Significance
LPL is an important enzyme for the maintenance of triglyceride levels in the blood. Hypertriglyceridemia is a risk factor for cardiovascular disease (CVD), which is the leading cause of death worldwide. LPL production, intracellular trafficking, and activity in the capillaries are all highly regulated. Here, we uncover the existence of a helical LPL polymer, which is an inactive state of LPL. This inactive state may represent a previously hypothesized form used to store LPL prior to release into the interstitial space. Our insight into the diversity of oligomeric states LPL can adopt may be key to understanding regulation of LPL trafficking.
Keywords: lipoprotein lipase, cryo-electron microscopy, helical reconstruction
Abstract
Lipases are enzymes necessary for the proper distribution and utilization of lipids in the human body. Lipoprotein lipase (LPL) is active in capillaries, where it plays a crucial role in preventing dyslipidemia by hydrolyzing triglycerides from packaged lipoproteins. Thirty years ago, the existence of a condensed and inactive LPL oligomer was proposed. Although recent work has shed light on the structure of the LPL monomer, the inactive oligomer remained opaque. Here we present a cryo-EM reconstruction of a helical LPL oligomer at 3.8-Å resolution. Helix formation is concentration-dependent, and helices are composed of inactive dihedral LPL dimers. Heparin binding stabilizes LPL helices, and the presence of substrate triggers helix disassembly. Superresolution fluorescent microscopy of endogenous LPL revealed that LPL adopts a filament-like distribution in vesicles. Mutation of one of the helical LPL interaction interfaces causes loss of the filament-like distribution. Taken together, this suggests that LPL is condensed into its inactive helical form for storage in intracellular vesicles.
All living organisms must direct nutrients for either utilization or storage. These decisions are highly regulated, as ensuring nutrient availability is essential for survival. Triglycerides are an especially energy-dense form of nutrients, and lipoprotein lipase (LPL) plays a key role in the spatiotemporal regulation of triglyceride utilization. LPL is a secreted lipase that hydrolyzes triglycerides from circulating lipoproteins, making free fatty acids (FFAs) available to tissues for utilization or storage (1). Improper partitioning of FFAs to the peripheral tissues can have major metabolic consequences. For example, elevated plasma triglyceride levels are a risk factor for coronary artery disease, underscoring the importance of understanding LPL function and regulation (2–6). Each step of LPL function is regulated, from production to intracellular trafficking to activity in the capillary endothelium (7–9).
LPL is a secreted glycoprotein that contains five disulfide bonds and requires an endoplasmic reticulum (ER) protein, lipase maturation factor 1 (LMF1), to successfully fold and traffic out of the ER to the Golgi (10). From the Golgi, LPL is sorted into vesicles in which LPL can be stored, targeted to the lysosomes for degradation, or secreted into the interstitial space (11, 12). In adipose tissue, the pool of stored LPL is thought to be available for secretion in response to insulin signaling (11, 13, 14). Interestingly, stored LPL was previously reported to exist in a “cryptic,” inactive state within membrane-bound compartments of the cell (15, 16). Recent work has also shown that LPL secretion is mediated by Syndecan-1 (SDC1), a heparan sulfate proteoglycan (HSPG) that facilitates LPL packaging into sphingomyelin-rich vesicles (17). The sorting of SDC1 is mediated by a transmembrane (TM) domain, which partitions SDC1 into distinct lipid domains in the trans-Golgi (17). Approximately two thirds of the total LPL is sequestered and packaged into partitioning SDC1 vesicles, and the remaining third of the LPL within the cell is constitutively trafficked via bulk flow in undefined secretory vesicles (18). It is unclear how LPL is preferentially sorted into SDC1 vesicles versus bulk flow, although the interaction of LPL with SDC1 HSPGs likely plays a role.
Stored LPL can be secreted into the interstitial space, where it interacts with HSPGs that bind to the multiple heparin binding sites on each LPL molecule (18). LPL is next bound by glycosylphosphatidylinositol-anchored high-density lipoprotein-binding protein 1 (GPIHBP1) and transported into the capillary (19), where it acts on chylomicrons and very-low-density lipoproteins (VLDLs) to hydrolyze packaged triglycerides and release FFAs. In addition to regulation at the secretion stage, LPL is also regulated by macromolecular inhibitors after it has reached the capillary. These inhibitors are members of the angiopoietin-like (ANGPTL) family of proteins and inhibit LPL in different tissues based on nutritional state (8). This facilitates directing FFAs postprandially to the white adipose tissue (WAT) for storage and during fasting to the oxidative tissues, such as the heart and muscles, to provide energy (8).
Recent studies of LPL in complex with GPIHBP1 have revealed that LPL may be active in a 1:1 complex with one molecule of each protein (20–22). This contradicts previous reports that the active form of LPL is a dimer (23–26). In the previous model, active LPL formed dimers, which could exchange subunits with other dimers (23, 24). Additionally, there was a separate catalytically inactive monomer form of LPL that could not dimerize (23, 24). Modeling of dimeric LPL was initially based on the crystallographic dimer of monomeric horse pancreatic lipase (PL) (27, 28). Recent modeling utilized single-molecule fluorescence resonance energy transfer (smFRET) to measure the distance between site-specific labels on two subunits of LPL. This latest dimer model flipped the dimerization interface to the opposite side of the protein as had been initially predicted (29). Based on these recent developments, it is necessary to revisit the function of monomeric and dimeric LPL. Reports on the cryptic state of LPL within the cell suggest that LPL also adopts a third oligomeric state, specifically one that is condensed or polymerized (15, 16). This condensed state was theorized based on the finding that LPL activity in cell lysate was lower than expected. When cell lysate was diluted, LPL activity was found to increase with increasing dilution, reaching the expected activity for the amount of LPL present (15). LPL was thus hypothesized to form an inactive, concentration-dependent oligomer in the cell. In this work, we report a cryo-EM structure of an inactive helical oligomer of LPL at 3.8-Å resolution.
Results
Purified LPL Forms Concentration-Dependent Helices.
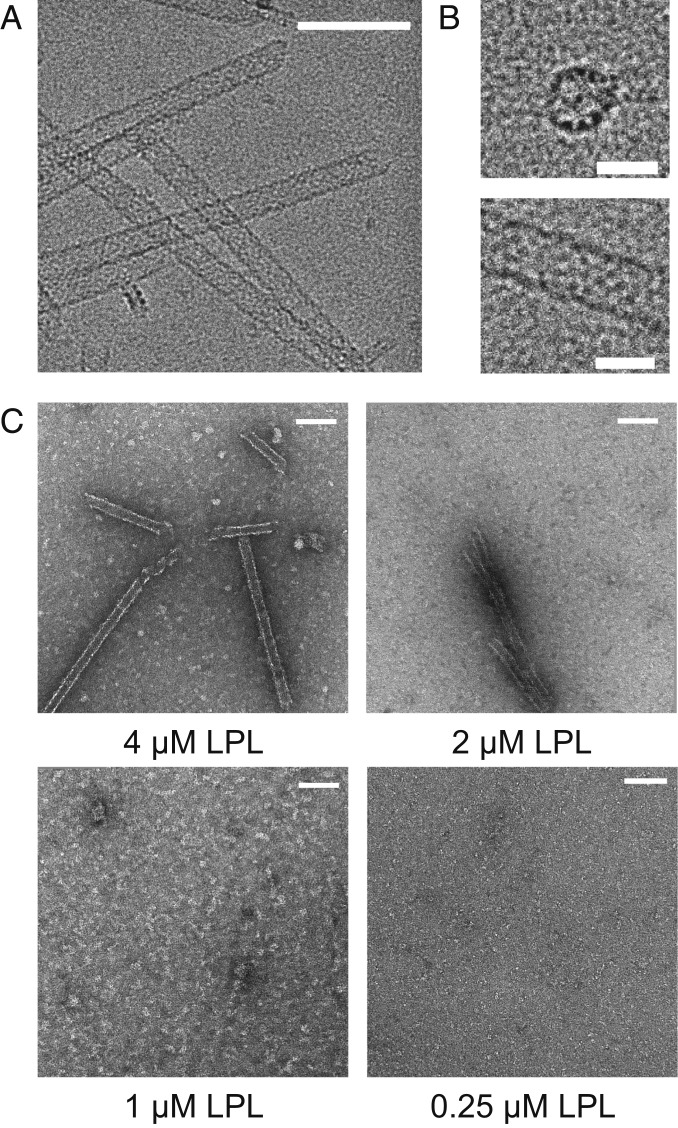
While preparing grids for cryo-EM, we unexpectedly discovered that bovine LPL formed helical structures under certain freezing conditions (Fig. 1 A and B). We confirmed the presence of helices using negative stain transmission electron microscopy (TEM) and performed a dilution series, which revealed that these helices were concentration-dependent and formed at concentrations above ∼2 µM (Fig. 1C). At the concentration used to create the cryo-grids (3.3 µM), LPL helices were common, but, as concentration was reduced, helices disappeared and single particles predominated. We utilized bovine LPL in these experiments to facilitate the purification of high concentrations of untagged, wild type LPL (SI Appendix, Fig. S1). It is difficult to achieve the high concentrations of LPL needed, in the absence of GPIHBP1, using human tissue culture (30). Following cleavage of the signal sequence, bovine LPL shares ∼94% sequence similarity with human LPL and displays similar activity to human LPL (31). To determine if the helices were dependent on the buffer used to dilute the sample, we tested a range of pHs and salt concentrations in the diluting buffer. We found that LPL helices were not affected by dilution in buffers ranging from pH 6.5 to 8.5 (SI Appendix, Fig. S2A). However, when LPL was diluted using buffers with salt concentrations below 100 mM NaCl (575 mM final NaCl concentration), we observed the formation of aggregates, with the aggregates becoming more prominent as the diluting buffer concentration dropped to 0 mM NaCl (SI Appendix, Fig. S2B). This may be a result of the buffer mismatch between the LPL storage buffer, which has 2 M NaCl, and the dilution buffer.
