Significance
The alarmone (p)ppGpp is a central signaling nucleotide that is synthesized by RelA-SpoT Homologue (RSH) enzymes, and rewires bacterial physiology in response to stress. It is an unanswered question why bacteria often carry multiple (p)ppGpp-synthesising RSHs in the same genome. We have answered that question for five subfamilies of small alarmone synthetases (SASs) by discovering that they are toxins of a novel toxin−antitoxin system, and that they act by synthesizing ppGpp and its unusual analogue—ppApp. These distinct toxic SAS (toxSAS) subfamilies are neutralized by six neighboring antitoxin genes. The toxSAS system and its weaponizing of nucleotide second messengers gives a new angle to the hotly debated and controversial topic of cross-talk between TAs and the alarmone-mediated stringent response.
Keywords: ppGpp, ppApp, toxin, antitoxin, alarmone
Abstract
Under stressful conditions, bacterial RelA-SpoT Homolog (RSH) enzymes synthesize the alarmone (p)ppGpp, a nucleotide second messenger. (p)ppGpp rewires bacterial transcription and metabolism to cope with stress, and, at high concentrations, inhibits the process of protein synthesis and bacterial growth to save and redirect resources until conditions improve. Single-domain small alarmone synthetases (SASs) are RSH family members that contain the (p)ppGpp synthesis (SYNTH) domain, but lack the hydrolysis (HD) domain and regulatory C-terminal domains of the long RSHs such as Rel, RelA, and SpoT. We asked whether analysis of the genomic context of SASs can indicate possible functional roles. Indeed, multiple SAS subfamilies are encoded in widespread conserved bicistronic operon architectures that are reminiscent of those typically seen in toxin−antitoxin (TA) operons. We have validated five of these SASs as being toxic (toxSASs), with neutralization by the protein products of six neighboring antitoxin genes. The toxicity of Cellulomonas marina toxSAS FaRel is mediated by the accumulation of alarmones ppGpp and ppApp, and an associated depletion of cellular guanosine triphosphate and adenosine triphosphate pools, and is counteracted by its HD domain-containing antitoxin. Thus, the ToxSAS–antiToxSAS system with its multiple different antitoxins exemplifies how ancient nucleotide-based signaling mechanisms can be repurposed as TA modules during evolution, potentially multiple times independently.
Bacteria encounter a variety of different environmental conditions during their life cycles, to which they need to respond and adapt in order to survive. This can include slowing down their growth and redirecting their metabolic resources during nutritional stress, until conditions improve and the growth rate can increase. One of the main ways that bacteria signal stress is through production of the alarmone nucleotides ppGpp and pppGpp, collectively referred to as (p)ppGpp (1). At high concentrations, (p)ppGpp is a potent inhibitor of bacterial growth (2), targeting transcription, translation, and ribosome assembly (1). The (p)ppGpp is produced and degraded by proteins of the RelA/SpoT homolog (RSH) superfamily, named after the two Escherichia coli representatives—multidomain “long” RSH factors RelA and SpoT (3). In addition to long RSHs, bacteria can encode single-domain RSHs: small alarmone synthetases (SAS) and small alarmone hydrolases (SAH).
It is currently unknown why some bacteria carry multiple SASs and SAHs, which can belong to many different subfamilies. Conservation of gene order through evolution can reveal potentially interacting proteins and shed light on the cellular role of proteins (4). Therefore, we developed a computational tool—FlaGs, standing for flanking genes (5)—for analyzing the conservation of genomic neighborhoods, and applied it to our updated database of RSH sequences classified into subfamilies. Surprisingly, we find that some subfamilies of SAS can be encoded in conserved and often overlapping two-gene (and sometimes three-gene) operon architectures that are reminiscent of toxin−antitoxin (TA) loci (6). The potential for SAS toxicity is supported by the observation that, when (p)ppGpp is overproduced—for example, if synthesis by RelA is not balanced by hydrolysis by SpoT—the alarmone becomes toxic and inhibits growth (7).
The first direct evidence that RSH toxicity per se might be a bona fide function of some SASs was provided by Dedrick et al. (8). They showed that gp29, an SAS encoded by the mycobacterial Cluster N bacteriophage Phrann, is exceedingly toxic to Mycobacterium smegmatis. This toxicity is countered by coexpression of its neighboring gene (gp30)—a proposed inhibitor of the SAS. Neither the molecular mechanism of gp29-mediated toxicity nor its neutralization by gp30 are known. The gp29-mediated abrogation of growth is proposed to be a defense mechanism against coinfection by other bacteriophages, such as Tweety and Gaia (8).
The regulatory interplay between gp29 and gp30 is typical of that seen in TA systems. The latter are diverse and ubiquitous small operons that usually encode two genes that are often overlapping. One gene encodes a toxic protein, and the other encodes an antitoxin (RNA or protein) that effectively neutralizes the toxic effect. Known toxins can act in a number of ways (6), commonly, by targeting translation by cutting or modifying the ribosome, translation factors, transfer RNAs (tRNAs), or messenger RNAs (mRNAs). Similarly, antitoxins counteract the toxins through different mechanisms (6): through base-pairing of the antitoxin RNA with the toxin mRNA (type I TA systems), direct protein−protein inhibition (type II), inhibition of the toxin by the antitoxin RNA (type III), or indirect nullification of the toxicity (type IV). Recently, Pseudomonas aeruginosa Tas1 has been identified as a secreted toxic effector of a type 6 secretion system (T6SS) that carries a proline−alanine−alanine−arginine (PAAR) domain for toxin delivery (9) fused to a divergent RSH domain. Instead of (p)ppGpp, the enzymatic domain produces (pp)pApp, that is, pApp, ppApp, and pppApp (10). In this case, the toxin and its immunity protein are encoded within a larger conserved T6SS cluster (not the two or three gene operons that are seen with TAs), and the toxic effect is directed to another cell, unlike with TA systems where the growth inhibitory effect is on the producing cell itself.
In this study, we have uncovered the evolutionary diversity of SAS-based toxin (toxSAS) TA systems using sensitive in silico sequence searching and gene neighborhood analysis. We have experimentally validated five SAS subfamilies as belonging to bona fide TA systems and demonstrated, through mutagenesis, that the toxicity of SASs is strictly dependent on a functional (p)ppGpp synthetase active site. Of our six identified antitoxins, five are strictly specific in counteracting only their cognate toxSAS, and one can universally neutralize all of the toxSASs. This latter antitoxin encodes a (p)ppGpp degrading enzyme—SAH—and acts as a type IV antitoxin degrading the molecular product of toxSAS synthetic activity.
Results
Updated RSH Phylogeny across the Tree of Life.
Our previous evolutionary analysis of the RSH protein family applied high-throughput sensitive sequence searching of 1,072 genomes from across the tree of life (3). Since the number of available genomes has grown dramatically in the last decade, we revisited the evolution of RSHs, taking advantage of our computational tool, FlaGs, to ask whether the conservation of gene neighborhoods might be indicative of functional associations (5). We identified and classified all of the RSHs in 24,072 genomes from across the tree of life using our previous Hidden Markov Model (HMM)-based method. We then carried out phylogenetic analysis to identify new subfamilies, generated new HMMs, and updated the classification in our database (Datasets S1 and S2). We have identified 30 subfamilies of SASs, 11 subfamilies of SAHs, and 13 subfamilies of long RSHs (Fig. 1). The nomenclature follows that of our previous analysis, where prefixes are used to indicate taxonomic distributions (3). As “Rel” is frequently used in the names of SASs such as RelP, RelQ, and RelV, we have continued this usage here, as in our previous analysis of RSH evolution (3), and similarly use the “Spo” suffix to denote hydrolases as SpoT is the only ppGpp hydrolase of E. coli.
