins are critical innate immunity proteins (e.g., apolipoprotein A-1 and complement factor H) that contribute to various immunity pathways (26, 27). There were obvious connections between functional proteins and recognition indexes in SI Appendix, Fig. S11. The dysopsonins (e.g., clusterin and apolipoproteins) or opsonins (e.g., immune proteins and complement protein) allowed similar patterns in functional motifs interacting with recognition receptors (measured by recognition indexes). The epitopes of the tested functional proteins enabled specific receptor recognition on macrophages (e.g., immune proteins in the corona activated the NF-κB pathway) (16, 26, 38). To verify the model practicality for different cell types, three cell lines (RAW264.7, human leukemic cell line [THP-1], and dendritic cell line [DC2.4]) were investigated in SI Appendix, Figs. S11 and S12. The prediction accuracy for most of important functional compositions was high with R2 > 0.8. The high prediction accuracy was probably because machine learning is a data-driven model and there are tight and steady relationships between cellular recognition and functional corona compositions. According to the prediction accuracy shown in Fig. 2 and SI Appendix, Figs. S11 and S12, the models enabled the prediction of the cell recognition of NPs in both fetal bovine serum (FBS) and human serum (HS). Unlike experiments alone (25, 27), a combination of integrated machine learning and experimentation supported the notion that corona functional compositions play a dominant role in mediating biomolecular recognition of NP–corona complexes (3, 39). Predicting biomolecular recognition provides a method for designing nanocarriers (especially actively targeted nanocarriers) and avoiding unintended biological outcomes (e.g., cellular uptake and immune responses) during clinical applications (1, 5).
The complex relationships among protein corona formation and numerous NP properties or cellular recognition in biological environments challenged the model prediction ability. With heterogeneous data in hand, the RF model was applied to explore unknown and complex relationships hidden in various quantitative and qualitative factors (21 factors overall). With robust learning capability, RF performed well on corona functional composition prediction on various NPs (e.g., most R2 > 0.75). Similarity analysis was also applied to analyze the heterogeneity distribution in RF, enabling the corona formation patterns to be extracted from the data-driven model. Because of the complexity in biological environments, machine learning with robust prediction capability would promote the applications of NPs in human healthcare.
Conclusion
The prediction of protein corona functional compositions is critical for the design of ideal NPs for clinical applications, but methods to quantitatively predict protein coronas have been unavailable to date (1, 5). The high-dimensional relationships (involving at least eight qualitative factors and 13 quantitative factors) between the protein corona and NP properties present challenges to traditional models and experimental methods (18, 19). By collecting knowledge from previous efforts, the present work assembled evidence to investigate protein corona formation behaviors, overcoming the limitations and uncertainty in distinct studies. Here, machine learning (i.e., RF) was used to learn the complex relationships between NP properties and corona composition and then to comprehensively and quantitatively predict the formation of protein coronas and the related cell responses. The most important factors (NP without modification and surface modification) determining corona formation were identified by the RF model. The similarity network was applied to visualize the heterogeneity distribution of the priority factors, illustrating that the same NPs shared unique protein binding patterns according to the factor-response dependence extracted from high-performing models. Experiments verified the functional and physicochemical compositions of predicted protein coronas. Moreover, the present work associated cellular recognition with the diverse functional compositions of the corona, as predicted by machine learning models. The predicted functional compositions of protein coronas were tightly correlated with cellular recognition. NPs with “stealth” properties induced unwanted immune responses and resulted in anaphylaxis after being coated with a complex protein corona (1). Quantitative prediction of the recognition mediation of protein coronas provides a method for designing nanocarriers (especially actively targeted nanocarriers) and avoiding unintended biological outcomes (e.g., cellular uptake and immune responses) during clinical applications.
Materials and Methods
Data Extraction.
The present work screened 56 papers (cited in the SI Appendix), and the details of the screening methods are provided in the SI Appendix. After reviewing the titles, abstracts, and full text, the present work mined the literature and extracted the data representing important factors describing the formation of the protein corona on NPs. Ten significant and independent factors (i.e., NP without modifications, modification, sizeTEM, zeta potential, protein source, plasma concentration, NP concentration, centrifugation speed, centrifugation time, and centrifugation temperature) were identified for further analyses. The details are provided in the SI Appendix. The RPAs of proteins were classified by physicochemical descriptions (pI, mass, length, GRAVY, aliphatic index, and cysteine content) of proteins. According to the biological functions of proteins identified from the UniProt database, the functional components were classified as immune proteins, apolipoproteins, complement proteins, coagulation proteins, clusterin, and other proteins. The RPAs of the 178 independent proteins extracted (with >100 RPA data pieces) were also selected to describe the compositions of the protein corona (the details are provided in SI Appendix). Finally, 567 and 652 data points were extracted for protein corona composition models and individual protein models, respectively.
