Significance
By ingesting apoptotic cells and other cellular debris, macrophages accumulate cholesterol. Some of the cholesterol is esterified and stored in cytosolic lipid droplets, mitigating the toxicity from free cholesterol. Eventually, however, macrophages must unload surplus cholesterol—a process often referred to as “cholesterol efflux.” Cholesterol efflux is an important physiologic process because it would be expected to retard the formation of cholesterol-rich macrophage foam cells in atherosclerotic plaques. Most studies of cholesterol efflux have focused on the ability of ABC transporters to export cholesterol onto high-density lipoproteins. The current study examines another mechanism. We found, using NanoSIMS imaging, that [13C]cholesterol-loaded macrophages unload cholesterol directly into adjacent smooth muscle cells. This mechanism is potentially relevant to cholesterol efflux by tissue macrophages.
Keywords: macrophages, smooth muscle cells, nanoSIMS imaging, cholesterol
Abstract
Cholesterol-laden macrophage foam cells are a hallmark of atherosclerosis. For that reason, cholesterol metabolism in macrophages has attracted considerable scrutiny, particularly the mechanisms by which macrophages unload surplus cholesterol (a process referred to as “cholesterol efflux”). Many studies of cholesterol efflux in macrophages have focused on the role of ABC transporters in moving cholesterol onto high-density lipoproteins (HDLs), but other mechanisms for cholesterol efflux likely exist. We hypothesized that macrophages have the capacity to unload cholesterol directly onto adjacent cells. To test this hypothesis, we used methyl-β-cyclodextrin (MβCD) to load mouse peritoneal macrophages with [13C]cholesterol. We then plated the macrophages (in the absence of serum or HDL) onto smooth muscle cells (SMCs) that had been metabolically labeled with [15N]choline. After incubating the cells overnight in the absence of HDL or serum, we visualized 13C and 15N distribution by nanoscale secondary ion mass spectrometry (NanoSIMS). We observed substantial 13C enrichment in SMCs that were adjacent to [13C]cholesterol-loaded macrophages—including in cytosolic lipid droplets of SMCs. In follow-up studies, we depleted “accessible cholesterol” from the plasma membrane of [13C]cholesterol-loaded macrophages with MβCD before plating the macrophages onto the SMCs. After an overnight incubation, we again observed substantial 13C enrichment in the SMCs adjacent to macrophages. Thus, macrophages transfer cholesterol to adjacent cells in the absence of serum or HDL. We suspect that macrophages within tissues transfer cholesterol to adjacent cells, thereby contributing to the ability to unload surplus cholesterol.
The presence of cholesterol ester-rich “macrophage foam cells” in early atherosclerotic lesions has prompted interest in mechanisms by which macrophages dispose of surplus cholesterol, a process referred to as “cholesterol efflux.” Studies from many laboratories established that ABC transporters play an important role in cholesterol efflux from macrophages. ABCA1 unloads cholesterol onto nascent HDL particles (1–4), and a deficiency of ABCA1 results in an accumulation of macrophage foam cells in tissues (5–7). ABCG1 unloads cholesterol onto mature HDL particles (4, 8). The expression of both transporters is regulated by the LXRα and LXRβ transcription factors, which are activated by oxysterol ligands in cholesterol-loaded cells (9, 10).
While many studies of cholesterol efflux by macrophages have focused on the capacity of ABC transporters to transfer cellular cholesterol onto HDL particles, a variety of observations have suggested that other mechanisms exist. For example, Babiker et al. (11) reported that cultured macrophages secrete oxygenated cholesterol metabolites and proposed that those metabolites contribute to cholesterol efflux. Also, Ho et al. (12) reported that intact human erythrocytes (in the absence of serum or HDL) unload cholesterol from peritoneal macrophages, implying that cholesterol unloading could be promoted by cell-to-cell interactions.
The laboratory of Howard Kruth reported that cultured macrophages export cholesterol from the plasma membrane in the form of “cholesterol microdomains” and proposed that the release of these microdomains serves to unload surplus cholesterol (13–16). More recently, He et al. (17) showed that cultured macrophages release 30-nm vesicular particles from the plasma membrane onto the surrounding substrate, and they went on to show that these particles are enriched in “accessible cholesterol” (a pool of cholesterol that is not sequestered by sphingolipids) (18). The accessible pool of cholesterol is mobile and important in regulating the SREBP transcription factors (18, 19) in the endoplasmic reticulum, as well as the hedgehog signaling pathway on the plasma membrane (20). The observation that cultured macrophages release plasma membrane-derived cholesterol-rich vesicles raised the possibility that macrophages might be capable of unloading cholesterol onto adjacent cells (21).
A key goal of the current study was to test whether cultured macrophages are capable of transferring cholesterol to adjacent cells in the absence of acceptors in the medium (e.g., serum, HDL). A second goal was actually to visualize the efflux of cholesterol, rather than simply recording the disappearance of radiolabeled cholesterol from cells. To address these goals, we loaded macrophages with uniformly labeled [13C]cholesterol and then plated the macrophages onto a monolayer of smooth muscle cells (SMCs) that had been metabolically labeled with [15N]choline. We then analyzed the movement of [13C]cholesterol from macrophages to the 15N-rich SMCs by nanoscale secondary ion mass spectrometry (NanoSIMS) imaging.
Results
Transfer of [13C]cholesterol from Macrophages to SMCs.
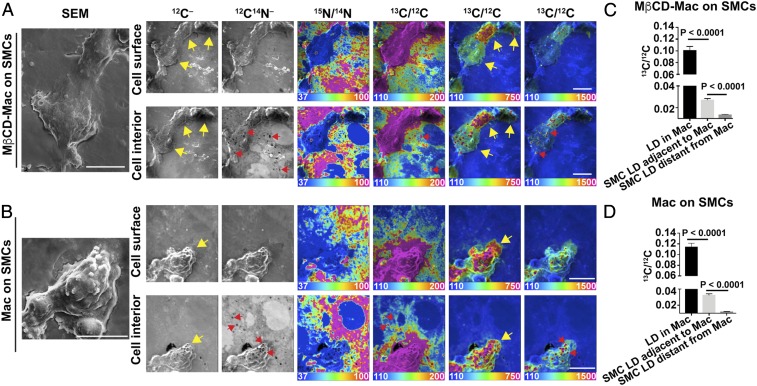
Mouse peritoneal macrophages were loaded with [13C]cholesterol complexed with methyl-β-cyclodextrin (MβCD), plated on a silicon wafer, and incubated overnight. Cell-surface and cell-interior NanoSIMS images revealed 13C enrichment in macrophages (SI Appendix, Fig. S1). The ratio of 13C to 12C in the cytosolic lipid droplets of macrophages (visible in 12C14N–, 32S–, and 13C/12C images) averaged 21%, ∼18-fold higher than 13C natural abundance (1.1%).