Fig. 1.
Purified LPL forms concentration-dependent helices. (A) Cryo-EM image of purified bovine LPL forming helices (3.3 μM LPL). (Scale bar, 100 nm.) (B) Zoomed view of LPL helix end-on (Upper) and side (Lower) views. (Scale bars, 25 nm.) (C) Representative negative stain EM images show that LPL helices form at high concentrations (4 μM and 2 μM), but do not form at lower concentrations (1 μM and 0.25 μM LPL). (Scale bars, 100 nm.)
Helical Reconstruction of LPL.
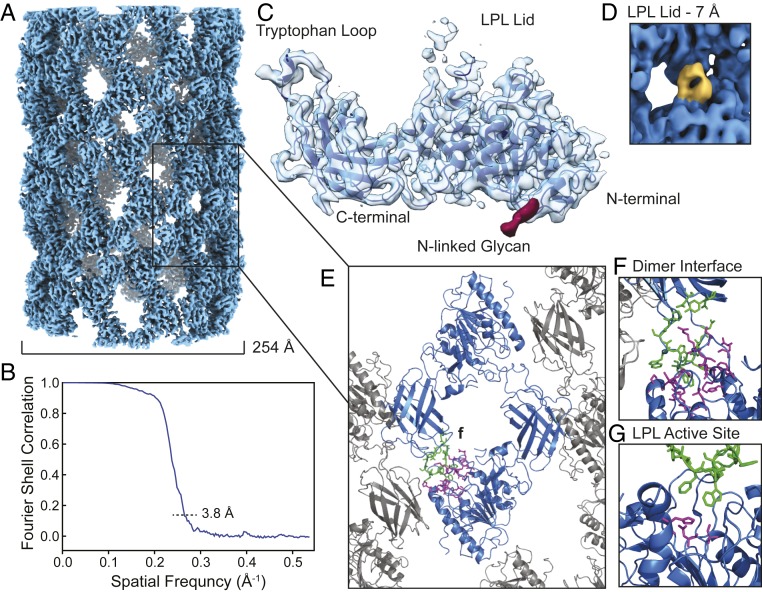
We collected a cryo-EM data set of the LPL helices and used an averaged power spectrum of nonoverlapping helical segments to empirically determine the initial helical parameters as C1 symmetry, rise = 10.7 Å, and rotation = 130.1° (yielding a pitch of 29.6 Å; SI Appendix, Fig. S3A) (32). These parameters were refined during helical reconstruction in Relion (33) to rise = 10.88 Å and rotation = 130.05° (yielding a pitch of 30.1 Å). During reconstruction, LPL helices were found to have dihedral symmetry, so, in later steps, D1 symmetry was applied to the reconstruction (Fig. 2A). Using the “gold standard” Fourier shell correlation (FSC) plot, the final resolution for the LPL helix reconstruction is 3.8 Å (Fig. 2B). We used I-TASSER to generate a homology model of a monomer of bovine LPL as the starting point for our atomic model (34–36). We refined this homology model using density for a single LPL subunit that was segmented out of the helical reconstruction (Fig. 2C). We generated the LPL dimer through dihedral symmetry, and then used the dimer to generate the helix structure through imposition of the helical symmetry parameters. The model was built in Coot and real-space refined in PHENIX (37–39). The map-to-model Fourier cross-resolution (FCR) confirmed a resolution of 3.8 Å (Table 1 and SI Appendix, Fig. S3 B and C) (40). We were able to identify all five of LPL’s disulfide bonds and one of the N-linked glycans in the structure. Residues within the lid region that covers LPL’s active site (N247–D264) were not interpretable when filtered to 3.8 Å (Fig. 2C). This is likely due to the mobility of the lid region, which gave rise to heterogeneity of the lid conformation within the helix. However, the cryo-EM density corresponding to the lid domain can be clearly seen when the reconstruction is filtered to 7-Å resolution (Fig. 2D).
Fig. 2.
Cryo-EM reconstruction of the LPL helix. (A) Surface of the helical reconstruction of LPL. (B) FSC plot showing 3.8-Å reconstruction resolution at the 0.143 cutoff. (C) Segmented density for a single LPL subunit (light blue) with the atomic model superimposed (dark blue) and the glycan at N73 highlighted (red). The broken density of LPL lid domain is seen in the top center of the segmented subunit density. (D) Surface of LPL helical reconstruction (blue) filtered to 7 Å to show the LPL lid density (yellow). (E) Zoomed model of LPL helix with a dihedral dimer shown in blue and the surrounding subunits in gray. LPL dimer interface residues are shown in green and magenta (SI Appendix, Table S1). (F) Zoomed view of the dimer interface of helical LPL in E. The density map of the dimer interface is shown in SI Appendix, Fig. S5A. (G) The LPL dimer interface viewed after a 180° rotation shows the LPL active site is buried in the dimer interface. The three active site residues are shown in magenta (S162, D186, H271), and tryptophan loop from the opposing subunit is shown in green (D415-W424).
Table 1.
Refinement statistics for the LPL helix model
| Data collection and processing | |
| Magnification | 45,000 |
| Voltage, kV | 200 |
| Pixel size, Å | 0.93 |
| Total Dose, e−/Å2 | 62.2 |
| Number of segments | 108,911 |
| B-factor | −101 |
| Map resolution, Å | |
| Model:map FCR, 0.38 | 3.8 |
| Map:map FSC, 0.143 | 3.8 |
| d99 | 3.9 |
| Helical symmetry | |
| Rise, Å | 10.88 |
| Rotation, ° | 130.05 |
| Symmetry | D1 |
| Model refinement | |
| Model to map CC | 0.80 |
| Clash score, all atoms | 1.56 |
| Ramachandran favored, % | 91.73 |
| Ramachandran outliers, % | 0.0 |
| Rotamer outliers, % | 0.27 |
| Cβ deviations > 0.25 Å | 0.0 |
| RMS deviations | |
| Bond, Å | 0.004 |
| Angles, ° | 0.775 |
| Deposition IDs | |
| PDB ID | 6U7M |
| EMDB ID | 20673 |
Dihedral LPL Dimer Interface Reveals an Occluded Active Site.
In the asymmetric subunit of the helix, two LPL molecules interact to form a head-to-tail dihedral dimer, in which the N-terminal and C-terminal domains of two opposing LPL subunits form the dimer interface (Fig. 2 E and F). Examination of the lid region reveals that the LPL lid is effectively open and displaced from the LPL active site. However, the active site is occluded by the tryptophan loop (D415–W424) of the other LPL subunit (Fig. 2G). Both the active site serine (S162) and histidine (H271) are buried in the dimer interface. Protein interfaces, surfaces and assemblies (PISA) at the European Bioinformatics Institute (PDBePISA, http://www.ebi.ac.uk/pdbe/prot_int/pistart.html) analysis of the LPL helix dimer revealed that the helix dimer interface buries 5.5% of each subunit’s surface area, with a favorable ∆G of −21.1 kcal/mol and a ∆G P value of 0.011 (SI Appendix, Table S1A) (41). In this instance, ∆G represents the solvation energy of the interface and the ∆G P value represents how surprising and specific the interface is in the context of the structure. The values for the helical dimer interface show it is strongly favorable and far more hydrophobic than the average surface on LPL. Additionally, PDBePISA identified the helical LPL dimer interface as playing an essential role in complex formation. Therefore, the dimer interface we observe in helical LPL is unlikely to be an artifact. The tryptophan loop in the helix dimer blocks access to the active site, and thus it is likely that the helix dimer is an inactive form of LPL.
This dimerization interface bears some similarities to a recently published structure of inhibitor bound LPL in complex with GPIHBP1 (SI Appendix, Fig. S4A) (22). In this structure, an inhibitor occupies the active site of LPL, which displaces the tryptophan loop from the dimerization interface seen in the helical structure (SI Appendix, Fig. S4B). The LPL lid domain is resolved in this structure, and alignment with our experimental density shows a different orientation relative to the LPL lid in the helix structure (SI Appendix, Fig. S4C). Using PDBePISA analysis, the authors found that 5.5% of LPL surface area was buried in the crystal dimer, with a ∆G of −6.5 kcal/mol and a ∆G P value of 0.377, which led to the conclusion that the interface was likely a crystal artifact (22, 41). The differences between the helical and crystal dimer interfaces emanate from the facts that the helix dimer does not include the LPL lid and the active site of the crystal dimer is blocked by an inhibitor, which prevents contacts in that vicinity (SI Appendix, Table S1B-C). Additionally, when one LPL subunit is used to align the helical and crystal structures, the other LPL subunit in the helix LPL dimer is tilted by an angle of 20.4° relative to the crystal dimer (SI Appendix, Fig. S4 D and E).
Helical Contacts within the LPL Filament.