RF Regression and Validation.
As a data-driven model, RF builds trees using a bootstrap sample from the overall data, and the best partitions in a subset of factors were selected randomly for each node of the trees. The predictions were performed by the RF algorithm aggregating the results of each tree, and the majority vote for classification analysis and the average for regression analysis were conducted. To quantify the relative importance of different factors, the increase in MSE and the mean decrease in the node impurity of each RF model were calculated by the R package randomForest. Because the two parameters (ntree and mtry) of RF cannot determine the predictive accuracy or model performance, the default values for the two parameters were set. To measure the performance and the predictive accuracy of the RF model, the R2 and RMSE values between the predictions and observations were calculated. RF used ∼63% of the raw data to construct the trees and validated the model performance with the remaining out-of-bag data in each RF bootstrap sample (40). Because of the out-of-bag validation, RF was robustly tolerant to overfitting (23). Moreover, 10-fold cross-validation was applied to avoid overfitting.
Visualization of the Heterogeneity Distribution of Priority Factors in Models.
To estimate the roles of key factors in the formation of the protein corona, a similarity network was applied to visualize the heterogeneity distribution of priority factors in functional composition models of the protein corona. The similarity network was drawn from the proximity matrix of models using the Kamada-Kawai layout algorithm by the R package igraph. Each node represented a data piece of the functional composition models. The nodes were colored according to the priority factors. From well-performed RF models, the values of connected nodes were more than four times higher than the average in each proximity matrix. The connecting nodes shared a similarity in the RF models. The clustering density was utilized to measure the tightness and heterogeneity of the network.
Characterization of NPs for Model Verification.
NH2- and COOH-coated Au NPs (G820971 and G820972) were obtained from Macklin Company, China. Fe3O4 NPs (MB9863) and CIT-modified Fe3O4 NPs (MB9866) were obtained from Meilunbio Company, China. Ag NPs (XFJ14) and TiO2 NPs (XFI02) were obtained from Nanjing XFNANO Materials Tech Co., Ltd., China. The morphology of the nanomaterials was examined using high-resolution TEM (JEM-2800, JEOL, Japan). The hydrodynamic size and zeta potential of the nanomaterials in water and phosphate-buffered saline (PBS) were measured using a ZetaSizer Nano-ZS instrument (Malvern Instruments, Worcestershire, UK).
Model Verification and Prediction of Unknown Protein Coronas.
Normal human plasma was obtained from Jiaozuo LFFBio Tech Co., Ltd., China, and centrifuged at 1,408 × g, and the supernatant was collected for interaction with NPs. Then, Fe3O4 and Fe3O4-CIT NPs (2.5 mg/mL, 850 µL) dispersed in PBS were incubated with human plasma (3,400 µL) in a shaker at 150 rpm and 37 °C for 1 h. Au-COOH and Au-NH2 NPs (2.5 mg/mL, 850 µL) were incubated with 3,400 µL of human plasma at 37 °C for 1 h. After incubation, the NP–plasma protein complexes were separated through centrifugation (21,913 × g, 4 °C) for 15 min, and the pellets were intensively washed with PBS and collected for further analysis. The protein was identified by mass spectrometry, and the details are provided in SI Appendix.
Cellular Uptake and Cytokine Analysis.