Fig. 1.
Maximum likelihood phylogenies of the (p)ppGpp (A) hydrolase and (B) synthetase domains. Trees were generated from RaxML (46) and IQ-TREE (47) analyses of alignments of representatives across the RSH family with (A) the (p)ppGpp hydrolase (HD) domain-containing dataset (698 amino acid positions, 519 sequences) and (B) the ppGpp synthetase (SYNTH) domain-containing dataset (699 amino acid positions, 722 sequences). Shading behind the branches indicates the boundary between multidomain type (long) RSHs and single-domain (small) RSHs. The long RSH groups also contain members that seem to have secondarily lost domains through evolution to become single domain (members of the RickSpo and RickRel groups). Inset box shows the legend for subfamily and intersubfamily support, and support values within subfamilies; those that are less than 60% MLB are not shown. Branch length is proportional to the number of substitutions per site (see scale bar). The red skull and crossbones symbol indicates those subfamilies of SASs that we have confirmed, with toxicity neutralization assays, to contain toxSASs. The SAH group PbcSpo that we have found contains an antitoxin is indicated with a green plus sign.
Putative toxSAS TA Modules Are Widespread in Actinobacteria, Firmicutes, and Proteobacteria.
We ran FlaGs on each of all of the subfamilies and discovered that SAS genes can be frequently found in conserved bicistronic (sometimes overlapping) loci that are characteristic of TA loci. Five SAS subfamilies displaying particularly well conserved TA-like arrangements—FaRel (which is actually a three gene operon), FaRel2, PhRel, PhRel2, and CapRel (Fig. 2 and Datasets S3 and S4)—were selected for further investigation. Among bacteria, PhRel [standing for Phage Rel, the group to which Gp29 (8) belongs] and FaRel are found in multiple species of Firmicutes and Actinobacteria (hence the “Fa” prefix), along with representatives of various Proteobacteria; FaRel2 is found in multiple Actinobacteria, and Firmicutes, while PhRel2 is found in Firmicutes, in addition to Bacillus phages. CapRel as a subfamily can be found in a wide diversity of bacteria (including Cyanobacteria, Actinobacteria, and Proteobacteria); hence the “Cap” prefix. The putative antitoxins are nonhomologous among cognate groups, with the exception of PhRel and CapRel, which share a homologous putative antitoxin (Fig. 2). PhRel and CapRel are sister groups in the RSH phylogeny with medium support (81% MLB RAxML, 96% UFB IQ-TREE, where MLB stands for maximum likelihood bootstrap [as per RaxML output] and UFB stands for ultra-fast bootstrap [as per IQ-Tree output]); Fig. 1 and Dataset S5), suggesting the TA arrangement has been conserved during the diversification of these groups from a common ancestor.
Fig. 2.
Conservation of gene neighborhood and maximum likelihood phylogenetic analysis of tested SAS proteins. Genes that encode proteins belonging to a homologous cluster in more than one genomic neighborhood are colored and numbered (see Dataset S4 for the identity of clusters with flanking gene accession numbers). The SAS gene is shown in black, and nonconserved genes are uncolored. Validated TAs have red taxon names. SASs that we have tested and are nontoxic have purple taxon names. Purple- and green-outlined gray genes are pseudogenes and RNA genes, respectively. Bacteriophage names are indicated with an icon. Numbers on branches are percentages of bootstrap support from 100 replicates.
The potential antitoxins are named with an “AT” prefix to the SAS name. ATfaRel is a predicted SAH of the PbcSpo family (Fig. 1), and ATphRel2 is a GepA (genetic element protein A) family homolog. GepA proteins, which carry the DUF4065 domain, have previously been associated with TA loci (11), and are related to the proteolysis-promoting SocA antitoxin of the SocB toxin (12). The other potential antitoxins (ATcapRel, ATfaRel2, AT2faRel, and ATPhRel) have no homology to proteins or domains of known function, as determined using the National Center for Biotechnology Information (NCBI) Conserved Domain Database search, which scans for NCBI-curated domains and domain models from other sources such as Pfam (9).
toxSAS-Anti-toxSAS Operons Encode Bona Fide Type II and Type IV TAs.
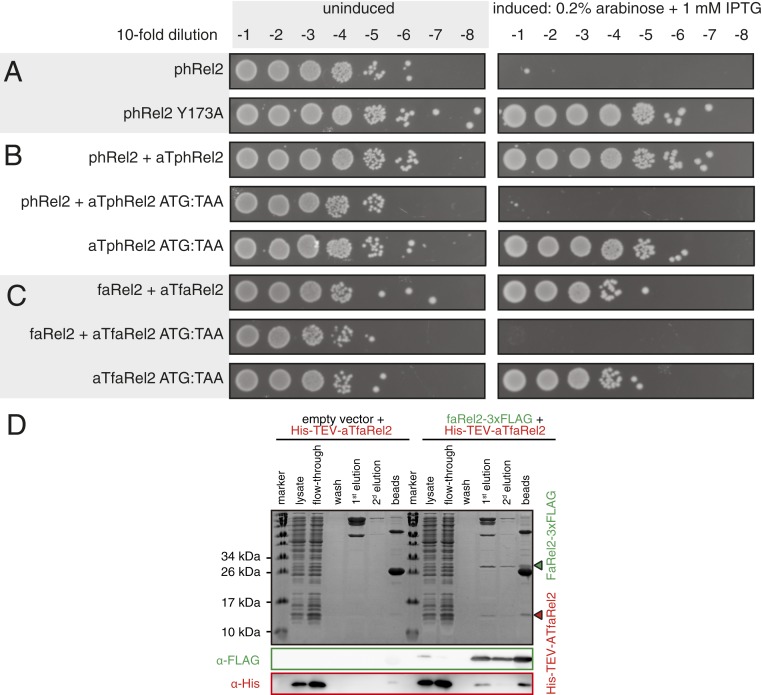
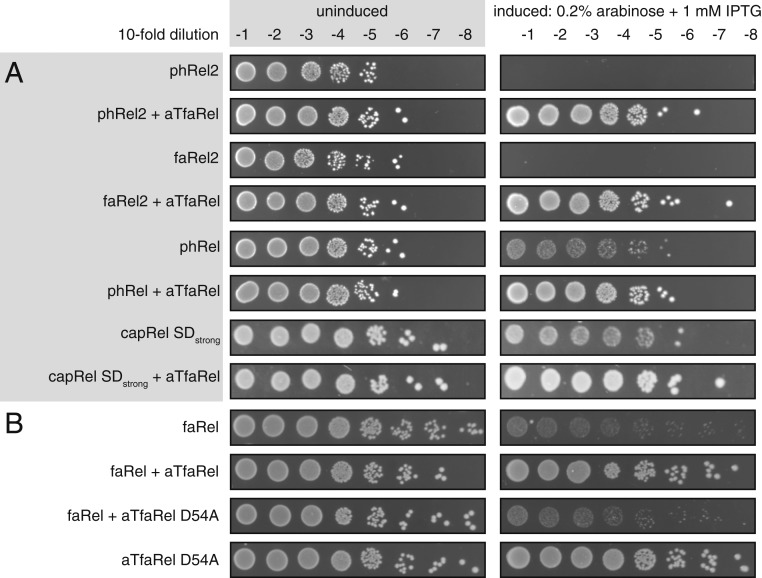
To demonstrate that a gene pair constitutes a genuine TA system, it is necessary to demonstrate both the toxicity of the putative toxin and the ability of the putative antitoxin to rescue the growth defect (thus the name “toxicity neutralization assay”). We carried out toxicity neutralization assays for SASs encoded in conserved TA-like architectures using E. coli strain BW25113 (13). Putative toxSAS and antitoxin genes were expressed under the control of arabinose- and isopropyl β-d-1-thiogalactopyranoside (IPTG)-inducible promoters, respectively (13). Using this approach, we have verified five toxSASs as toxic components of bona fide TA systems: Bacillus subtilis la1a PhRel2 (Fig. 3A), Coprobacillus sp. D7 FaRel2 (Fig. 3B), Mycobacterium phage Phrann PhRel (gp29) (Fig. 3C), Cellulomonas marina FaRel (Fig. 3D), and Mycobacterium tuberculosis AB308 CapRel (Fig. 3E). Importantly, coexpression of putative antitoxins restored growth in all of the cases. C. marina FaRel is encoded as the central gene in a conserved three-gene operon (Fig. 2), and its toxic effect can be neutralized by expression of either the upstream or, to a lesser extent, the downstream flanking gene (Fig. 3D).