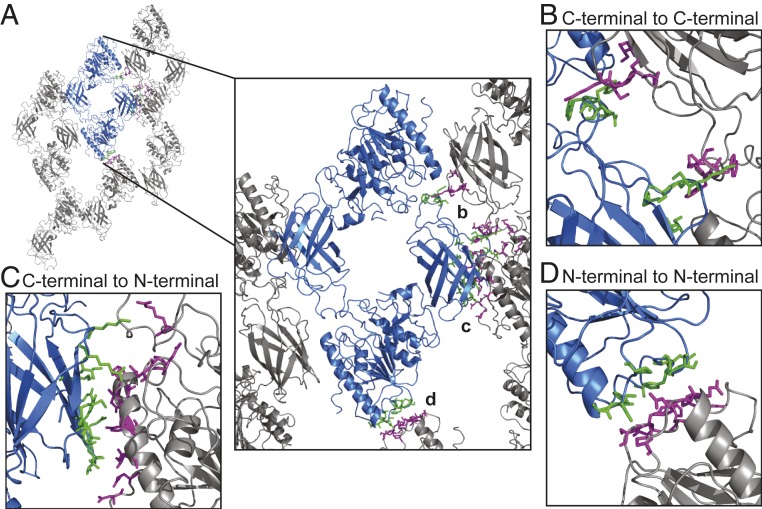
The asymmetric subunit of the LPL helix is a dihedral dimer, and each LPL molecule makes multiple contacts with other LPL molecules in the adjacent asymmetric subunits (Fig. 3A). We identified a C-terminal to C-terminal helical interface, which represents 1.4% of the surface area (Fig. 3B and SI Appendix, Fig. S5B and Table S1D). PDBePISA analysis found that this interface had a ∆G of −5.9 kcal/mol with a ∆G P value of 0.101 (41). Each LPL molecule also has two interfaces to the adjacent asymmetric subunits between the N-terminal domain and C-terminal domain (Fig. 3C and SI Appendix, Fig. S5C and Table S1D). Each N-terminal to C-terminal helical interface accounts for 2.1% of the subunit surface area, for a total of 4.2% per subunit. This interface had a ∆G of −1.6 kcal/mol with a ∆G P value of 0.564 (41). Finally, we found an N-terminal to N-terminal helical interface, which covers 1.4% of the subunit surface area (Fig. 3D and SI Appendix, Fig. S5D and Table S1D). The N-terminal to N-terminal interface had a ∆G of −2.9 kcal/mol with a ∆G P value of 0.390 (41). Interestingly, this interface also contains the density for the N-linked glycosylation at N73. It is likely that the location in the interface stabilized the glycan, allowing resolution in the reconstruction. Including both dimerization and helical interfaces, 12.6% of the surface area (2,709 Å2) of each LPL subunit is buried. Although the helical interfaces have higher ∆G P values than the LPL dimerization interface, taken together, they show how helix formation is energetically favorable.
Fig. 3.
LPL helices form contacts between dimers. (A) Model of a section of the LPL helix with a dihedral dimer shown in blue and the surrounding subunits in gray. Zoomed view of the helical dimer with helical interface residues shown in green and magenta for (B) the C-terminal to C-terminal interface, (C) the C-terminal to N-terminal interface, and (D) the N-terminal to N-terminal interface (SI Appendix, Table S1). Density map for the helical interfaces can be seen in SI Appendix, Fig. S5 B–D, including the N-linked glycan at N73 in the N-terminal to N-terminal interface.
Heparin Stabilizes LPL Helices.
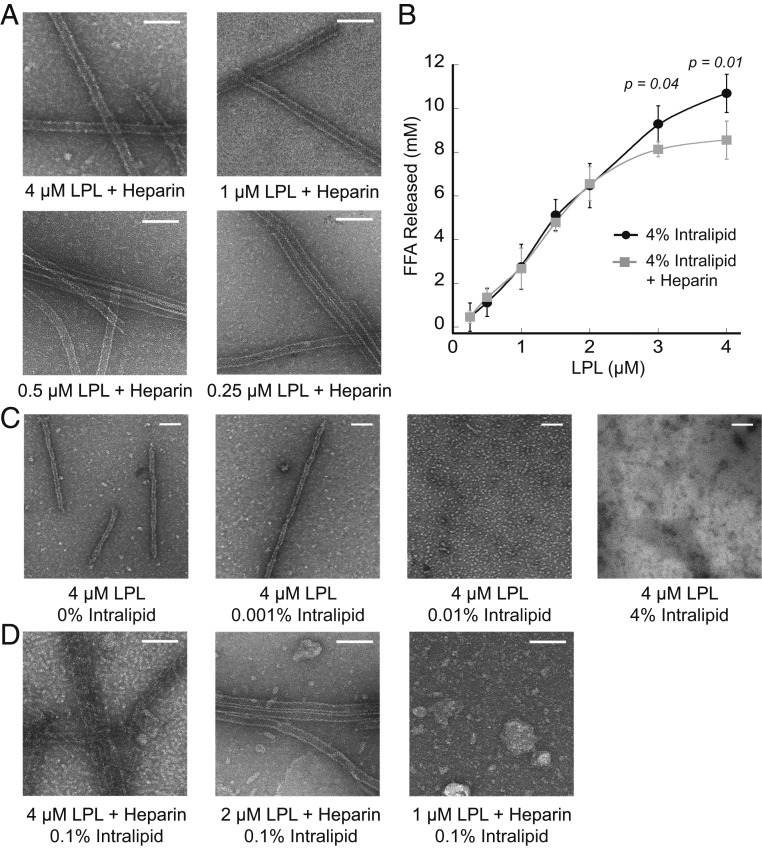
We tested the effects of small molecules known to interact with LPL, including sodium deoxycholate and heparin, on helix structure. By negative stain TEM, we found that deoxycholate disrupted the formation of LPL helices, whereas helices were stable in the presence of heparin (SI Appendix, Fig. S6A). LPL has several known heparin binding sites (29, 42), which are located on the outside of the helical structure (SI Appendix, Fig. S6B). Due to recent work showing that SDC1, an HSPG, interacts with LPL in vesicles (17), we further explored the effect of heparin on the helical form of LPL. We performed a dilution series of LPL in the presence of heparin using negative stain TEM. This revealed that heparin stabilizes LPL helices through dilution to concentrations as low as 31 nM LPL (Fig. 4A and SI Appendix, Fig. S6C), whereas helices do not form when LPL alone is diluted to these lower concentrations (Fig. 1C).
Fig. 4.
Heparin stabilizes inactive LPL helices. (A) Representative negative stain TEM images show that dilution of LPL in the presence of 0.07 U/μL heparin stabilizes LPL helices at an LPL concentration range from 4 μM to 0.25 μM. Heparin-stabilized LPL helices were observed down to 31 nM (SI Appendix, Fig. S6D). (B) FFAs released by LPL acting on intralipid substrate were assessed with (gray square) and without (black circle) 0.07 U/μL heparin. With heparin stabilization, inactive LPL helices form at higher LPL concentrations (4 μM and 3 μM), resulting in a significantly lower release of FFA. Error bars are SD of four replicates, and significance was determined by two-tailed Student’s t test. (C) Negative stain TEM images show 4 μM LPL alone and with increasing intralipid concentrations. LPL helices disassemble in the presence of 0.01% intralipid. (D) Negative stain TEM images of heparin-stabilized LPL in the presence of 0.1% intralipid. As LPL concentration decreases, intralipid triggers helix disassembly even when heparin is present. (Scale bars, 100 nm.)
LPL Helices Are Inactive and Can Disassemble in the Presence of Substrate.
Because LPL’s active site is occluded in the helices, we next set out to understand the interplay between LPL helix formation and catalytic activity. We performed LPL activity assays on intralipid at LPL concentrations spanning those needed for helix formation. Intralipid is comprised of triglycerides with a small amount of phospholipids and bears similarity to native LPL substrates (43). We incubated increasing concentrations of LPL with intralipid, stopped the reaction with Triton X-100, and analyzed free fatty acid (FFA) release. We found that LPL incubated with intralipid displayed a linear increase in released FFA compared to LPL concentration (Fig. 4B, black circles). However, when LPL was tested in the presence of heparin, which stabilizes helical formation, the LPL activity did not increase linearly, but rather leveled off at LPL concentrations of 3 and 4 μM (Fig. 4B, gray squares).
To visualize the relationship between helix formation and LPL activity, we performed negative stain TEM of LPL mixed with intralipid. LPL alone dissociates in the presence of very small amounts of intralipid (0.01%; Fig. 4C). Thus, in the activity assay conditions (4% intralipid), LPL is not in its helical form, explaining the linear increase in LPL activity with increasing LPL concentration. We also performed negative stain TEM of LPL in the presence of heparin and intralipid and found that heparin allows LPL to maintain its helical form even when intralipid is present. For example, at 4 and 3 μM, LPL forms heparin-stabilized, inactive helices despite the presence of 4% intralipid (SI Appendix, Fig. S6D). This helix stabilization explains the nonlinear relationship between LPL concentration and activity (Fig. 4B). However, heparin did not stabilize LPL helices at lower LPL concentrations in the presence of intralipid. For example, at 0.1% intralipid and 1 μM LPL (Fig. 4D), heparin-stabilized helices disassembled. At 4% intralipid, heparin did not stabilize helices at LPL concentrations of 2 μM and below. This allowed LPL to adopt its active form, resulting in a linear relationship between FFA released and LPL concentration (Fig. 4B). These data illustrate that LPL helices are not an active form of LPL.
Endogenous LPL Has a Unique Arrangement in Vesicles.