The murine macrophage cell line RAW264.7 was obtained from the Shanghai Cell Bank of the Type Culture Collection of China. Cells were grown in Dulbecco's modified Eagle medium (high glucose, Ding Guo, China) supplemented with 10% FBS (AusGeneX, Australia) and a final concentration of 100 units/mL penicillin/streptomycin in a humidified incubator with 5% CO2 at 37 °C. Macrophages were seeded on 24-well plates at a density of 5 × 104 cells/well for 12 h and then incubated in fresh serum-free medium for 2 h. The NPs (Fe3O4, Fe3O4-CIT, Ag, TiO2, Au-NH2, and Au-COOH NPs) at 50 mg/L with or without protein coronas were incubated with the macrophages in serum-free medium. After 4 h of exposure, the cells were washed twice with PBS, lysed in cell lysis buffer (Beyotime Biotechnology, China), and then centrifuged at 11,180 × g for 5 min. The supernatant (20 μL) was used to determine the protein concentration by using a BCA Kit (Beyotime Biotechnology, China), and the rest was digested using HNO3 until no color was observed. After the digestive solution was filtered through a 0.22-µm micropore membrane, the concentrations of metal elements were measured by inductively coupled plasma mass spectrometry (Elan drc-e, PerkinElmer). The intracellular ion content was normalized to the total protein content and is represented as a percentage with respect to the control group. To analyze the effects of cell types and culture medium on models, another two cell types (dendritic cell line [DC2.4] and human leukemic cell line [THP-1]) in both FBS and HS were tested. The cytokines (tumor necrosis factor-α and interleukin 6) were measured using ELISA kits (Dakewe, Shenzhen, China), and the details are provided in the SI Appendix.
Data Availability.
Code in the paper is available at https://github.com/BanZhan/RF-and-PC.
Supplementary Material
Acknowledgments
The authors thank Zhixiao Liu, Caijiao He, and Na Wong from Nankai University for literature collection and data extraction, and Ying Zhang and Han Zhang from Nankai University for suggestions. This work was financially supported by the National Natural Science Foundation of China (grant no. 21722703).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: Code in the paper is available at GitHub (https://github.com/BanZhan/RF-and-PC).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1919755117/-/DCSupplemental.
References
- 1.Lundqvist M., et al. , Nanoparticle size and surface properties determine the protein corona with possible implications for biological impacts. Proc. Natl. Acad. Sci. U.S.A. 105, 14265–14270 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schöttler S., et al. , Protein adsorption is required for stealth effect of poly(ethylene glycol)- and poly(phosphoester)-coated nanocarriers. Nat. Nanotechnol. 11, 372–377 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Castagnola V., et al. , Biological recognition of graphene nanoflakes. Nat. Commun. 9, 1577 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pilkington E. H., et al. , Profiling the serum protein corona of fibrillar human islet amyloid polypeptide. ACS Nano 12, 6066–6078 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y., Cai R., Chen C., The Nano–bio interactions of nanomedicines: Understanding the biochemical driving forces and redox reactions. Acc. Chem. Res. 52, 1507–1518 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Oh J. Y., et al. , Cloaking nanoparticles with protein corona shield for targeted drug delivery. Nat. Commun. 9, 4548 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kelly P. M., et al. , Mapping protein binding sites on the biomolecular corona of nanoparticles. Nat. Nanotechnol. 10, 472–479 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Zhou Q., Yue Z., Li Q., Zhou R., Liu L., Exposure to PbSe nanoparticles and male reproductive damage in a rat model. Environ. Sci. Technol. 53, 13408–13416 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Vilanova O., et al. , Understanding the kinetics of protein-nanoparticle corona formation. ACS Nano 10, 10842–10850 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chandran P., Riviere J. E., Monteiro-Riviere N. A., Surface chemistry of gold nanoparticles determines the biocorona composition impacting cellular uptake, toxicity and gene expression profiles in human endothelial cells. Nanotoxicology 11, 507–519 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Monopoli M. P., et al. , Physical-chemical aspects of protein corona: Relevance to in vitro and in vivo biological impacts of nanoparticles. J. Am. Chem. Soc. 133, 2525–2534 (2011). [DOI] [PubMed] [Google Scholar]
- 12.Tenzer S., et al. , Nanoparticle size is a critical physicochemical determinant of the human blood plasma corona: A comprehensive quantitative proteomic analysis. ACS Nano 5, 7155–7167 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Pozzi D., et al. , Surface chemistry and serum type both determine the nanoparticle-protein corona. J. Proteomics 119, 209–217 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Talamini L., et al. , Influence of size and shape on the anatomical distribution of endotoxin-free gold nanoparticles. ACS Nano 11, 5519–5529 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Ban Z., Zhou Q., Sun A., Mu L., Hu X., Screening priority factors determining and predicting the reproductive toxicity of various nanoparticles. Environ. Sci. Technol. 52, 9666–9676 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Qiu B., et al. , Fabrication of nickel–cobalt bimetal phosphide nanocages for enhanced oxygen evolution catalysis. Adv. Funct. Mater. 28, 1706008 (2018). [Google Scholar]