Fig. 3.
The two- and three-gene toxSAS operons encode bona fide TA systems. Representatives of groups of toxSAS are validated as TAs: (A) B. subtilis la1a phRel2:aTphRel2, (B) Coprobacillus sp. D7 faRel2:aTfaRel2, (C) Mycobacterium phage Phrann phRel:aTphRel (gp29:gp30), (D) C. marina faRel:aTfaRel and faRel:aT2faRel, and (E) Mycobacterium sp. AB308 capRel:aTcapRel. To perform the toxicity neutralization assays on LB plates, overnight cultures of E. coli strains transformed with pBAD33 and pKK223-3 vectors or derivatives expressing putative toxSAS toxins and antitoxins, correspondingly, were serially diluted from 101- to 108-fold and spotted on LB medium supplemented with appropriate antibiotics as well as either 1% glucose (repression conditions; Left) or 0.2% arabinose and 1 mM IPTG (induction conditions; Right). To assay the toxicity in liquid media, bacteria were grown at 37 °C in Mops minimal media supplemented with 0.5% glycerol, 0.2% arabinose, and 1 mM IPTG (induction conditions). The growth curves represent the geometric mean of three biological replicates, and shading represents the SE; µ2 is the growth rate (±SE) either upon induction of the toxin (in red) or in the absence of the toxin (in black, vector control).
The validated toxSAS toxins differ in the strength of the toxic effect in our system (Fig. 3). FaRel2 and PhRel2 are exceedingly potent, and no overnight bacterial growth is detected upon expression of these toxins from the original pBAD33 vector. FaRel and PhRel are significantly weaker, and small colonies are readily visible upon overnight incubation. Despite the well-conserved bicistronic organization, M. tuberculosis AB308 CapRel initially displayed no detectable toxicity. Thus, we added a strong Shine−Dalgarno motif (SDstrong, 5′ AGGAGG 3′ located six nucleotides upstream of the AUG start codon) to increase the translation initiation efficiency and drive up its protein levels. With this approach, the protein is toxic, and, importantly, the toxicity is readily counteracted by the antitoxin ATcapRel (Fig. 3E). Similarly, moderately toxic phRel and faRel become significantly more toxic when their protein production levels are increased by introduction of SDstrong (SI Appendix, Fig. S1A), or when bacteria are grown on relatively poor M9 minimal media instead of lysogeny broth (LB) (SI Appendix, Fig. S1B). We have validated the observed toxicity by following bacterial growth in liquid culture (Fig. 3). This assay is more sensitive than plating on solid media, with both growth inhibition by toxSAS and neutralization by the antitoxin being clear even in the case of moderately toxic phRel, faRel, and capRel. Mycobacterium phage Squirty PhRel (8) did not display significant toxicity even when the expression was driven with a strong Shine−Dalgarno sequence (SI Appendix, Fig. S2A). The reason for this seems to be a large deletion in the synthetase active site in Squirty PhRel (SI Appendix, Fig. S3). We also tested the well-studied bacterial SASs that are not encoded in TA-like arrangements [Staphylococcus aureus RelP (14, 15) and Enterococcus faecalis RelQ (16, 17)]. We detected no toxicity, even when expression is driven by a strong Shine−Dalgarno sequence (SI Appendix, Fig. S2B). All of the 11 verified toxins and antitoxin sequences and NCBI accession numbers are listed in SI Appendix, Table S1.
Next, we tested whether enzymatic activity is responsible for the toxicity of toxSASs. To do so, we substituted a conserved tyrosine in the so-called G loop for alanine (SI Appendix, Fig. S3). This residue is critical for binding the nucleotide substrate and is highly conserved in (p)ppGpp synthetases (18). All of the tested mutants—PhRel2 Y173A (Fig. 4A), FaRel2 Y128A, PhRel Y143A, and FaRel Y175A—are nontoxic (SI Appendix, Fig. S4). Therefore, we conclude that production of a toxic alarmone is, indeed, the universal causative agent of growth inhibition by toxSASs. Finally, the toxicity does not rely on the functionality of the host RSH machinery, since the toxicity phenotype is identical in a ΔrelA ΔspoT (ppGpp0) BW25113 E. coli strain (SI Appendix, Fig. S5).
Fig. 4.
Active site mutations abrogate toxicity of toxSASs, and toxSAS antitoxins work as proteins, not RNA. (A) Active site mutation Y173A renders phRel2 toxSAS nontoxic. Analogous experiments with all other identified toxSAS support the essentiality of the enzyme function for toxicity (SI Appendix, Fig. S4). (B) Mutation of the start codon to stop renders the aTphRel2 antitoxin ORF unable to protect from the phRel2 toxin. (C) Mutation of the start codon to stop renders the aTfaRel2 antitoxin ORF unable to protect from the faRel2 toxin. Equivalent experiments with aTphRel are presented in SI Appendix, Fig. S6. (D) A pull-down assay demonstrates complex formation between Coprobacillus sp. D7 FaRel2 and ATfaRel2. When coexpressed with 3xFLAG-tagged FaRel2, His-TEV−tagged ATfaRel2 is absorbed from the cellular lysate on ANTI-FLAG M2 Affinity Gel and is eluted with the FLAG peptide. When His-TEV−tagged ATfaRel2 is expressed alone, it neither sorbs nor is eluted from the resin. As an empty vector, pBAD33 was used. Proteins immunoprecipitated with the anti-FLAG antibody were eluted two times and detected by SDS/PAGE and Western blotting (loading order: first and second elution, followed by ANTI-FLAG M2 Affinity Gel beads postelution). An equivalent experiment with aTphRel is presented in SI Appendix, Fig. S8.