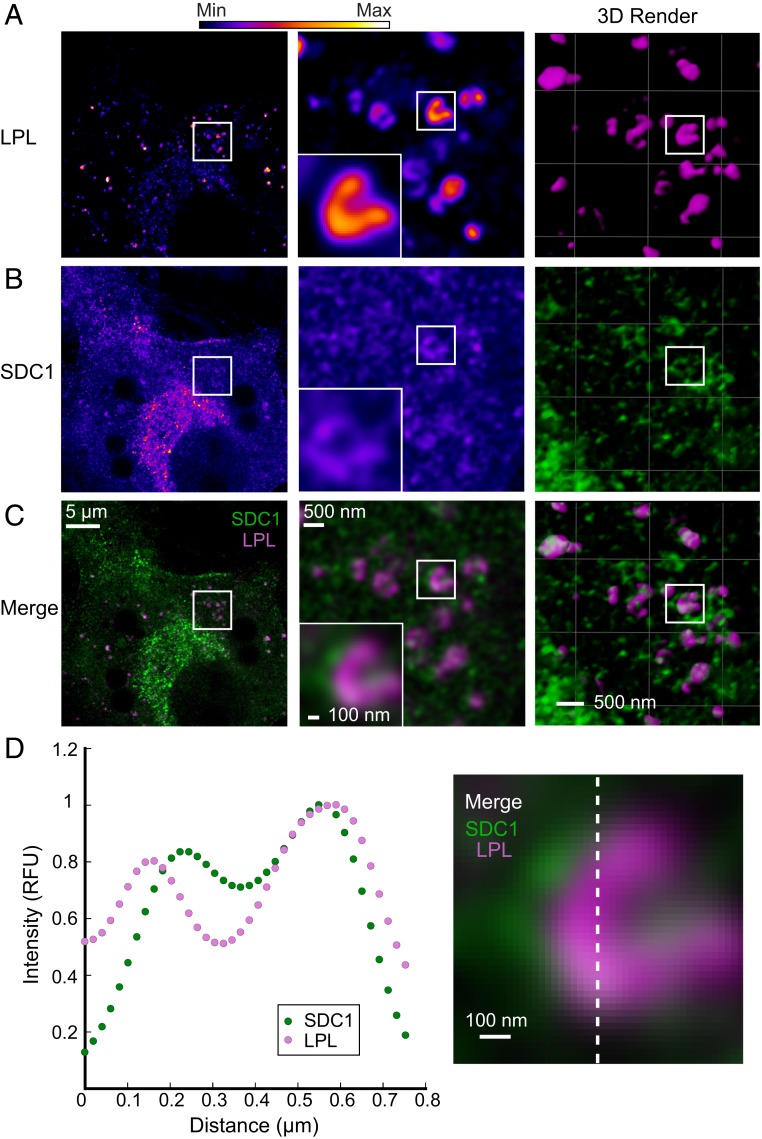
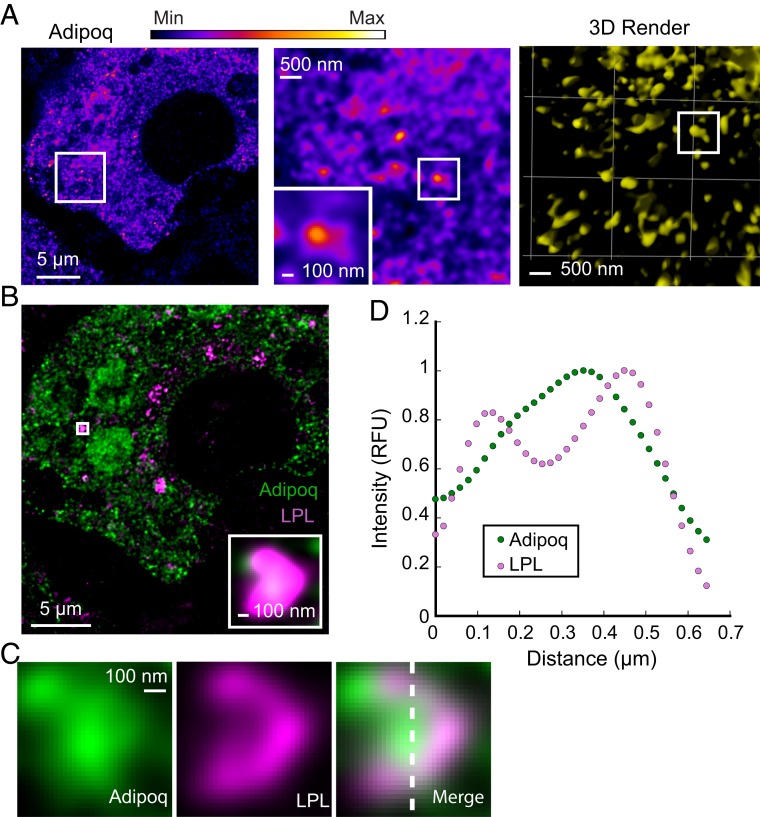
In order to better understand the potential role of helical LPL in vivo, we examined the distribution of endogenous LPL in differentiated 3T3-L1 mouse adipocytes (14). Adipocytes were immunolabeled with antibodies to fluorescently detect both LPL (SI Appendix, Fig. S7) and SDC1, which have been previously shown to traffic through the cell together (17). The cells were examined using stimulated emission depletion (STED) microscopy, a super-resolution technique with a resolution of ∼50 nm (44). We found that LPL was not evenly distributed inside of intracellular vesicles, as would be expected for a soluble cargo protein. Imaging using the enhanced resolution of STED reveals that LPL does not occupy the center of vesicles, but creates a compacted structure that follows the vesicular membrane (Fig. 5A). This distribution of LPL suggests it adopts a filament-like structure in vesicles. SDC1 is a TM protein that marks the vesicular membrane, and can be seen surrounding LPL (Fig. 5 B–D). By comparison, adiponectin (Adipoq), another high-volume adipocyte cargo, was distributed uniformly throughout the vesicles (Fig. 6A), which differs from the distribution seen with LPL (Fig. 6 B–D). Although STED microscopy does not provide sufficient resolution to unambiguously identify the LPL helical oligomer (due to its small 25-nm diameter), it provides compelling evidence that LPL does not adopt a typical soluble protein distribution in vesicles (compare Figs. 5D and 6D). It is likely that LPL’s interaction with SDC1, an integral membrane protein, results in the LPL distribution we observe along the membrane. Given that the LPL/SDC1 interaction is mediated by HSPGs (17) and that heparin stabilizes LPL helices, it is likely that LPL is a helical oligomer inside of storage vesicles.
Fig. 5.
LPL adopts a filament-like distribution in vesicles in adipocytes. Representative STED micrographs of endogenous LPL and SDC1 in 3T3-L1 adipocytes. Immunolabeled endogenous LPL (A), endogenous SDC1 (B), and merged micrographs (C) show that SDC1 colocalizes with LPL vesicles. LPL adopts a condensed and filament-like distribution on the interior face of the vesicles. The 3D renderings further illustrate the filament-like distribution of LPL and show the colocalization of SDC1 and LPL. Fluorescence is colored using a fire gradient (A and B, Left and Middle, scale at top). In the merged micrographs (C) and 3D renderings (A–C, Right), SDC1 is in green and LPL is in magenta. Zoomed views of the left micrographs are seen in the middle panels, with an additional zoom of a single vesicle (Insets). (Scale bars are as labeled.) (D) To illustrate the distribution of LPL and SDC1, the intensity of fluorescence from a vertical slice of the micrograph (indicated by a dashed white line) was graphed. LPL and SDC1 intensity track together, and both have fluorescence peaks that correspond to the sides of the vesicle. The highest intensities of LPL and SDC1 were independently normalized to 1.
Fig. 6.
Unlike LPL, adiponectin is uniformly distributed in vesicles. (A) STED microscopy of immunolabeled endogenous adiponectin (Adipoq) in 3T3-L1 adipocytes shows that Adipoq is uniformly distributed within vesicles. This differs from the distinctive distribution seen with LPL (Fig. 5), although both Adipoq and LPL are soluble, high-volume vesicle cargos in adipocytes. Fluorescence is colored using a fire gradient (A, Left and Middle, scale at top). A zoomed view of the micrograph (Left) is shown (Middle), with an additional zoom of a single vesicle (Inset). Adipoq is in yellow in the 3D render. (B) A merge of immunolabeled, endogenous Adipoq (green) and LPL (magenta) in adipocytes shows the distinct distributions of the two cargos. (C) Zoomed views of the Adipoq, LPL, and merged fluorescence of a single vesicle are shown. (D) A graph of the intensity of fluorescence from a vertical slice of the merged single vesicle (indicated by a dashed white line). This illustrates the different intensity distributions of LPL and Adipoq within a single vesicle. Adipoq fluorescence is evenly distributed through the center of the vesicle, and LPL fluorescence seen as two peaks on the side of the vesicle. The highest intensities of LPL and Adipoq were independently normalized to 1. (Scale bars are as labeled.)
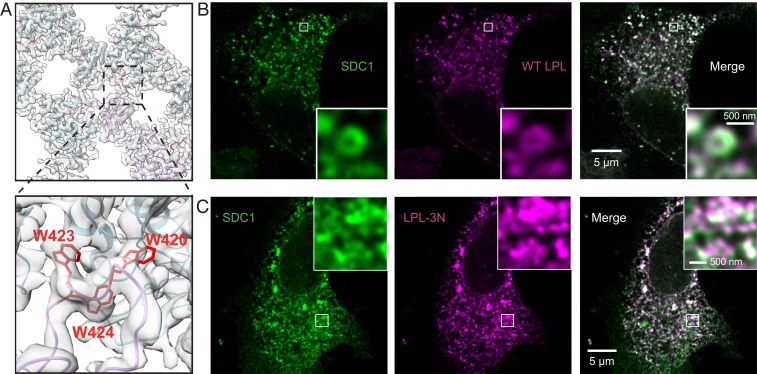
Mutations at the LPL Helical Interface Disrupt the Filament-Like Distribution of LPL.