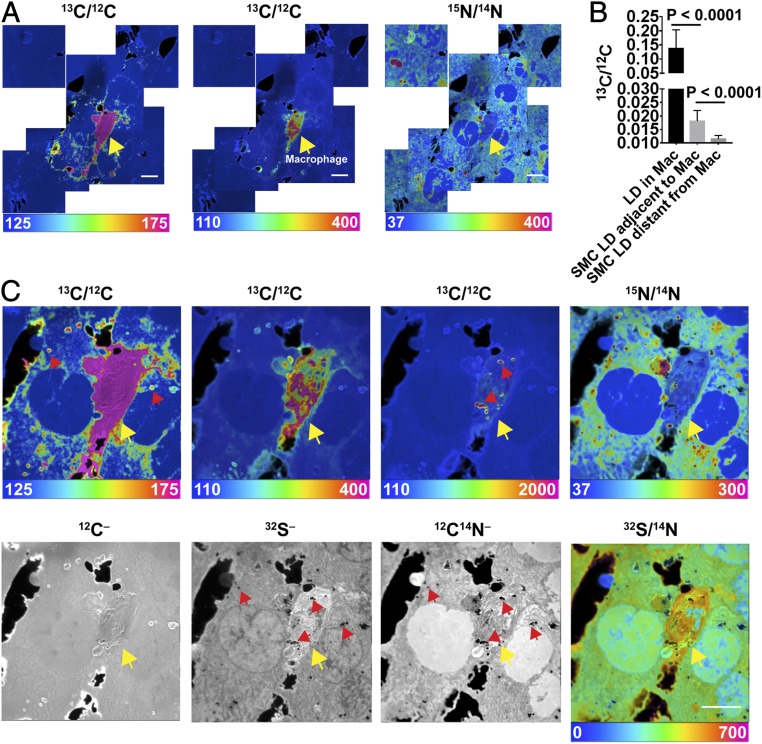
To determine if macrophages transfer [13C]cholesterol to adjacent cells, we plated [13C]cholesterol-loaded macrophages onto a monolayer of SMCs that had been metabolically labeled with [15N]choline (which is incorporated into phosphatidylcholine and sphingolipids). Labeling macrophages and SMCs with different isotopes made it possible to distinguish the two cell types by NanoSIMS imaging. After incubating the cocultured cells overnight at 37 °C, the cells were prepared for NanoSIMS analyses. NanoSIMS imaging revealed transfer of the [13C]cholesterol from macrophages onto adjacent SMCs, but not more distant SMCs (Fig. 1A). 13C enrichment of adjacent SMCs was evident in cytosolic lipid droplets and in the abutting plasma membranes of adjacent SMCs within the monolayer (Fig. 1 A and C). The cytosolic lipid droplets were identified as dark regions in 12C14N– and 32S– NanoSIMS images (regions with negligible 14N or 32S) (Fig. 1C and SI Appendix, Fig. S2). The 13C/12C ratio in cytosolic lipid droplets of SMCs adjacent to the macrophage was substantial, ∼13% as high as the 13C/12C ratio in macrophage lipid droplets (Fig. 1B). 13C enrichment in cytosolic lipid droplets of distant SMCs was negligible (Fig. 1B).
Fig. 1.
Transfer of [13C]cholesterol from [13C]cholesterol-loaded macrophages into adjacent SMCs. The SMCs had been grown for 21 d in medium containing [15N]choline. [13C]cholesterol-loaded macrophages were plated onto a ∼90 to 95% confluent monolayer of SMCs and incubated overnight at 37 °C in serum-free medium. NanoSIMS images were recorded from the interior of cells (note the presence of nuclei with negligible amounts of 15N enrichment). (A) A mosaic of 13C/12C and 15N/14N NanoSIMS images centered on a [13C]cholesterol-loaded macrophage. The macrophage is noted with a yellow arrow. The large black holes in the NanoSIMS images represent regions of the silicon wafer that were not covered by cells. (Scale bars: 10 μm.) Ratio scales were multiplied by 10,000. The natural abundance of 13C is 1.1%. 12C–, 32S–, and 12C14N– NanoSIMS images of the same field are shown in SI Appendix, Fig. S2. (B) 13C/12C ratio (mean ± SD) in macrophage (Mac) cytosolic lipid droplets (LD) (n = 23 LDs in five regions of 45 μm × 45 μm), LDs in SMCs immediately adjacent to the macrophage (n = 52 LDs in five regions of 45 μm × 45 μm), and LDs in more distant SMCs (n = 42 LDs in four regions of 45 μm × 45 μm). The cytosolic LDs in SMCs and macrophages were identified in the 32S– and 12C14N– NanoSIMS images (SI Appendix, Fig. S2). (C) High magnification image of the [13C]cholesterol-loaded macrophage shown in A. Ratio scales were multiplied by 10,000. The ratio scale of 125–175 shows the transfer of [13C]cholesterol from Macs to SMCs; the ratio scale of 110–400 shows [13C]cholesterol enrichment in Macs; and the ratio scale of 110–2,000 shows [13C]cholesterol enrichment in cytosolic LDs in Macs. Note that the macrophage is relatively enriched in 32S. Red arrows show a few examples of cytosolic LDs and yellow arrow points to the macrophage. (Scale bar: 10 μm.)
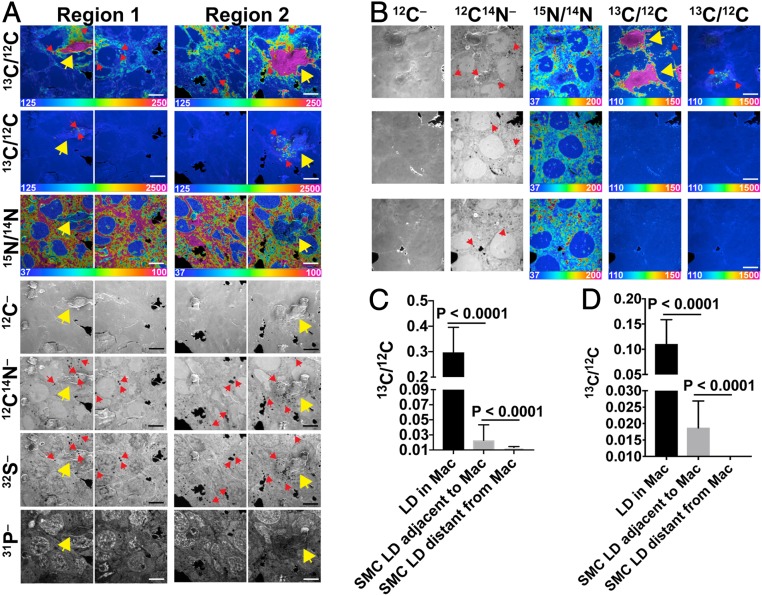
Transfer of [13C]cholesterol from [13C]cholesterol-loaded macrophages to SMCs was observed in two additional experiments (Fig. 2 and SI Appendix, Fig. S3). Substantial 13C enrichment was observed in the lipid droplets of SMCs adjacent to [13C]cholesterol-loaded macrophages (Fig. 2 A and B). The 13C/12C ratio in lipid droplets of adjacent SMCs in the experiment shown in Fig. 2A averaged 7.6% of the 13C/12C ratio in macrophage lipid droplets (Fig. 2C). The 13C/12C ratio in lipid droplets of adjacent SMCs in the experiment shown in Fig. 2B averaged 17% as high as the ratio in macrophage lipid droplets (Fig. 2D).
Fig. 2.
Two additional experiments demonstrating transfer of [13C]cholesterol from [13C]cholesterol-loaded macrophages to SMCs. In each experiment, we recorded NanoSIMS images of [13C]cholesterol-loaded macrophages that had been plated onto an ∼90 to 95% confluent layer of [15N]choline-labeled SMCs. In the first experiment (A), 50,000 macrophages were plated onto the SMC monolayer. Each “region” contains one macrophage (yellow arrows). A thin white line indicates a small gap between individual NanoSIMS images. In the second experiment (B) 30,000 macrophages were plated onto the SMC monolayer. One region (Top Row) contains two macrophages (yellow arrows); two other regions (Middle and Lower Rows) show nearby areas containing SMCs but no macrophages. For both experiments, the large black areas are regions of the silicon wafer without cells. Small black holes in the 12C14N– (A and B) and 32S– (A) images are cytosolic lipid droplets (LDs), which contain negligible amounts of 14N and 32S. Red arrows show examples of cytosolic LDs. NanoSIMS images were recorded from the cell interior (note the negligible amounts of 15N enrichment in A and B and the high 31P– signal in SMC nuclei in A). Ratio scales were multiplied by 10,000. (Scale bars: 10 μm.) (C) 13C/12C ratios (mean ± SD) in macrophage cytosolic LDs (n = 92 LDs in nine regions of 48 μm × 48 μm), LDs in SMCs immediately adjacent to the macrophage (n = 137 LDs in 15 regions of 48 μm × 48 μm), and LDs in SMCs distant from the macrophage (n = 101 LDs in six regions of 48 μm × 48 μm) for the experiment shown in A. Quantification includes data from the NanoSIMS image shown in SI Appendix, Fig. S3. (D) 13C/12C ratios (mean ± SD) in macrophage cytosolic LDs (n = 33 LDs in five regions of 45 μm × 45 μm), LDs in SMCs immediately adjacent to the macrophage (n = 28 LDs in five regions of 45 μm × 45 μm), and LDs in SMCs distant from the macrophage (n = 86 LDs in two regions of 45 μm × 45 μm) for the experiment shown in B.