We then investigated whether toxSAS antitoxins inhibit toxSASs on the level of RNA (as in types I and III TA systems) or protein (as in types II and IV TA systems). The former scenario is theoretically possible, since, as we have shown earlier, E. faecalis SAS RelQ binds single-stranded RNA and is inhibited in a sequence-specific manner (16). To discriminate between the two alternatives, we mutated the start codon of the aTphRel2, aTfaRel2, and aTphRel antitoxin open reading frames (ORFs) to a stop codon, TAA. Since all of these mutants fail to protect from the cognate toxSAS (Fig. 4 B and C and SI Appendix, Fig. S6), we conclude that they act as proteins.
The proteinaceous nature of antitoxins suggests that they act as type II or IV TA pairs. Type II TAs rely on complex formation, and the interaction can be probed using pull-down assays. We characterized the Coprobacillus sp. D7 FaRel2 and ATfaRel2 TA system, where the large size difference between the toxin and antitoxin allows efficient separation of the two with sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS/PAGE) (Fig. 4D). For specific detection by Western blotting, we used C-terminally 3xFLAG-tagged FaRel2 and N-terminally His-TEV−tagged ATfaRel2. The functionality of the tagged constructs was validated by toxicity neutralization assays (SI Appendix, Fig. S7). Indeed, we observed interaction between the tagged FaRel2 and ATfaRel2 proteins (Fig. 4D), and pull-down experiments with the Mycobacterium phage Phrann PhRel and ATphRel TA pair yielded similar results (SI Appendix, Fig. S8).
The C. marina ATfaRel SAH Hydrolase Antitoxin Cross-Inhibits All Identified toxSAS SASs.
The antitoxin ATfaRel is a member of the PbcSpo subfamily of SAH hydrolases (Fig. 1A). This suggests it acts via degradation of the alarmone nucleotide produced by the toxSAS (and thus as a type IV TA system that does not require direct physical interaction of the TA pair). Therefore, we hypothesized that ATfaRel is able to mitigate the toxicity of all of the identified toxSAS classes through alarmone degradation. This is indeed the case (Fig. 5A). Similarly, coexpression of human SAH MESH1 (19) universally counteracts the toxicity of toxSASs (SI Appendix, Fig. S9). To test whether the hydrolysis activity is strictly necessary for antitoxin function, we generated a point mutant of ATfaRel (D54A). Mutation of the homologous active site residue of Rel from Streptococcus dysgalactiae subsp. equisimilis (RelSeq) abolishes (p)ppGpp hydrolysis (20). As expected, the D54A mutant is unable to counteract the toxicity from FaRel (Fig. 5B).
Fig. 5.
C. marina ATfaRel SAH universally counteracts all identified toxSASs. (A) C. marina aTfaRel neutralizes the toxicity of all identified toxSAS toxins. (B) Toxicity neutralization by C. marina aTfaRel is abolished by the D54A mutation that inactivates the hydrolytic activity of aTfaRel.
FaRel Toxicity Is Mediated by Accumulation of ppGpp and ppApp Alarmones and Depletion of ATP and GTP.
To gain first indications for the mechanism of toxSAS-mediated growth inhibition, we assessed the effects of C. marina FaRel expression in E. coli on overall cell morphology (phase contrast microscopy and FM 5-95 outer membrane staining) and nucleoid appearance (DAPI staining) (Fig. 6A and SI Appendix, Fig. S10). While no change in cell morphology was evident, a rapid decondensation of the nucleoid caused by faRel induction was observed (Fig. 6A and SI Appendix, Fig. S10). This is reminiscent of the decondensation caused by the transcriptional inhibitor rifampicin (Fig. 6A and SI Appendix, Fig. S10), and also by acute RelA-mediated stringent response (21), suggesting that transcription might be affected upon induction of FaRel.
Fig. 6.
Expression of the C. marina FaRel RSH enzyme leads to overproduction of the ppGpp and ppApp alarmones and depletion of intracellular GTP and ATP. (A) Induction of FaRel triggers nucleoid decondensation in E. coli. Depicted are phase-contrast (Upper) and fluorescence images (Upper Middle, Lower Middle, and Lower) of E. coli cells costained with DNA dye DAPI and outer membrane dye FM 5-95. The representative cells carry either an empty or FaRel-expressing vector, and are imaged under uninducing (Mops−glucose medium) or inducing (15 min in Mops−glycerol−arabinose medium) conditions. Note the loss of visible nucleoid structure upon induction of FaRel. As a positive control, cells containing empty vector (Mops−glucose medium) were incubated with rifampicin, which triggers nucleoid decondensation through inhibition of transcription. See SI Appendix, Fig. S10 for a larger field of view with more cells. (B) Pulse-labeling assays following incorporation of 3H-uridine (black traces), 35S-methionine (red traces), and 3H-thymidine (blue traces). E. coli BW25113 cells transformed with empty vector control plasmid pBAD33 were treated with 300 µg/mL kanamycin, 100 µg/mL rifampicin, and 30 µg/mL nalidixic acid as controls for specific inhibition of translation, transcription, and replication, respectively (Left). Expression of FaRel from the pBAD33-faRel plasmid was induced with 0.2% l-arabinose (Right). (C) The expression of C. marina FaRel leads to the accumulation of the alarmone ppGpp as detected by TLC. Alarmone accumulation is efficiently counteracted by wild-type aTfaRel but not its enzymatically compromised D54A mutant. Autoradiograms of a representative TLC plate and a biological replicate (SI Appendix, Fig. S12) are presented. (D and E) Nucleotide pools in E. coli BW25113 expressing C. marina faRel alone. Cell cultures were grown in defined minimal Mops medium supplemented with 0.5% glycerol at 37 °C with vigorous aeration. Expression of C. marina faRel was induced with 0.2% l-arabinose at OD600 0.5. Intracellular nucleotides are expressed in pmol per OD600 per milliliter of culture as per the key in each plot. Error bars indicate the SE of the arithmetic mean of three biological replicates. (F) The 32P-ppApp synthesis assays with either 0.5 µM C. marina faRel Y175A or 30 nM E. coli RelA supplemented with starved ribosomal complexes (0.25 µM 70S IC(MV) + 2 µM tRNAVal) using 0.5 mM 32P ATP and 1 mM ADP as substrates. Experiments were performed in Hepes:Polymix buffer, pH 7.5 at 37 °C, in the presence of 5 mM Mg2+. (G) Neutralization of C. marina faRel toxicity by overexpression of E. coli SpoT. Expression of SpoT from pMG25 was driven by 50 µM IPTG, and expression of faRel from pBAD33 was driven by 0.2% arabinose.
To test whether C. marina FaRel expression indeed inhibits transcription, we assayed macromolecular synthesis rates by following incorporation of 35S-methionine in proteins, 3H-uridine in RNA, and 3H-thymidine in DNA (Fig. 6B; see SI Appendix, Fig. S11 for method validation). Kanamycin, rifampicin, and nalidixic acid were used as controls for specific inhibition of translation, transcription, and replication, respectively. The addition of antibiotics causes rapid (within 2 min) inhibition of the corresponding target process (Fig. 6 B, Left). While expression of FaRel was inhibitory to transcription, translation, and replication, the first process to be affected was transcription: The kinetics of inhibition is similar to that of rifampicin (Fig. 6 B, Right). While the result is in good agreement with (p)ppGpp targeting all of the three processes, the swiftness of the effect on 3H-uridine incorporation is still surprising. A possible explanation is that 3H-uridine uptake is affected, which is a well-documented effect of (p)ppGpp accumulation (22).