It was previously observed that mutations to LPL’s tryptophan loop disrupt the SDC1/HSPG mediated targeting of LPL to sphingomyelin-rich vesicles for secretion (17). Based on analysis of our EM density map and LPL model, we know that the tryptophan loop residues (W417, W420, and W421 in human LPL) are a part of the dimer interface critical to helix formation (Figs. 2A and 7A). Without this highly favorable interface, we hypothesized that formation of the LPL helix would be disfavored. When these tryptophan residues are mutated to alanine, both LPL secretion and LPL’s ability to recognize substrates decreases; however, the LPL active site is still functional (27, 45). Therefore, we mutated all three tryptophan residues to asparagine (LPL-3N), as a structurally homologous monomeric lipase, pancreatic lipase, utilizes an asparagine at the position equivalent to W417 (28). We transfected WT LPL and LPL-3N along with SDC1 into HEK-293 cells that were stably expressing LMF1 (to enhance LPL production) (30). Western blots and LPL activity assays showed that both WT LPL and LPL-3N were secreted into the media as active enzymes (SI Appendix, Fig. S8), although we did observe lower secretion of LPL-3N.
Fig. 7.
Mutations at the LPL dimer interface disrupt its filament-like distribution. (A) View of the LPL dihedral dimer interface from inside the helical reconstruction (transparent gray) with LPL models fit into the map. The zoomed view highlights the tryptophan loop that plays an integral role in forming the repeating subunit of the LPL helix. The three homologous tryptophan residues from bovine LPL (W420, W423, W424) were mutated to asparagine in human LPL (W417N, W420N, W421N) to create the LPL-3N mutant. SDC1 and either (B) WT human LPL or (C) LPL-3N were transfected into HEK-293 cells and immunolabeled. Representative STED micrographs of SDC1 (green) and WT LPL or LPL-3N (magenta) show that WT LPL adopts a filament-like distribution within HEK-293 vesicles. This can be seen clearly in the zoomed images (Insets). The LPL-3N mutant is differently distributed, without distinct large puncta and a lack of the filament-like distribution seen within WT LPL vesicles. (Scale bars are as labeled.)
Examining these transfected cells using STED microscopy, we observed that WT LPL adopts a filament-like distribution in the HEK-293 cells as LPL does endogenously in 3T3 adipocytes (Fig. 7B). The LPL-3N mutant displays a qualitatively different distribution compared to WT LPL and did not appear filament-like (Fig. 7C). To better understand this difference, we performed colocalization analysis of WT LPL and LPL-3N with SDC1 and calnexin (CNX), which serves as a marker for the ER, using confocal microscopy. We found that LPL-3N colocalizes significantly more with CNX than WT LPL (P = 0.006; SI Appendix, Fig. S9 A and B). However, LPL-3N colocalizes significantly less with SDC1 compared to WT LPL (P = 0.009; SI Appendix, Fig. S9 C and D). This confirms our qualitative observation that LPL-3N does not collect in large puncta with filament-like distribution in SDC1 vesicles. The mutation of the three tryptophan residues results in an increased ER colocalization and a lack of the SDC1 vesicle filament-like LPL distribution. This corresponds with the observations of Sundberg et al., who examined the effect of mutating LPL aromatic residues (Y417A, W420A, W423A, and W424A) on trafficking of LPL (17). LPL with mutated aromatic residues was no longer preferentially targeted to the sphingomyelin secretion pathway; rather, equivalent secretion of LPL by both bulk flow and the sphingomyelin pathway was observed (17). We propose that reducing the ability of LPL to form the energetically favorable helical LPL oligomer leads to loss of helical oligomerization, resulting in LPL no longer being preferentially targeted to the SDC1/HSPG sphingomyelin secretion pathway. This in turn causes the loss of the filament-like distribution seen in vesicles for LPL-3N.
Discussion
The field of LPL research has recently been expanded by the first structures of LPL (21, 22). However, these structures also called into question the conventional thinking that LPL forms an active dimer, because these structures show LPL in complex with GBIHBP1 may be a monomer. It was previously believed that monomeric LPL only represented an inactive form of LPL (23, 24). Given the ample evidence for the existence of dimeric LPL and the evidence for the 1:1 LPL:GPIHBP1 complex, LPL clearly has a diversity of conformational forms. We have identified a helical form of LPL created by dihedral LPL dimers, which broadens our understanding of LPL oligomerization. We have shown that the LPL helix structure is concentration-dependent and inactive.
This helical structure illustrates a mode of LPL interaction, wherein LPL can form an inactive dimer that assembles into a larger structure. In the helix, 12.6% of each LPL molecule’s surface area is buried. The dimer interface found in helical LPL is energetically favorable and buries the active site serine and histidine in LPL, taking the place of the LPL lid that is thought to cover the active site in the active form of LPL. The structure of LPL with the lid on top of the active site has not yet been solved. This research is hampered by the inherent flexibility of the lid region, which must move to allow substrates access to the LPL active site. Our reconstruction has low-resolution data for the lid density, which shows that it has been displaced from its likely closed conformation.
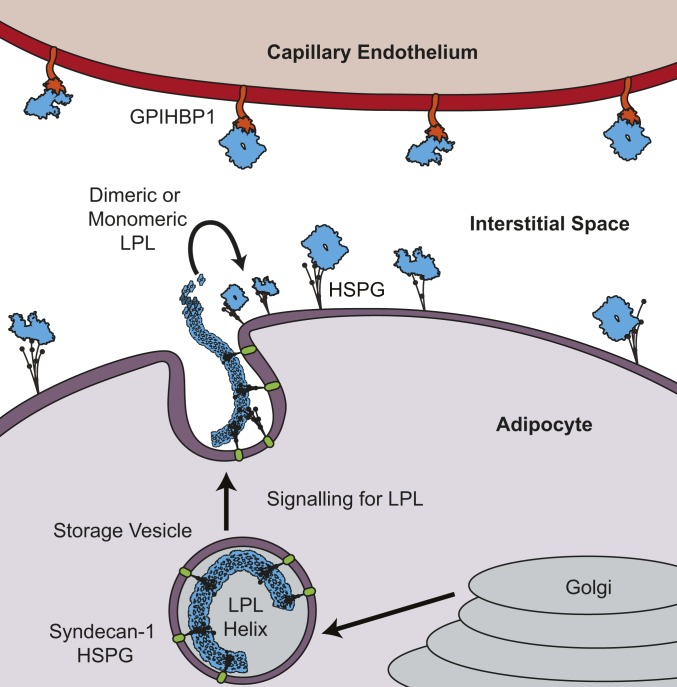
An inactive form of LPL is likely physiologically important for storage during trafficking. LPL has been reported to have a cryptic state in cell lysate that is inactive and concentration-dependent (15, 16). This cryptic state shares many similarities to the helical LPL that we identified. Cells store pools of LPL that can be released in response to nutritional signaling. However, it is not ideal to have active LPL inside the cell, where it could potentially hydrolyze needed lipids. LPL in an inactive conformation would provide an appealing solution to the issue and give the cell flexibility to store LPL without concern for unwanted hydrolysis (Fig. 8). A helical structure is also a stable way to pack an aggregation-prone protein, such as LPL, for efficient storage in a small compact space (46).
Fig. 8.
Model for storage of inactive LPL in adipocytes. Following LPL folding in the ER and maturation in the Golgi apparatus, LPL is concentrated and stored in an inactive form in vesicles, likely the helical form identified in this work. SDC1 mediates sequestration of LPL into vesicles, and binding to HSPGs stabilizes the LPL helix. Upon signaling, LPL vesicles move to the membrane and LPL is secreted into the interstitial space. LPL is diluted in the interstitial space, and helices dissociate into monomeric or dimeric LPL. The LPL monomers or dimers are bound by HSPGs and transferred to GPIHBP1 on the capillary endothelium. Diagram is not to scale.
Other metabolic enzymes form helices and filaments in the cell (46–49). In some cases, the formation of helices leads to the inactivation of enzymes, such as Glutamine synthetase (Gln1), which forms inactive filaments in low pH to facilitate storage during cellular starvation (50). Inosine monophosphate dehydrogenase (IMPDH) also has an inactive helical form in the presence of GTP (51). For both Gln1 and IMPDH, formation of inactive filaments helps regulate metabolic processes in the cell. Similarly, helical LPL could provide an additional layer of regulation to facilitate control of triglyceride hydrolysis. CTP synthase (CTPS) also forms filaments comprised of inactive dimers and tetramers, as well as an active filament of CTPS tetramers (52, 53). This illustrates that diverse oligomeric states can be an effective way of controlling protein activity. LPL, which has now been identified in three oligomeric forms, could similarly utilize oligomeric state to control its activity.