- 17.Walkey C. D., et al. , Protein corona fingerprinting predicts the cellular interaction of gold and silver nanoparticles. ACS Nano 8, 2439–2455 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Gajewicz A., How to judge whether QSAR/read-across predictions can be trusted: A novel approach for establishing a model’s applicability domain. Environ. Sci. Nano 5, 408–421 (2018). [Google Scholar]
- 19.Wang D., et al. , A QSAR-based mechanistic study on the combined toxicity of antibiotics and quorum sensing inhibitors against Escherichia coli. J. Hazard. Mater. 341, 438–447 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Breiman L., Random forest. Mach. Learn. 45, 5–32 (2001). [Google Scholar]
- 21.Jordan M. I., Mitchell T. M., Machine learning: Trends, perspectives, and prospects. Science 349, 255–260 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Ahneman D. T., Estrada J. G., Lin S., Dreher S. D., Doyle A. G., Predicting reaction performance in C-N cross-coupling using machine learning. Science 360, 186–190 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Oh E., et al. , Meta-analysis of cellular toxicity for cadmium-containing quantum dots. Nat. Nanotechnol. 11, 479–486 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Findlay M. R., Freitas D. N., Mobed-Miremadi M., Wheeler K. E., Machine learning provides predictive analysis into silver nanoparticle protein corona formation from physicochemical properties. Environ. Sci. Nano 5, 64–71 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Haute D., Liu A. T., Berlin J. M., Coating metal nanoparticle surfaces with small organic molecules can reduce nonspecific cell uptake. ACS Nano 12, 117–127 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guan J., et al. , Enhanced immunocompatibility of ligand-targeted liposomes by attenuating natural IgM absorption. Nat. Commun. 9, 2982 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simon J., et al. , Hydrophilicity regulates the stealth properties of polyphosphoester-coated nanocarriers. Angew. Chem. Int. Ed. Engl. 57, 5548–5553 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Labouta H. I., Asgarian N., Rinker K., Cramb D. T., Meta-analysis of nanoparticle cytotoxicity via data-mining the literature. ACS Nano 13, 1583–1594 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Pisani C., et al. , The species origin of the serum in the culture medium influences the in vitro toxicity of silica nanoparticles to HepG2 cells. PLoS One 12, e0182906 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Butler K. T., Davies D. W., Cartwright H., Isayev O., Walsh A., Machine learning for molecular and materials science. Nature 559, 547–555 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Sakulkhu U., et al. , Significance of surface charge and shell material of superparamagnetic iron oxide nanoparticle (SPION) based core/shell nanoparticles on the composition of the protein corona. Biomater. Sci. 3, 265–278 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Ghosal S., et al. , An explainable deep machine vision framework for plant stress phenotyping. Proc. Natl. Acad. Sci. U.S.A. 115, 4613–4618 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee J., Morita M., Takemura K., Park E. Y., A multi-functional gold/iron-oxide nanoparticle-CNT hybrid nanomaterial as virus DNA sensing platform. Biosens. Bioelectron. 102, 425–431 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Hu Y., Mignani S., Majoral J. P., Shen M., Shi X., Construction of iron oxide nanoparticle-based hybrid platforms for tumor imaging and therapy. Chem. Soc. Rev. 47, 1874–1900 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Andersen A. J., et al. , Single-walled carbon nanotube surface control of complement recognition and activation. ACS Nano 7, 1108–1119 (2013). [DOI] [PubMed] [Google Scholar]
- 36.Boraschi D., et al. , Nanoparticles and innate immunity: New perspectives on host defence. Semin. Immunol. 34, 33–51 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Mo J., Xie Q., Wei W., Zhao J., Revealing the immune perturbation of black phosphorus nanomaterials to macrophages by understanding the protein corona. Nat. Commun. 9, 2480 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lara S., et al. , Identification of receptor binding to the biomolecular corona of nanoparticles. ACS Nano 11, 1884–1893 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Corbo C., et al. , Unveiling the in vivo protein corona of circulating leukocyte-like carriers. ACS Nano 11, 3262–3273 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Bylander T., Estimating generalization error on two-class datasets using out-of-bag estimates. Mach. Learn. 48, 287–297 (2002). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Code in the paper is available at https://github.com/BanZhan/RF-and-PC.