We next proceeded to assessing the effects on the intracellular nucleotide pools, with a special focus on (p)ppGpp. First, we used metabolic labeling with 32P-orthophosphoric acid combined with one-dimensional thin-layer chromatography (TLC) separation and autoradiography to assess the accumulation and degradation of nucleotide alarmones upon expression of the C. marina FaRel toxSAS and ATfaRel SAH (Fig. 6C and SI Appendix, Fig. S12). The expression of FaRel results in accumulation of 32P-ppGpp, which is counteracted by wild-type—but not D54A substituted—ATfaRel. While the TLC-based approach is efficient, allowing simultaneous analysis of multiple samples, rather than quantifying the absolute concentrations of nucleotides, the approach relies on the metabolic conversions for 32P labeling of nucleotide species—which could, in principle, be affected by the toxins, since, as was shown for P. aeruginosa Tas1, (pp)pApp produced by this effector binds to PurF and inhibits de novo purine biosynthesis (10). Moreover, one-dimensional TLC is ill-suited for resolving ppApp in complex nucleotide mixtures, since it can comigrate with guanosine triphosphate (GTP) (23). Therefore, we next applied the more quantitative and direct high-pressure liquid chromatography (HPLC)-based approach (24). Our approach utilizes strong anion exchange (SAX) for detection and quantification of ppGpp and pppGpp, while all of the other nucleotide species are analyzed by ion-paired reverse-phase (IPRP) chromatography (24).
We analyzed the kinetics of nucleotide pool changes upon expression of either FaRel alone (Fig. 6 D and E) or coexpressed with ATfaRel (SI Appendix, Fig. S13). Expression of FaRel dramatically perturbs both guanosine (Fig. 6D) and adenosine (Fig. 6E) pools. While both GTP and adenosine triphosphate (ATP) are rapidly depleted, uridine triphosphate (UTP) and cytidine triphosphate (CTP) levels, after the initial drop at 2 min, remain stable (SI Appendix, Fig. S13). The result is consistent with neither UTP nor CTP serving as substrates for RSH enzymes. The ppGpp levels peak at 5 min and drop at 10 min. The likely explanation is exhaustion of ATP and GTP that serve as substrates for the RSH enzymes. The efficient depletion of ATP, which is approximately two times more abundant in E. coli than GTP (2.2 mM vs. 900 µM) (24), is surprising given that RSH-catalyzed pppGpp synthesis is expected to consume guanosines and adenosines in a one-to-one ratio. A possible explanation is that FaRel catalyzes synthesis of (p)ppApp, similarly to a Streptomyces morookaensis SAS homolog (25), and a divergent RelV subfamily homolog Tas1 that is a fusion protein delivered as a toxic effector protein as part of the T6SS of some P. aeruginosa strains (10). While the inspection of SAX traces used for quantification of ppGpp failed to detect ppApp produced by FaRel, we noticed the appearance of a strong peak on IPRP upon FaRel expression (SI Appendix, Fig. S14 A and B). We chemically synthesized pure ppApp using a modification of the synthetic scheme that was originally developed for ppGpp (26) (SI Appendix, Fig. S15). Spike-in experiments and ultraviolet spectroscopy establish the identity of the novel peak as ppApp (SI Appendix, Fig. S14 C–G). Quantification of ppApp accumulation over time upon FaRel expression shows that, at 5 min, ppApp becomes the dominant adenosine species, superseding ATP (Fig. 6E). It was shown earlier that ppApp binds to PurF and inhibits de novo purine biosynthesis (10), thus explaining the drop in ATP and GTP levels: Upon inhibition of synthesis, the two nucleotides are expected to be rapidly consumed by transcription and translation. In principle, the reduced nucleotide pools could also be caused either by FaRel-dependent rapid inhibition of cell metabolism or by triggered leakage of cytoplasmic content. However, we can rule out these alternative scenarios, since, as judged by our microscopy experiments using the membrane potential-sensitive dye DiSC3(5) (27) and the membrane permeability indicator Sytox Green (28), the cells remained both intact and well energized upon expression of FaRel (SI Appendix, Fig. S16). We do not see the membrane hyperpolarization that was speculated to be triggered by (pp)pApp accumulation in the case of Tas1 (10).
The next logical step was to characterize the enzyme biochemically. Despite our best efforts, we failed to express and purify wild-type FaRel to homogeneity, even when coexpressed with ATfaRel. We could, however, purify the enzymatically compromised Y175A mutant. Importantly, when overexpressed, FaRel Y175A potently inhibits bacterial growth, and this toxicity is counteracted by ATfaRel (SI Appendix, Fig. S17), indicating that Y175A and wild-type FaRel are likely to share the same mechanism of toxicity. We directly validated, biochemically, that while catalytically compromised FaRel Y175A can indeed synthesize 32P-ppApp using 32P-ATP and ADP as substrates, this activity is not a general feature of RSH enzymes, since we detect no 32P-ppApp when we test wild-type E. coli RelA activated by starved ribosomal complexes (Fig. 6F). Finally, we tested whether the housekeeping E. coli (p)ppGpp hydrolase SpoT can neutralize the toxicity of FaRel. Surprisingly, while P. aeruginosa SpoT was reported to be unable to counteract the toxicity of Tas1 (10), overexpression of E. coli SpoT from the high-copy vector pMG25 (29) efficiently rescues the growth defect caused by FaRel expression (Fig. 6G). This could be a result of SpoT levels being higher in our study, or species-specific differences in the hydrolytic activities of the housekeeping RSH machinery. Alternatively, it could reflect potential differences between the synthetic activities of FaRel and Tas1; for example, the latter was shown to produce pApp and pppApp in addition to ppApp (10).
Type II Antitoxins Protect Only from Cognate toxSAS Toxins.
The gp29-mediated abrogation of growth is employed by the Phrann phage as a defense mechanism against superinfection by other phages (8). This raises the question of cross-inhibition between toxSAS TA systems: Do all of the identified antitoxins inhibit all of the toxSASs (similarly to how the type IV antitoxin SAH ATfaRel protects from all of the tested toxSASs; see Fig. 5A and Table 1), or is the inhibition specific to toxSAS subfamilies TAs? Therefore, we exhaustively tested pairwise combinations of all of the toxSASs with all of the antitoxins (Table 1 and SI Appendix, Fig. S18). ATphRel2, ATfaRel2, ATphRel, ATcapRel, and AT2faRel antitoxins could not counteract noncognate toxSASs, demonstrating that different classes provide specific discrimination of self from nonself.
Table 1.
Cross-talk among identified toxSAS and their antitoxins as well as standalone phage-encoded SAHs
| Mycobacterium sp. AB308 ATcapRel | B. subtilis la1a ATphRel2 | Coprobacillus sp. D7 ATfaRel2 | Mycobacterium phage Phrann gp29 ATphRel | C. marina ATfaRel SAH | C. marina AT2faRel | Salmonella phage PVP-SE1 SAH (PbcSpo) | Salmonella phage SSU5 SAH (PaSpo) | |
| CapRel | + | – | – | – | + | – | + | + |
| PhRel2 | – | + | – | – | + | – | + | + |
| FaRel2 | – | – | + | – | + | – | + | + |
| PhRel | – | – | – | + | + | – | + | + |
| FaRel | – | – | – | – | + | + | + | + |
Toxicity neutralization assays are presented in Fig. 5A and SI Appendix, Figs. S18 and S19. Plus (+) and minus (–) symbols indicate the ability and inability, respectively, of the antitoxin to neutralize toxicity.