Given the possibility that helical LPL is a storage conformation for LPL in the cell, the ability of heparin to bind and stabilize helices takes on further significance. LPL interacts with the HSPG SDC1 in vesicles (17). This likely leads to LPL being tethered along the membrane, as we observed using STED microscopy. This atypical distribution of a soluble protein in vesicles likely indicates the presence of helical LPL. In vitro, we observe that heparin binds LPL and stabilizes its helical structure at low LPL concentrations, and heparin is structurally similar to the HSPGs LPL binds in vivo (54). HS chains range in length from 20 to 150 nm, indicating there are multiple ways for the HS chains to interact with the outward-facing heparin binding sites on the 25-nm-diameter LPL helix (55). LPL is known to follow both SDC1-marked and bulk flow pathways (18). These data raise the possibility that LPL may be sorted into different secretory routes depending on its protein–protein interactions or structural state. Mutations to LPL in its tryptophan loop are shown to prevent SDC1-dependent trafficking and are likely to interfere with LPL oligomerization. It is possible that one mechanism for secretory control of bulky protein oligomers, such as those formed by LPL, is sorting into specific secretory pathways depending on oligomeric state.
Following secretion of LPL to the interstitial spaces, LPL would be diluted and thus capable of adopting an active dimer or monomer form (Fig. 8). We observed that the presence of LPL substrate also led to the disassembly of helices, which represents another safety mechanism, ensuring that LPL will preferentially act on its substrate rather than remain sequestered in its inactive form. We would not expect LPL substrates to be found in vesicles, but, given that heparin stabilized LPL has increased resistance to substrate-induced helical disassembly, helical LPL in vesicles would be able to tolerate the presence of low levels of substrates due to binding of HSPGs.
Studies on LPL structure and trafficking will continue to shed light on the triggers dictating LPL conformation in the cell. The structure of an inactive LPL adds insight to our understanding of the many LPL interactions that take place during the intricate process of preparing LPL for secretion. Further elucidation of this complex process is crucial for understanding how LPL’s quaternary structure can regulate its cellular function.
Materials and Methods
Bovine LPL Purification.
Bovine LPL was purified from raw cow’s milk using heparin Sepharose at 4 °C as previously described (56), with minor modifications (SI Appendix, Fig. S1).
Negative Stain Transmission Electron Microscopy (TEM).
Continuous carbon grids (CF300-Cu-UL; Electron Microscopy Sciences) were glow-discharged using a Harrick Plasma Cleaner for 30 s. The grids were suspended from reverse tweezers, and 5 μL of each sample was applied to the grid. Each sample was incubated on the grid for 30 s. Grids were first washed with 100 μL of dilution buffer (500 mM NaCl, 20 mM Tris HCl, pH 8) and then washed with 100 μL of 1% uranyl acetate stain. Grids were incubated for 1 min in stain before being blotted and air-dried. Negative stain grids were imaged using a ThermoFisher (FEI) TECNAI T12 G2 transmission electron microscope equipped with either a Gatan 794 1k × 1k CCD camera with Digital Montage software or a Gatan Rio CMOS camera with Gatan Microscopy Suite software.
Negative Stain TEM Sample Preparation.
To generate samples for the LPL concentration gradient, the first LPL concentration was created by diluting LPL and stored in heparin buffer B at 1:4 ratio using dilution buffer (500 mM NaCl, 20 mM Tris HCl, pH 8) to reach 4 μM LPL. For the remaining serial dilutions, LPL was diluted in a 1:4 mix of heparin buffer B and dilution buffer. To test the effect of diluting LPL in different salt concentrations and pHs, LPL in heparin buffer B was mixed with the tested buffers to reach 4 μM LPL. Buffers for testing pH all contained 500 mM NaCl and either 10 mM Bis-Tris, pH 6.5, 20 mM Tris⋅HCl, pH 7.5, 20 mM Tris⋅HCl, pH 8, or 20 mM Tris⋅HCl, pH 8.5. We confirmed that the final pH matched the diluting buffer using pH strips. Buffers for testing salt concentrations all contained 20 mM Tris⋅HCl, pH 8, and either 500, 400, 300, 200, 150, 100, 50, or 0 mM NaCl. This resulted in the final salt concentrations of 875 mM, 800 mM, 725 mM, 650 mM, 612.5 mM, 575 mM, 537.5 mM, and 500 mM, respectively. To test the effect of heparin and deoxycholate, the additives were included in the dilution buffer, which was then used to dilute LPL to 4 μM. Dilution buffer was supplemented with either 0.5 to 0.1 U/μL heparin (AC411210010; Fisher) or 1 mM sodium deoxycholate (D6750; Sigma) for final concentrations of 0.1 to 0.07 U/μL heparin and 0.7 mM deoxycholate, respectively. The effects of dimethylsulfoxide (DMSO) and intralipid emulsion (I141-100ML; Sigma) were assessed by diluting LPL stored in heparin buffer B with dilution buffer. Additives were diluted with phosphate buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4) and then added to the LPL mixture to reach the final LPL concentration.
Cryo-Grid Preparation.
Cryo-grids were prepared with a Leica EM GP set to 98% humidity and 25 °C. Immediately prior to preparing TEM grids, they were glow-discharged for 30 s using a PELCO easiGlow plasma cleaner (with 15 mA current and under 0.38 mbar vacuum). A total of 3 μL of sample (LPL in heparin buffer B was diluted 1:4 using dilution buffer to a final concentration of 3.3 μM) was applied to the carbon side of the TEM grids, preblotted for 15 s, and then blotted for 3 s with Whatman no. 1 filter paper. Cryo-grids were plunge frozen in liquid ethane and stored under liquid nitrogen.
Cryo-EM Data Collection.
Cryo-grids were imaged with a Thermo Fisher 200 kV Talos Arctica TEM and Gatan K2 summit direct electron detector operated in counting mode. Data were collected over a 3-d period using Latitude in Digital Micrograph. The microscope was aligned in nano-probe mode at spot size 5 and 70 μm C2 aperture (57). Coma-free alignment was carried out in SerialEM (58). Data were collected at 45,000× nominal magnification corresponding to a pixel size of 0.93 Å/pixel and a measured flux of 4.2 e−/pixel/s. Movies were collected with a 12.8-s exposure at 0.4 s/frame for 32 total frames, corresponding to a total dose of 62.2 e−/Å2.
Helical Reconstruction.
A total of 1,764 micrographs were processed using MotionCor2 and CTFFIND4 (59, 60). A total of 4,192 LPL helices were manually picked using both EMAN2 and Relion 3.0 (61, 62). To determine the helical parameters of the LPL helix, we extracted helix images from EMAN2 and transferred the data into Spider (63), where we cut nonoverlapping 1,200-pixel boxes from the helices (64). We calculated the power spectra of each box and averaged the power spectra together to create a high-resolution power spectrum, which contained the layer lines needed to derive the helical parameters for LPL (65). The helical symmetry was determined as a 10.7-Å rise and 130.1° rotation after searching through a number of possible symmetries by trial and error. Only the helical symmetry scheme n = −11, n = +3 yielded interpretable density (SI Appendix, Fig. S3A). The correct handedness was confirmed by examination of alpha helices in the reconstruction density, which can be clearly seen at this resolution. We imported this information into Relion 3.0 to further improve the resolution (33). We cut overlapping 384 × 384-pixel boxes from LPL helices with a 19-Å offset from dose-weighted micrographs using all frames, which yielded 157,128 segments in the initial data set. We performed two-dimensional (2D) classification and selected the best classes, leaving 136,324 particles (86%). We then proceeded to three-dimensional (3D) classification using a featureless cylinder with 127-Å radius as an initial model, refining both helical parameters during the classification. The best class contained 108,911 particles (69%), which we used for 3D refinement. The helical parameters refined to rise = 10.88 Å and pitch = 130.05°. Following postprocessing, the estimated resolution was 4.5 Å. Using Relion 3.0, we performed CTF refinement and particle polishing (61). We took these polished particles and repeated 3D refinement. Postprocessing was performed in Relion using automated B-factor map sharpening, yielding a B-factor of −101. The final resolution for the LPL helix was 3.8 Å using the gold-standard FSC 0.143 criterion (66).
LPL Helix Model.
I-TASSER (34–36) was used to generate a series of homology models for a monomer of LPL using the bovine LPL sequence. The resulting model with the highest C-score, which was modeled on the LPL/GPIHBP1 crystal structure (6E7K) (21), was used as the initial model for model building. The helical reconstruction map was segmented using Chimera (67) to isolate the density representing a single monomer of LPL. All residues can be successfully traced and built into the map except for the lid region (N247–D264). We identified five-disulfide bonds, C57–C50, C246–C269, C308–C313, C448–C468, and one glycan at N73. Density for the glycan at N389 was not seen. The LPL monomer structure was refined using PHENIX real-space refinement and Coot (37–39). Subsequently, a filament model was generated from the helical symmetry and refined against the full cryo-EM map in PHENIX and Coot (37–39). The model-to-map FCR and the per-residue real space correlation coefficient (SI Appendix, Fig. S3 B and C) were calculated to measure the accuracy of the model to the experimental map. The model-to-map FCR confirmed a resolution of 3.8 Å, as determined by the spatial frequency at 0.38 (the square root of 0.143) FCR (68). In total, the resolution was confirmed using three different approaches: the map-to-map FSC, the map-to-model FCR, and the d99 (40). The refinement statistics are shown in Table 1. PDBePISA was used for the helical interface analysis.
Intralipid Non-Esterified Fatty Acid (NEFA) Assay.