We also imaged macrophages and SMCs after culturing the cells overnight at 4 °C (SI Appendix, Fig. S4). Small amounts of 13C enrichment were detectable in the SMCs of cocultures that were incubated at 4 °C, but, in contrast to cocultured cells incubated at 37 °C, we observed little or no 13C enrichment in SMC cytosolic lipid droplets or in the plasma membranes of abutting SMCs. Very low levels of 15N enrichment (∼13% more than natural abundance) were observed in macrophages when the cocultures were incubated at 4 °C. Somewhat higher levels of 15N enrichment (∼44% more than natural abundance) were observed in macrophages when the cocultures were incubated at 37 °C. 15N enrichment in macrophages was detectable by NanoSIMS in cocultures incubated at 37 °C (Figs. 1 and 2 and SI Appendix, Figs. S3, S9, and S10).
Depleting Cholesterol from the Plasma Membrane of [13C]cholesterol-Loaded Macrophages.
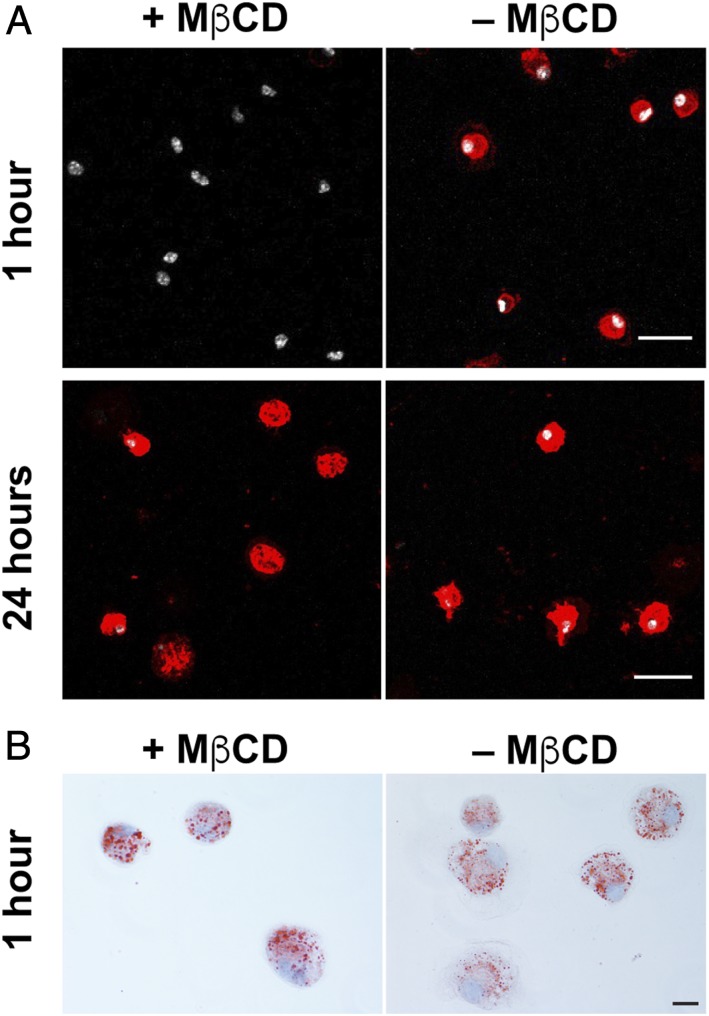
In the studies shown in Figs. 1 and 2 and SI Appendix, Fig. S3, macrophages were loaded overnight with [13C]cholesterol, washed extensively, and then plated onto SMC monolayers. In each experiment, we observed substantial [13C]cholesterol transfer from macrophages to adjacent SMCs. After considering these findings, we wanted to make sure that the cholesterol transfer from macrophages to SMCs did not result from nonphysiologic overloading of the macrophage plasma membrane with cholesterol (as a result of loading cells with [13C]cholesterol–MβCD complexes). In subsequent studies, we examined cholesterol transfer from [13C]cholesterol-loaded macrophages in which we had depleted the accessible cholesterol pool (17) from the plasma membrane by incubating the cells for 30 min with 10 mM MβCD. As expected, the MβCD incubation markedly depleted “accessible cholesterol” from the macrophage plasma membrane, as judged by the binding of fluorescently labeled ALO-D4 (21) (Fig. 3A). Those macrophages—loaded with [13C]cholesterol but depleted in plasma membrane accessible cholesterol—were used in subsequent coculture studies.
Fig. 3.
“Accessible cholesterol” from the plasma membrane was depleted with MβCD. [13C]Cholesterol-loaded macrophages were incubated with or without MβCD for 30 min. (A) Binding of Alexa 594-labeled ALO-D4 (red) to [13C]cholesterol-loaded macrophages was assessed 1 h and 24 h later. ALO-D4 binds to the accessible pool of cholesterol (18). After 1 h, ALO-D4 binding to MβCD-treated macrophages was negligible whereas binding to untreated cells was robust. After 24 h, ALO-D4 binding to the surface of cells that had been treated with MβCD had recovered. Cell nuclei were stained with DAPI (white). Images of 1-h and 24-h samples were recorded with different settings, but both of the 1-h samples were imaged with the same setting, and both of the 24-h samples were imaged with the same setting. (B) Oil red O staining 1 h after cholesterol-loaded macrophages were treated with or without MβCD. Cell nuclei were stained with hematoxylin (light blue). (Scale bars: 25 μm.)
As expected, the depletion of accessible cholesterol from the plasma membrane of the MβCD-treated [13C]cholesterol-loaded macrophages was transient. After 24 h, ALO-D4 binding to the macrophages was robust (Fig. 3A), indicating that the accessible cholesterol pool in the plasma membrane had been replenished. The incubation of macrophages with MβCD did not reduce cell viability (SI Appendix, Fig. S5). Also, the MβCD incubation had little impact on the size or numbers of cytosolic lipid droplets, as judged by Oil Red O staining (Fig. 3B). Also, the incubation of [13C]cholesterol-loaded macrophages with MβCD did not significantly reduce their level of 13C enrichment, as judged by NanoSIMS analyses (SI Appendix, Fig. S6).
Transfer of [13C]cholesterol from “MβCD-Pretreated” Macrophages to Adjacent SMCs.