Numerous SASs and SAHs Are Encoded in Prophage-Derived Regions of Bacterial Genomes.
Our initial search has identified 13 SASs in bacteriophage genomes, 2 of which we have confirmed as toxSASs (Figs. 2 and 3). However, this is likely to be an underestimate for two reasons. Firstly, the currently sequenced phage genomes are a small sample of their entire diversity (30), and, secondly, as prophages reside in bacterial genomes, their genes may not be identified as phage in origin. Therefore, we used the tool PHASTER (Phage Search Tool Enhanced Release) (31) to identify phage-like regions of bacterial genomes around SAS genes. In addition to the already identified phage-encoded CapRel, PhRel, and PhRel2, we find 63 prophage regions around representatives in groups belonging to 12 different SASs (Dataset S6). SAHs are found in many more prophages and prophage-like regions than SASs (90 versus 63 instances; Dataset S6). We tested SAHs encoded by Salmonella phages PVP-SE1 (32) (PbcSpo subfamily) and SSU5 (33) (PaSpo subfamily) in toxicity neutralization assays against validated toxSASs. Like the C. marina SAH ATfaRel, both of these stand-alone phage-encoded SAHs efficiently mitigate the toxicity of all of the tested toxSASs (Table 1 and SI Appendix, Fig. S19).
Discussion
Using our tool FlaGs, we have made the surprising discovery that multiple SAS subfamilies can be encoded in TA-like genetic architectures. Through subsequent experimental validation, we have found that the organization of SAS genes into conserved TA-like two- or three-gene arrangements is an indicator of toxicity (Fig. 7). Identification of bicistronic architectures has previously been used as a starting point for prediction of TAs (34, 35). However, these studies focused on species that do not encode toxSASs, and, therefore, these TA systems were not detected. By being associated with novel antitoxins, toxSASs have also escaped identification in “guilt by association” analysis of thousands of genomes (36). This long-term obscurity is despite toxSAS-containing subfamilies being broadly distributed, present in 239 genera belonging to 15 Gram-positive and -negative phyla of bacterial genomes sampled in this study. Thus, it is likely that there are other previously unknown TA systems to be found that are identifiable through searching for conservation of gene neighborhoods across disparate lineages, as we have done with FlaGs.
Fig. 7.
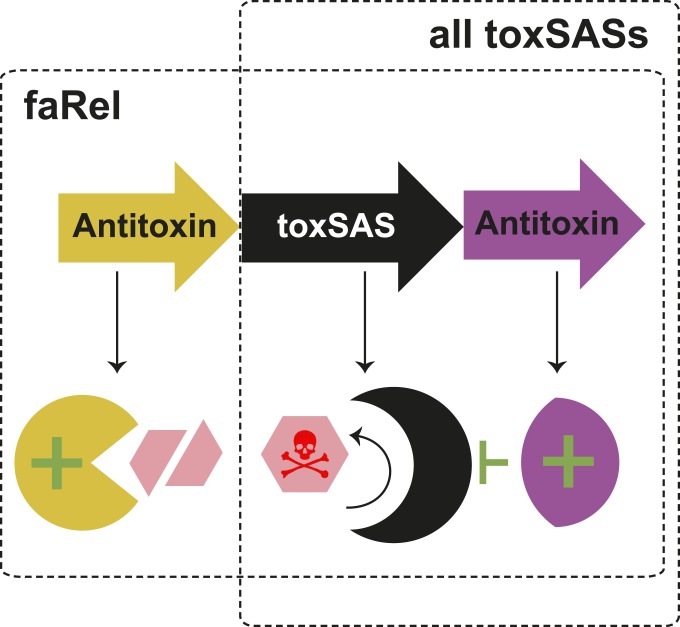
A graphical overview of the toxSAS TA system. ToxSASs exert their toxicity through production of toxin nucleotide alarmones ppGpp and ppApp. In the faRel system, the toxSAS gene is flanked by two neighboring antitoxin genes, each of which is sufficient to counteract the toxicity. One antitoxin (purple) acts as a type II antitoxin working through protein:protein interactions, while the other (yellow) acts as a type IV antitoxin degrading the molecular product of toxSAS synthetic activity. The other identified toxSASs function as a two-gene TA pair, as typically seen in type II TA loci.
As we have found RSH proteins as widespread representatives of most phyla (Dataset S2), they were most likely present in the last common ancestor of bacteria. Thus, RSHs have likely been used for billions of years by bacteria to regulate their growth rate in response to their environment by synthesizing and hydrolyzing nucleotide alarmones. Paradoxically, the very ability of an alarmone to down-regulate growth for continued survival is also what gives it toxic potential. We have identified 30 subfamilies of SASs, 5 of which we have validated as containing toxins, and 2 of which we have validated as nontoxic (RelP and RelQ). It is likely that SASs exist on a continuum in terms of toxicity, with an antitoxin only being required at a certain level of toxicity. This is supported by the observation that not all toxSASs have the same level of toxicity, with one (M. tuberculosis AB308 CapRel) requiring a strong Shine−Dalgarno in order to observe any toxicity in our system. For our five validated toxSAS systems, there are five different homologous groups of antitoxins. This—and the lack of a multisubfamily toxSAS-specific clade in phylogenetic analysis—suggests toxic SASs could have evolved independently multiple times from nontoxic SASs. In the evolution of a ToxSAS−antiToxSAS module from a nontoxic SAS, it is unlikely that the toxic component evolved before the regulatory antitoxin, as this would be detrimental to fitness. Rather, it is more likely that an SAS became regulated by a neighboring gene, which relaxed enzymatic constraints on the SAS, allowing it to relax the precision of enzymatic catalysis, resulting in production of both the “normal” alarmone ppGpp and its toxic analog ppApp. While depletion of ATP and GTP pools is expected to contribute to the inhibition of transcription, the fact that the SAH antitoxin efficiently counteracts the toxicity of all ToxSAS SAS enzymes clearly points to accumulation of the alarmones as the cause of the toxicity. We hypothesize that the depletion of the ATP and GTP substrates is responsible for the decrease in ppGpp levels after the initial spike at around 5 min after the induction of FaRel expression. The ppApp levels remain high, suggesting that the hydrolysis activity of E. coli SpoT expressed at wild-type levels is insufficient for degradation, thus ensuring the efficient shutdown of bacterial growth.
The specific biological role of most of the toxSASs is unclear, with the exception of the phage PhRel−ATphRel (Gp29−Gp30) toxSAS TA pair, which seems to have a role in inhibition of superinfection (8). In this system, PhRel encoded by a prophage protects Mycobacteria from infection by a second phage. Phage infection has previously been linked to alarmone accumulation and stringent response (37–39), although this may not be a universal response to infection (40). Presumably, alarmone accumulation is an example of a so-called abortive infection mechanism (41), where infected hosts are metabolically restricted, but the larger population is protected. A corollary of alarmone-mediated phage inhibition is that incoming phages could bypass this defense system by encoding alarmone hydrolases. Indeed, we have found a variety of different SAHs in different phage genomes and prophage-like regions of bacterial genomes, suggesting there could be cross-talk between ToxSASs and SAHs during infection and superinfection. With the exception of some Arthrobacter representatives that are found on large so-called “megaplasmids,” toxSASs are not plasmid-localized. Thus, they do not seem to have a role as addiction modules for plasmid maintenance.