LPL stored in heparin buffer B was diluted 5:16 using dilution buffer. Subsequent dilutions were performed using 5:16 heparin buffer B:dilution buffer. For samples with heparin, heparin was added to the dilution buffer, for a final concentration of 0.07 U/μL. Intralipid 20% (suspended in PBS) was added to each sample to create a final concentration of 4% intralipid. Intralipid was added at 22 °C and incubated for 30 s, and the reaction was stopped with a final concentration of 2.5% Triton X-100. Samples were diluted an additional 1.5× in PBS. A total of 5 μL of each sample was moved into three separate wells in a clear 96-well plate to create technical triplicates. FFAs released were quantified as has previously been reported (69). Briefly, each sample was incubated for 15 min at 37 °C with 45 μL of reagent A (133 mM KPO4 pH 7.5, 3.3 mM MgCl2, 4.4 mM adenosine triphosphate, 1 mM coenzyme A [CoA], 0.055 U/mL acetyl-CoA synthetase [ACS], 0.5% Triton X-100). Next, each sample was incubated for 10 min at 37 °C with 50 μL of reagent B (133 mM KPO4 pH 7.5, 0.6 mM N-ethyl-N-[2-hydroxy-3-sulfopropyl]-3-methylaniline [TOOS], 6 U/mL horseradish peroxidase, 1.73 mM 4-aminoantipyrine, 10 mM N-ethylmaleimide [NEM], 5 U/mL acetyl-CoA oxidase [ACO], 0.5% Triton X-100). Endpoint absorption was assessed at 555 nm using a M5 Spectramax plate reader. Sodium palmitate (P9767-5G; Sigma) was used to create an FFA standard curve for each assay. Four biological replicates were conducted for each sample. Data were graphed in KaleidaGraph, and significance was assessed using a two-tailed Student’s t test.
Cell Culture.
For immunofluorescence studies, 3T3-L1 adipocytes were differentiated and maintained as described previously (14). Flp-In T-REx HEK 293 cells were grown and maintained in M1 media. HEK 293-LMF1 cells stably expressing LMF1 were described previously (30). HEK 293-LMF1 cells were transiently transfected 24 h before assaying using JetPrime transfection regent per manufacturer instructions. Cells were induced to express LMF1 at the time of transfection with tetracycline for 24 h.
Immunofluorescence and Microscopy.
3T3-L1 adipocytes were grown on no. 1.5 glass coverslips coated with poly-D-lysine and rat tail collagen. Cells were fixed with 4% paraformaldehyde for 10 min at 22 °C and washed with PBS. Next, cells were permeabilized with 0.3% Triton-X 100 in 5% bovine serum albumin (BSA)–PBS for 15 min at 22 °C. Cells were then blocked for 30 min at 22 °C in 5% BSA–PBS plus normal donkey serum. Cells were then stained overnight with anti-LPL antibody (1:40; R&D AF7197) and either anti-SDC1 (1:500) or anti-Adipoq (1:500; Invitrogen PA1-054). Cells were then stained with Alexa Fluor 594 (1:1,000) and Atto647N (1:500) and mounted in ProLong Diamond antifade (Invitrogen).
HEK cells were grown on no. 1.5 glass coverslips coated with poly-D-lysine and fibronectin. Cells were treated as indicated for the 3T3-L1 adipocytes, then stained overnight with anti-LPL antibody (1:40; R&D AF7197) and either anti-HA (1:1,500; CST C29F4) or anti-calnexin (1:50; CST 2433). Cells were then stained with Alexa Fluor 594 (1:1,000) and Atto647N (STED; 1:500) or Alexa Fluor 488 (Confocal; 1:1,000) and mounted in ProLong Diamond antifade.
For STED microscopy, cells were imaged using STED on a Leica SP8 equipped using a 93× NA 1.3 glycerol lens. Z-stack images were deconvolved using Huygens software (https://www.svi.nl). Additional image analysis was done using the Fiji suite (70) and Imaris (https://imaris.oxinst.com). For confocal microscopy, cells were imaged using a Zeiss 710 laser scanning confocal microscope with 40×/1.4 plan apo lens, and images were deconvolved with Bitplane Autoquant 3. For colocalization analyses in Fiji, single-cell Z-stacks were selected and thresholded for the top 2% of pixel intensities, and colocalization was computed as the Mander’s coefficient using the JACop plugin (71). The Wilcoxon signed-rank test was used to compute significance.
Immunoblotting.
For Western blotting, cells were transfected and induced to express LMF1 as detailed earlier. After 24 h, media was replaced with 500 μL of 1% FBS DMEM with 10 U/mL heparin for 1 h. Media was collected, centrifuged for 10 min at 1,000 × g to remove debris, and moved to fresh tubes. Cells were lysed in radioimmunoprecipitation assay (RIPA) buffer with 1 mM phenylmethylsulfonyl fluoride (PMSF), rotated at 4 °C for 30 min, and cleared at 16,000 × g for 10 min, and protein concentrations were measured by BCA assay. An equal volume of media or 25 μg of cell lysate was used for Western blots. Samples were separated on a 12% sodium dodecyl sulfate/polyacrylamide gel electrophoresis (SDS/PAGE) gel, transferred to PVDF, and probed with anti-LPL (R&D; 1:200) and HRP anti-goat (R&D; 1:1,000).
LPL Media Activity Assay.
LPL activity in conditioned media was assayed as previously described (72).
Data Availability.
Data for structural studies were deposited in the PDB (ID code 6U7M) and the EMDB (accession no. 20673).
Supplementary Material
Acknowledgments
We thank members of the S.B.N. and E.H.E. labs for discussions and assistance. We thank the University of North Carolina at Chapel Hill (UNC) Hooker Imaging Core; the UNC Microscopy Services Lab; UNC Information Technology Services Research Computing, which assisted with computing services on Google Cloud platform; and the Duke Light Microscopy Core Facility. The authors acknowledge support from the National Institutes of Health (Grant nos. R01-HL12565 to S.B.N., R35-GM122510 to E.H.E., 1F31-DK12272801A1 to B.S.R.) and the American Heart Association (Grant no. 20POST35210031 to K.H.G.). This work was supported in part by the Intramural Research Program of the NIH; National Institute of Environmental Health Sciences (ZIC ES103326 to M.J.B.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The data for structural studies in this paper have been deposited in the Protein Data Bank (PDB; https://www.rcsb.org/) under ID code 6U7M and in the Electron Microscopy Data Bank (EMDB; https://www.ebi.ac.uk/pdbe/emdb/) under ID no. 20673.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1916555117/-/DCSupplemental.
References
- 1.Kersten S., Physiological regulation of lipoprotein lipase. Biochim. Biophys. Acta 1841, 919–933 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Miller M., et al. ; American Heart Association Clinical Lipidology, Thrombosis, and Prevention Committee of the Council on Nutrition, Physical Activity, and Metabolism; Council on Arteriosclerosis, Thrombosis and Vascular Biology; Council on Cardiovascular Nursing; Council on the Kidney in Cardiovascular Disease , Triglycerides and cardiovascular disease: A scientific statement from the American heart association. Circulation 123, 2292–2333 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Castelli W. P., Epidemiology of triglycerides: A view from framingham. Am. J. Cardiol. 70, 3H–9H (1992). [DOI] [PubMed] [Google Scholar]
- 4.Le N. A., Walter M. F., The role of hypertriglyceridemia in atherosclerosis. Curr. Atheroscler. Rep. 9, 110–115 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Sathiyakumar V., et al. , Novel therapeutic targets for managing dyslipidemia. Trends Pharmacol. Sci. 39, 733–747 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Stahel P., Xiao C., Hegele R. A., Lewis G. F., The atherogenic dyslipidemia complex and novel approaches to cardiovascular disease prevention in diabetes. Can. J. Cardiol. 34, 595–604 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Farese R. V. Jr, Yost T. J., Eckel R. H., Tissue-specific regulation of lipoprotein lipase activity by insulin/glucose in normal-weight humans. Metabolism 40, 214–216 (1991). [DOI] [PubMed] [Google Scholar]
- 8.Zhang R., The ANGPTL3-4-8 model, a molecular mechanism for triglyceride trafficking. Open Biol. 6, 150272 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Preiss-Landl K., Zimmermann R., Hämmerle G., Zechner R., Lipoprotein lipase: The regulation of tissue specific expression and its role in lipid and energy metabolism. Curr. Opin. Lipidol. 13, 471–481 (2002). [DOI] [PubMed] [Google Scholar]
- 10.Roberts B. S., Babilonia-Rosa M. A., Broadwell L. J., Wu M. J., Neher S. B., Lipase maturation factor 1 affects redox homeostasis in the endoplasmic reticulum. EMBO J. 37, e97379 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vannier C., Ailhaud G., Biosynthesis of lipoprotein lipase in cultured mouse adipocytes. II. Processing, subunit assembly, and intracellular transport. J. Biol. Chem. 264, 13206–13216 (1989). [PubMed] [Google Scholar]
- 12.Klinger S. C., et al. , SorLA regulates the activity of lipoprotein lipase by intracellular trafficking. J. Cell Sci. 124, 1095–1105 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Pradines-Figueres A., Vannier C., Ailhaud G., Short-term stimulation by insulin of lipoprotein lipase secretion in adipose cells. Biochem. Biophys. Res. Commun. 154, 982–990 (1988). [DOI] [PubMed] [Google Scholar]
- 14.Semenkovich C. F., Wims M., Noe L., Etienne J., Chan L., Insulin regulation of lipoprotein lipase activity in 3T3-L1 adipocytes is mediated at posttranscriptional and posttranslational levels. J. Biol. Chem. 264, 9030–9038 (1989). [PubMed] [Google Scholar]
- 15.Pradines-Figuères A., Vannier C., Ailhaud G., Lipoprotein lipase stored in adipocytes and muscle cells is a cryptic enzyme. J. Lipid Res. 31, 1467–1476 (1990). [PubMed] [Google Scholar]
- 16.Vannier C., Deslex S., Pradines-Figuères A., Ailhaud G., Biosynthesis of lipoprotein lipase in cultured mouse adipocytes. I. Characterization of a specific antibody and relationships between the intracellular and secreted pools of the enzyme. J. Biol. Chem. 264, 13199–13205 (1989). [PubMed] [Google Scholar]