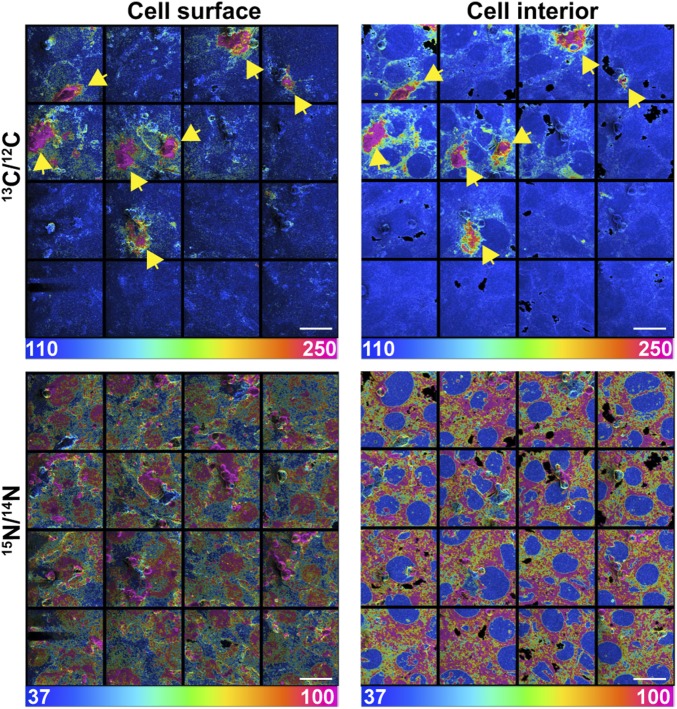
Depleting accessible cholesterol from [13C]cholesterol-loaded macrophages with MβCD had little impact on the transfer of [13C]cholesterol from macrophages to adjacent SMCs during an overnight incubation at 37 °C. The transfer of cholesterol to adjacent SMCs was evident in 13C/12C and 15N/14N NanoSIMS mosaic images—both images of the cell surface and cell interior (Fig. 4). Additional NanoSIMS images of the same mosaics are shown in SI Appendix, Figs. S7 and S8, but the size of the mosaics is larger (showing a larger area containing additional macrophages). Also, in addition to 13C/12C and 15N/14N images, SI Appendix, Figs. S7 and S8 provide 12C14N–, 12C–, and 31P– images (SI Appendix, Figs. S7 and S8).
Fig. 4.
Transfer of [13C]cholesterol from MβCD-treated [13C]cholesterol-loaded macrophages to adjacent SMCs. In this experiment, “accessible cholesterol” was removed from the plasma membrane of the [13C]cholesterol-loaded macrophages by incubating the cells with MβCD for 30 min. The macrophages were then washed extensively and plated onto a monolayer of [15N]choline-labeled SMCs (∼90 to 95% confluency). NanoSIMS images were recorded from the cell surface (Left) and the cell interior (Right). Note the negligible amounts of 15N enrichment in SMC nuclei. Mosaics of 13C/12C and 15N/14N NanoSIMS images (16 images, each 48 µm by 48 µm) show seven 13C-enriched macrophages (yellow arrows) on [15N]choline-enriched SMCs. A larger NanoSIMS mosaic containing this region is shown in SI Appendix, Figs. S7 and S8. Ratio scales were multiplied by 10,000. (Scale bars: 20 μm.)
In an independent experiment, we examined [13C]cholesterol transfer to SMCs with [13C]cholesterol-loaded macrophages that had not been pretreated with MβCD and [13C]cholesterol-loaded macrophages that had been preincubated with MβCD (Fig. 5 A and B and SI Appendix, Figs. S9 and S10). Regardless of whether the macrophages were preincubated with MβCD, [13C]cholesterol transfer to adjacent SMCs was obvious, with [13C]cholesterol enrichment in the cytosolic lipid droplets of SMCs adjacent to macrophages (Fig. 5 C and D). The 13C/12C ratio in lipid droplets of SMCs adjacent to macrophages in the experiment depicted in Fig. 5A averaged 26% of the 13C/12C ratio in the macrophage lipid droplets (Fig. 5C). The 13C/12C ratio lipid droplets of SMCs adjacent to macrophages in the experiment depicted in Fig. 5B averaged 27% as high as the ratio in macrophage lipid droplets (Fig. 5D).
Fig. 5.
Transfer of [13C]cholesterol from [13C]cholesterol-loaded macrophages to adjacent SMCs. [13C]cholesterol-loaded macrophages were plated onto a monolayer of [15N]choline-labeled SMCs (∼90 to 95% confluency). (A) NanoSIMS images (from the cell surface and the cell interior) of [13C]cholesterol-loaded macrophages that had been incubated with MβCD for 30 min before plating onto the SMCs. In this image, there are three macrophages with varying amounts of 13C enrichment (yellow arrows). (B) NanoSIMS images (from the cell surface and the cell interior) of a [13C]cholesterol-loaded macrophage that had not been preincubated with MβCD (yellow arrows). Cytosolic lipid droplets (LDs) (red arrows) are visible in the 12C14N– images. Ratio scales were multiplied by 10,000. SEM images of the macrophages are shown to the Far Left. (Scale bars: 10 µm.) Additional images of macrophages (both MβCD-untreated and treated)) from this experiment are shown in SI Appendix, Figs. S9 and S10. (C) 13C/12C ratios (mean ± SD) in macrophage cytosolic LDs (n = 17 droplets in five regions of 40 μm × 40 μm), LDs in SMCs immediately adjacent to the macrophage (n = 158 droplets in nine regions of 40 μm × 40 μm), and LDs in distant SMCs (n = 8 droplets in one region of 40 μm × 40 μm) for images containing MβCD-treated macrophages. (D) 13C/12C ratios (mean ± SD) in macrophage cytosolic LDs (n = 179 droplets in six regions of 40 μm × 40 μm), LDs in SMCs immediately adjacent to the macrophage (n = 1,167 droplets in 9 regions of 40 μm × 40 μm), and LDs in distant SMCs (n = 368 droplets in three regions of 40 μm × 40 μm) for images containing macrophages that were not pretreated with MβCD.
The transfer of [13C]cholesterol from macrophages to SMCs was substantial. In experiments involving [13C]cholesterol-loaded macrophages that had not been pretreated with MβCD (Fig. 5A and SI Appendix, Fig. S9), 13C secondary ions (above natural abundance and normalized to 12C) in the SMCs immediately adjacent to macrophages were 8.9% of the 13C secondary ions in macrophages (n = 6 macrophages and adjacent SMCs quantified). In [13C]cholesterol-loaded macrophages that had been preincubated with MβCD (Fig. 5B and SI Appendix, Fig. S10), 13C secondary ions (above natural abundance and normalized to 12C) were 12.9% of 13C secondary ions in macrophages (n = 11 macrophages and adjacent SMCs quantified).
In independent studies with [13C]cholesterol-loaded macrophages that had not been preincubated with MβCD (Fig. 2A and SI Appendix, Fig. S3), 13C secondary ions (above natural abundance and normalized to 12C) in SMCs adjacent to macrophages were 11.7% of the 13C secondary ions in macrophages (n = 25 macrophages and adjacent SMCs quantified). In parallel studies (Fig. 4 and SI Appendix, Figs. S7 and S8) involving [13C]cholesterol-loaded macrophages that had been preincubated with MβCD, 13C secondary ions (above natural abundance and normalized to 12C) were 8.1% of the 13C secondary ions in macrophages (n = 22 macrophages and adjacent SMCs quantified). Our results are summarized in SI Appendix, Table S1.
In the coculture experiment illustrated in SI Appendix, Fig. S8 the total area covered by SMCs was far greater than the area covered by macrophages. In that study, the total number of 13C atoms (above natural abundance) in SMCs within the entire mosaic was 1.7-fold greater than the number of 13C atoms (above natural abundance) in all of the macrophages combined.