The (p)ppGpp-mediated signaling has previously—and controversially—been suggested to be implicated in induction of TA-mediated growth inhibition through antitoxin degradation (42, 43). Here, we have discovered a direct connection between these systems, with a subset of TAs themselves producing alarmones to inhibit growth. Importantly, we show that overexpression of the core genome E. coli RSH SpoT can neutralize the toxicity mediated by ppGpp and ppApp synthesized by FaRel toxSAS. This is indicative of a spectrum of RSH functions ranging from growth rate control (housekeeping RSH enzymes), to dramatic, but potentially reversible growth inhibition (toxSAS enzymes discovered in the current report), to, finally, dedicated cytotoxic effectors [secretion system toxins such as Tas1 (10)].
Materials and Methods
Bioinformatic Methods for Identification and Classification of RSH Sequences across the Tree of Life.
RSH sequences from 24,072 genomes were identified and classified with HMM sequence searching and maximum likelihood phylogenetic analysis, as described in SI Appendix, Supplementary Materials and Methods. The alignments used for phylogenetic analysis, and trees with all branch support values, are available in Dataset S5. All sequences and their classification can be found in SI Appendix, Table S1, and subfamily distributions across taxonomy are recorded in SI Appendix, Table S2. Gene neighborhood analysis was carried out with our tool FlaGs, and the output for ToxSASs as well as RelP, RelQ, and RelV can be found in Datasets S3 and S4. To detect SAS genes that may be phage in origin but reside in bacterial genomes, we used the tool PHASTER (31), taking a region of DNA equivalent to four upstream and four downstream genes around each SAS and SAH gene (one representative strain per bacterial species).
Toxicity Neutralization Assays.
Toxicity neutralization assays were performed on LB medium (Lennox) plates (VWR). E. coli BW25113 strains cotransformed with pBAD33 [medium copy number, p15A origin of replication, CmlR, toxins expressed under the control of PBAD promoter (44)], and either pKK223-3 harboring antitoxin genes [medium copy number, ColE1 origin of replication, AmpR, antitoxins expressed under the control of PTac promoter (45)] or pMG25 [high copy number, pUC origin of replication, AmpR, E. coli spoT expressed under the control of PA1/04/03 promoter (29)] were grown in liquid LB medium (BD) supplemented with 100 µg/mL carbenicillin (AppliChem) and 20 µg/mL chloramphenicol (AppliChem) as well as 1% glucose (repression conditions). Serial 10-fold dilutions were spotted (5 µL per spot) on solid LB plates containing carbenicillin and chloramphenicol in addition to either 1% glucose (repressive conditions), or 0.2% arabinose combined with 1 mM IPTG (induction conditions). Plates were scored after an overnight incubation at 37 °C. Sequences were codon-optimized for expression in E. coli.
Growth Assays.
Unless stated otherwise, growth assays were performed in liquid Mops minimal medium (1× Mops mixture [AppliChem], 1.32 mM K2HPO4 [VWR Lifesciences], 0.1 mg/mL thiamine [Sigma], 0.1% casamino acids [VWR Lifesciences], and the carbon source—either 0.5% glycerol [VWR Chemicals] or 1% glucose). The media was supplemented with carbenicillin and chloramphenicol. Overnight cultures were grown in Mops medium supplemented with 1% glucose at 37 °C. The cultures were diluted to a final optical density 600 (OD600) of 0.01 in Mops medium supplemented with 0.5% glycerol, 0.2% arabinose, and 1 mM IPTG. Growth was then monitored using a Bioscreen C Analyzer (Oy Growth Curves Ab Ltd) at 37 °C for 10 h.
The experimental procedures for construction of plasmids, microscopy, quantification of nucleotide pools by TLC and HPLC, protein expression and purification, and enzymatic assays are described in detail in SI Appendix, Supplementary Materials and Methods.
Data Availability.
All data and protocols are available in the main text, SI Appendix, and Datasets S1–S6, with descriptions in SI Appendix. The Python code of our tool FlaGs is available from https://github.com/GCA-VH-lab/FlaGs.
Supplementary Material
Acknowledgments
We are grateful to the Protein Expertise Platform Umeå University and Mikael Lindberg for constructing the plasmids used in this work, Mohammad Roghanian for purifying E. coli RelA, Victoriia Murina for assistance with setting up macromolecular labelling assays, and Anaïs Poirier for help with toxicity neutralization experiments. This work was supported by the Swedish Research Council (Vetenskapsrådet) (Grant 2017-03783 to V.H., and Grants 2015-04746 and 2019-01085 to G.C.A.); Molecular Infection Medicine Sweden (MIMS) (V.H.); Ragnar Söderbergs Stiftelse (V.H.); Kempestiftelserna (Grant SMK-1858.3 to G.C.A.); Carl Tryggers Stiftelse för Vetenskaplig Forskning (Grant 19-24 to G.C.A.); Jeanssons Stiftelser grant to G.C.A.; Umeå Universitet Insamlingsstiftelsen för medicinsk forskning (G.C.A. and V.H.); Umeå Centre for Microbial Research (UCMR) gender policy program grant to G.C.A.; Biotechnology and Biological Sciences Research Council (BBSRC) New Investigator Award BB/S00257X/1 to H.S.; Czech Ministry of Education and Sport via the Joint Programming Initiative on Antimicrobial Resistance (JPIAMR) (Grant 8F19006 to D.R. and V.H.); Fonds National de Recherche Scientifique (grants FRFS-WELBIO CR-2017S-03, FNRS CDR J.0068.19, and FNRS-PDR T.0066.18 to A.G.-P.); The European Union from the European Regional Development Fund through the Centre of Excellence in Molecular Cell Engineering (award 2014-2020.4.01.15-0013 to T.T. and V.H.); and the Estonian Research Council (Grants PRG335 and IUT2-22 to T.T. and V.H.). Funding for open access charge is from Swedish Research Council (Grant 2019-01085 to G.C.A.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The Python code of our tool FlaGs is available on GitHub at https://github.com/GCA-VH-lab/FlaGs.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1916617117/-/DCSupplemental.
References
- 1.Hauryliuk V., Atkinson G. C., Murakami K. S., Tenson T., Gerdes K., Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat. Rev. Microbiol. 13, 298–309 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ryals J., Little R., Bremer H., Control of rRNA and tRNA syntheses in Escherichia coli by guanosine tetraphosphate. J. Bacteriol. 151, 1261–1268 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atkinson G. C., Tenson T., Hauryliuk V., The RelA/SpoT homolog (RSH) superfamily: Distribution and functional evolution of ppGpp synthetases and hydrolases across the tree of life. PLoS One 6, e23479 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dandekar T., Snel B., Huynen M., Bork P., Conservation of gene order: A fingerprint of proteins that physically interact. Trends Biochem. Sci. 23, 324–328 (1998). [DOI] [PubMed] [Google Scholar]
- 5.Saha C. K., Sanchez Pires R., Brolin H., Atkinson G. C., Predicting functional associations using flanking genes (FlaGs). bioRxiv:10.1101/362095 (4 July 2018).