- 17.Sundberg E. L., Deng Y., Burd C. G., Syndecan-1 mediates sorting of soluble lipoprotein lipase with sphingomyelin-rich membrane in the Golgi apparatus. Dev. Cell 51, 387–398.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pillarisetti S., et al. , Endothelial cell heparanase modulation of lipoprotein lipase activity. Evidence that heparan sulfate oligosaccharide is an extracellular chaperone. J. Biol. Chem. 272, 15753–15759 (1997). [DOI] [PubMed] [Google Scholar]
- 19.Davies B. S., et al. , GPIHBP1 is responsible for the entry of lipoprotein lipase into capillaries. Cell Metab. 12, 42–52 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beigneux A. P., et al. , Lipoprotein lipase is active as a monomer. Proc. Natl. Acad. Sci. U.S.A. 116, 6319–6328 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Birrane G., et al. , Structure of the lipoprotein lipase-GPIHBP1 complex that mediates plasma triglyceride hydrolysis. Proc. Natl. Acad. Sci. U.S.A. 116, 1723–1732 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arora R., et al. , Structure of lipoprotein lipase in complex with GPIHBP1. Proc. Natl. Acad. Sci. U.S.A. 116, 10360–10365 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lookene A., Zhang L., Hultin M., Olivecrona G., Rapid subunit exchange in dimeric lipoprotein lipase and properties of the inactive monomer. J. Biol. Chem. 279, 49964–49972 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Osborne J. C. Jr, Bengtsson-Olivecrona G., Lee N. S., Olivecrona T., Studies on inactivation of lipoprotein lipase: Role of the dimer to monomer dissociation. Biochemistry 24, 5606–5611 (1985). [DOI] [PubMed] [Google Scholar]
- 25.Iverius P. H., Ostlund-Lindqvist A. M., Lipoprotein lipase from bovine milk. Isolation procedure, chemical characterization, and molecular weight analysis. J. Biol. Chem. 251, 7791–7795 (1976). [PubMed] [Google Scholar]
- 26.Garfinkel A. S., et al. , Lipoprotein lipase: Size of the functional unit determined by radiation inactivation. J. Lipid Res. 24, 775–780 (1983). [PubMed] [Google Scholar]
- 27.Kobayashi Y., Nakajima T., Inoue I., Molecular modeling of the dimeric structure of human lipoprotein lipase and functional studies of the carboxyl-terminal domain. Eur. J. Biochem. 269, 4701–4710 (2002). [DOI] [PubMed] [Google Scholar]
- 28.van Tilbeurgh H., Roussel A., Lalouel J. M., Cambillau C., Lipoprotein lipase. Molecular model based on the pancreatic lipase x-ray structure: Consequences for heparin binding and catalysis. J. Biol. Chem. 269, 4626–4633 (1994). [PubMed] [Google Scholar]
- 29.Hayne C. K., et al. , We FRET so you don’t have to: New models of the lipoprotein lipase dimer. Biochemistry 57, 241–254 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu M. J., et al. , Coexpression of novel furin-resistant LPL variants with lipase maturation factor 1 enhances LPL secretion and activity. J. Lipid Res. 59, 2456–2465 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hayashi R., Tajima S., Yamamoto A., Purification and characterization of lipoprotein lipase from human postheparin plasma and its comparison with purified bovine milk lipoprotein lipase. J. Biochem. 100, 319–331 (1986). [DOI] [PubMed] [Google Scholar]
- 32.Egelman E. H., The iterative helical real space reconstruction method: Surmounting the problems posed by real polymers. J. Struct. Biol. 157, 83–94 (2007). [DOI] [PubMed] [Google Scholar]
- 33.He S., Scheres S. H. W., Helical reconstruction in RELION. J. Struct. Biol. 198, 163–176 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roy A., Kucukural A., Zhang Y., I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang J., et al. , The I-TASSER suite: Protein structure and function prediction. Nat. Methods 12, 7–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y., I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9, 40 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adams P. D., et al. , PHENIX: A comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Afonine P. V., et al. , Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D Struct. Biol. 74, 531–544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Afonine P. V., et al. , New tools for the analysis and validation of cryo-EM maps and atomic models. Acta Crystallogr. D Struct. Biol. 74, 814–840 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krissinel E., Henrick K., Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Lookene A., Chevreuil O., Ostergaard P., Olivecrona G., Interaction of lipoprotein lipase with heparin fragments and with heparan sulfate: Stoichiometry, stabilization, and kinetics. Biochemistry 35, 12155–12163 (1996). [DOI] [PubMed] [Google Scholar]
- 43.Lutz O., et al. , Fat emulsion particle size: Influence on the clearance rate and the tissue lipolytic activity. Am. J. Clin. Nutr. 50, 1370–1381 (1989). [DOI] [PubMed] [Google Scholar]
- 44.Vicidomini G., Bianchini P., Diaspro A., STED super-resolved microscopy. Nat. Methods 15, 173–182 (2018). [DOI] [PubMed] [Google Scholar]
- 45.Lookene A., Groot N. B., Kastelein J. J., Olivecrona G., Bruin T., Mutation of tryptophan residues in lipoprotein lipase. Effects on stability, immunoreactivity, and catalytic properties. J. Biol. Chem. 272, 766–772 (1997). [DOI] [PubMed] [Google Scholar]
- 46.O’Connell J. D., Zhao A., Ellington A. D., Marcotte E. M., Dynamic reorganization of metabolic enzymes into intracellular bodies. Annu. Rev. Cell Dev. Biol. 28, 89–111 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shen Q. J., et al. , Filamentation of metabolic enzymes in Saccharomyces cerevisiae. J. Genet. Genomics 43, 393–404 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Selwood T., Jaffe E. K., Dynamic dissociating homo-oligomers and the control of protein function. Arch. Biochem. Biophys. 519, 131–143 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Park C. K., Horton N. C., Structures, functions, and mechanisms of filament forming enzymes: A renaissance of enzyme filamentation. Biophys. Rev. 11, 927–994 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Petrovska I., et al. , Filament formation by metabolic enzymes is a specific adaptation to an advanced state of cellular starvation. eLife 3, e02409 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Anthony S. A., et al. , Reconstituted IMPDH polymers accommodate both catalytically active and inactive conformations. Mol. Biol. Cell 28, 2600–2608 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Noree C., Monfort E., Shiau A. K., Wilhelm J. E., Common regulatory control of CTP synthase enzyme activity and filament formation. Mol. Biol. Cell 25, 2282–2290 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lynch E. M., Kollman J. M., Coupled structural transitions enable highly cooperative regulation of human CTPS2 filaments. Nat. Struct. Mol. Biol. 27, 42–48 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meneghetti M. C., et al. , Heparan sulfate and heparin interactions with proteins. J. R. Soc. Interface 12, 0589 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sarrazin S., Lamanna W. C., Esko J. D., Heparan sulfate proteoglycans. Cold Spring Harbor Perspect. Biol. 3, a004952 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bengtsson-Olivecrona G., Olivecrona T., Phospholipase activity of milk lipoprotein lipase. Methods Enzymol. 197, 345–356 (1991). [DOI] [PubMed] [Google Scholar]
- 57.Herzik M. A. Jr, Wu M., Lander G. C., Achieving better-than-3-Å resolution by single-particle cryo-EM at 200 keV. Nat. Methods 14, 1075–1078 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schorb M., Haberbosch I., Hagen W. J. H., Schwab Y., Mastronarde D. N., Software tools for automated transmission electron microscopy. Nat. Methods 16, 471–477 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zheng S. Q., et al. , MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rohou A., Grigorieff N., CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zivanov J., et al. , New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tang G., et al. , EMAN2: An extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 (2007). [DOI] [PubMed] [Google Scholar]
- 63.Shaikh T. R., et al. , SPIDER image processing for single-particle reconstruction of biological macromolecules from electron micrographs. Nat. Protoc. 3, 1941–1974 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Egelman E. H., A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy 85, 225–234 (2000). [DOI] [PubMed] [Google Scholar]
- 65.Egelman E. H., Reconstruction of helical filaments and tubes. Methods Enzymol. 482, 167–183 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Scheres S. H., Chen S., Prevention of overfitting in cryo-EM structure determination. Nat. Methods 9, 853–854 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pettersen E. F., et al. , UCSF Chimera–A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 68.Penczek P. A., Resolution measures in molecular electron microscopy. Methods Enzymol. 482, 73–100 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Garrett C. K., Broadwell L. J., Hayne C. K., Neher S. B., Modulation of the activity of Mycobacterium tuberculosis LipY by its PE domain. PLoS One 10, e0135447 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schindelin J., et al. , Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bolte S., Cordelières F. P., A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 224, 213–232 (2006). [DOI] [PubMed] [Google Scholar]
- 72.Hayne C. K., Lafferty M. J., Eglinger B. J., Kane J. P., Neher S. B., Biochemical analysis of the lipoprotein lipase truncation variant, LPLS447X, reveals increased lipoprotein uptake. Biochemistry 56, 525–533 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data for structural studies were deposited in the PDB (ID code 6U7M) and the EMDB (accession no. 20673).