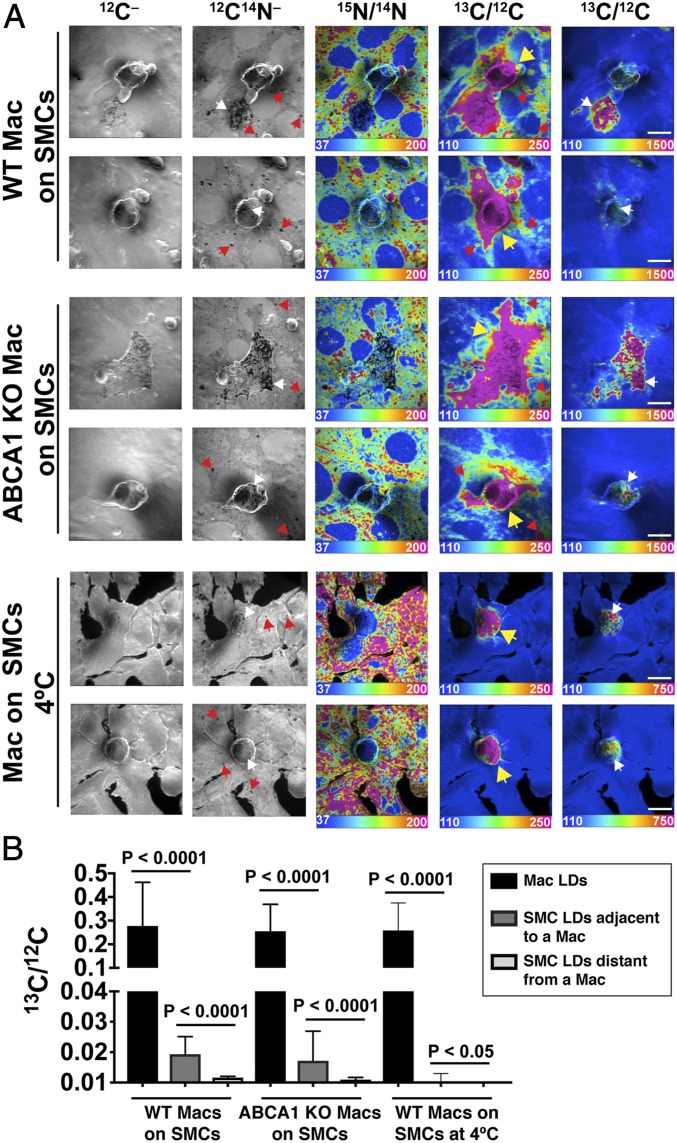
Next, we tested whether cholesterol is transferred from ABCA1 knockout mouse peritoneal macrophages to adjacent SMCs. Wild-type (WT) or ABCA1 knockout macrophages were loaded with [13C]cholesterol, resulting in an accumulation of [13C]cholesterol ester-rich cytosolic lipid droplets (SI Appendix, Fig. S11). The [13C]cholesterol-loaded macrophages were then incubated with 10 mM MβCD for 30 min, transiently depleting “accessible cholesterol” from the plasma membrane (SI Appendix, Fig. S11). Next, the [13C]cholesterol-loaded macrophages were plated onto SMCs that had been metabolically labeled with [15N]choline, and the cocultures were incubated overnight at 37 °C. NanoSIMS analyses revealed that WT and ABCA1 KO macrophages transferred similar amounts of [13C]cholesterol to the 15N-enriched SMCs (Fig. 6 A and B and SI Appendix, Table S1). The 13C/12C ratio in lipid droplets of SMCs adjacent to WT macrophages averaged 7.0% of the 13C/12C ratio in lipid droplets of the WT macrophages; the 13C/12C ratio in lipid droplets of SMCs adjacent to ABCA1 KO macrophages averaged 6.7% of the 13C/12C ratio in lipid droplets of the ABCA1 KO macrophages (Fig. 6B).
Fig. 6.
Transfer of [13C]cholesterol from [13C]cholesterol-loaded WT or ABCA1 knockout (KO) macrophages to SMCs. [13C]cholesterol-loaded WT or ABCA1 KO macrophages were plated onto a monolayer of [15N]choline-labeled SMCs (∼90 to 95% confluency). (A) NanoSIMS images of the interior of [13C]cholesterol-loaded WT or ABCA1 KO macrophages (Mac) that had been plated onto SMCs and incubated overnight at 37 °C. As a control, NanoSIMS images were also obtained on WT macrophages that had been plated onto SMCs and then incubated overnight at 4 °C. In all of these studies, the [13C]cholesterol-loaded macrophages were preincubated for 30 min with 10 mM MβCD before being plated onto the SMCs. The preincubation with MβCD markedly reduced levels of “accessible cholesterol” in the plasma membrane of the [13C]cholesterol-loaded macrophages, as judged by the binding of fluorescently labeled ALO-D4 (SI Appendix, Fig. S11). In these NanoSIMS images, [13C]cholesterol-loaded macrophages are indicated with a yellow arrow. Cytosolic lipid droplets (LDs) in SMCs (red arrows) were detectable as nitrogen-poor black “holes” in the 12C14N– NanoSIMS images. SMC LDs were also visible in the 13C/12C NanoSIMS images when the cells were incubated at 37 °C. LDs in macrophages (white arrows) were visible in both 12C14N– and 13C/12C NanoSIMS images. (B) 13C/12C ratios (mean ± SD) in LDs of WT macrophages that were incubated overnight at 37 °C (n = 188), LDs in SMCs immediately adjacent to a macrophage (n = 73), and LDs in distant SMCs (n = 8). The bar graph also shows 13C/12C ratios in ABCA1 KO macrophage LDs (n = 264), LDs in SMCs immediately adjacent to a macrophage (n = 141), and LDs in distant SMCs (n = 5). As an experimental control, we measured 13C/12C LD ratios in LDs of macrophages and SMCs when the cocultured cells were incubated overnight at 4 °C. 13C/12C ratios were measured in macrophage LDs (n = 141), SMC LDs immediately adjacent to a macrophage (n = 127), and more distant SMC LDs (n = 18) (P < 0.0001).
Consistent with the studies shown in SI Appendix, Fig. S4 , there was negligible transfer of [13C]cholesterol from macrophages to SMCs when the cocultured cells were incubated overnight at 4 °C (Fig. 6 A and B).
Discussion
In the current studies, we took advantage of NanoSIMS imaging to determine if [13C]cholesterol-loaded macrophages unload cholesterol onto adjacent cells. Multiple coculture studies, performed in the absence of serum or HDL, revealed that the [13C]cholesterol in [13C]cholesterol-loaded macrophages moves into adjacent SMCs. By NanoSIMS imaging, the transfer of [13C]cholesterol into adjacent SMCs was easily detectable after an overnight incubation at 37 °C, visible in both SMC cytosolic lipid droplets and along the abutting plasma membranes of adjacent SMCs.
The amount of [13C]cholesterol transferred from macrophages into adjacent SMCs during an overnight incubation was substantial. The mean 13C/12C ratio in cytosolic lipid droplets of SMCs adjacent to macrophages was ∼8 to 26% of the 13C/12C ratio in lipid droplets of macrophages (Figs. 1, 2, and 5). Initially, we were concerned that the transfer of [13C]cholesterol from macrophages to adjacent SMCs might have been due to nonphysiologic amounts of [13C]cholesterol in the plasma membrane of macrophages (perhaps as a consequence of cholesterol loading with MβCD), but this was not the case. In follow-up studies, we preincubated [13C]cholesterol-loaded macrophages with MβCD (thereby transiently depleting the plasma membrane of cholesterol) before plating the macrophages onto the SMC monolayer. The MβCD preincubation had little or no impact on the transfer of [13C]cholesterol into adjacent SMCs. The mean 13C/12C ratio in the cytosolic lipid droplets of SMCs adjacent to the “pre-incubated macrophages” was ∼7 to 27% of that in the cytosolic lipid droplets of macrophages (Figs. 5 and 6). We observed consistent findings when we quantified 13C atoms in macrophages and in the SMC monolayer. After an overnight incubation, the total number of 13C atoms in SMCs (above natural abundance) was 1.7-fold greater than in the macrophages (SI Appendix, Fig. S8). In coculture studies involving WT and ABCA1-deficient macrophages, we observed similar amounts of [13C]cholesterol transfer to SMCs (Fig. 6). Thus, the transfer of cholesterol from macrophages to SMCs did not depend on ABCA1 expression.