- 6.Harms A., Brodersen D. E., Mitarai N., Gerdes K., Toxins, targets, and triggers: An overview of toxin-antitoxin biology. Mol. Cell 70, 768–784 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Xiao H., et al. , Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J. Biol. Chem. 266, 5980–5990 (1991). [PubMed] [Google Scholar]
- 8.Dedrick R. M., et al. , Prophage-mediated defence against viral attack and viral counter-defence. Nat. Microbiol. 2, 16251 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marchler-Bauer A., et al. , CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 45, D200–D203 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ahmad S., et al. , An interbacterial toxin inhibits target cell growth by synthesizing (p)ppApp. Nature 575, 674–678 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaldalu N., Kasari V., Atkinson G., Tenson T., “Type II toxin-antitoxin loci: The unusual mqsRA locus” in Prokaryotic Toxin-Antitoxins, Gerdes K., Ed. (Springer, Berlin, Germany, 2013), pp. 93–105. [Google Scholar]
- 12.Aakre C. D., Phung T. N., Huang D., Laub M. T., A bacterial toxin inhibits DNA replication elongation through a direct interaction with the β sliding clamp. Mol. Cell 52, 617–628 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jurėnas D., et al. , AtaT blocks translation initiation by N-acetylation of the initiator tRNAfMet. Nat. Chem. Biol. 13, 640–646 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Manav M. C., et al. , Structural basis for (p)ppGpp synthesis by the Staphylococcus aureus small alarmone synthetase RelP. J. Biol. Chem. 293, 3254–3264 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geiger T., Kästle B., Gratani F. L., Goerke C., Wolz C., Two small (p)ppGpp synthases in Staphylococcus aureus mediate tolerance against cell envelope stress conditions. J. Bacteriol. 196, 894–902 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beljantseva J., et al. , Negative allosteric regulation of Enterococcus faecalis small alarmone synthetase RelQ by single-stranded RNA. Proc. Natl. Acad. Sci. U.S.A. 114, 3726–3731 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abranches J., et al. , The molecular alarmone (p)ppGpp mediates stress responses, vancomycin tolerance, and virulence in Enterococcus faecalis. J. Bacteriol. 191, 2248–2256 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Steinchen W., et al. , Structural and mechanistic divergence of the small (p)ppGpp synthetases RelP and RelQ. Sci. Rep. 8, 2195 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun D., et al. , A metazoan ortholog of SpoT hydrolyzes ppGpp and functions in starvation responses. Nat. Struct. Mol. Biol. 17, 1188–1194 (2010). [DOI] [PubMed] [Google Scholar]
- 20.Hogg T., Mechold U., Malke H., Cashel M., Hilgenfeld R., Conformational antagonism between opposing active sites in a bifunctional RelA/SpoT homolog modulates (p)ppGpp metabolism during the stringent response [corrected]. Cell 117, 57–68 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Ferullo D. J., Lovett S. T., The stringent response and cell cycle arrest in Escherichia coli. PLoS Genet. 4, e1000300 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Winslow R. M., Lazzarini R. A., Amino acid regulation of the rates of synthesis and chain elongation of ribonucleic acid in Escherichia coli. J. Biol. Chem. 244, 3387–3392 (1969). [PubMed] [Google Scholar]
- 23.Sobala M., Bruhn-Olszewska B., Cashel M., Potrykus K., Methylobacterium extorquens RSH enzyme synthesizes (p)ppGpp and pppApp in vitro and in vivo, and leads to discovery of pppApp synthesis in Escherichia coli. Front. Microbiol. 10, 859 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Varik V., Oliveira S. R. A., Hauryliuk V., Tenson T., HPLC-based quantification of bacterial housekeeping nucleotides and alarmone messengers ppGpp and pppGpp. Sci. Rep. 7, 11022 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oki T., Yoshimoto A., Sato S., Takamatsu A., Purine nucleotide pyrophosphotransferase from Streptomyces morookaensis, capable of synthesizing pppApp and pppGpp. Biochim. Biophys. Acta 410, 262–272 (1975). [DOI] [PubMed] [Google Scholar]
- 26.Schattenkerk C., Wreesmann C. T., van der Marel G. A., van Boom J. H., Synthesis of riboguanosine pentaphosphate ppprGpp (Magic Spot II) via a phosphotriester approach. Nucleic Acids Res. 13, 3635–3649 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Te Winkel J. D., Gray D. A., Seistrup K. H., Hamoen L. W., Strahl H., Analysis of antimicrobial-triggered membrane depolarization using voltage sensitive dyes. Front. Cell Dev. Biol. 4, 29 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roth B. L., Poot M., Yue S. T., Millard P. J., Bacterial viability and antibiotic susceptibility testing with SYTOX green nucleic acid stain. Appl. Environ. Microbiol. 63, 2421–2431 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takada H., et al. , The C-terminal RRM/ACT domain is crucial for fine-tuning the activation of ‘long’ RelA-SpoT Homolog enzymes by ribosomal complexes. Front. Microbiol. 11, 277 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hatfull G. F., Hendrix R. W., Bacteriophages and their genomes. Curr. Opin. Virol. 1, 298–303 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arndt D., et al. , PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44, W16-21 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Santos S. B., et al. , Genomic and proteomic characterization of the broad-host-range Salmonella phage PVP-SE1: Creation of a new phage genus. J. Virol. 85, 11265–11273 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim M., Kim S., Ryu S., Complete genome sequence of bacteriophage SSU5 specific for Salmonella enterica serovar Typhimurium rough strains. J. Virol. 86, 10894 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Horesh G., et al. , SLING: A tool to search for linked genes in bacterial datasets. Nucleic Acids Res. 46, e128 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wei Y. Q., Bi D. X., Wei D. Q., Ou H. Y., Prediction of type II toxin-antitoxin loci in Klebsiella pneumoniae genome sequences. Interdiscip. Sci. 8, 143–149 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Leplae R., et al. , Diversity of bacterial type II toxin-antitoxin systems: A comprehensive search and functional analysis of novel families. Nucleic Acids Res. 39, 5513–5525 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fernández L., et al. , Low-level predation by lytic phage phiIPLA-RODI promotes biofilm formation and triggers the stringent response in Staphylococcus aureus. Sci. Rep. 7, 40965 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Slomińska M., Neubauer P., Wegrzyn G., Regulation of bacteriophage lambda development by guanosine 5′-diphosphate-3′-diphosphate. Virology 262, 431–441 (1999). [DOI] [PubMed] [Google Scholar]
- 39.Tabib-Salazar A., et al. , T7 phage factor required for managing RpoS in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 115, E5353–E5362 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patterson-West J., et al. , The E. coli global regulator DksA reduces transcription during T4 infection. Viruses 10, E308 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blower T. R., et al. , A processed noncoding RNA regulates an altruistic bacterial antiviral system. Nat. Struct. Mol. Biol. 18, 185–190 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goormaghtigh F., et al. , Reassessing the role of type II toxin-antitoxin systems in formation of Escherichia coli type II persister cells. MBio 9, e00640-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harms A., Fino C., Sørensen M. A., Semsey S., Gerdes K., Prophages and growth dynamics confound experimental results with antibiotic-tolerant persister cells. MBio 8, e01964-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guzman L. M., Belin D., Carson M. J., Beckwith J., Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177, 4121–4130 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brosius J., Holy A., Regulation of ribosomal RNA promoters with a synthetic lac operator. Proc. Natl. Acad. Sci. U.S.A. 81, 6929–6933 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stamatakis A., RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nguyen L. T., Schmidt H. A., von Haeseler A., Minh B. Q., IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data and protocols are available in the main text, SI Appendix, and Datasets S1–S6, with descriptions in SI Appendix. The Python code of our tool FlaGs is available from https://github.com/GCA-VH-lab/FlaGs.