The NanoSIMS instrument accurately quantifies numbers of 12C– and 13C– ions released by the primary Cs+ beam. Thus, the observation that there were greater numbers of 13C atoms (above natural abundance) in SMCs than that in macrophages in a large NanoSIMS mosaic (SI Appendix, Fig. S8) implies that substantial amounts of cholesterol transfer had occurred. However, there are caveats in drawing conclusions about the absolute amount of cholesterol transfer. For example, NanoSIMS images are created from secondary ions released from a very thin slice of the cocultured cells (a depth of only ∼5 to 10 nm). Thus, quantitative analyses of secondary ions could be misleading if the cytosolic lipid droplets were distributed unevenly. In theory, it would be possible to record numbers of secondary ions from hundreds of successive NanoSIMS images spanning the entire depth of cocultured cells. Unfortunately, NanoSIMS imaging is very time-consuming and very expensive, making those sorts of studies impractical.
We showed that ABCA1-deficient macrophages transfer cholesterol to adjacent SMCs. In the future, it should be possible to use our experimental approaches to assess the relevance of other cell-surface proteins to cholesterol transfer. The fact that [13C]cholesterol transfer was largely confined to adjacent SMCs implies that the movement of cholesterol depends on direct interactions between the plasma membranes of macrophages and SMCs (as opposed to being mediated by a protein secreted by macrophages into the cell culture medium). Given that the accessible pool of cholesterol is known to be mobile (18, 19), we speculate that much of the cholesterol transferred to SMCs was from the accessible pool within the macrophage plasma membrane, but it is certainly possible that sphingolipid- or phospholipid-sequestered pools contribute to the transfer of cholesterol. It is conceivable that cholesterol moves from macrophages into SMCs via tunneling nanotubes (22), membranous connections between different cells. These structures are known to exist in cultured macrophages (22).
In a recent paper, Hu et al. (21) reported that cultured macrophages, when incubated in either fetal bovine serum (FBS) or lipoprotein-depleted serum, release large numbers of 30-nm “accessible cholesterol”-rich vesicles onto the surrounding substrate. They presented evidence that these particles represent fragments of the macrophage plasma membrane that are torn away and left behind during the movement of filopodia and lamellipodia (21). Scanning electron microscopy studies revealed that macrophages also release these vesicular particles onto a collagen substrate and onto the surface of dead cells (21). However, whether the cholesterol transfer to SMCs in the current studies involves the release of plasma membrane-derived vesicles by macrophages and their subsequent assimilation into abutting SMCs is unknown. On a practical level, documenting the release of particles by macrophages and their assimilation into the plasma membranes of living SMCs would be extremely challenging. As we look to the future, a reasonable next step would be to use NanoSIMS imaging to quantify cholesterol transfer from [13C]cholesterol-loaded macrophages to the intimal endothelial cells or the medial SMCs of explanted mouse aortas.
We suspect that tissue macrophages, when overloaded with cholesterol, transfer some of their cholesterol directly to adjacent cells. By internalizing senescent erythrocytes, splenic macrophages take up large amounts of cholesterol. We suspect that some of that cholesterol moves directly from macrophages into adjacent lymphocytes, fibroblasts, and endothelial cells of the spleen. Passing off surplus cholesterol to adjacent cells would be expected to protect macrophages from excessive amounts of free cholesterol and limit the accumulation of cholesterol ester droplets. We suspect that cholesterol transfer from tissue macrophages to adjacent cells could actually augment the efficiency of cholesterol movement to HDL, simply because the surplus cellular cholesterol would be dispersed among greater numbers of cells, thereby increasing opportunities for interactions with HDL. We further suspect that cell-to-cell transfer of cholesterol might be ineffective in certain settings. For example, we suspect that macrophages in the necrotic cores of atherosclerotic plaques would have limited opportunities to off-load cholesterol onto adjacent cells.
Materials and Methods
Mouse Strains.
WT mice (C57BL/6) were purchased from The Jackson Laboratory.
Cell Lines.
Mouse SMCs were purchased from ATCC (MOVAS ATCC CRL-2797) and were grown in monolayer cultures at 37 °C with 8 to 9% CO2. SMCs were maintained in SMC medium containing 1× minimum essential medium nonessential amino acids solution, 1 mM sodium pyruvate, 10% (vol/vol) FBS (HyClone), and 0.2 mg/mL G418 sulfate (Thermo Fisher Scientific) in Dulbecco’s modified Eagle’s medium (DMEM) (Thermo Fisher Scientific) (23).
Mouse Peritoneal Macrophages.
Macrophages were elicited and isolated as described in He et al. (17). WT macrophages were elicited by injecting mice intraperitoneally with 1 mL of 3% (wt/vol) Difco Fluid Thioglycollate Medium (Becton, Dickinson & Co.). Peritoneal macrophages were collected 3 d after the injection with 10 mL of cold Dulbecco’s phosphate-buffered saline (PBS) without Ca2+ and Mg2+. After pelleting cells by centrifugation (400 × g for 5 min at 4 °C), we used red blood cell lysing buffer Hybri-Max (Sigma) to remove red blood cells. Next, macrophages were washed two times with cold PBS and then plated onto FBS-coated Petri dishes (8 million cells per dish). Macrophages were incubated in medium containing 2 mM glutamine, 1 mM sodium pyruvate, and 10% (vol/vol) FBS (HyClone) in DMEM (Thermo Fisher Scientific) overnight before initiating any treatments (e.g., cholesterol loading).
[13C]cholesterol Loading of Macrophages and [15N]choline Loading of SMCs.
[13C]cholesterol was produced as described (24) using a Saccharomyces cerevisiae strain (RH6829) engineered to produce cholesterol rather than ergosterol (17, 25, 26). The cholesterol contained 94% 13C, as judged by mass spectrometry. Peritoneal macrophages from C57BL/6 WT or Abca1-deficient macrophages (27) were loaded with [13C]cholesterol/MβCD as described (17). Briefly, macrophages were incubated with 20 μL/mL [13C]cholesterol/MβCD (final concentration, 50 μM [13C]cholesterol in 500 μM MβCD) in DMEM containing 2 mM glutamine, 1 mM sodium pyruvate, 5% lipoprotein-deficient serum, 50 μM mevastatin (Calbiochem), and 50 μM mevalonolactone (Sigma) for 24 h at 37 °C. In some cases, we pretreated the [13C]cholesterol-loaded macrophages with 10 mM MβCD for 30 min at 37 °C before plating them onto monolayers of SMCs. The MβCD pretreatment of the [13C]cholesterol-loaded macrophages virtually abolished the accessible pool of cholesterol from the plasma membrane, as judged by binding studies with fluorescently labeled ALO-D4 (21). The 30-min time point and the 10 mM MβCD concentration were chosen because this method depleted the accessible cholesterol pool without affecting cell viability. SMCs were loaded with 2 mg/mL [15N]choline chloride (Sigma-Aldrich) in SMC medium without G418 sulfate at 37 °C for 21 d.
Immunocytochemistry.
ALO-D4 was purified and labeled with Alexa Fluor 594 as described (17). Adherent macrophages were washed three times (10 min each) in PBS/Ca/Mg containing 0.2% (wt/vol) bovine serum albumin (BSA). Cells were then incubated with 20 μg/mL Alexa Fluor 594-labeled ALO-D4 in PBS/Ca/Mg containing 0.2% (wt/vol) BSA for 2 h at 4 °C and then rinsed two times with PBS. The macrophages were then fixed with 3% (wt/vol) paraformaldehyde (PFA) for 15 min at room temperature and washed three times with PBS/Ca/Mg. After staining DNA with 10 μg/mL DAPI in PBS/Ca/Mg, the cells were mounted with Prolong Gold anti-fade reagent (Life Technologies). Images were recorded with a Zeiss LSM700 confocal fluorescence microscope with an Axiovert 200 M stand and processed with Zen 2010 (Zeiss).
Oil Red O Staining.
Adherent macrophages were washed three times (10 min each) in PBS/Ca/Mg containing 0.2% (wt/vol) BSA. Cells were then fixed with 4% (wt/vol) PFA for 10 min at room temperature and rinsed three times with PBS/Ca/Mg. The macrophages were rinsed once with 60% (vol/vol) isopropanol followed by incubation with 0.24% (wt/vol) Oil Red O in 60% isopropanol for 10 min at room temperature. Cells were rinsed once with 60% (vol/vol) isopropanol and once with distilled water. Cell nuclei were stained with Gill’s Hematoxylin (Vector laboratories) for 4 min and then rinsed with distilled water three times. Cells were incubated in saturated sodium bicarbonate for 1 min and then rinsed with distilled water before mounting with glycerol. Images were recorded with a Nikon eclipse E600 microscope and processed with NIS elements software.
Coculture of Macrophages and SMCs.
After [13C]cholesterol loading, macrophages were detached from the Petri dish by incubating in cold Dulbecco’s PBS without Ca2+ and Mg2+ (Gibco) containing 5% FBS and 5 mM ethylenediaminetetraacetic acid for 45 min at 4 °C. Macrophages were washed three times in DMEM by centrifugation at 400 × g for 5 min. Dead cells were removed with a Dead Cell Removal Kit according to the manufacturer’s instructions (Miltenyi Biotec). After dead cell removal, 100,000 macrophages were plated in a 24-well plate. Cell viability was tested 1 h after plating with the fluorescence-based LIVE/DEAD Cell Imaging Kit (ThermoFisher). After mixing the LIVE/DEAD dyes, the mixture was diluted 1:1 in DMEM, and cells were incubated in the diluted mixture for 15 min at room temperature. Images were recorded by confocal fluorescence microscopy as described earlier.
At the same time, 1,000 to 100,000 macrophages were plated on a 0.5-cm2 silicon wafer coated with 0.1 mg/mL poly-d-lysine hydrobromide (Sigma-Aldrich) (macrophage-only control) or on a poly-d-lysine–coated silicon wafer with a monolayer of 90 to 95% confluent SMCs that were metabolically labeled with [15N]choline chloride. Macrophage-only samples and macrophage–SMC cocultures were incubated in serum-free media containing 2 mM glutamine and 1 mM sodium pyruvate in DMEM for 1 h at 37 °C, followed by washing once with fresh serum-free media to remove cells that did not adhere. Next, samples were incubated in serum-free media for an additional 23 h at 37 °C. In some cases, macrophage–SMC cocultures were incubated in DMEM (with 4,500 mg/L glucose and l-glutamine, without sodium bicarbonate [Millipore Sigma]) for 1 h at 4 °C, followed by washing once with fresh medium and incubating for an additional 23 h at 4 °C.
Sample Preparation for NanoSIMS and Scanning Electron Microscopy.
Cells were fixed with 2.5% glutaraldehyde in 0.1 M sodium cacodylate for 1 h, washed with 0.1 M sodium cacodylate, incubated with 2% osmium tetroxide in 0.1 M sodium cacodylate for 30 min, washed with ice-cold water, and air dried (17). Samples were then coated with 4 nm of iridium with an ion beam sputtering system (South Bay Technologies). Samples were imaged with a Zeiss Supra 40VP scanning electron microscope with a 3-KeV incident beam.
NanoSIMS Analyses.
A NanoSIMS 50L or 50 instrument (CAMECA) was used to analyze samples as previously described (21). Briefly, a focused 133Cs+ primary beam was used to bombard cells that had been coated with 4 nm of iridium; the secondary ions and secondary electrons (SEs) released by the primary beam (e.g., 16O–,12C–, 13C–, 12C2–, 12C13C–, 12C14N–, 12C15N–, 31P–, and 32S–) were collected and quantified.
For “cell surface” imaging in Fig. 4 and SI Appendix, Figs. S1 and S7, a 133Cs+ primary beam (primary aperture D1 = 1) was used to scan an area 50 μm × 50 μm with a total dose of ∼3.3 × 1015 ions per square centimeter to remove the iridium coating and implant 133Cs+. In the same region, images (48 μm × 48 μm) were obtained with an ∼3-pA beam current (primary aperture D1 = 2), a dwell time of ∼100 μs per pixel, and multiple frames of 512 × 512 pixels. For the “cell interior” imaging in Figs. 2A and 4 and SI Appendix, Figs. S1–S4 and S8, a 133Cs+ primary beam (∼1-nA current) was used to scan an area 50 μm × 50 μm with a total dose >1 × 1017 ions per square centimeter, and implant 133Cs+. In the same region, images (48 μm × 48 μm) were obtained with an ∼3-pA beam current (primary aperture D1 = 2), a dwell time of 1.5 ms per pixel, and multiple frames of 512 × 512 pixels. For the “cell interior” imaging in Fig. 2B, regions were scanned as described earlier; images (45 μm × 45 μm) were obtained with an ∼8-pA beam current (primary aperture D1 = 2), a dwell time of 3 ms per pixel, and multiple frames of 512 × 512 pixels. For the “cell interior” imaging in Fig. 1 and SI Appendix, Fig. S2, regions were scanned as described; images (45 μm × 45 μm) were obtained with an ∼3-pA beam current (primary aperture D1 = 2), a dwell time of 7.5 ms per pixel or 5 ms per pixel, and one frame of 512 × 512 pixels.
For the “cell surface” imaging in Fig. 5 and SI Appendix, Figs. S9 and S10, a 133Cs+ primary beam (primary aperture D1 = 1) was used to scan an area 50 μm × 50 μm with a total dose of ∼4.5 × 1015 ions per square centimeter to remove the iridium coating and implant 133Cs+. In the same region, images (40 μm × 40 μm) were obtained with an ∼4-pA beam current (primary aperture D1 = 3), a dwell time of ∼500 μs per pixel, and multiple frames of 512 × 512 pixels. For cell interior imaging in Fig. 5 and SI Appendix, Figs. S9 and S10, a 133Cs+ primary beam current (∼1.3-nA; primary aperture D1 = 0) was used to scan an area 50 μm × 50 μm with a total dose >1 × 1017 ions per square centimeter, and implant 133Cs+. In the same region, images (40 μm × 40 μm) were obtained with an ∼4-pA beam current (primary aperture D1 = 3), a dwell time of 500 μs per pixel, and multiple frames of 512 × 512 pixels. The imaging in Fig. 6 (from the cell interior) used a 133Cs+ primary beam current (∼2 nA; primary aperture D1 = 1) to scan an area 50 μm × 50 μm with a total dose >1 × 1017 ions per square centimeter, and to implant 133Cs+. In the same region, 12C2– and 12C13C images (40 μm × 40 μm) were obtained with an ∼3-pA beam current (primary aperture D1 = 2), a dwell time of 500 μs per pixel, and multiple frames of 512 × 512 pixels. 12C14N– and 12C15N– images (40 μm × 40 μm) were obtained with an ∼3-pA beam current (primary aperture D1 = 3), a dwell time of 500 μs per pixel, and multiple frames of 512 × 512 pixels.
To quantify the 15N/14N ratio or the ratio of (13C sum)/(12C sum) in macrophages or SMCs, we identified macrophages (enriched in [13C]cholesterol and low in [15N]choline) and SMCs (enriched in [15N]choline) from the 13C–, 12C15N–, or 15N/14N NanoSIMS images. Regions of interest (ROIs) were defined with ImageJ. ROIs were drawn on the [13C]cholesterol-loaded macrophages based on the low 15N/14N ratio and high 12C13C– signal. ROIs on adjacent SMCs were drawn over the cytoplasm of 1 to 2 SMCs adjacent to the [13C]cholesterol-loaded macrophages (identified by a high 15N/14N ratio), avoiding SMC nuclei (low 15N/14N ratio). 15N/14N ratios, 12C13C– and 12C2– secondary ion signals, and areas of the ROIs were extracted by the OpenMIMS plugin in ImageJ, and the ratios of (13C sum)/(12C sum) were calculated.
To quantify the 13C/12C ratio in lipid droplets of macrophages and SMCs, we identified lipid droplets (enriched in 12C [lipids] and low in 14N and 32S [proteins]) by 12C– and 12C14N– and 32S– NanoSIMS images. ROIs encircling lipid droplets were defined using ImageJ. The mean 15N/14N and 13C/12C ratios of the ROI were measured with the OpenMIMS plugin in ImageJ and processed by Prism 8.0.2. A Student’s t test with Welch’s correction was used to assess differences.
Data Availability.
Data are included in the figures of this paper.
Supplementary Material
Acknowledgments
This work was supported by National Heart, Lung, and Blood Institute Grants HL090553, HL087228, and HL125335; the Swiss National Centre for Competence in Research in Chemical Biology; the Swiss National Science Foundation (H.R.); and Transatlantic Network Grants from the Fondation Leducq (12CVD04 and 19CVD04). H.J. was supported by an Australian Research Council Discovery Early Career Researcher Award and a Cancer Council Western Australia Early Career Investigator Grant. We thank Anne P. Beigneux for proofing and editing the manuscript.
Footnotes
The authors declare no competing interest.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1922879117/-/DCSupplemental.
References
- 1.Cuchel M., Rader D. J., Macrophage reverse cholesterol transport: Key to the regression of atherosclerosis? Circulation 113, 2548–2555 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Ouimet M., Barrett T. J., Fisher E. A., HDL and reverse cholesterol transport. Circ. Res. 124, 1505–1518 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Phillips M. C., Molecular mechanisms of cellular cholesterol efflux. J. Biol. Chem. 289, 24020–24029 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yvan-Charvet L., Wang N., Tall A. R., Role of HDL, ABCA1, and ABCG1 transporters in cholesterol efflux and immune responses. Arterioscler. Thromb. Vasc. Biol. 30, 139–143 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brooks-Wilson A. et al., Mutations in ABC1 in Tangier disease and familial high-density lipoprotein deficiency. Nat. Genet. 22, 336–345 (1999). [DOI] [PubMed] [Google Scholar]
- 6.Bodzioch M. et al., The gene encoding ATP-binding cassette transporter 1 is mutated in Tangier disease. Nat. Genet. 22, 347–351 (1999). [DOI] [PubMed] [Google Scholar]
- 7.Fredrickson D. S., Altrocchi P. H., Avioli L. V., Goodman D. S., Goodman H. C., Tangier disease. Combined clinical staff conference at the National Institutes of Health. Ann. Intern. Med. 55, 1016–1031 (1961). [Google Scholar]
- 8.Kennedy M. A. et al., ABCG1 has a critical role in mediating cholesterol efflux to HDL and preventing cellular lipid accumulation. Cell Metab. 1, 121–131 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Venkateswaran A. et al., Control of cellular cholesterol efflux by the nuclear oxysterol receptor LXR alpha. Proc. Natl. Acad. Sci. U.S.A. 97, 12097–12102 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chawla A. et al., A PPAR gamma-LXR-ABCA1 pathway in macrophages is involved in cholesterol efflux and atherogenesis. Mol. Cell 7, 161–171 (2001). [DOI] [PubMed] [Google Scholar]
- 11.Babiker A. et al., Elimination of cholesterol in macrophages and endothelial cells by the sterol 27-hydroxylase mechanism. Comparison with high density lipoprotein-mediated reverse cholesterol transport. J. Biol. Chem. 272, 26253–26261 (1997). [DOI] [PubMed] [Google Scholar]
- 12.Ho Y. K., Brown M. S., Goldstein J. L., Hydrolysis and excretion of cytoplasmic cholesteryl esters by macrophages: Stimulation by high density lipoprotein and other agents. J. Lipid Res. 21, 391–398 (1980). [PubMed] [Google Scholar]
- 13.Ong D. S. et al., Extracellular cholesterol-rich microdomains generated by human macrophages and their potential function in reverse cholesterol transport. J. Lipid Res. 51, 2303–2313 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Freeman S. R. et al., ABCG1-mediated generation of extracellular cholesterol microdomains. J. Lipid Res. 55, 115–127 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jin X. et al., ABCA1 contributes to macrophage deposition of extracellular cholesterol. J. Lipid Res. 56, 1720–1726 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin X. et al., ABCA1 (ATP-binding cassette transporter A1) mediates ApoA-I (Apolipoprotein A-I) and ApoA-I mimetic peptide mobilization of extracellular cholesterol microdomains deposited by macrophages. Arterioscler. Thromb. Vasc. Biol. 36, 2283–2291 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He C. et al., Macrophages release plasma membrane-derived particles rich in accessible cholesterol. Proc. Natl. Acad. Sci. U.S.A. 115, E8499–E8508 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Das A., Brown M. S., Anderson D. D., Goldstein J. L., Radhakrishnan A., Three pools of plasma membrane cholesterol and their relation to cholesterol homeostasis. eLife 3, e02882 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Infante R. E., Radhakrishnan A., Continuous transport of a small fraction of plasma membrane cholesterol to endoplasmic reticulum regulates total cellular cholesterol. eLife 6, e25466 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kinnebrew M. et al., Cholesterol accessibility at the ciliary membrane controls hedgehog signaling. eLife 8, e50051 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu X. et al., Release of cholesterol-rich particles from the macrophage plasma membrane during movement of filopodia and lamellipodia. eLife 8, e50231 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dupont M., Souriant S., Lugo-Villarino G., Maridonneau-Parini I., Vérollet C., Tunneling nanotubes: Intimate communication between myeloid cells. Front. Immunol. 9, 43 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim P. H. et al., Disrupting the LINC complex in smooth muscle cells reduces aortic disease in a mouse model of Hutchinson-Gilford progeria syndrome. Sci. Transl. Med. 10, eaat7163 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alfonso-García A., Pfisterer S. G., Riezman H., Ikonen E., Potma E. O., D38-cholesterol as a Raman active probe for imaging intracellular cholesterol storage. J. Biomed. Opt. 21, 61003 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Souza C. M. et al., A stable yeast strain efficiently producing cholesterol instead of ergosterol is functional for tryptophan uptake, but not weak organic acid resistance. Metab. Eng. 13, 555–569 (2011). [DOI] [PubMed] [Google Scholar]
- 26.Shivapurkar R., Souza C. M., Jeannerat D., Riezman H., An efficient method for the production of isotopically enriched cholesterol for NMR. J. Lipid Res. 52, 1062–1065 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Timmins J. M. et al., Targeted inactivation of hepatic Abca1 causes profound hypoalphalipoproteinemia and kidney hypercatabolism of apoA-I. J. Clin. Invest. 115, 1333–1342 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are included in the figures of this paper.